Summary
The fossil record is widely informative about evolution, but fossils are not systematically used to study the evolution of stem cell-driven renewal. Here, we examined evolution of the continuous growth (hypselodonty) of rodent molar teeth, which is fuelled by the presence of dental stem cells. We studied occurrences of 3500 North American rodent fossils, ranging from 50 million years ago (mya) to 2 mya. We examined changes in molar height to determine if evolution of hypselodonty shows distinct patterns in the fossil record, and we found that hypselodont taxa emerged through intermediate forms of increasing crown height. Next, we designed a Markov simulation model, which replicated molar height increases throughout the Cenozoic, and, moreover, evolution of hypselodonty. Thus, by extension, the retention of the adult stem-cell niche appears to be a predictable quantitative rather than a stochastic qualitative process. Our analyses predict that hypselodonty will eventually become the dominant phenotype.
Introduction
The relationship between the environment and morphology is a classical question in evolutionary biology. Novel traits enable species to adapt to changes within their ecological niches (Heard and Hauser, 1995; Vermeij, 2006), and this is especially evident in the diverse forms of mammalian teeth. One key trait of teeth is crown height. The evolutionarily basal mammalian molars are characterized by low, or brachydont, crowns that do not extend beyond the jawbone, following eruption (Figure 1A). Derived mammalian species, in particular herbivorous mammals with abrasive diets, have evolved teeth with a taller mesodont or hypsodont crown, a portion of which is reserved within the jawbone following completion of development, and that continues to erupt as it is abraded (Figure 1B,C). However, the limited extent to which tall teeth can fit in the jaw has led lagomorph (rabbits and hares) and some rodent lineages to evolve continuously growing, hypselodont molars (Figure 1D). Hypselodont molars, like continuously growing rodent incisors, have stem cells at the base of the tooth in niches called cervical loops; these continuously produce cells that mineralize the tooth (Tummers and Thesleff, 2003). Thus, hypselodonty allows the continual replenishment of worn tooth structures throughout the lifetime of the animal (Tummers and Thesleff, 2003), whereas even extremely hypsodont teeth are of finite duration. Several genes important for stem cell-driven dental renewal in extant mammals have been identified (Harada et al., 1999; Klein et al., 2008; Wang et al., 2007), and comparison of voles and mice suggested that prolonged expression of some of these genes could result in taller crowns (Yokohama-Tamaki et al., 2006). In addition, the fossil record has provided extensive information about the evolution of tooth height in large and small mammals (Damuth and Janis, 2011; Janis, 1988; Jardine et al., 2012; Jernvall and Fortelius, 2002). However, to date no studies have addressed the evolution of hypselodonty in the fossil record as it relates to the evolution of stem cell niches.
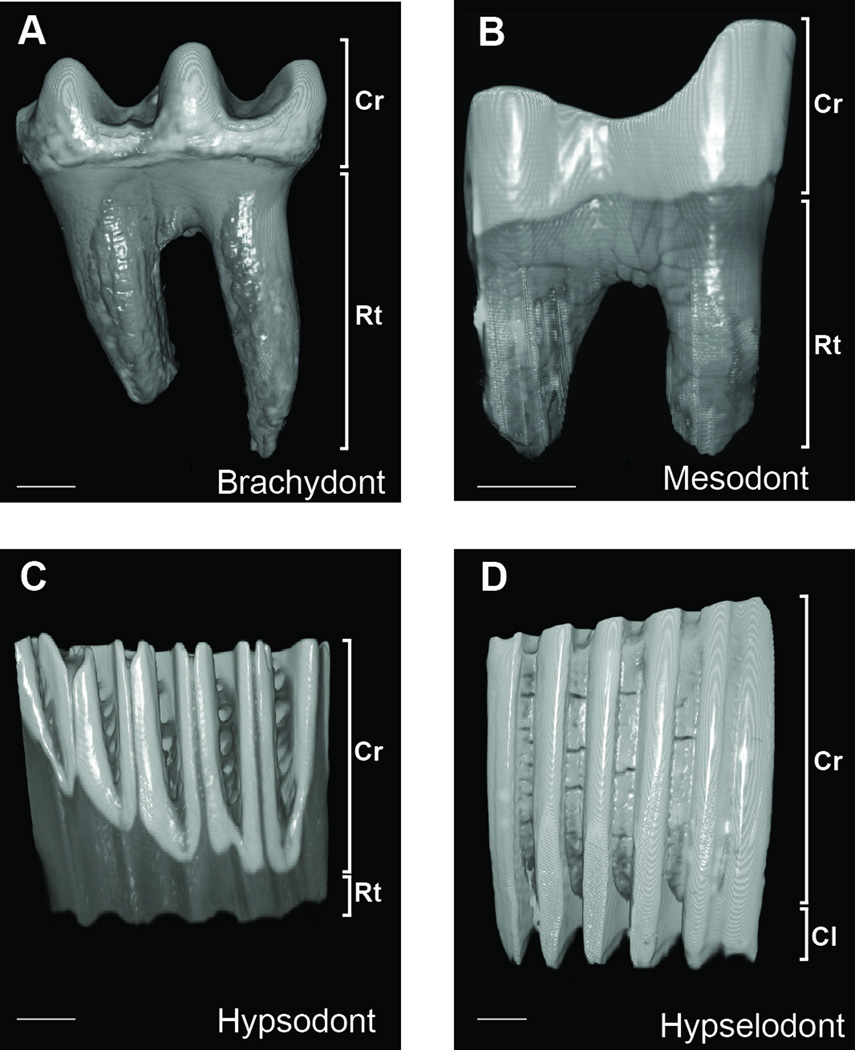
Figure 1. Extant rodent molar crown phenotypes.
