Abstract
The development of ESR methods that measure long-range distance distributions has advanced biophysical research. However, the spin labels commonly employed are highly flexible, which leads to ambiguity in relating ESR measurements to protein-backbone structure. Herein we present the double-histidine (dHis) Cu2+-binding motif as a rigid spin probe for double electron–electron resonance (DEER) distance measurements. The spin label is assembled in situ from natural amino acid residues and a metal salt, requires no postexpression synthetic modification, and provides distance distributions that are dramatically narrower than those found with the commonly used protein spin label. Simple molecular modeling based on an X-ray crystal structure of an unlabeled protein led to a predicted most probable distance within 0.5 of the experimental value. Cu2+ DEER with the dHis motif shows great promise for the resolution of precise, unambiguous distance constraints that relate directly to protein-backbone structure and flexibility.
Keywords: copper, DEER, EPR spectroscopy, proteins, spin labeling
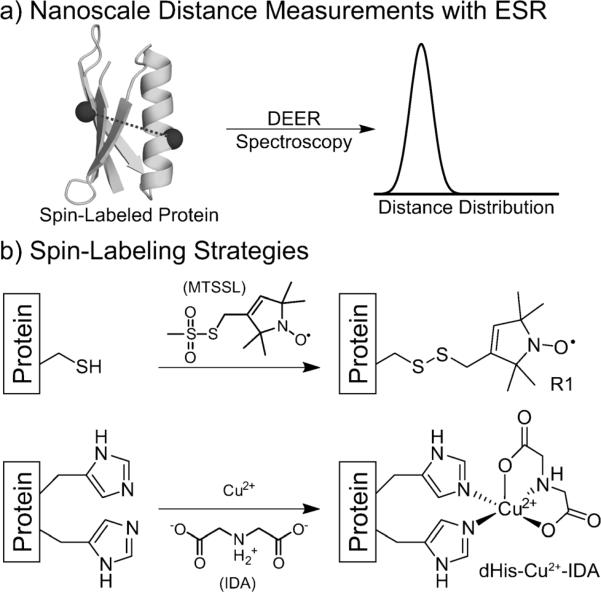
In recent years, the measurement of 1–10 nm distances between paramagnetic species in proteins by pulsed electron spin resonance (ESR) spectroscopic methods has greatly advanced biophysical research.[1] Most current applications of these methods rely on covalent modification of cysteine residues with thiol-reactive spin labels.[1a] The most commonly utilized spin-labeling reagent reacts to form the nitroxidefunctionalized residue R1 (Figure 1b), and the nitroxide is the reporter for ESR distance measurements. Site-directed spin labeling (SDSL), and the Cys-derived R1 residue in particular, has found widespread use in protein ESR as well as NMR spectroscopy.[2] Despite its promise in these applications, SDSL suffers from a significant limitation: R1 measurements are dominated by the conformational dynamics of its flexible side chain rather than local protein-backbone fluctuations.
Figure 1.
a) Overview of the DEER experiment. b) The mutations and resulting side chains of the most common spin label (R1) and the paramagnetic metal label reported herein (dHis–Cu2+–IDA). The widths of the resultant distance distributions are heavily influenced by the flexibility of the side chain.
Significant efforts to investigate R1 conformational preferences through X-ray crystallography[3] and computational techniques[4] have helped to address this limitation in part, though the accurate prediction of interspin distances in proteins remains challenging. Much work has also been devoted to the development of alternate protein spin labels that are more rigid than R1. Slight changes to the basic R1 residue through modification of the nitroxide ring[5] or its replacement with alternate heterocycles[6] can rigidify its structure. To date, the most promising label for proteins appears to be the bifunctional RX side chain, in which the nitroxide ring is covalently attached to two neighboring cysteine residues.[7] This added rigidity comes at the price of a more complex synthetic scheme necessary to introduce RX into expressed proteins. Thus, despite significant progress, there remains a need for a protein-spin-labeling method that provides narrow, readily interpretable distances and minimizes postexpression synthetic manipulations.
