Abstract
Recently, we identified Glut4 as a palmitoylated protein in adipocytes. To understand the role of Glut4 palmitoylation in Glut4 membrane trafficking, a process that is essential for maintenance of whole body glucose homeostasis, we have characterized Glut4 palmitoylation. We found that Glut4 is palmitoylated at Cys223 and Glut4 palmitoylation at Cys223 is essential for insulin dependent Glut4 membrane translocation as substitution of Cys223 with a serine residue in Glut4 (C223S Glut4) diminished Glut4 responsiveness to insulin in membrane translocation in both adipocytes and CHO-IR cells. We have examined C223S Glut4 subcellular localization and observed that it was absence from tubular-vesicle structure, where insulin responsive Glut4 vesicles were presented. Together, our studies uncover a novel mechanism under which Glut4 palmitoylation regulates Glut4 sorting to insulin responsive vesicles, thereby insulin-dependent Glut4 membrane translocation.
Keywords: Fatty acid, adipocytes, palmitoylation, membrane, insulin, Glucose transporter
1. INTRODUCTION
Glut4 (Glucose transporter 4), predominantly expressed in adipose and muscle tissues, is a member of facilitative glucose transporter family that consists of 12 members [1]. Unlike other glucose transporter which constitutively resides on plasma membrane, Glut4 is stored in intracellular compartments in the forms of a small membrane vesicles (termed Glut4 vesicle) under basal condition and only distributed to following insulin stimulation [2,3]. As insulin regulated glucose transporter, Glut4 plays a critical role in the regulation of whole body glucose homeostasis and peripheral insulin sensitivity. In mice and human impaired Glut4 function results in hyperglycemia and insulin resistance, the hallmark of type II diabetes [4,5,6]
Glut4 vesicles are presented in various subcellular compartments including Trans Golgi network (TGN), tubular-vesicular structure, endosomes and cytoplasm. However, only ones in TGN and tubular-vesicular structure are distributed to plasma membrane following insulin stimulation (for a review see reference [7]). It is therefore, to be effectively distributed to plasma membrane following insulin stimulation, Glut4 vesicles have to be sorted to these insulin responsive compartments.
Protein palmitoylation is a reversible posttranslational modification, through which palmitate (a 16 carbon fatty acid) is attached onto cysteine residues [8]. Palmitoylation equips the protein with a hydrophobic moiety. This moiety serves as an anchor to facilitate the association of targeting proteins with cellular membranes [9], thereby regulating protein and cellular membrane interaction, a process that is essential for protein trafficking, vesicle and membrane trafficking, protein subcellular membrane localization and cell signaling [10,11].
We have been interested in the role of protein palmitoylation in adipose tissue as adipose tissue is a vital organ that regulates body glucose homeostasis and peripheral insulin sensitivity [12,13]. Toward this, we carried out a global proteomic analysis of adipose palmitoylated proteins and identified more than 800 putative palmitoylated protein in adipose tissue including Glut4 [14]. In the present study, we have characterized Glut4 palmitoylation and found that Glut4 palmitoylation plays a critical role for Glut4 membrane translocation.
2. MATERIAL AND METHODS
2.1 Reagents
Insulin, dexamethasone, 3-Isobutyl-1-methylxanthine (IBMX), Methyl methanethiosulfonate (MMTS), saponin, hydroxylamine hydrochloride, anti-syntaxin 4, anti-GFP, anti-Glut1, and anti-Flag antibodies were from Sigma. Thiopropyl Sepharose 6B was from GE Healthcare Life Science. Streptavidin Agarose was from Pierce. 17-octadecynoic acid (17-ODYA) was from Caymen. Biotin-Azide was from Invitrogen. Anti-HA antibody was from Covance. Anti-Glut4 (1F8), anti-EEA1 (early endosome antigen 1), anti-IRAP (insulin responsive aminopeptidase), anti-AS160, anti-Akt and anti-phospho-Akt antibodies were from Cell Signaling.
2.2 Plasmid construction
Glut4 expression vectors have been described previously [15]. HA-Glut4-GFP and HA-Glut4-mCherry were a gift from Dr. Joshua J Zimmerberg (NICHD, NIH) [16]. The C223S, C361S, C363S and C361/363S Glut4 were generated through site-directed mutagenesis kit from Stratagene according to manufacturer’s instruction with the following primers. C223S Glut4, 5′-GTCCTGCTGCCCTTCTCTCCCGAGAGCCCCCGCT-3′ (forward) and 5′-AGCGGGGGCTCTCGGGAGAGAAGGGCAGCAGGAC-3′ (reverse); C361S Glut4, 5′-GGCCTGGCGGGCATGTCTGGCTGTGCCATCCTGATGAC-3′ (forward) and 5′-GTCATCAGGATGGCACAGCCAGACATGCCCGCCAGGCC-3′ (reverse); C363S Glu4, 5′-GGCCTGGGGGCATGTGTGGCTCTGCCATCCTGATGAC-3′ (forward) and 5′-GTCATCAGGATGGCAGAGCCACACATGCCCGCCAGGCC-3′ (reverse); and C361/363S Glut4, 5′-GGCCTGGCGGGCATGTCTGGCTCTGCCATCCTGATGAC-3′ (forward) and 5′-GTCATCAGGATGGCAGAGCCAGACATGCCCGCCAGGCC-3′ (reverse). All mutations were verified by sequencing. The adenovirus generation and production have been described previously [17].
