Abstract
Nonsense mutations across the whole coding sequence of Syne1/ Nesprin1 have been linked to Autosomal Recessive Cerebellar Ataxia Type I (ARCA1). However, nothing is known about the molecular etiology of this late-onset debilitating pathology. In this work, we report that Nesprin1 giant is specifically expressed in CNS tissues. We also identified a CNS-specific splicing event that leads to the abundant expression of a KASH-LESS variant of Nesprin1giant (KLNes1g) in the cerebellum. KLNes1g displayed a noncanonical localization at glomeruli of cerebellar mossy fibers whereas Nesprin2 exclusively decorated the nuclear envelope of all cerebellar neurons. In immunogold electron microscopy, KLNes1g colocalized both with synaptic vesicles within mossy fibers and with dendritic membranes of cerebellar granule neurons. We further identified vesicle- and membrane-associated proteins in KLNes1g immunoprecipitates. Together, our results suggest that the loss of function of KLNes1g resulting from Nesprin1 nonsense mutations underlie the molecular etiology of ARCA1.
Keywords: ARCA1, Autosomal recessive cerebellar ataxia TypeI, cerebellar granule neuron, cerebellar mossy fiber, Cerebellum, KASH, KLNes1g, Nesprin, Spinocerebellar ataxia
INTRODUCTION
Autosomal recessive cerebellar ataxia type I (ARCA1) has been linked to nonsense mutations scattered along the gene encoding Syne1 (Synaptic nuclear envelope 1) also known as Nesprin1 (Nuclear envelope spectrin 1) (Figure 1A). These mutations were initially reported in populations from the Quebec region of Beauce and more recently in French, Japanese and Brazilian patients (Dupre et al., 1993; Gros-Louis et al., 2007; Izumi et al., 2013; Noreau et al., 2013). This late onset pathology, which belongs to the complex spectrum of hereditary ataxias (Mancuso et al., 2014), is characterized by cerebellar dysarthria, limb and gait ataxia and diffuse cerebellar atrophy. Nothing is currently known about the molecular etiology of that pathology.
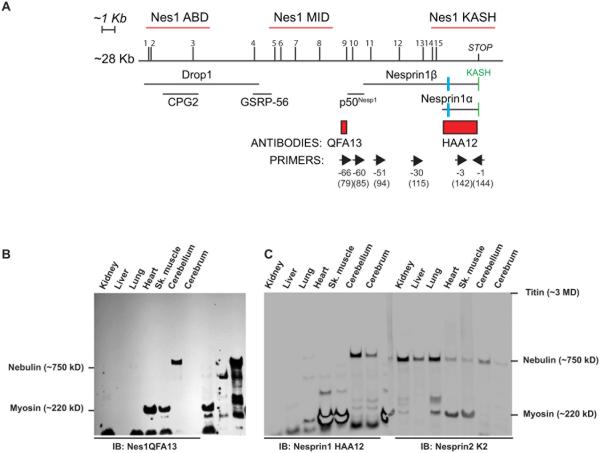
Figure 1.
A) Localization of unique ARCA1 mutations (1 to 15), hybridization probes (red lines), antibody epitopes (red rectangles), floxed exon in ActinCreNesprin1Δ/Δ mice (blue rectangle), primers (forward and reverse arrows above corresponding exon number) and known Nesprin1 isoforms relative to Nesprin1 giant transcript XM_006512463.1 used as a reference in this work. The reverse exon numbering system used in (Zhang et al., 2009a) that simplifies exon labeling was adopted in this work. The classical numbering of corresponding exons of reference transcript XM_006512463.1 is also indicated between brackets. B, C) Autoradiogram of mouse tissue lysates (40µg) immunobloted with Nesprin1 QFA13 (B). Note the high molecular weight band expressed in the cerebellum. A single membrane with duplicate loading of the same tissues was immunobloted with Nesprin1 HAA12 (C, left panel) and Nesprin2K2 (C, right panel) and imaged quantitatively. Note the CNS-specific expression pattern of Nesprin1giant by comparison to Nesprin2 giant. Molecular weight markers correspond to large proteins abundantly expressed skeletal muscle used as molecular weight rulers (Fig.S2).
Human and mouse Nesprin1 are exceptionally large genes composed of more than 140 exons assembled within a predicted ~28kb transcript (Zhang et al., 2002; Zhang et al., 2001) encoding a large protein called Nesprin1 giant (1MDa). Nesprin2 giant (~750kDa), which is encoded by a distinct gene, displays the same structural organization than Nesprin1giant (Fig.S1) (Rajgor et al., 2012; Zhen et al., 2002). The latter consists of an N-terminal calponin homology domain that binds to actin, a main core flanked by multiple spectrin repeats and a C-terminal KASH (Klarsicht/Anc1/Syne1 homology) domain. This domain (~ 60 amino acids) includes a transmembrane domain followed by a polypeptide protruding within the perinuclear space that separates the inner from the outer membrane of the NE. This is where the KASH domain directly interacts with the SUN domain of Sun proteins, a family of transmembrane proteins residing within the inner nuclear membrane (Fig.S1) (Sosa et al., 2012; Zhou et al., 2012). As such, SUN/KASH interactions within the perinuclear space mediate the formation of macromolecular assemblies called LINC (Linkers or the Nucleoskeleton to the Cytoskeleton) complexes (Crisp et al., 2006; Padmakumar et al., 2005) that span the nuclear envelope and underlie nuclear migration and anchorage within developing tissues and syncytia (Lombardi et al., 2011; Luxton and Starr, 2014; Razafsky and Hodzic, 2009; Starr and Fischer, 2005).
Nesprin1 also encodes smaller C-terminal isoforms such as Nesprin1α (120kDa) and Nesprin1β (350kDa) that interact with Sun proteins via their KASH domain (Apel et al., 2000; Padmakumar et al., 2004; Rajgor et al., 2012; Zhang et al., 2001). Additional N-terminal isoforms such as Drop1, CPG2, GSRP-56 and p50Nesp1 have also been identified (Fig.1A)(Cottrell et al., 2004; Kobayashi et al., 2006; Marme et al., 2008; Rajgor et al., 2012; Rajgor et al., 2014) .
ARCA1 mutations are scattered across the whole coding sequence of Nesprin1 and underlie a very homogenous set of cerebellar clinical symptoms. Because all ARCA1 nonsense mutations are predicted to affect Nesprin1giant (Fig.1A), we hypothesized that this isoform exerts essential cerebellum-specific functions that may not be compensated by Nesprin2giant, its structural homolog. In agreement with these hypotheses, results presented in this work uncovered a KASH-LESS variant of Nesprin1giant we named KLNes1g. This variant is specifically expressed in the CNS and most abundant in the cerebellum where it may be involved in vesicular trafficking and/or in dendritic membranes structural organization.
RESULTS
Nesprin1 giant is specifically expressed in the CNS
We first sought to unequivocally detect and examine the tissue distribution of Nesprin1giant. Classical SDS-PAGE does not allow for the efficient resolution and transfer onto membranes of such a high molecular weight protein. We turned to vertical agarose gel electrophoresis that palliate to these issues (Warren et al., 2003) . Two antisera raised against distant epitopes located either on the C-terminal side (Nes1HAA12) or in the middle region (Nes1QFA13) of Nesprin1 giant were used in this study (Fig.1A, see Material and Methods). Nes1QFA13 detected a large protein specifically in the cerebellum (Fig.1B) and Nes1HAA12 detected the same molecular weight protein in the cerebellum and, to a lesser extent, the cerebrum (Fig.1C, left). The molecular weight of that immunoreactive band was estimated at 980 kDa by comparison to molecular weights “rulers” provided by titin, nebulin and myosin heavy chain that are abundantly expressed in skeletal muscle lysates (Fig.S2). This molecular weight corresponds to the predicted molecular weight of the giant isoform of Nesprin1 (~1MDa). Immunoblotting with Nesprin2 K2, whose epitope is located on the C-terminal side of Nesprin2 (Khatau et al., 2012), detected a 750 kDa protein that corresponds to the predicted molecular weight of Nesprin2 giant (Fig.1C, right panel). Strikingly, by contrast to Nesprin1 giant that was specifically detected in CNS tissues (cerebellum, cerebrum and retina, data not shown), Nesprin2 giant was ubiquitously expressed.