Paramagnetic metal centers can be used as an alternative to nitroxides for spin labeling biological systems.[8] These probes have been introduced into proteins by the use of both native metal-binding sites[9] and unnatural chelating side chains introduced by the SDSL-like modification of cysteine.[10] Natural metal-binding sites can provide excellent rigidity,[9b,c,10b,11] but can only be used when the system of interest natively binds metals. The best unnatural metal-chelating tag shows distance-distribution widths comparable to those of R1, thus suggesting similar flexibility.[12] Para-magnetic metals can alternatively be incorporated into a protein through the creation of artificial metal-binding sites.[11b,13] One such structure that has been utilized for a variety of applications is the double-histidine (dHis) motif (Figure 1b),[13a,14] in which two strategically placed His sides chains are used to chelate Cu2+. The dHis motif has been used to chelate Cu2+ in the context of an α-helix in T4-lysozyme to measure short-range, relaxation-based, average distances at room temperature.[14c]
Presented herein is the first use of the dHis–Cu2+ motif as a spin probe for distance measurements. We show that this readily introduced motif is ideal as a highly rigid spin label in both α-helix and β-sheet environments. Individual histidine residues contain only two rotatable bonds (as compared to five in R1), and the simultaneous coordination of Cu2+ by two histidine residues highly restricts the movement of the metal center relative to the protein backbone.
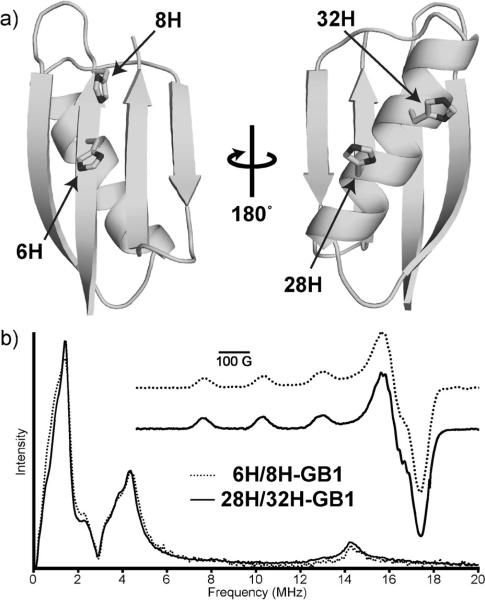
To investigate the use of the dHis–Cu2+ motif, we incorporated it into the B1 immunoglobulin-binding domain of protein G (GB1). Two Cu2+-binding dHis sites were introduced into GB1—one in the α-helix and one in the β-sheet. For the helix site, K28H and Q32H mutations were performed to yield 28H/32H-GB1. The i and i + 4 placement of the histidine residues accounts for the turn of the helix and produces a known metal-binding environment.[14a] In the sheet, I6H and N8H mutations were performed to yield 6H/8H-GB1. The i and i + 2 residue placement creates a dHis site by the placement of both histidines on the solvent-exposed face of the sheet. Unlike the dHis helix site, to the best of our knowledge, the β-sheet design has yet to be demonstrated experimentally. For Cu2+ DEER measurements, the helix and sheet mutations described above were combined into a single 6H/8H/28H/32H-GB1 variant. Finally, a 6C/28C double mutant of GB1 was created and spin-labeled to yield 6R1/28R1-GB1. Plasmid preparation, protein expression, purification, and SDSL were performed by standard methods (see the Supporting Information). Circular dichroism data (see the Supporting Information) indicated that all mutant proteins retained a native folded structure. Furthermore, we determined the crystal structure of 6H/8H/28H/32H-GB1 (Figure 2a), which showed a tertiary fold identical to that of wild-type GB1 (PDB: 4WH4).
Figure 2.
a) Crystal structure of 6H/8H/28H/32H-GB1 (PDB: 4WH4). b) ESEEM and CW (inset) data for each of the individual dHis sites complexed with Cu2+–IDA. All data are indicative of coordination by two histidine residues and IDA for both the α-helix (28H/32H) and β-sheet (6H/8H) sites.