2.3 Cell culture, transient transfection and subcellular fractionation
Differentiation of 3T3-L1 preadipocytes into adipocytes and subcellular fractionation have been described previously [15]. CHO-IR cells were cultured in RPMI-1640 supplemented with 10% (v/v) FBS, 2 mM L-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). L6 rat skeletal myoblasts were cultured in DMEM supplemented with 15% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. When the cells were infected with viruses, at least 75% of cells were transduced. The transient transfection was carried out with Lipofectamine 2000 according to the manufacturer’s instruction.
2.4 Protein palmitoylation assay
Thiopropyl captivation (TPC) of S-palmitoylated protein assay [14] and 17-ODYA metabolic labeling and Click chemistry have been described previously [18].
2.5 Immunocytochemistry and cell imaging
The cells grown on coverslips were fixed and incubated with the indicated primary antibodies followed by Cy3-conjugated goat anti-mouse/rabbit secondary antibody. The fluorescence imaging was captured with confocal microscopy (Olympus) using a 60x objective lens. The total internal reflection fluorescence (TIRF) experiments were carried out on a Nikon TIRF microscope using a 60x objective lens with a 1.49 NA, 488nm excitation and a standard GFP emission filter. Quantification of fluorescence density was performed with Images J software.
2.6 Western blotting
The cellular proteins extracted from cells were prepared and separated on SDS PAGE and transferred to nitrocellulose membranes (Biorad). Then, the membranes were incubated with each of the indicated primary antibody, followed by incubation with a horseradish peroxidase-conjugated secondary antibody (Bio-Rad). The protein bands were visualized using an ECL detection system (Biorad). For quantification, the densities of bands were measured with Image J software.
2.7 Statistical analysis
Means ± S.D./S.E.M. were calculated and statistically significant differences among groups were determined by one-way ANOVA analysis followed by post hoc comparisons, or by two-tailed unpaired Student’s t test between two groups as appropriate, with significance at p < 0.05.
3 RESULTS
3.1 Glut4 is palmitoylated at Cysteine 223
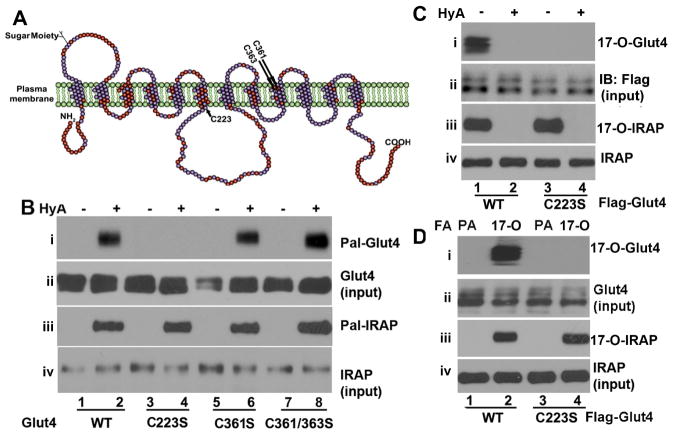
There are three cysteine residues in Glut4 including Cys223, Cys361 and Cys363 (Fig. 1A). Substitution of Cys361 and Cys363 individually or in combination with serine residues had no significant impact on Glut4 palmitoylation as judged by the amount of Glut4 captured by thiopropyl sepharose (panel i) in TPC assay. However, substitution of Cys223 with a serine residue (designated as C223S Glut4) abolished Glut4 captivation by thiopropyl sepharose (Fig. 1B, panel i), arguing that Cys223 is the cysteine residue that undergoes palmitoylation in Glut4. The diminished C223S Glut4 palmitoylation could not be ascribed to the failure of experiments as IRAP palmitoylation was detected in all samples (Fig. 1B, panel iii. Panel iv showed the input levels of IRAP in each sample.), nor to sample variation as comparable cellular levels of wildtype and C223S Glut4 were observed in the corresponding samples (panel ii).
Figure 1.