Nesprin1 transcripts are abundantly expressed in mouse cerebellum
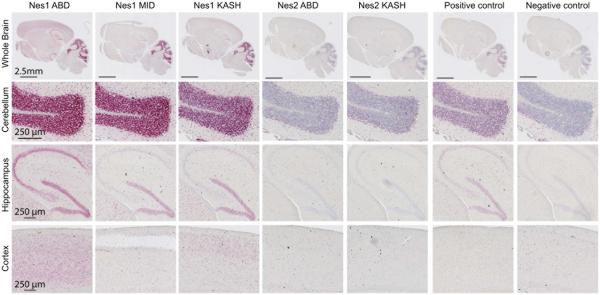
We next examined the relative distribution and expression levels of Nesprin1 and Nesprin2 transcripts in mouse brain. In Situ hybridization (ISH) was performed on whole consecutive brain slices from adult C57/Bl6 mice. Because of the large size of Nesprin1 (~28 Kb, XM_006512463.1) and Nesprin2 (~21 Kb, NM_001005510.2) transcripts, we used probes covering ~5 Kb of either the N-terminal (ABD) or C-terminal (KASH) ends of Nesprin1 and Nesprin2. An additional central 5kb probe was used for Nesprin1 (Nes1MID) (Fig.1A). As shown in Figure 2, Nesprin1 transcripts were most abundant in the cerebellar granule cell layer (GCL). The ISH signal was attributable to cerebellar granule neurons (CGN) that overwhelmingly populate the GCL. By contrast, Nesprin2 transcripts expression was significantly lower in the GCL. Similar hybridization patterns observed in the GCL with Nes1ABD, Nes1MID or Nes1KASH probes was consistent with the expression of large Nesprin1 transcripts encoding the giant isoform in the GCL.
Figure 2.
Localization and relative expression level of Nesprin1 and Nesprin2 transcripts in mouse brain. In situ hybridization (RNAscope, see Material and Methods) with indicated probes (see Fig.1A) was performed on consecutive adult brain slices of C57Bl\6 mice. Upper panels: large scans of whole brain slices. Lower panels: zoomed in pictures of indicated brain structures.
In the hippocampus, Nesprin1 hybridization patterns were rather probe-specific; Nes1ABD probes, which overlap with Drop1 and CPG2 (Fig.1A), labelled the hippocampal formation and dentate gyrus with comparable intensities whereas Nes1MID and Nes1KASH probes preferentially labeled the dentate gyrus. Nes2ABD and Nes2KASH probes marginally hybridized in the hippocampus.
In the cerebral cortex, Nes1ABD probes homogenously labeled all cortical layers whereas Nes1MID and Nes1KASH probes emphasized more discrete cortical layers. Nes2 probes displayed weaker and more diffuse hybridization signals across the cerebral cortex. Together, these results indicate that the mouse brain displays significant difference in abundance and relative distribution of Nesprin1 and Nesprin2 transcripts.
Immunolocalization of Nesprin1 and Nesprin2 in the cerebellum
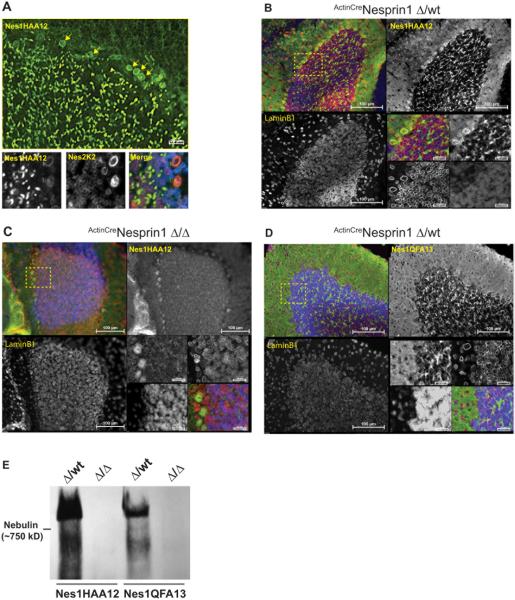
In immunofluorescence microscopy, Nes1HAA12 detected nuclear rims corresponding to the NE of Purkinje cells (PCs) (Fig.3A, upper panel). However, despite the abundant expression of Nesprin1 transcripts in the GCL (Fig.2), we could not detect any nuclear rims in CGN. Instead, we observed a non-canonical distribution of Nesprin1 immunoreactivity in the form of bright and large (~5µm) speckles scattered across the whole GCL. Nesprin2 also localized at the NE of PCs. However, by contrast to Nesprin1, Nesprin2 exclusively displayed a canonical localization at the NE of CGN (Fig.3A, lower panels). To confirm the specificity of noncanonical Nesprin1 immunoreactivity in GCL speckles, we repeated the same experiment on P15 cerebellar slices from ActinCreNes1Δ/Δ mice (see Material and Methods) in which exon-16 is floxed (Zhang et al., 2009a) (Fig.1A). Nesprin1 immunoreactivity was readily detected at the NE of PCs and in GCL speckles from Actin-CreNes1Δ/wt cerebella (Fig.3B) but not in cerebellar slices from ActinCreNes1Δ/Δ littermates (Fig.3C). Consistent with the expression of Nesprin1giant in GCL speckles, Nes1QFA13 also recognized GCL speckles (Fig.3D). Furthermore, Nesprin1 giant was undetectable in cerebellar lysates from ActinCreNes1Δ/Δ mice immunoblotted with Nes1HAA12 or Nes1QFA13 (Fig.3E). Together, these results implied that the noncanonical Nesprin1 immunoreactivity in GCL speckles corresponds to Nesprin1giant.
Figure 3.
Noncanonical localization of Nesprin1 in the GCL. A) Main panel: Representative immunofluorescence pattern of adult C57/Bl6 cerebellar slices processed with Nes1HAA12. Note the strong Nesprin1 immunoreactive “speckles” within the GCL in addition to Nesprin1 rims at the NE of PCs (arrows). Scale bar: 20 µm. Lower panels: Close up view of the PC-CGN interface colabeled with mouse Nes1HAA12 and rabbit Nes2K2 antibodies. Scale bars: 10 µm. B, C) Nes1HAA12 immunoreactivity of P15 cerebellar slices isolated either from ActinCreNes1Δ/wt mice (B) or from ActinCreNes1Δ/Δ mice (C). LaminB1 was used as a control to ensure epitope accessibility. Note the absence of Nesprin1 speckles and NE rims within the GCL and PCs of ActinCreNes1Δ/Δ cerebellum, respectively. Scale Bars: 100 µm and 20 µm (insets). D) Representative immunofluorescence pattern of P15 cerebellar slices from ActinCreNes1Δ/wt mice processed with Nes1QFA13. Note that whereas Nes1QFA13 recognizes GCL speckles, it does not detect any NE signal in PCs. Scale Bars: 100 µm and 20 µm (insets). E) Nes1HAA12 and Nes1QFA13 immunoblots of cerebellar lysates prepared either from P15 ActinCreNes1Δ/wt or from ActinCreNes1Δ/Δ mice. Note the lack of any detectable expression of Nesprin1 giant in ActinCreNes1Δ/Δ samples in agreement with the lack of speckles detection in the corresponding cerebellar slices (C).