One potential difficulty associated with the dHis motif is its relatively low Cu2+-binding affinity. Apparent dissociation constants ranging from 200 to 2 mm have been observed for various α-helix sites.[13b] At the Cu2+ concentrations necessary to fully populate these sites, dHis–Cu2+ binding can be accompanied by nonspecific binding of the metal elsewhere in the protein. Previous NMR spectroscopic studies have suggested the existence of a nonspecific Cu2+-binding site in GB1.[15] We validated this observation by ESR spectroscopy (see the Supporting Information). As a means to prevent nonspecific binding, Cu2+ was introduced to the dHis sites as a complex with iminodiacetate (IDA; Figure 1b).[13a] On the basis of a crystal structure of Cu2+ coordinated to two imidazole ligands and an IDA derivative,[16] the Cu2+–IDA complex was expected to have one equatorial nitrogen ligand, one equatorial oxygen ligand, and one axial oxygen ligand (Figure 1b). The IDA–Cu2+ complex retains two open binding sites that enable cis coordination by two histidine residues, thus promoting Cu2+ selectivity for the dHis sites. Electron spin echo envelope modulation (ESEEM) data of 28H/32HGB1 with Cu2+–IDA in D2O suggested that the sixth open site in the complex is occupied by solvent water (see the Supporting Information). Importantly, the use of Cu2+–IDA abolished nonspecific binding to wild-type GB1 at concentrations at which the dHis sites are occupied (see the Supporting Information).
To investigate the binding environment of the dHis–Cu2+ sites, we carried out continuous wave (CW) and ESEEM experiments on 6H/8H-GB1 (β-sheet site) and 28H/32H-GB1 (α-helix site) in the presence of Cu2+–IDA (Figure 2b). CW spectra are sensitive to atoms coordinated equatorially to the Cu2+ center. The remarkably similar spectra and corresponding fits (see the Supporting Information) suggest a consistent binding environment involving three nitrogen atoms and one oxygen atom (see Figure 1b)[17] for both sites. The normalized Cu2+ ESEEM spectrum for each dHis mutant (Figure 2b) displayed features common for histidine coordination,[18] and the similarity of the spectra suggests similar binding environments for the two sites. In particular, the appearance of the slight feature at approximately 8 MHz suggests multiple histidine coordination.[19] Additionally, the ratio of the nuclear-quadrupole-interaction (ca. 2 MHz) feature and the double-quantum (ca. 4 MHz) feature is similar in both complexes, thus indicating that the number of histidine residues that coordinate to Cu2+ is the same in the two mutants.[19a,20] The ratio also matches well with comparable data for a model complex containing two histidine residues (see the Supporting Information).[20b] Taken together, the ESEEM data and the CW data support the assembly of the binding environment as shown in Figure 1b for both sites.
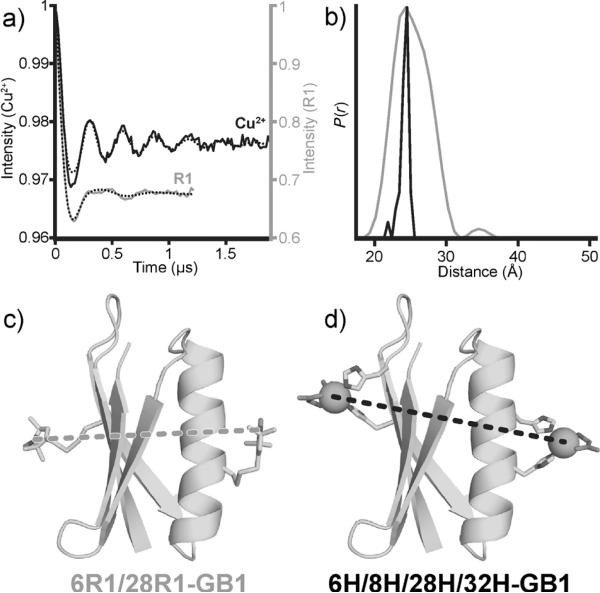
Cu2+ and nitroxide DEER experiments were performed on 6H/8H/28H/32H-GB1 and 6R1/28R1-GB1, respectively. For the Cu2+ measurement, orientational effects were probed but found to be negligible (see the Supporting Information). Inspection of the baseline-subtracted DEER signals and the resultant distance distributions (Figure 3a,b) enables direct comparison of the behavior of the two different spin labels. The modulation depths of the DEER measurements were 2.2% for Cu2+, and 33% for R1. Nevertheless, a good signal-to-noise ratio was achieved in a reasonable collection time (ca. 16 h) for the Cu2+ measurement. A dramatic increase in the number of modulation periods was observed for dHis–Cu2+ relative to the use of R1 (Figure 3a), thus suggesting a much narrower distance distribution in the former case. The data were analyzed with Tikhonov regularization[21] (Figure 3b), which revealed a distance-distribution width (between 16 and 84% probability) for dHis that was remarkably narrower (1.0Å) than that found for the analogous R1 sample (5.2Å). This narrow distribution suggests the bound Cu2+ centers are highly localized in space, thus providing a very precise distance measurement.