TPC assay to show that Glut4 is palmitoylated at Cys223. A. The diagrammatic presentation of human Glut4 peptide. The transmembrane domains and all three cysteine residues are indicated. B. TPC assay showing Glut4 palmitoylation at Cys223. Flag-tagged-Glut4 or its mutants were transiently expressed in HEK293 cells, and subject to TPC assay as described under “Materials and Methods.” Panels i and iii palmitoylated Glut4 (Pal-Glut4) and IRAP (Pal-IRAP), respectively Panels ii and iv: input levels of Glut4 and IRAP, respectively. WT, wildtype; C223S, C361S and C363S, Glut4 in which Cys223, Cys361 and Cys363 were substituted with a serine residue, respectively. C361/363S, Glut4 in which both Cys361 and Cys363 were substituted with serine residues. HyA, hydroxylamine hydrochloride. These experiments were repeated at least three times. C. 17-ODYA metabolic labeling and Click Chemistry to show that Glut4 is palmitoylated at Cys223. Panel i, 17-ODYA labeled Glut4 (17-O-Glut4). Panel ii, Glut4 input. HyA, hydroxylamine hydroxylchloride. The + and − indicate the samples treated with or without HyA. Panal iii and iv are 17-ODYA labeled IRAP and IRAP input, respectively. D. Similar to except that the cells were also treated with palmitate (PA). FA: fatty acid. These experiments were repeated at least three times with similar results.
To further verify that Glut4 is indeed modified by palmitate at Cys223, 17-ODYA metabolic labeling of wildtype and C223S Glut4 were performed in HEK293 cells. 17-ODYA metabolic labeling is an assay to specifically detect cysteine palmitoylation [18]. As shown in Figure 1C, wildtype Flag-Glut4 was readily isolated from 17-ODYA metabolically labeled HEK293 cells with streptavidin agarose (panel i), which was blocked by HyA treatment (panel i), a process that removes palmitoylate from cysteine residues. In contrast, no C223S Glut4 was isolated from the 17-ODYA labeled cells under all circumstances (Figure 1C, panel i). 17-ODYA had no impact on Glut4 expression, as comparable levels of wildtype and C223S Glut4 were seen in the corresponding samples (Fig. 1C, panel ii). As a control, we also examined the 17-ODYA labeled IRAP and observed comparable levels of 17-ODYA labeled IRAP in both wildtype and C223S Glut4 expressed cells (Fig. 1C, panel iii).
Next, we carried out the labeling experiments with 17-ODYA and palmitate in parallel. As shown in Fig. 1D, in agreement with the notion that Click Chemistry only occurs on 17-ODYA, no Glut4 was captured in palmitate treated cells although wildtype Glut4 and IRAP were readily captured by streptavidin agarose in the 17-ODYA labeled cells (panel i and iii). Taking all together, we conclude that Glut4 is palmitoylated at Cys223.
3.2 Glut4 palmitoylation at Cys223 is required for Glut4 membrane translocation
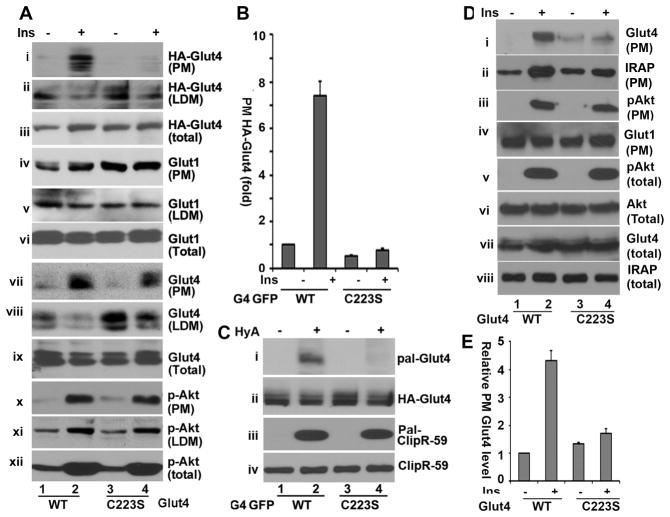
To determine the role of Glut4 palmitoylation in Glut4 membrane translocation, wildtype and C223S HA-Glut4-GFP were introduced into 3T3-L1 adipocytes via adenoviral gene transfer, respectively. Then, PM and low density microsomes (LDM), two fractions where Glut4 are mainly presented in adipocytes, were isolated following insulin (10nM) treatment for 30 min, and the distribution of wildtype or C223S HA-Glut4-GFP in each fraction was assessed. As shown in Fig. 2A, insulin treatment increased the amount of HA-Glut4-GFP in PM roughly by 7-fold (panel i, and Fig. 2B for quantification) and decreased that in LDM (panel ii), as previously observed [19]. The amount of C223S HA-Glut4-GFP in the PM was low under basal condition and only marginally increased following insulin stimulation (Figure 2A, panel i, and Figure 2B). Accordingly, its levels in LDM showed a minimum change following insulin stimulation (Figure 3A1, panel ii). The differences of HA-Glut4-GFP in different fractions observed here could not be ascribed to HA-Glut4-GFP expression and sample variations, as comparable levels of wildtype and C223S HA-Glut4-GFP in total cell homogenates (Figure 2A, panel iii) and of Glut1 in PM (panel iv), LDM (panel v) and total cell homogenates (vi) were seen.