The cerebellum expresses a significant pool of Nesprin1 giant devoid of KASH domain
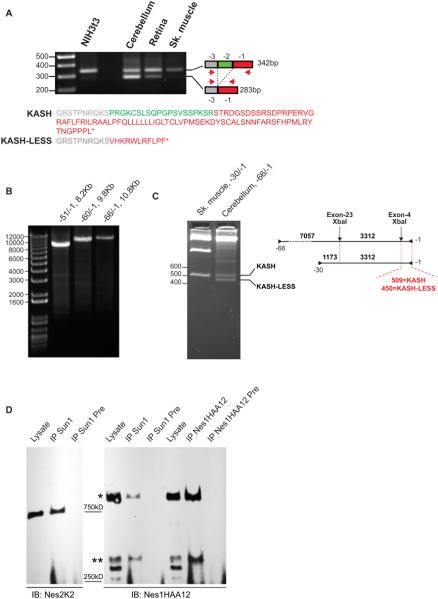
The non-canonical localization of Nesprin1 giant within GCL speckles was intriguing since the KASH domain dictates the localization of Nesprins at the NE (Crisp et al., 2006; Padmakumar et al., 2005; Stewart-Hutchinson et al., 2008). However, Nesprin1 transcripts lacking the penultimate exon (called exon-2 hereafter) predict the synthesis of Nesprin1 variants devoid of KASH domains (Rajgor et al., 2012; Razafsky et al., 2013; Zhang et al., 2009a). Interestingly, this splicing preferentially occurs in CNS tissues and is especially frequent in the cerebellum (Fig. 4A and (Razafsky et al., 2013)). As a result, the KASH domain is swapped for a short stretch of 11 amino acids followed by an early termination STOP codon (Fig.4A). We therefore examined the splicing status of exon-2 in large Nesprin1 transcripts isolated from cerebellum. As shown in Figure 4B, long Nesprin1 transcripts could be amplified using a common reverse primer from exon-1 combined with forward primers annealing to exon −51, −60 and −66 (Fig.1A). Because − 66/−1 amplicons (10.8kb) include the coding sequence of Nes1QFA13 and Nes1HAA12 epitopes (Fig.1A), they must originate from transcripts encoding Nesprin1 giant. To probe the splicing status of exon-2 within −66/−1 amplicons, we took advantage of an XbaI restriction site located within exon-4. As shown in Figure 4C, XbaI restriction products indicated that exon-2 was spliced in about 50% of transcripts encoding Nesprin1 giant in the cerebellum. As predicted by the lack of exon-2 splicing in muscle tissues (Fig.4A), an XbaI control digest performed on −30/−1 amplicons from muscle tissues only generated a single restriction band of 509 bp confirming that muscle tissues only express KASH-containing isoforms of Nesprin1(Fig.4C). Similar results were obtained by performing BssSI restriction digests (Fig.S3). These results show that a significant pool of cerebellar transcripts encode a variant of Nesprin1 giant devoid of KASH domain. More importantly, these results provide a mechanistic explanation as to how the bulk of Nesprin1 giant immunoreactivity observed in the GCL does not localize at the NE of CGN. Hereafter, the KASH-LESS variant of Nesprin1 giant is called KLNes1g.
Figure 4.
A) KLNes1g is synthesized in the CNS through the alternative splicing of exon-2 of Nesprin1 giant transcripts. RT-PCR on total RNA from indicated tissues with primers −1 and −3 emphasizes the abundant splicing of exon-2 in the cerebellum. Sizes are in bp. The lower panel shows the translation of PCR products. Colors refer to encoding exons. B) Long RT-PCR amplification of Nesprin1 cDNA from total cerebellum RNA carried out with primers pairs depicted in Fig.1A. Sizes are in bp. C) Large cerebellar Nesprin1 transcripts encode KASH-LESS variants. XbaI digestion of cerebellar amplicons −66/−1 reveals the splicing of exon-2 through the generation of a 450 bp restriction fragment on 2.5% agarose gels. In agreement with the lack of exon-2 splicing in skeletal muscle (A), control XbaI digest of −30/−1 amplicon from skeletal muscle generates a single 509bp fragment. Right: depiction of XbaI restriction sites within RT-PCR amplicons. Sizes are in bp. D) A significant pool of Nesprin1giant is not immunoprecipitated by Sun1 (see text for details). Cerebellar lysates were immunoprecipitated with the indicate antibodies and their respective preimmune sera (Pre). *: Nesprin1 giant; **: 350kDa band that most likely corresponds to Nesprin1β.
Distinguishing KASH-containing Nesprin1 giant from KLNes1g at the protein level constitutes an obvious challenge. However, immunoprecipitation experiments shown in Figure 4D were informative. Indeed, Nes1HAA12, which pulls down all Nesprins disregarding the status of their KASH domain, immunoprecipitated both Nesprin1 giant (Fig.4D, right panel, *) and a ~350kDa protein, which likely corresponds to Nesprin1β at the NE of PCs (Fig.4D, right panel, **), in a ~3/1 ratio. By contrast, in Sun1 immunoprecipitations that specifically pulls down KASH-containing isoforms of Nesprins, this ratio was ~1/1. This less efficient coimmunoprecipitation with Sun1 was not due to the large size of Nesprin1giant since Nesprin2 giant was efficiently pulled-down by Sun1 in the same conditions (Figure 4D, left panel). Even though they only provide a crude and indirect biochemical approach, these immunoprecipitations results combined with immunofluorescence microscopy and RT-PCR experiments together support the idea that a significant pool of KLNes1gis expressed in the cerebellum.
KLNes1g associates with glomeruli vesicles and CGN dendritic membranes
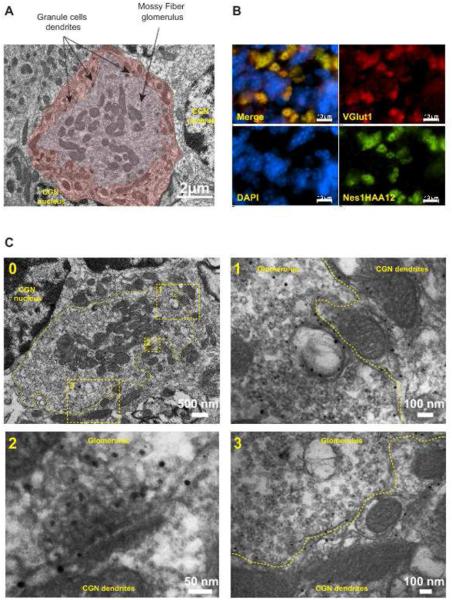
The size and GCL distribution of Nesprin1 immunoreactive speckles hinted at mossy fiber glomeruli that correspond to synaptic “hubs” between mossy fibers and CGN (Fig.5A and (Xu-Friedman and Regehr, 2003)). Accordingly, KLNes1g immunoreactivity significantly overlapped with Vglut1 as well as with other pre- and post-synaptic markers (Fig.5B and data not shown). Immunogold labelling further revealed KLNes1g colocalization both with glomeruli vesicles and CGN dendritic membranes (Fig.5C).
Figure 5.
A) Transmission electron microscopy of a mossy fiber glomerulus within the GCL of a P30 cerebellum from a C57/Bl6 mouse. The glomerulus is pseudocolored in light brown and the adjacent CGN dendrites are pseudocolored in dark brown. B) Coimmunolabeling of the GCL with Vglut1 and Nes1HAA12. C) Immunogold electron microscopy of cerebellar glomeruli and surrounding CGN dendrites. A single glomerulus is delineated by a dashed line in the low magnification image (0). 1 and 3: higher magnification of two regions revealing the colocalization of KLNes1g with a significant pool of synaptic vesicles and with dendritic membranes. 2: Higher magnification showing KLNes1g immunoreactivity within synaptic vesicles at a release site.