Figure 3.
a) Baseline-subtracted time-domain DEER data for both the R1 (gray) and the Cu2+ samples (black) as well as the best fit (dotted black lines) from Tikhonov regularization; modulation depths have been scaled for comparison. b) The distance distributions for each sample. c,d) Structural models of 6R1/28R1-GB1 and 6H/8H/28H/32H-GB1 complexed with Cu2+–IDA with measured interspin distances.
We rationalized the interspin distances observed in the DEER experiments by a combination of X-ray crystallography and molecular modeling. For the nitroxide sample, we modeled R1-modified Cys residues into a published crystal structure of wild-type GB1; rotamers were based on a previous crystal structure (Figure 3c).[3e] The observed Cu2+– Cu2+ distance was interpreted on the basis of the X-ray crystal structure of 6H/8H/28H/32H-GB1 crystallized in the absence of copper (Figure 2a). All efforts to derivatize these crystals with copper were unsuccessful; however, force-field minimization at both the helix and sheet sites provided similar histidine rotamers and coordination geometries to those observed in a previously reported crystal structure of Cu2+ bound to an IDA derivative and two imidazole ligands.[16] The predicted Cu2+–Cu2+ distance in this model (Figure 3d) was 25 Å, in excellent agreement with the measured most probable distance by DEER of 24.5 Å. The ease with which this distance was precisely predicated is remarkable. Despite significant efforts, it is still difficult to predict R1-based distances by modeling techniques.[3e,4b,d,22]
The results reported herein on dHis–Cu2+ as a protein spin label for DEER are noteworthy in light of the many successful applications of the bifunctional RX label.[23] Although RX shows greatly increased rigidity as compared to that of R1, our findings suggest dHis–Cu2+ is superior in terms of the width of distance distributions and the simplicity of structural interpretation. In the first use of RX, distribution widths of 2.8 and 2.0 Å were reported in two different model proteins, in which analogous R1 measurements displayed widths of 6.5 and 4.7 Å respectively.[7] Additionally, one of the distances was modeled by using a crystal structure of the RX-modified protein, and the predicted distance was 1.4 Å longer than the DEER distance.[7] The distance distribution between the two dHis–Cu2+ sites in this study is considerably narrower (1.0 Å), and the predicted interspin distance was within 0.5 Å of the experiment value. Overall, the dHis–Cu2+–IDA motif displays distinct advantages over RX as a highly precise, site-specific spin probe. These advantages are multiplied by the operational simplicity of introducing multiple dHis sites in a single chain and modifying those sites by the addition of a metal salt and ligand to the solution prior to ESR spectroscopic measurement.
In summary, we have shown herein that the dHis–Cu2+–IDA motif is a highly rigid, site-specific spin label useful for ESR distance measurements in proteins. The label can be incorporated in both α-helix and β-sheet contexts by the use of natural amino acids and requires no postexpression synthetic manipulations. The distance distribution observed by DEER between dHis sites in a small protein is significantly narrower than that observed for an analogous mutant with R1 labels. The most probable distances are readily interpreted through simple modeling. Together, these results demonstrate that Cu2+ DEER with dHis motifs is a simple yet highly effective means of overcoming the inherent limitations of the commonly used R1 label. Furthermore, this histidine-dependent motif is an ideal alternative in proteins in which cysteine incorporation and/or labeling is problematic. Given that dHis coordination greatly restricts the motion of the Cu2+ ion, the resultant distance measurements can provide structural constraints that can be more readily related to backbone flexibility. With further development, it may be possible to elucidate Cα–Cα distance distributions from these measurements. Such Cu2+ DEER measurements will also be important for NMR spectroscopic structure determination based on paramagnetic relaxation enhancement,[24] and could possibly be combined with crystallography and/or NMR spectroscopy for the accurate structure determination of very large, multicomponent biological systems.[25] Finally, this labeling may be also be suitable for in-cell metal–metal ESR measurements[26] or the triangulation of locations of paramagnetic-metal binding.[27]
Supplementary Material
Footnotes
This research was supported by a grant from the National Science Foundation (MCB-1157712 to S.S.), and the Bruker E680 instrument used was purchased with funds from the National Institutes of Health (grant 1S10RR028701).
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.201501968.