Figure 2.
Glut4 palmitoylation at Cys223 is required for Glut4 membrane translocation in 3T3-L1 adipocytes. A. Subcellular fractionation assay to examine Glut4 subcellular localization in 3T3-L1 adipocytes that were transduced with adenoviral vectors expressing either wildtype or C223S HA-Glut4-GFP and were treated with or without insulin for 30 min. The PM, LDM and total cell homogenates (total) were analyzed in Western blot with an anti-GFP antibody to detect HA-Glut4-GFP (i, ii and iii), anti-Glut1 (iv, v and vi). Ins, insulin. (A2) Western blot analysis of factions in (A1) to examine endogenous Glut4 (vii, viii and ix) and phospho-Akt (x, xi and xi) in PM, LDM and total, respectively. B. Densitometry analysis of HA-Glut4-GFP in PM from Western blots in (A1) The level of HA-Glut4-GFP without insulin treatment was set as 1 after normalized to total cellular Glut4 and Glut1 in PM. Bar graphs represent means ± S.D. (n = 3). The Glut4 level without insulin treatment was set as 1 after normalized to total cellular Glut4. Bar graphs represent means ± S.D. (n = 3). C. TPC assay to examine wildtype and C223S HA-Glut4-GFP palmitoylation in 3T3-L1 adipocytes with ClipR-59 palmitoylation as a control. D. Subcellular fractionation of wildtype and C223S Glut4 in CHO-IR cells. CHO-IR cells were transduced with wildtype or C223S HA-Glut4-GFP adenovirus and treated with or without insulin for 30 min. The PM fractions were prepared and analyzed in Western blot with anti-GFP antibody (i), anti-IRAP (ii), anti-phospho-Akt (iii) and anti-Glut1 antibodies (iv), respectively. The total cellular levels of phospho-Akt (v), Akt (vi), HA-Glut4-GFP (vii) and IRAP (viii) were also shown. Glut4, HA-Glut4-GFP. (B) Densitometry analysis of HA-Glut4-GFP in PM from Western blots in (A). The level of HA-Glut4-GFP was set as 1 after normalized to Glut1 in PM and total cellular HA-Glut4-GFP. Bar graphs represent means ± S.E.M from two independent experiments. In all cases, P<0.025
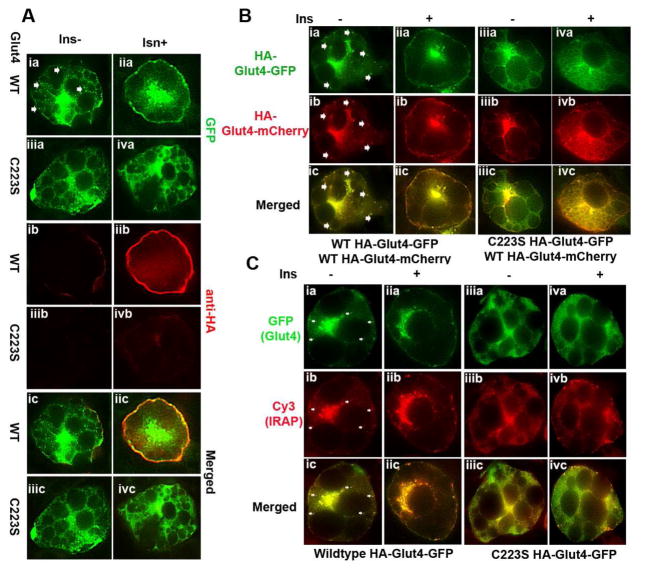
Figure 3.
Subcellular localization of C223S Glut4 in 3T3-L1 adipocytes. A. 3T3-L1 adipocytes that were transduced with adenoviral vectors expressing either wildtype or C223S HA-Glut4-GFP and were treated with or without insulin for 30 min. Then, the cells were fixed with 3.7% paraformaldehyde and immunostained with anti-HA antibody Red) without permeablization and fluorescence images were captured with confocal fluorescence microscope. The arrow indicates that vesicular structures. B. 3T3-L1 adipocytes were co-transduced with adenoviral vectors that express wildtype HA-Glut4-mCherry together with wildtype HA-Glut4-GFP or C223S HA-Glut4-GFP. The cells were treated with or without insulin for 30 min and then fixed with 3.7% paraformaldehyde. The fluorescence images were captured with confocal fluorescence microscope. Panels i and ii are the cells expressing HA-Glut4-mCherry and wildtype HA-Glut4-GFP. Panel iii and iv are the cells expressing HA-Glut4-mCherry and C223S HA-Glut4-GFP. (C) Immunocytochemistry of 3T3-L1 adipocytes transduced with adenoviral vectors that express wildtype or C223S HA-Glut4-GFP with an anti-IRAP antibody. The cells were incubated with rabbit monoclonal anti-IRAP antibody followed by Cy3-conjugated anti-rabbit IgG antibody.