Vesicular and membrane-associated proteins coimmunoprecipitate with KLNes1g
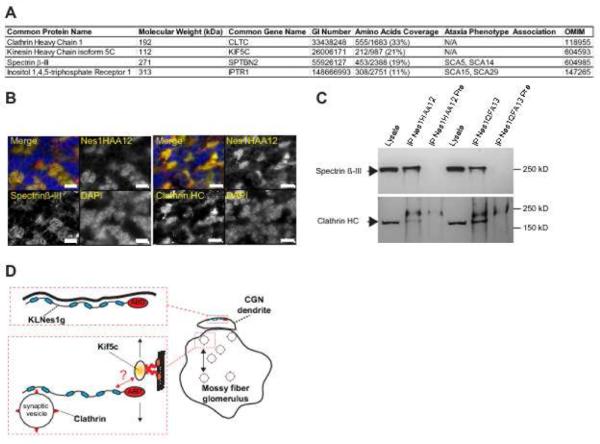
To provide preliminary insights into the biological function of KLNes1g in the cerebellum, silver-stained proteins specifically detected in Nes1HAA12 and Nes1QFA13 but not in their respective preimmune immunoprecipitates were identified by mass spectrometry. This approach identified the heavy chain of Clathrin, the heavy chain of Kinesin 5C, SpectrinβIII, and the Inositol 1, 4, 5-triphosphate receptor with high confidence as judged by their extensive amino acid coverage (Fig.6A). Accordingly, Spectrin-βIII and Clathrin colocalized with Nesprin1 at cerebellar glomeruli (Fig.6B) and were immunodetected in Nes1HAA12 and Nes1QFA13 immunoprecipitates (Fig.6C). Consistent with immunogold labelling data (Fig.5C), these preliminary experiments supported a functional association of KLNes1g with vesicles and dendritic membranes (Fig.5D).
Figure 6.
Identification of proteins coimmunoprecipitating with KLNes1g . A) List of proteins identified in Nes1QFA13 and Nes1HAA12 immunoprecipitates. B) Colocalization of Spectrin β-III and Clathrin with KLNes1g at glomeruli. Scale bars: 10 µm. C) Immunobloting of immunoprecipitations performed with Nes1HAA12, QFA13 and their respective preimmune sera (Pre) with Spectrin β-III and Clathrin antibodies. D) Functional model of KLNes1g acting in vesicular trafficking within cerebellar glomeruli and as a membrane scaffold for CGN dendrites. Blue ovals: spectrin repeats; ABD: Actin-binding domain. The question mark indicates unknown binding modalities.
Disruption of LINC complexes at the NE of PCs does not induce any obvious cerebellar phenotype
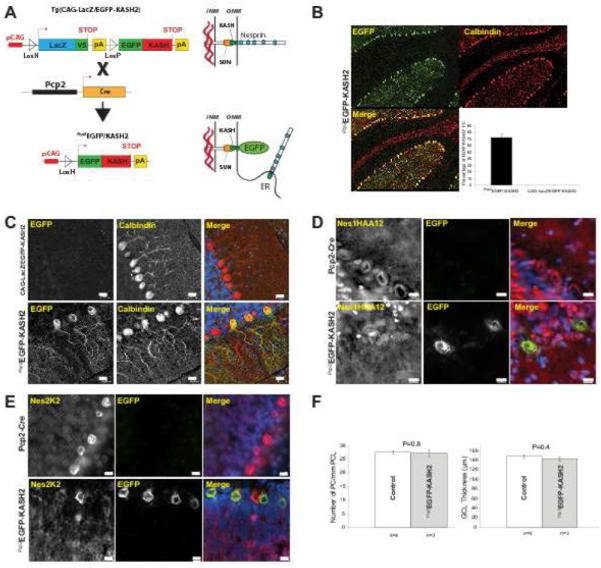
Since a NE localization of Nesprin1 was only observed in PCs, we examined the consequences of its functional inactivation in that compartment using Tg(CAG-LacZ/EGFP-KASH2), a dominant negative mouse model allowing for the spatiotemporal control of EGFP-KASH2 expression in specific tissue/cell types (Razafsky and Hodzic, 2014). As a result of the saturation of available SUN domains by EGFP-KASH2, this approach allows for the displacement of endogenous Nesprins from the NE of targeted cells (Fig.7A). Tg(CAG-LacZ/EGFP-KASH2) were bred to Tg(Pcp2-Cre) mice (Barski et al., 2000) that initiate Cre recombinase expression in PCs at around postnatal day6 (P6). As shown in Figure 7B and 7C, a robust and widespread pattern consisting of EGFP-KASH2 nuclear rims was specifically observed in ~70% of PCs from recombinant transgenic offspring (called Pcp2EGFP-KASH2 hereafter). As expected, endogenous Nesprin1 and Nesprin2 were undetectable at the NE of PCs expressing EGFP-KASH2 whereas they localized at the NE of non-recombinant PCs (Fig.7D and 7E). ARCA1 is a late-onset disease whose most common histological feature consists of cerebellar atrophy. However, despite the displacement, i.e. the functional inactivation, of both Nesprin1 and Nesprin2 from the NE of PCs, 10 month-old Pcp2EGFP-KASH2 mice did not display any obvious behavioral difference by comparison to their control littermates. In addition, the positioning and density of PCs as well as GCL thickness measured in Pcp2EGFP-KASH2 cerebellar slices were not affected (Fig.7F). These results suggest that the functional impairment of Nesprin1 at the NE of PCs does not induce any obvious cerebellar phenotype further reinforcing the idea that ARCA1 phenotypes originates from KLNes1g dysfunction.
Figure 7.
Disruption of LINC complexes at the NE of PCs does not induce any obvious cerebellar phenotype. A) Depiction of the genetic strategy to induce EGFP-KASH2 expression in mouse cerebellum using Pcp2-Cre, a strain that initiates the expression of Cre recombinase at P6. B) Representative paraffin-embedded sections from 10 month-old Pcp2EGFP-KASH2 cerebella processed for calbindin and EGFP immunofluorescence microscopy. EGFP-KASH2 expression was homogenously distributed within ~70% of the PCs population. C) Higher magnification of similar staining emphasizing the rim-like pattern of EGFP-KASH2 in recombinant PCs. Tg(CAG-LacZ/EGFP-KASH2) littermates did not display any EGFP-KASH2 signal. Scale bars: 20 µm. D, E) Endogenous Nesprins are displaced from the NE of EGFP-KASH2+ PCs. Immunofluorescence microscopy of cerebella from indicated genotypes either with Nes1HAA12 (D) or Nesprin2K2 (E). Note that whereas Nesprin rims are present in all PCs of Pcp2-Cre cerebella, Nesprin1 and Nesprin2 perinuclear rims are absent from the NE of EGFP-KASH2+ PCs. Scale bars: 10 µm. F) Disruption of LINC complexes in PCs does not affect cerebellar organization. Average PCs counts and GCL thickness were measured from cerebella of 4 control and 3 recombinant mice from two distinct 10 month-old litters.
DISCUSSION
In this work, we uncovered surprising aspects of Nesprin1 biology that provide new hypotheses related to the molecular etiology of ARCA1. Indeed, Nesprin1 giant was specifically expressed in CNS tissues with highest expression levels in the cerebellum whereas Nesprin2 giant was detected in all tissues. The localization of Nesprin1 and Nesprin2 in cerebellar slices also displayed essential differences; whereas Nesprin2 exclusively localized at the NE of all cerebellar neurons, the bulk of Nesprin1 immunoreactivity colocalized with mossy fiber glomeruli. We provide evidence that this immunoreactivity corresponds to KLNes1g, a variant of Nesprin1 giant devoid of KASH domain. The canonical NE localization of Nesprin1 was only observed in PCs. We anticipate that this signal corresponds to Nesprin1β since QFA13 failed to detect the NE of PCs (Fig.3D) and Nesprin1α is not expressed in the cerebellum (data not shown). For the first time, these data emphasize divergent biological properties of Nesprin1giant vs. Nesprin2 giant as well as a new biological function for Nesprin1 through the synthesis of KLNes1g.