References
- 1.a Hubbell W, López C, Altenbach C, Yang Z. Curr. Opin. Struct. Biol. 2013;23:725–733. doi: 10.1016/j.sbi.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Jeschke G. Annu. Rev. Phys. Chem. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 2.Jeschke G. Prog. Nucl. Magn. Reson. Spectrosc. 2013;72:42–60. doi: 10.1016/j.pnmrs.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 3.a Langen R, Oh KJ, Cascio D, Hubbell WL. Biochemistry. 2000;39:8396–8405. doi: 10.1021/bi000604f. [DOI] [PubMed] [Google Scholar]; b Fleissner MR, Cascio D, Hubbell WL. Protein Sci. 2009;18:893–908. doi: 10.1002/pro.96. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Kroncke BM, Horanyi PS, Columbus L. Biochemistry. 2010;49:10045–10060. doi: 10.1021/bi101148w. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Freed DM, Khan AK, Horanyi PS, Cafiso DS. Biochemistry. 2011;50:8792–8803. doi: 10.1021/bi200971x. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Cunningham TF, McGoff MS, Sengupta I, Jaroniec CP, Horne WS, Saxena S. Biochemistry. 2012;51:6350–6359. doi: 10.1021/bi300328w. [DOI] [PubMed] [Google Scholar]
- 4.a Fajer MI, Li H, Yang W, Fajer PG. J. Am. Chem. Soc. 2007;129:13840–13846. doi: 10.1021/ja071404v. [DOI] [PubMed] [Google Scholar]; b Polyhach Y, Bordignon E, Jeschke G. Phys. Chem. Chem. Phys. 2011;13:2356–2366. doi: 10.1039/c0cp01865a. [DOI] [PubMed] [Google Scholar]; c Hatmal MM, Li Y, Hegde BG, Hegde PB, Jao CC, Langen R, Haworth IS. Biopolymers. 2012;97:35–44. doi: 10.1002/bip.21699. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Sarver J, Townsend J, Rajapakse G, Jen-Jacobson L, Saxena S. J. Phys. Chem. B. 2012;116:4024–4033. doi: 10.1021/jp211094n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a Columbus L, Kalai T, Jeko J, Hideg K, Hubbell WL. Biochemistry. 2001;40:3828–3846. doi: 10.1021/bi002645h. [DOI] [PubMed] [Google Scholar]; b Fawzi NL, Fleissner MR, Anthis NJ, Kalai T, Hideg K, Hubbell WL, Clore GM. J. Biomol. NMR. 2011;51:105–114. doi: 10.1007/s10858-011-9545-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toledo Warshaviak D, Khramtsov VV, Cascio D, Alten-bach C, Hubbell WL. J. Magn. Reson. 2013;232:53–61. doi: 10.1016/j.jmr.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleissner MR, Bridges MD, Brooks EK, Cascio D, Kalai T, Hideg K, Hubbell WL. Proc. Natl. Acad. Sci. USA. 2011;108:16241–16246. doi: 10.1073/pnas.1111420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a Narr E, Godt A, Jeschke G. Angew. Chem. Int. Ed. 2002;41:3907–3910. doi: 10.1002/1521-3773(20021018)41:20<3907::AID-ANIE3907>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2002;114:4063–4066. [Google Scholar]; b Becker JS, Saxena S. Chem. Phys. Lett. 2005;414:248–252. [Google Scholar]; c Yang Z, Becker J, Saxena S. J. Magn. Reson. 2007;188:337–343. doi: 10.1016/j.jmr.2007.08.006. [DOI] [PubMed] [Google Scholar]; d Bode BE, Plackmeyer J, Prisner TF, Schiemann O. J. Phys. Chem. A. 2008;112:5064–5073. doi: 10.1021/jp710504k. [DOI] [PubMed] [Google Scholar]; e Yang Z, Ji M, Saxena S. Appl. Magn. Reson. 2010;39:487–500. [Google Scholar]