Next, we evaluated the levels of endogenous Glut4 in each fraction. As shown in Fig. 2A, insulin increased the levels of Glut4 in PM (panel vii) and decreased that in LDM (panel viii) in wildtype HA-Glut4-GFP expressed cells. In C223S HA-Glut4-GFP expressed adipocytes, insulin also increased the levels of endogenous Glut4 in PM (Fig. 2A, panel vii) and deceased that in LDM (panel viii). However, a modest but significant decrease in PM Glut4 and an increase in LDM Glut4 were seen in C223S HA-Glut4-GFP expressed adipocytes compared to wildtype HA-Glut4-GFP expressed cells, (Figure 2A, panels vii), implying that C223S HA-Glut4-GFP expression might exert a negative effect on endogenous Glut4 membrane translocation. These differences were not because of Glut4 expression, as comparable levels of Glut4 were observed in total cell lysates (Fig. 2A, panel ix). In these experiments, we also examined the status of Akt activation, an essential event for insulin to promote Glut4 membrane translocation [20]. Comparable levels of Akt phosphorylation were observed in PM, LDM and total cell lysates from both wildtype HA-Glut4-GFP and C223S HA-Glut4-GFP expressed cells (Fig. 2A, panels x, xi and xi). This indicates that C223S HA-Glut4-GFP expression has no adverse effects on Akt activation.
Next, we examined the palmitoylation status of HA-Glut4-GFP. In agreement with the notion that Cys223 is the palmitoylated cysteine residue in Glut4, substitution of Cys223 with a serine residue in HA-Glut4-GFP also blocked Glut4 palmitoylation (Fig. 2C, panel i). Fig. 2C, panel iii showed the palmitoylation status of ClipR-59, a known palmitoylated protein [21], and panels ii and iv showed the input levels of HA-Glut4-GFP and ClipR-59, respectively.
To further substantiate the impact of palmitoylation of Glut4 on its membrane translocation, we next examined C223S Glut4 membrane translocation in CHO-IR cells. CHO-IR cells are the fibroblast-like Chinese hamster ovary cells stably transfected with a human insulin receptor (IR) [22]. They not express Glut4 endogenously, but consist of specialized insulin-responsive vesicles. When ectopically expressed in CHO-IR cells, Glut4 is targeted to these specialized vesicles and exhibits insulin-dependent membrane translocation. Thus, CHO-IR cells constitute a complementary system to adipocytes in the study of Glut4 membrane translocation [23,24]. As shown in Fig. 2D, insulin stimulation resulted in ~4-fold increase in PM wildtype HA-Glut4-GFP (panel i and Figure 2E), in agreement with the view that CHO-IR cells are responsive to insulin in Glut4 membrane translocation. In marked contrast, C223S HA-Glut4-GFP exhibited only a modest insulin response in its membrane translocation (Fig. 2D, panel i, and Figure 2E). We have also examined IRAP and phospho-Akt in PM and observed that insulin stimulation increased the levels of both proteins in wildtype Glut4 expressed CHO-IR cells, as expected; However, the levels of IRAP induced by insulin in C223S Glut4 expressed cells were less than that in wildtype Glut4 expressed cells (Fig. 2D, panel ii). The expression of C223S Glut4 had no impact on Akt activation in PM and total cell lysates (Fig. 2D, panels iii and v). We have also examined PM Glut1 (Fig 2D, panel iv) and total cellular Akt (panel vi), HA-Glut4-GFP (panel vii), and IRAP (panel viii). No alterations in these proteins were observed, ruling out sample variations.
3.3 C223S Glut4 is differentially colocalized from wildtype Glut4 and IRAP
By modulating protein-membrane interaction, palmitoylation regulates protein membrane subcellular localization [9]. Toward this, we examined the subcellular localization of C223S HA-Glut4-GFP in 3T3-L1 adipocytes. Shown in Fig. 3A, wildtype HA-Glut4-GFP exhibits typical Glut4 subcellular localizations, i.e. on cell surface as well as presumably tubular and vesicular structures indicated by arrows. On the other hand, C223S-HA-Glut4-GFP showed more diffused subcellular localization (compare panel i and iii).
Next, we performed a cytohistochemistry with anti-HA antibodies on these cells under nonpermeablized condition. The HA epitope of HA-Glut4-GFP is inserted into the exofacial of Glut4. When stained with anti-HA antibody under nonpermeablized condition, only is cell surface HA-Glut4-GFP detected, thereby measuring membrane associated Glut4 [25]. Shown in Fig. 3A, 3T3-L1 adipocytes expressing wildtype HA-Glut4-GFP showed a cell peripheral surface staining (panel ib), the intensity of which strongly increased following insulin stimulation (panel iib). In contrast, the ones that expressed C223S HA-Glut4-GFP showed a minimum anti-HA staining (panel ivb), further demonstrating that C223S HA-Glut4-GFP lacks insulin responsiveness in membrane translocation.