We propose that dysfunction(s) of KLNes1g underlie ARCA1 for the following reasons. First, all ARCA1 nonsense mutations would lead to KLNes1g truncations. Second, because KLNes1g is specifically expressed in CNS tissues and most abundant in the cerebellum, it potentially explains why ARCA1 mutations mostly targets cerebellar function and integrity. Third, by contrast to the well-established redundancy of Nesprin1 and Nesprin2 functions at the NE, a loss of function of the KLNes1g cannot be rescued by Nesprin2 since both proteins localize to very distinct cellular compartments. These considerations may further explain why ARCA1 mutations are exclusively associated to Nesprin1 mutations. Fourth, despite the in vivo functional inactivation of Nesprins at the NE of PCs, Pcp2EGFP-KASH2 mice do not develop any cerebellar phenotype suggesting that ARCA1 is not clinically related to a loss of function of Nesprin1 at the NE of PCs. Finally, the genetic strategy used to generate Nesprin1KO mice (Zhang et al., 2009b) is not predicted to affect the coding sequence of KLNes1g. In agreement with our hypothesis, these mice do not develop any cerebellar ataxia phenotype further reinforcing the idea that Nesprin1 function at the NE does not underlie ARCA1. By contrast, two additional Nesprin1 KO models raised using genetic strategies that affect the coding sequence of KLNes1g display severe phenotypes (Puckelwartz et al., 2009; Zhang et al., 2009a). Whether cerebellar defects contribute to these phenotypes, which are currently attributed to skeletal muscle pathologies, is currently unknown. However, it is important to note that none of the ARCA1 mutations reported to date fall into the coding sequence of Nesprin1α (Fig.1A) suggesting that a protein encoded by these exons may play vital developmental functions in mice and underlie distinct pathologies in humans. Together, these results support that the molecular etiology of ARCA1 originates from a loss of function of KLNes1g.
Interestingly, the functional duality of Nesprin1 may be evolutionary-conserved since a similar alternative splicing of Klarsicht is associated with the synthesis of KASH-containing and KASH-less variants in Drosophila. Whereas the former is involved in nuclear positioning in photoreceptors, the latter mediates lipid droplets movement in embryos (Guo et al., 2005; Mosley-Bishop et al., 1999; Razafsky et al., 2012; Welte et al., 1998; Yu et al., 2011). To that regard, immunogold labeling experiments and the coimmunoprecipitation of Kif5C and Clathrin with KLNes1g together suggest the involvement of KLNes1g in vesicular trafficking (Fig.6D). By analogy to the role of Klarsicht in molecular motor coordination during lipid droplets movements (Welte et al., 1998), KLNes1g may possibly coordinate molecular motors in vesicular transport. This model is overall consistent with our data and with a possible evolutionary-conserved role of KLNes1g and KASH-LESS variants of Klarsicht in vesicular trafficking. Because CGN dendrites were significantly labelled with gold particles, KLNes1g may also act as a scaffold for CGN dendritic membrane (Fig.6D). This function for KLNes1g is further supported by the Golgi localization of recombinant Nesprin1 fragment corresponding to the central region of Nesprin1 giant (Gough et al., 2003). According to these models, ARCA1 mutations may alter synaptic transmission through synaptic vesicles trafficking and/or CGN dendrites ultrastructural defects.
Mutations of Spectrin-βIII and Inositol 1, 4, 5-triphosphate Receptor proteins underlie late-onset spinocerebellar ataxia phenotypes (Ikeda et al., 2006; van de Leemput et al., 2007) (Fig.6A). Whereas these interactions remain to be further examined, the identification of these proteins in KLNes1g immunoprecipitates suggests that, together, these proteins belong to a common pathway acting at cerebellar glomeruli. Alternatively, because Spectrin-βIII and Inositol 1, 4, 5-triphosphate Receptor are abundantly expressed in PCs dendrites (Sugawara et al., 2013), we cannot exclude that, beside a role at the interface between mossy fibers and CGN, KLNes1g may also exert its non-canonical functions at CGN/PCs synapses.
MATERIAL AND METHODS
Animals
Animal protocols used in this study strictly adhered to the ethical and sensitive care and use of animals in research and were approved by the Washington University School of Medicine Animal Studies Committee (Animal Welfare Insurance Permit #A-3381-01, protocol#20130225). Tg(CAG-LacZ/EGFP-KASH2) were recently described (Razafsky and Hodzic, 2014). Tg(Pcp2-Cre) were purchased from the Jackson Laboratories (#004146, B6.129-Tg(Pcp2-cre)2Mpin/J)(Barski et al., 2000). Actin-Cre and Nesprin1 conditional knockout mice were previously described (Zhang et al., 2009a). Mouse colonies were maintained and genotyped at the Mouse Genetics Core (Washington University School of Medicine).
In Situ hybridization
ISH was performed on mouse brain sections using the RNAscope 2.0 Red Kit (Advanced Cell Diagnostics, #310036) according to the manufacturer’s instructions. Briefly, whole brains from 1 month-old C57B\l6 mice were fixed in 4% PFA/PBS overnight at 4°C and embeded in paraffin. Five µm-thick microtome sections were deparaffinized in xylene, followed by dehydration in an ethanol series. Sections were then incubated in a boiling citrate buffer, rinsed with water and immediately treated with protease. Hybridization with target probes, preamplifier and amplifier were carried out at 40°C followed by development using the supplied Fast Red reagents. Control hybridizations were carried out in parallel with all test hybridizations. Samples were couterstained with Hematoxylin and imaged with an Olympus NanoZoomer Whole-Slide Imaging System.
Antibodies
Anti-LaminB1 (Santa Cruz Biotechnology, #SC-6210), anti-EGFP (AbCam, #ab3970), anti-Calbindin (Sigma, #C9848), anti-VGLUT1 (Synaptic Systems, #135302), anti-Clathrin HC (Santa Cruz Biotechnology, #SC-6579) and anti-Spectrin βIII (Santa Cruz Biotechnology, #SC-9660) were used in this study. Nesprin1 (Nes1HAA12) and Nesprin2 (Nes2K2) antisera specificity was previously reported (Khatau et al., 2012; Razafsky et al., 2013). HRP- and Alexa-conjugated secondary antibodies were obtained from Santa Cruz Biotechnology and Invitrogen, respectively. To generate Nes1QFA13, total RNA of mouse cerebellum was reverse transcribed and PCR amplified using primers annealing to exons −66 and −64. Amplicons were subcloned in pGEX-4T1 to produce a GST fusion protein. Bacterial synthesis of recombinant protein was induced with IPTG in BL21 E. coli and the fusion protein was purified from inclusion bodies in urea. Polyclonal antibodies were produced by immunizing mice and rabbits with the purified GST fusion protein. Half the rabbit serum was affinity purified on a CNBr sepharose column (PRIMM Biotech, Cambridge, MA, USA). The specificity of Nes1QFA13 used in this study is shown in Figure 3.
Immunoprecipitation
Freshly dissected cerebella were minced and added to RIPA buffer (150 mM NaCl, 50 mM Tris pH 7.6, 0.5% Sodium Deoxycholate, 0.1% SDS, 1% Triton X-100) supplemented with protease inhibitors (Roche, #11697498001) and Zirconium oxide beads (0.5 mm, Next Advance). The tissue was then grinded in a Bullet Blender (Next Advance) for 5 min and centrifuged at 12,000 g for 10 min at 4°C to collect the supernatant. The latter was pre-cleared for at least one hour with Protein A/G beads (Pierce, #20421) and incubated with fresh Protein A/G and ~3 µg of immunoprecipitating antibodies on a rotary shaker overnight at 4°C. Beads were washed three times with RIPA buffer and resuspended in gel loading buffer. For silver staining, 20% of immunoprecipitates were loaded on SDS-PAGE gels that were further processed according to the manufacturer recommendation (Silverquest, Invitrogen, #LC6070). Silver stained bands detected in immune but not in preimmune immunoprecipitations were processed for tryptic digest and Mass spectrometry analyses (Donald Danforth Plant Sciences Center Proteomics and Mass Spectrometry Facility (St. Louis, MO, USA)).