- 9.a van Amsterdam IM, Ubbink M, Canters GW, Huber M. Angew. Chem. Int. Ed. 2003;42:62–64. [PubMed] [Google Scholar]; Angew. Chem. 2003;115:64–67. [Google Scholar]; b Merz GE, Borbat PP, Pratt AJ, Getzoff ED, Freed JH, Crane BR. Biophys. J. 2014;107:1669–1674. doi: 10.1016/j.bpj.2014.07.068. [DOI] [PMC free article] [PubMed] [Google Scholar]; c van Wonderen JH, Kostrz DN, Dennison C, MacMillan F. Angew. Chem. Int. Ed. 2013;52:1990–1993. doi: 10.1002/anie.201208166. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2013;125:2044–2047. [Google Scholar]
- 10.a Goldfarb D. In: Structural Information from Spin-Labels and Intrinsic Paramagnetic Centres in the Biosciences. Timmel CR, Harmer JR, editors. Springer; Berlin: 2012. pp. 163–204. [Google Scholar]; b Ji M, Ruthstein S, Saxena S. Acc. Chem. Res. 2014;47:688–695. doi: 10.1021/ar400245z. [DOI] [PubMed] [Google Scholar]; c Goldfarb D. Phys. Chem. Chem. Phys. 2014;16:9685–9699. doi: 10.1039/c3cp53822b. [DOI] [PubMed] [Google Scholar]
- 11.a Yang Z, Kurpiewski MR, Ji M, Townsend JE, Mehta P, Jen-Jacobson L, Saxena S. Proc. Natl. Acad. Sci. USA. 2012;109:E993–1000. doi: 10.1073/pnas.1200733109. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Yang Z, Jiménez-Osés G, López CJ, Bridges MD, Houk KN, Hubbell WL. J. Am. Chem. Soc. 2014;136:15356–15365. doi: 10.1021/ja5083206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cunningham TF, Shannon MD, Putterman MR, Arachchige R, Sengupta I, Gao M, Jaroniec CP, Saxena S. J. Phys. Chem. B. 2014;119:2839–2843. doi: 10.1021/jp5103143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a Arnold FH, Haymore BL. Science. 1991;252:1796–1797. doi: 10.1126/science.1648261. [DOI] [PubMed] [Google Scholar]; b Regan L. Annu. Rev. Biophys. Biomol. Struct. 1993;22:257–287. doi: 10.1146/annurev.bb.22.060193.001353. [DOI] [PubMed] [Google Scholar]
- 14.a Todd RJ, Van Dam ME, Casimiro D, Haymore BL, Arnold FH. Proteins Struct. Funct. Genet. 1991;10:156–161. doi: 10.1002/prot.340100209. [DOI] [PubMed] [Google Scholar]; b Higaki JN, Fletterick RJ, Craik CS. Trends Biochem. Sci. 1992;17:100–104. doi: 10.1016/0968-0004(92)90245-5. [DOI] [PubMed] [Google Scholar]; c Voss J, Salwinski L, Kaback HR, Hubbell WL. Proc. Natl. Acad. Sci. USA. 1995;92:12295–12299. doi: 10.1073/pnas.92.26.12295. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Jung K, Voss J, He M, Hubbell WL, Kaback HR. Biochemistry. 1995;34:6272–6277. doi: 10.1021/bi00019a003. [DOI] [PubMed] [Google Scholar]; e Nicoll AJ, Miller DJ, Futterer K, Ravelli R, Allemann RK. J. Am. Chem. Soc. 2006;128:9187–9193. doi: 10.1021/ja061513u. [DOI] [PubMed] [Google Scholar]
- 15.Nadaud PS, Sengupta I, Helmus JJ, Jaroniec CP, Biomol J. NMR. 2011;51:293–302. doi: 10.1007/s10858-011-9536-y. [DOI] [PubMed] [Google Scholar]
- 16.Brandi-Blanco MP, de Benavides-Gim nez MM, Gonz lez-P rez JM, Choquesillo-Lazarte D. Acta Crystallogr. Sect. E. 2007;63:m1678–m1679. [Google Scholar]
- 17.Peisach J, Blumberg WE. Arch. Biochem. Biophys. 1974;165:691–708. doi: 10.1016/0003-9861(74)90298-7. [DOI] [PubMed] [Google Scholar]
- 18.Mims WB, Peisach J. J. Chem. Phys. 1978;69:4921–4930. [Google Scholar]
- 19.a Goldfarb D, Fauth J, Tor Y, Shanzer A. J. Am. Chem. Soc. 1991;113:1941–1948. [Google Scholar]; b Hernández-Guzmán J, Sun L, Mehta AK, Dong J, Lynn DG, Warncke K. ChemBioChem. 2013;14:1762–1771. doi: 10.1002/cbic.201300236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.a McCracken J, Pember S, Benkovic SJ. J. Am. Chem. Soc. 1988;110:1069. [Google Scholar]; b Silva KI, Michael BC, Geib SJ, Saxena S. J. Phys. Chem. B. 2014;118:8935–8944. doi: 10.1021/jp500767n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeschke G, Chechik V, Ionita P, Godt A, Zimmermann H, Banham J, Timmel CR, Hilger D, Jung H. Appl. Magn. Reson. 2006;30:473–498. [Google Scholar]