To further examine C223S HA-Glut4 subcellular localization, we compared co-localization of C223S HA-Glut4-GFP with HA-Glut4-mCherry which similar to wildtype HA-Glut4-GFP, exhibits identical subcellular localization to endogenous Glut4 [16]. As shown in Fig. 3B, HA-Glut4-mCherry and wildtype HA-Glut4-GFP exhibited identical subcellular localization (panel ia–c) and both were distributed to plasma membrane following insulin stimulation (panel iia–c). Surprisingly, C223S HA-Glut4-GFP also showed subcellular co-localization with HA-Glut4-mCherry (Figure 3A, panel iiia–c). However, C223S HA-Glut4-GFP expression not only altered HA-Glut4-mCherry subcellular localization as HA-Glut4-mCherry disappeared from presumably vesicular structures regardless of insulin stimulation (Fig. 3B, compare panels iib and ivb), but also reducing the levels of HA-Glut4-mcherry on plasma membrane, suggesting that C223S Glut4 expression affects wildtype Glu4 subcellular localization and membrane translocation as shown in Fig. 2A.
Next, we assessed subcellular co-localization of C223S HA-Glut4-GFP with IRAP, a maker of major cargo of insulin-responsive Glut4 vesicles [26,27]. We observed that C223S-Glut4-GFP expression affected IRAP subcellular localization as well as IRAP membrane translocation (Fig. 3C) just as it did to HA-Glut4 mCherry.
4 DISCUSSION
In the present report, we have characterized Glut4 palmitoylation and its role in Glut4 membrane translocation. Despite that there are three cysteine residues in Glut4, our studies indicate that Cys223 is the cysteine residue that is palmitoylated, as substitution of Cys223 but not the other two cysteine residues with a serine residue abolished Glut4 palmitoylation (Fig 1D). Cysteine residues can be acylated by different fatty acids [8]. Our data from 17-ODYA metabolic labeling and Click Chemistry experiments (Fig. 1C) demonstrated that Cys223 in Glut4 is modified by palmitate.
To determine the role of Glut4 palmitoylation, we examined how palmitoylation defective C223S Glut4 responds to insulin in its membrane translocation. In 3T3-L1 adipocytes and CHO-IR cells, C223S Glut4 showed a diminished responsiveness to insulin in membrane translocation (Fig. 2), arguing that Glut4 palmitoylation at Cys223 is critical for insulin-dependent membrane translocation.
To get insight into the mechanism by which Glut4 palmitoylation is involved in Glut4 membrane translocation, we examined the subcellular localization of palmitoylation defective C223S Glut4 in 3T3-L1 adipocytes and observed that C223S HA-Glut4 exhibited a different subcellular cellular localization from willdtype one (Fig. 3A), most notably, in vesicular and tubular structures. Based on these data, we believed that Glut4 palmitoylation likely affect Glut4 sorting to Glut4 storage vesicles.
In our studies, we also observed that C223S Glut4 expression interferes with the membrane translocation of endogenous Glut4 (Figure 2A), as well as co-expressed wildtype Glut4 (Fig 3B). At present, why C223S Glut4 interferes with membrane translocation of wildtype Glut4 remains unexplored. Studies of protein palmitoylation revealed that protein palmitoylation defective mutant usually has no dominant effect on wildtype protein palmitoylation as the rate limiting step of protein palmtioylaiton is the autopalmitoylation of palmitoyltransferase [28,29]. Thus, it is unlikely that C223S Glut4 expression suppresses endogenous Glut4 palmitoylation. Supporting this view, we have also observed that C223S Glut4 expression reduced IRAP membrane translocation (Fig. 4). Each Glut4 vesicles consists of multiple copies of Glut4 [27,30]. Therefore, it is plausible that the impact of C223S Glut4 on IRAP and Glut4 membrane translocation is because C223S Glut4 is present in the same vesicles with endogenous Glut4 and IRAP, thereby interfering the membrane translocation of endogenous Glut4 and IRAP. Regardless, the finding that Glut4 palmitoylation regulates Glut4 membrane trafficking is in the line with the notion that palmitoylation regulates proteins trafficking exampled by Ras protein [31].
In summary, we have showed that Glut4 palmitoylation at Cys223, which is essential for insulin-dependent Glut4 membrane translocation. To our knowledge, this is the first study showing that lipid modification is involved in Glut4 membrane translocation. Glut4 membrane translocation is essential for maintenance of whole body glucose homeostasis [1] and protein palmitoylation is often influenced by the cellular levels of palmitate and glucose, which is often become aberrant under obesity and insulin resistance [32,33,34]. We believe that our studies not only provides novel insight into the regulation of Glut4 vesicle trafficking, but also aid us understanding. In mammals, protein palmitoylation is mediated by a family of DHHC palmitoyltransferases [35,36]. Identifying the DHHC proteins that palmitoylate Glut4 will shed more light into the regulation of Glut4 membrane translocation by palmitoylation.