RT-PCR
Mouse tissues were isolated and resuspended in Trizol (Invitrogen, #15596020), grinded with a Bullet Blender (Next Advance, Averill Park, NY, USA) and centrifuged. Supernatants were used for RNA purification according to the manufacturer’s specifications. Total RNA was reverse transcribed with Superscript II (Invitrogen, #18064014) following manufacturer recommendations. Ten percent of RT reactions were used for PCR amplification either with Taq Hi-FI polymerase (Invitrogen, #1130411) or the Expanded Long Template PCR System (Roche #11681834001). Amplification of Nesprin1 transcripts was performed using the following primers (See Figure 1A): −3 (5’-GTCCCACATCCGGAAGAAGTACCCC-3’), −1 (5’-CTTCAGAGTGGAGGACCGTTGG-3’), −30 (5’-GGAGCAATCACAAAGCAGCCTGTGACG-3’), −51 (5’-CCACCATCAGGATGAAAGCAGCTGGCAAGC-3’), −60 (5’-GCTCGAGACTGTTCGGAAATGGATCGAGAAAGC-3’), −66 (5’-GCATCACAACTAGAGCAAACCGGAAACGATACAAGC-3’). Ethidium bromide-stained agarose gels were imaged with a G:Box HR16 imaging system (Syngene, Fredrick. ND, USA). Sequencing reactions were performed at our in-house facility (Protein and Nucleic Acid Chemistry Laboratory, Washington University School of Medicine).
Vertical gel electrophoresis (VAGE) and immunoblotting procedures
We followed the procedure described in (Warren et al., 2003). Briefly, mouse tissues were grinded in gel loading buffer (8M Urea, 2M thiourea, 3% SDS, 75mM DTT, 0.03% bromophenol blue, 50mM Tris ( pH 6.8)) using a Bullet Blender and centrifuged at 12,000g for 10 min. Supernatants were heated at 60°C for 15 min and loaded on 1.5% agarose gels. VAGE was performed at 65V OVN at 4°C in 50mM Tris/385mM Glycine/0.1% SDS (Fig.S2). Proteins were then transferred to Optitran nitrocellulose membranes (GE Healthcare) for 2h20min at 4°C at 40V in 50mM Tris/40mM Glycine/0.04% SDS/20% methanol (pH 8.3). Membranes were stained with PonceauS to label Titin, Nebulin and Myosin from the skeletal muscle lysates (Fig.S2), blocked with 5% milk in TBST (10mM Tris, 150mM NaCl, 0.1% Tween 20 ( pH 7.3) ) for 1 hour at room temperature and incubated overnight at 4°C with the primary antibodies. After washing with TBST, membranes were incubated with the appropriate HRP-conjugated secondary antibodies. Signals were detected using SuperSignal® West Pico solutions (Thermo, #1856135) and exposed on x-ray film and/or imaged for quantification purposes on a G:Box HR16 imaging system (Syngene). Bands were quantified in non-saturated images using the GeneTools software package.
Preparation of mouse cerebella for immunofluorescence microscopy
Mice were anesthetized with a ketamine/xylazine cocktail and transcardially perfused with 30% sucrose/1XPBS followed by 4% PFA/PBS. After an OVN incubation in 30% sucrose/1XPBS at 4°C, cerebella were rinsed in PBS and embedded in OCT compound (Tissue-TEK). Cryosections (15µm) on Superfrost Plus slides (VWR) were fixed for 5 minutes in 4% PFA in PBS, rinsed three times in PBS, permeabilized with 0.5% Triton X-100/PBS and incubated with primary antibodies diluted in 10% donkey serum/0.5% Triton X-100 in PBS. Alexa conjugated secondary antibodies (Invitrogen) were incubated in the same conditions. Sections were then counterstained with DAPI and mounted with fluorescent mounting media (DAKO).
For quantification of PCs number and GCL thickness, mice were injected with a ketamine/xylazine cocktail and transcardially perfused with 30% sucrose in PBS followed by 4% paraformaldehyde in PBS. Dissected cerebella were incubated in 4% PFA/1XPBS OVN at 4°C, dehydrated for 30 minutes each in 70% ethanol and 80% ethanol, followed by two 30 minute cycles each of 95% ethanol, 100% ethanol, histological grade xylenes and four 30 minute cycles of Tissue-Prep 2 paraffin. 4 µm-thick slices were cut on a rotary microtome, floated out on a 49°C water bath, collected on poly-L-lysine coated slides and air dried overnight. Slides were deparaffinized and antigen retrieval performed in citrate buffer pH 6.0 in a pressure cooker for three minutes. Sections were then blocked with 20% donkey serum then treated with primary antibodies at 4°C overnight. Sections were rinsed in PBS, treated with appropriate Alexa conjugated secondary antibodies for one hour, rinsed again with PBS and mounted with Vectashield Hard Set with DAPI (Vector Laboratories, #H-1500).
Image acquisition and quantification
Image acquisition (single, large scans and Z-stacks) was performed with a Nikon Eclipse Ti coupled to a Coolsnap HQ2 (Photometrics) and an LED light source (Lumencor) with the NIS element software package (Nikon) with either 20× (n.a 1.0) or 40× (n.a. 1.4) objectives. PCs counts were determined with the quantification tools from NIS-Elements (Nikon). PCs labeled with Calbindin were counted across entire cerebellar sections from a minimum of three mice per genotype. The total number of PCs was divided by the length of a line drawn through the PCs layer on each image. The thickness of the GCL was measured at five random locations throughout each image and averaged. A standard t-test was used to compare statistical significance.
TEM and Immunogold TEM
For ultrastructural analysis, cerebella were fixed in 2% paraformaldehyde/2.5% glutaraldehyde in 100 mM cacodylate buffer, pH 7.2 overnight at 4°C. Samples were washed in cacodylate buffer and postfixed in 1% osmium tetroxide (Polysciences Inc.) for 1 hr. Samples were then rinsed extensively in dH20 prior to en bloc staining with 1% aqueous uranyl acetate (Ted Pella Inc.) for 1 hr. Following several rinses in dH20, samples were dehydrated in a graded series of ethanol and embedded in Eponate 12 resin (Ted Pella Inc.). Sections of 95 nm were cut with a Leica Ultracut UCT ultramicrotome (Leica Microsystems Inc.), stained with uranyl acetate and lead citrate, and viewed on a JEOL 1200 EX transmission electron microscope (JEOL USA Inc.,) equipped with an AMT 8 megapixel digital camera (Advanced Microscopy Techniques, Woburn, MA). For immunogold TEM, mice were injected with a ketamine/xylazine cocktail and transcardially perfused with 30% sucrose in PBS followed by 4% paraformaldehyde/2.5% glutaraldehyde in PBS. The cerebellum was isolated and permeabilized in 50% methanol at −20°C for five minutes, thawed and immediately embedded in 4% low meltpoint agarose in ACSF. Sagittal sections were sliced on a Leica VT1000S Vibratome (Leica BioSystems). Immunoreactions were carried out essentially as described above with affinity purified HAA12 antibody, except no Triton X-100 was present to maintain membrane ultrastructural integrity. FluorNanogold anti-rabbit Fab’ AlexaFluor 488 secondary antibody was used (Nanoprobes). Tissue slices were confirmed positive by fluorescence microscopy prior to proceeding with enhancement of nanogold and processing for transmission electron microscopy. Samples were then fixed in 4% paraformaldehyde/2.5% glutaraldehyde overnight and the 1.4 nm nanogold secondary antibody was metallographically enhanced using GoldEnhance EM Plus (Nanoprobes) according to the manufacturers’ instructions. Samples were then washed, postfixed with osmium, and processed for resin embedding and sectioning as described above.
Supplementary Material
Figure S1: Depiction of LINC complexes that assemble through direct interactions (dashed arrows) between evolutionary-conserved SUN and KASH domains, the molecular signature of Sun proteins and Nesprins, respectively. See text for details. INM, ONM: inner and outer nuclear membrane, respectively. PNS: perinuclear space.
Figure S2: Vertical agarose gel electrophoresis. Mouse skeletal muscle and cell culture lysates were run in duplicate on 1.5% agarose gels. Coomassie staining shows the resolution over several centimeters of well-characterized skeletal muscle proteins Titin (3 and 4MDa isoforms), Nebulin (750kDa) and myosin heavy chain (250kDa) (right panel, Coomassie before transfer). After transfer onto nitrocellulose membrane, the second gel was stained with Coomassie. As shown on the left panel (Coomassie after transfer), skeletal muscle proteins are fully transferred to nitrocellulose membranes. Ponceau S staining labels these proteins used as molecular rulers (not shown).