- 22.a Jeschke G, Bender A, Schweikardt T, Panek G, Decker H, Paulsen H. J. Biol. Chem. 2005;280:18623–18630. doi: 10.1074/jbc.M501171200. [DOI] [PubMed] [Google Scholar]; b Finiguerra MG, Prudencio M, Ubbink M, Huber M. Magn. Reson. Chem. 2008;46:1096–1101. doi: 10.1002/mrc.2290. [DOI] [PubMed] [Google Scholar]; c Swanson MA, Kathirvelu V, Majtan T, Frerman FE, Eaton GR, Eaton SS. Protein Sci. 2011;20:610–620. doi: 10.1002/pro.595. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Klose D, Klare JP, Grohmann D, Kay CW, Werner F, Steinhoff HJ. PLoS One. 2012;7:e39492. doi: 10.1371/journal.pone.0039492. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Hagelueken G, Ward R, Naismith JH, Schiemann O. Appl. Magn. Reson. 2012;42:377–391. doi: 10.1007/s00723-012-0314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; f Roux B, Islam SM. J. Phys. Chem. B. 2013;117:4733–4739. doi: 10.1021/jp3110369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.a Vanea E, Gruian C, Rickert C, Steinhoff HJ, Simon V. Biomacromolecules. 2013;14:2582–2592. doi: 10.1021/bm4003893. [DOI] [PubMed] [Google Scholar]; b Sahu ID, McCarrick RM, Troxel KR, Zhang R, Smith HJ, Dunagan MM, Swartz MS, Rajan PV, Kroncke BM, Sanders CR, Lorigan GA. Biochemistry. 2013;52:6627–6632. doi: 10.1021/bi4009984. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Bowman A, Hammond CM, Stirling A, Ward R, Shang W, El-Mkami H, Robinson DA, Svergun DI, Norman DG, Owen-Hughes T. Nucleic Acids Res. 2014;42:6038–6051. doi: 10.1093/nar/gku232. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Vendome J, Felsovalyi K, Song H, Yang Z, Jin X, Brasch J, Harrison OJ, Ahlsen G, Bahna F, Kaczynska A, Katsamba PS, Edmond D, Hubbell WL, Shapiro L, Honig B. Proc. Natl. Acad. Sci. USA. 2014;111:E4175–4184. doi: 10.1073/pnas.1416737111. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Sahu ID, Kroncke BM, Zhang R, Dunagan MM, Smith HJ, Craig A, McCarrick RM, Sanders CR, Lorigan GA. Biochemistry. 2014;53:6392–6401. doi: 10.1021/bi500943p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sengupta I, Nadaud PS, Helmus JJ, Schwieters CD, Jaroniec CP. Nat. Chem. 2012;4:410–417. doi: 10.1038/nchem.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.a Park SY, Borbat PP, Gonzalez-Bonet G, Bhatnagar J, Pollard AM, Freed JH, Bilwes AM, Crane BR. Nat. Struct. Mol. Biol. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]; b Duss O, Yulikov M, Jeschke G, Allain FH. Nat. Commun. 2014;5:3669. doi: 10.1038/ncomms4669. [DOI] [PubMed] [Google Scholar]; c Duss O, Michel E, Yulikov M, Schubert M, Jeschke G, Allain FH. Nature. 2014;509:588–592. doi: 10.1038/nature13271. [DOI] [PubMed] [Google Scholar]
- 26.Qi M, Gross A, Jeschke G, Godt A, Drescher M. J. Am. Chem. Soc. 2014;136:15366–15378. doi: 10.1021/ja508274d. [DOI] [PubMed] [Google Scholar]
- 27.Abdullin D, Florin N, Hagelueken G, Schiemann O. Angew. Chem. Int. Ed. 2015;54:1827–1831. doi: 10.1002/anie.201410396. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2015;127:1847–1851. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.