Supplementary Material
Highlights.
We have characterized palmitoylation of Glut4, an insulin responsive glucose transporter.
Glut4 is palmitoylated at Cys223.
Glut4 palmitoylation at Cys223 is required for insulin dependent Glut4 membrane translocation.
Glut4 palmitoylation at Cys223 is involved in Glut4 vesicle sorting.
Acknowledgments
This work was supported by NIH grant RO1 DK084319. We are especially grateful to Dr. Joshua J. Zimmerberg for the internal HA-tagged-Glut4 expression vector.
Footnotes
Conflict of interest
The manuscript authors confirm that they have no commercial associations that pose a conflict of interest in connection with publication of this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thorens B, Mueckler M. Glucose transporters in the 21st Century. Am J Physiol Endocrinol Metab. 2010;298:E141–145. doi: 10.1152/ajpendo.00712.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adekola K, Rosen ST, Shanmugam M. Glucose transporters in cancer metabolism. Curr Opin Oncol. 2012;24:650–654. doi: 10.1097/CCO.0b013e328356da72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leto D, Saltiel AR. Regulation of glucose transport by insulin: traffic control of GLUT4. Nat Rev Mol Cell Biol. 2012;13:383–396. doi: 10.1038/nrm3351. [DOI] [PubMed] [Google Scholar]
- 4.Graham TE, Kahn BB. Tissue-specific alterations of glucose transport and molecular mechanisms of intertissue communication in obesity and type 2 diabetes. Horm Metab Res. 2007;39:717–721. doi: 10.1055/s-2007-985879. [DOI] [PubMed] [Google Scholar]
- 5.Shulman GI. Cellular mechanisms of insulin resistance. J Clin Invest. 2000;106:171–176. doi: 10.1172/JCI10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nandi A, Kitamura Y, Kahn CR, Accili D. Mouse models of insulin resistance. Physiol Rev. 2004;84:623–647. doi: 10.1152/physrev.00032.2003. [DOI] [PubMed] [Google Scholar]
- 7.Bryant NJ, Govers R, James DE. Regulated transport of the glucose transporter GLUT4. Nat Rev Mol Cell Biol. 2002;3:267–277. doi: 10.1038/nrm782. [DOI] [PubMed] [Google Scholar]
- 8.Nadolski MJ, Linder ME. Protein lipidation. FEBS J. 2007;274:5202–5210. doi: 10.1111/j.1742-4658.2007.06056.x. [DOI] [PubMed] [Google Scholar]
- 9.Charollais J, Van Der Goot FG. Palmitoylation of membrane proteins (Review) Mol Membr Biol. 2009;26:55–66. doi: 10.1080/09687680802620369. [DOI] [PubMed] [Google Scholar]
- 10.Resh MD. Palmitoylation of ligands, receptors, and intracellular signaling molecules. Sci STKE. 2006;2006:re14. doi: 10.1126/stke.3592006re14. [DOI] [PubMed] [Google Scholar]
- 11.Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- 12.Galic S, Oakhill JS, Steinberg GR. Adipose tissue as an endocrine organ. Mol Cell Endocrinol. 2010;316:129–139. doi: 10.1016/j.mce.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 13.Kajimura S, Saito M. A new era in brown adipose tissue biology: molecular control of brown fat development and energy homeostasis. Annu Rev Physiol. 2014;76:225–249. doi: 10.1146/annurev-physiol-021113-170252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren W, Jhala US, Du D. Proteomic analysis of protein palmitoylation in adipocytes. Adipocytes. 2013;2:12–27. doi: 10.4161/adip.22117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ren W, Cheema S, Du K. The association of ClipR-59 protein with AS160 modulates AS160 protein phosphorylation and adipocyte Glut4 protein membrane translocation. J Biol Chem. 2012;287:26890–26900. doi: 10.1074/jbc.M112.357699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stenkula KG, Lizunov VA, Cushman SW, Zimmerberg J. Insulin controls the spatial distribution of GLUT4 on the cell surface through regulation of its postfusion dispersal. Cell Metab. 2010;12:250–259. doi: 10.1016/j.cmet.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du K, Herzig S, Kulkarni RN, Montminy M. TRB3: a tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science. 2003;300:1574–1577. doi: 10.1126/science.1079817. [DOI] [PubMed] [Google Scholar]