Figure S3: BssSI digest were performed as an additional way to analyze the splicing status of large Nesprin1 amplicons amplified by long RT-PCR. This approach takes advantage of a BssSI site located within exon-2. The presence of a 2545bp and of a 2779bp fragment is indicative of large Nesprin1 transcripts encoding KASH-containing and KASH-LESS variants of the protein, respectively. Sizes are in bp.
Highlights.
The giant isoform of Nesprin1 (980kDa) is specifically expressed in CNS tissues
A CNS-specific alternative splicing of Nesprin1 generates KLNes1g, a KASH-LESS variant of Nesprin1 giant
KLNes1g coimmunoprecipitates with proteins whose mutations underlie spinocerebellar ataxia
ARCA1 mutations may act through abnormal synaptic defects at mossy fibers/CGN interface
ACKNOWLEDGEMENTS
The authors are grateful to Dr. Ju Chen (UC San Diego) for sharing Nesprinf/f mice and to Dr. Wandy Beatty (Washington University) for excellent technical help with immunogold TEM. We thank Belinda McMahan from our in-house Morphology and Imaging Core, the team at the Mouse Genetics Core for handling mice breeding and genotyping. This work was funded by the small grant program from The McDonnell Center for Cellular and Molecular Neurobiology and supported by the Alafi Neuroimaging Laboratory, the Hope Center for Neurological disorders, and NIH Shared Instrumentation Grant (S10 RR0227552) to Washington University. The authors are supported by the National Eye Institute (#R01EY022632 to D.H.), a National Eye Institute Center Core Grant (#P30EY002687) and an unrestricted grant from Research to Prevent Blindness to the Department of Ophthalmology and Visual Sciences. The authors declare no competing financial interests.
ABBREVIATIONS
- ARCA1
Autosomal Recessive Cerebellar Ataxia Type I
- CGN
Cerebellar Granule Neurons
- CNS
Central Nervous System
- GCL
Granule Cell Layer
- KASH
Klarsicht/Anc1/Syne1 Homology
- ISH
In Situ Hybridization
- KLNes1g
KASH-LESS Nesprin1 giant
- LINC
LInkers of the Nucleoskeleton to the Cytoskeleton
- NE
Nuclear Envelope
- SUN
Sad1/UNc84
- PCs
Purkinje Cells
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- Apel ED, Lewis RM, Grady RM, Sanes JR. Syne-1, a dystrophin- and Klarsicht-related protein associated with synaptic nuclei at the neuromuscular junction. J Biol Chem. 2000;275:31986–31995. doi: 10.1074/jbc.M004775200. [DOI] [PubMed] [Google Scholar]
- Barski JJ, Dethleffsen K, Meyer M. Cre recombinase expression in cerebellar Purkinje cells. Genesis. 2000;28:93–98. [PubMed] [Google Scholar]
- Cottrell JR, Borok E, Horvath TL, Nedivi E. CPG2: a brain- and synapse-specific protein that regulates the endocytosis of glutamate receptors. Neuron. 2004;44:677–690. doi: 10.1016/j.neuron.2004.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupre N, Gros-Louis F, Bouchard JP, Noreau A, Rouleau GA. SYNE1-Related Autosomal Recessive Cerebellar Ataxia. In: Pagon RA, Adam MP, Ardinger HH, Bird TD, Dolan CR, Fong CT, Smith RJH, Stephens K, editors. GeneReviews(R) Seattle (WA): 1993. [Google Scholar]
- Gough LL, Fan J, Chu S, Winnick S, Beck KA. Golgi localization of Syne-1. Mol Biol Cell. 2003;14:2410–2424. doi: 10.1091/mbc.E02-07-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros-Louis F, Dupre N, Dion P, Fox MA, Laurent S, Verreault S, Sanes JR, Bouchard JP, Rouleau GA. Mutations in SYNE1 lead to a newly discovered form of autosomal recessive cerebellar ataxia. Nat Genet. 2007;39:80–85. doi: 10.1038/ng1927. [DOI] [PubMed] [Google Scholar]
- Guo Y, Jangi S, Welte MA. Organelle-specific control of intracellular transport: distinctly targeted isoforms of the regulator Klar. Mol Biol Cell. 2005;16:1406–1416. doi: 10.1091/mbc.E04-10-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y, Dick KA, Weatherspoon MR, Gincel D, Armbrust KR, Dalton JC, Stevanin G, Durr A, Zuhlke C, Burk K, Clark HB, Brice A, Rothstein JD, Schut LJ, Day JW, Ranum LP. Spectrin mutations cause spinocerebellar ataxia type 5. Nat Genet. 2006;38:184–190. doi: 10.1038/ng1728. [DOI] [PubMed] [Google Scholar]
- Izumi Y, Miyamoto R, Morino H, Yoshizawa A, Nishinaka K, Udaka F, Kameyama M, Maruyama H, Kawakami H. Cerebellar ataxia with SYNE1 mutation accompanying motor neuron disease. Neurology. 2013;80:600–601. doi: 10.1212/WNL.0b013e3182815529. [DOI] [PubMed] [Google Scholar]
- Khatau SB, Bloom RJ, Bajpai S, Razafsky D, Zang S, Giri A, Wu PH, Marchand J, Celedon A, Hale CM, Sun SX, Hodzic D, Wirtz D. The distinct roles of the nucleus and nucleus-cytoskeleton connections in three-dimensional cell migration. Sci Rep. 2012;2:488. doi: 10.1038/srep00488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Katanosaka Y, Iwata Y, Matsuoka M, Shigekawa M, Wakabayashi S. Identification and characterization of GSRP-56, a novel Golgi-localized spectrin repeat-containing protein. Exp Cell Res. 2006;312:3152–3164. doi: 10.1016/j.yexcr.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Lombardi ML, Jaalouk DE, Shanahan CM, Burke B, Roux KJ, Lammerding J. The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J Biol Chem. 2011;286:26743–26753. doi: 10.1074/jbc.M111.233700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton GW, Starr DA. KASHing up with the nucleus: novel functional roles of KASH proteins at the cytoplasmic surface of the nucleus. Curr Opin Cell Biol. 2014;28:69–75. doi: 10.1016/j.ceb.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso M, Orsucci D, Siciliano G, Bonuccelli U. The genetics of ataxia: through the labyrinth of the Minotaur, looking for Ariadne's thread. J Neurol. 2014;261(Suppl 2):S528–541. doi: 10.1007/s00415-014-7387-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marme A, Zimmermann HP, Moldenhauer G, Schorpp-Kistner M, Muller C, Keberlein O, Giersch A, Kretschmer J, Seib B, Spiess E, Hunziker A, Merchan F, Moller P, Hahn U, Kurek R, Marme F, Bastert G, Wallwiener D, Ponstingl H. Loss of Drop1 expression already at early tumor stages in a wide range of human carcinomas. Int J Cancer. 2008;123:2048–2056. doi: 10.1002/ijc.23763. [DOI] [PubMed] [Google Scholar]
- Mosley-Bishop KL, Li Q, Patterson L, Fischer JA. Molecular analysis of the klarsicht gene and its role in nuclear migration within differentiating cells of the Drosophila eye. Curr Biol. 1999;9:1211–1220. doi: 10.1016/s0960-9822(99)80501-6. [DOI] [PubMed] [Google Scholar]
- Noreau A, Bourassa CV, Szuto A, Levert A, Dobrzeniecka S, Gauthier J, Forlani S, Durr A, Anheim M, Stevanin G, Brice A, Bouchard JP, Dion PA, Dupre N, Rouleau GA. SYNE1 mutations in autosomal recessive cerebellar ataxia. JAMA neurology. 2013;70:1296–1231. doi: 10.1001/jamaneurol.2013.3268. [DOI] [PubMed] [Google Scholar]
- Padmakumar VC, Abraham S, Braune S, Noegel AA, Tunggal B, Karakesisoglou I, Korenbaum E. Enaptin, a giant actin-binding protein, is an element of the nuclear membrane and the actin cytoskeleton. Exp Cell Res. 2004;295:330–339. doi: 10.1016/j.yexcr.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Padmakumar VC, Libotte T, Lu W, Zaim H, Abraham S, Noegel AA, Gotzmann J, Foisner R, Karakesisoglou I. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J Cell Sci. 2005;118:3419–3430. doi: 10.1242/jcs.02471. [DOI] [PubMed] [Google Scholar]