- 18.Martin BR, Cravatt BF. Large-scale profiling of protein palmitoylation in mammalian cells. Nat Methods. 2009;6:135–138. doi: 10.1038/nmeth.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang S, Czech MP. The GLUT4 glucose transporter. Cell Metab. 2007;5:237–252. doi: 10.1016/j.cmet.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 20.Sakamoto K, Holman GD. Emerging role for AS160/TBC1D4 and TBC1D1 in the regulation of GLUT4 traffic. Am J Physiol Endocrinol Metab. 2008;295:E29–37. doi: 10.1152/ajpendo.90331.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren W, Sun Y, Du K. DHHC17 palmitoylates ClipR-59 and modulates ClipR-59 association with the plasma membrane. Mol Cell Biol. 2013;33:4255–4265. doi: 10.1128/MCB.00527-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ebina Y, Edery M, Ellis L, Standring D, Beaudoin J, Roth RA, Rutter WJ. Expression of a functional human insulin receptor from a cloned cDNA in Chinese hamster ovary cells. Proc Natl Acad Sci U S A. 1985;82:8014–8018. doi: 10.1073/pnas.82.23.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bogan JS, Hendon N, McKee AE, Tsao TS, Lodish HF. Functional cloning of TUG as a regulator of GLUT4 glucose transporter trafficking. Nature. 2003;425:727–733. doi: 10.1038/nature01989. [DOI] [PubMed] [Google Scholar]
- 24.Omata W, Shibata H, Suzuki Y, Tanaka S, Suzuki T, Takata K, Kojima I. Subcellular distribution of GLUT4 in Chinese hamster ovary cells overexpressing mutant dynamin: evidence that dynamin is a regulatory GTPase in GLUT4 endocytosis. Biochem Biophys Res Commun. 1997;241:401–406. doi: 10.1006/bbrc.1997.7810. [DOI] [PubMed] [Google Scholar]
- 25.Blot V, McGraw TE. Use of quantitative immunofluorescence microscopy to study intracellular trafficking: studies of the GLUT4 glucose transporter. Methods Mol Biol. 2008;457:347–366. doi: 10.1007/978-1-59745-261-8_26. [DOI] [PubMed] [Google Scholar]
- 26.Thoidis G, Kandror KV. A Glut4-vesicle marker protein, insulin-responsive aminopeptidase, is localized in a novel vesicular compartment in PC12 cells. Traffic. 2001;2:577–587. doi: 10.1034/j.1600-0854.2001.20807.x. [DOI] [PubMed] [Google Scholar]
- 27.Kandror KV, Pilch PF. The sugar is sIRVed: sorting Glut4 and its fellow travelers. Traffic. 2011;12:665–671. doi: 10.1111/j.1600-0854.2011.01175.x. [DOI] [PubMed] [Google Scholar]
- 28.Jennings BC, Linder ME. DHHC protein S-acyltransferases use similar ping-pong kinetic mechanisms but display different acyl-CoA specificities. J Biol Chem. 2012;287:7236–7245. doi: 10.1074/jbc.M111.337246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell DA, Mitchell G, Ling Y, Budde C, Deschenes RJ. Mutational analysis of Saccharomyces cerevisiae Erf2 reveals a two-step reaction mechanism for protein palmitoylation by DHHC enzymes. J Biol Chem. 2010;285:38104–38114. doi: 10.1074/jbc.M110.169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rowland AF, Fazakerley DJ, James DE. Mapping insulin/GLUT4 circuitry. Traffic. 2011;12:672–681. doi: 10.1111/j.1600-0854.2011.01178.x. [DOI] [PubMed] [Google Scholar]
- 31.Eisenberg S, Laude AJ, Beckett AJ, Mageean CJ, Aran V, Hernandez-Valladares M, Henis YI, Prior IA. The role of palmitoylation in regulating Ras localization and function. Biochemical Society transactions. 2013;41:79–83. doi: 10.1042/BST20120268. [DOI] [PubMed] [Google Scholar]
- 32.Baldwin AC, Green CD, Olson LK, Moxley MA, Corbett JA. A role for aberrant protein palmitoylation in FFA-induced ER stress and beta-cell death. Am J Physiol Endocrinol Metab. 2012;302:E1390–1398. doi: 10.1152/ajpendo.00519.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren W, Ulupi JS, Du K. Proteomic analysis of protein palmitoylation in adipocytes. Adipocyte. 2013;2:17–28. doi: 10.4161/adip.22117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pandey NR, Zhou X, Qin Z, Zaman T, Gomez-Smith M, Keyhanian K, Anisman H, Brunel JM, Stewart AF, Chen HH. The LIM domain only 4 protein is a metabolic responsive inhibitor of protein tyrosine phosphatase 1B that controls hypothalamic leptin signaling. J Neurosci. 2013;33:12647–12655. doi: 10.1523/JNEUROSCI.0746-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korycka J, Lach A, Heger E, Boguslawska DM, Wolny M, Toporkiewicz M, Augoff K, Korzeniewski J, Sikorski AF. Human DHHC proteins: a spotlight on the hidden player of palmitoylation. Eur J Cell Biol. 2012;91:107–117. doi: 10.1016/j.ejcb.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 36.Greaves J, Chamberlain LH. DHHC palmitoyl transferases: substrate interactions and (patho)physiology. Trends Biochem Sci. 2011;36:245–253. doi: 10.1016/j.tibs.2011.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.