- Puckelwartz MJ, Kessler E, Zhang Y, Hodzic D, Randles KN, Morris G, Earley JU, Hadhazy M, Holaska JM, Mewborn SK, Pytel P, McNally EM. Disruption of nesprin-1 produces an Emery Dreifuss muscular dystrophy-like phenotype in mice. Hum Mol Genet. 2009;18:607–620. doi: 10.1093/hmg/ddn386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajgor D, Mellad JA, Autore F, Zhang Q, Shanahan CM. Multiple novel nesprin-1 and nesprin-2 variants act as versatile tissue-specific intracellular scaffolds. PLoS One. 2012;7:e40098. doi: 10.1371/journal.pone.0040098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajgor D, Mellad JA, Soong D, Rattner JB, Fritzler MJ, Shanahan CM. Mammalian microtubule P-body dynamics are mediated by nesprin-1. J Cell Biol. 2014;205:457–475. doi: 10.1083/jcb.201306076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razafsky D, Blecher N, Markov A, Stewart-Hutchinson PJ, Hodzic D. LINC complexes mediate the positioning of cone photoreceptor nuclei in mouse retina. PLoS One. 2012;7:e47180. doi: 10.1371/journal.pone.0047180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razafsky D, Hodzic D. Bringing KASH under the SUN: the many faces of nucleo-cytoskeletal connections. J Cell Biol. 2009;186:461–472. doi: 10.1083/jcb.200906068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razafsky D, Hodzic D. Temporal and tissue-specific disruption of LINC complexes in vivo. Genesis. 2014;52:359–365. doi: 10.1002/dvg.22755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razafsky DS, Ward CL, Kolb T, Hodzic D. Developmental regulation of linkers of the nucleoskeleton to the cytoskeleton during mouse postnatal retinogenesis. Nucleus. 2013;4:399–409. doi: 10.4161/nucl.26244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa BA, Rothballer A, Kutay U, Schwartz TU. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell. 2012;149:1035–1047. doi: 10.1016/j.cell.2012.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Fischer JA. KASH 'n Karry: the KASH domain family of cargo-specific cytoskeletal adaptor proteins. Bioessays. 2005;27:1136–1146. doi: 10.1002/bies.20312. [DOI] [PubMed] [Google Scholar]
- Stewart-Hutchinson PJ, Hale CM, Wirtz D, Hodzic D. Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp Cell Res. 2008;314:1892–1905. doi: 10.1016/j.yexcr.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara T, Hisatsune C, Le TD, Hashikawa T, Hirono M, Hattori M, Nagao S, Mikoshiba K. Type 1 inositol trisphosphate receptor regulates cerebellar circuits by maintaining the spine morphology of purkinje cells in adult mice. J Neurosci. 2013;33:12186–12196. doi: 10.1523/JNEUROSCI.0545-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Leemput J, Chandran J, Knight MA, Holtzclaw LA, Scholz S, Cookson MR, Houlden H, Gwinn-Hardy K, Fung HC, Lin X, Hernandez D, Simon-Sanchez J, Wood NW, Giunti P, Rafferty I, Hardy J, Storey E, Gardner RJ, Forrest SM, Fisher EM, Russell JT, Cai H, Singleton AB. Deletion at ITPR1 underlies ataxia in mice and spinocerebellar ataxia 15 in humans. PLoS Genet. 2007;3:e108. doi: 10.1371/journal.pgen.0030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren CM, Krzesinski PR, Greaser ML. Vertical agarose gel electrophoresis and electroblotting of high-molecular-weight proteins. Electrophoresis. 2003;24:1695–1702. doi: 10.1002/elps.200305392. [DOI] [PubMed] [Google Scholar]
- Welte MA, Gross SP, Postner M, Block SM, Wieschaus EF. Developmental regulation of vesicle transport in Drosophila embryos: forces and kinetics. Cell. 1998;92:547–557. doi: 10.1016/s0092-8674(00)80947-2. [DOI] [PubMed] [Google Scholar]
- Xu-Friedman MA, Regehr WG. Ultrastructural contributions to desensitization at cerebellar mossy fiber to granule cell synapses. J Neurosci. 2003;23:2182–2192. doi: 10.1523/JNEUROSCI.23-06-02182.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YV, Li Z, Rizzo NP, Einstein J, Welte MA. Targeting the motor regulator Klar to lipid droplets. BMC Cell Biol. 2011;12:9. doi: 10.1186/1471-2121-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Felder A, Liu Y, Guo LT, Lange S, Dalton ND, Gu Y, Peterson KL, Mizisin AP, Shelton GD, Lieber RL, Chen J. Nesprin 1 is critical for nuclear positioning and anchorage. Hum Mol Genet. 2009a;19:329–341. doi: 10.1093/hmg/ddp499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Ragnauth C, Greener MJ, Shanahan CM, Roberts RG. The nesprins are giant actin-binding proteins, orthologous to Drosophila melanogaster muscle protein MSP-300. Genomics. 2002;80:473–481. [PubMed] [Google Scholar]
- Zhang Q, Skepper JN, Yang F, Davies JD, Hegyi L, Roberts RG, Weissberg PL, Ellis JA, Shanahan CM. Nesprins: a novel family of spectrin-repeat-containing proteins that localize to the nuclear membrane in multiple tissues. J Cell Sci. 2001;114:4485–4498. doi: 10.1242/jcs.114.24.4485. [DOI] [PubMed] [Google Scholar]
- Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, Xu R, Han M. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009b;64:173–187. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen YY, Libotte T, Munck M, Noegel AA, Korenbaum E. NUANCE, a giant protein connecting the nucleus and actin cytoskeleton. J Cell Sci. 2002;115:3207–3222. doi: 10.1242/jcs.115.15.3207. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Du X, Cai Z, Song X, Zhang H, Mizuno T, Suzuki E, Yee MR, Berezov A, Murali R, Wu SL, Karger BL, Greene MI, Wang Q. Structure of Sad1-UNC84 homology (SUN) domain defines features of molecular bridge in nuclear envelope. J Biol Chem. 2012;287:5317–5326. doi: 10.1074/jbc.M111.304543. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Depiction of LINC complexes that assemble through direct interactions (dashed arrows) between evolutionary-conserved SUN and KASH domains, the molecular signature of Sun proteins and Nesprins, respectively. See text for details. INM, ONM: inner and outer nuclear membrane, respectively. PNS: perinuclear space.
Figure S2: Vertical agarose gel electrophoresis. Mouse skeletal muscle and cell culture lysates were run in duplicate on 1.5% agarose gels. Coomassie staining shows the resolution over several centimeters of well-characterized skeletal muscle proteins Titin (3 and 4MDa isoforms), Nebulin (750kDa) and myosin heavy chain (250kDa) (right panel, Coomassie before transfer). After transfer onto nitrocellulose membrane, the second gel was stained with Coomassie. As shown on the left panel (Coomassie after transfer), skeletal muscle proteins are fully transferred to nitrocellulose membranes. Ponceau S staining labels these proteins used as molecular rulers (not shown).
Figure S3: BssSI digest were performed as an additional way to analyze the splicing status of large Nesprin1 amplicons amplified by long RT-PCR. This approach takes advantage of a BssSI site located within exon-2. The presence of a 2545bp and of a 2779bp fragment is indicative of large Nesprin1 transcripts encoding KASH-containing and KASH-LESS variants of the protein, respectively. Sizes are in bp.