(A) Brachydont phenotype is characterized by a crown/root proportion (crp) of 0.3–0.9. (B) Mesodont phenotype is characterized by crp of 0.9–1.5. (C) Hypsodont phenotype is characterized by crp of >1.5. (D) Hypselodont phenotype is characterized by continuous growth, as roots are replaced by active stem cell niches. Cr-crown; Rtroot; Cl-cervical loop. Scale line = 1mm.
Utilizing paleontological data, we set out to determine to what extent the evolution of rodent molars, and more generally the evolution of a complex system such as an adult stem cell niche, could be explained by a simple and quantifiable process of change in tooth height. Specifically, we asked whether the evolution of continuously growing molars can be considered a predictable quantitative evolutionary step no different than simple elongation of the crown over time or whether hypselodonty might be a more difficult step, perhaps requiring more time and additional evolutionary pressure parameters to appear in the fossil record. We utilized rodent fossil data found in localities across the entire North American continent from the New and Old World (NOW) database (http://helsinki.fi/science/now). Four classes of tooth crown height recorded in the database were assigned the values 1, 2, 3, and 4, respectively (Figure 1): brachydont (low), mesodont (medium high), hypsodont (high), and hypselodont (high with no root formation). These categories are considered relatively robust (Eronen et al., 2010a, b), and crown/root proportions for extant brachydont, mesodont, and hypsodont taxa are <0.9, 0.9–1.5, and >1.5, respectively. Dental stem cell niches cannot be themselves identified in fossils; however, as all extant hypselodont mammals lack roots, such characteristics in fossils are the best way of inferring the hypselodont phenotype.
Results
Fossil record and molecular clock analysis show morphological and taxonomic dynamics leading to the emergence of hypselodonty in North American Palaeogene and Neogene rodents
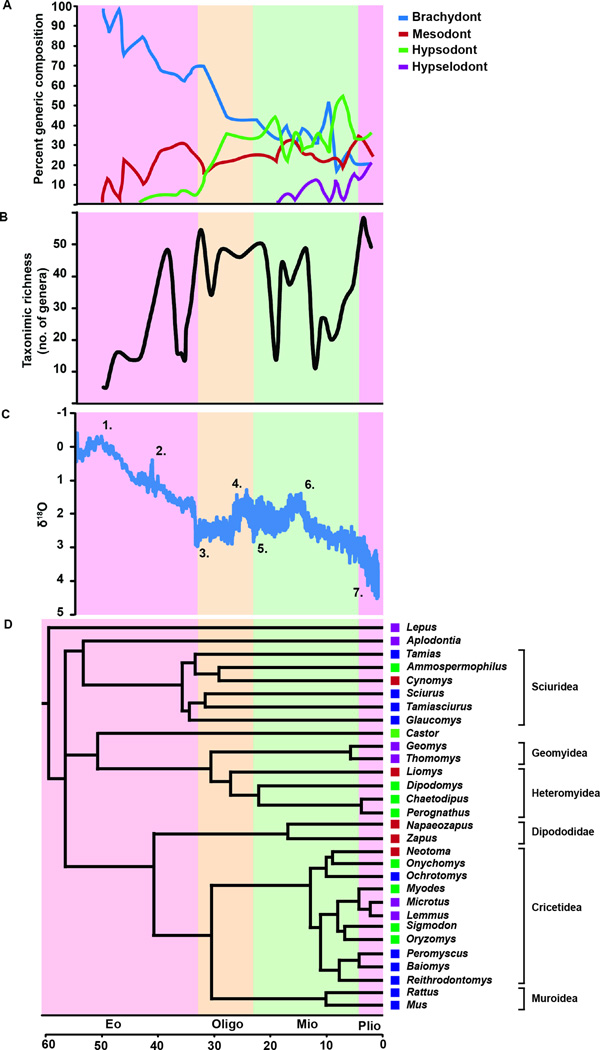
Analysis of the prevalence of four different categories of molar height in 3550 fossils from 50 mya to about 2 mya revealed that brachydont rodents dominated the North American rodent diversity from 50 mya until ~25 mya (Figure 2A and Table S1 and S2) and confirmed some of the findings that used earlier, albeit smaller, iterations of the rodent fossil datasets (Jardine et al., 2012). As the Mesodont as well as hypsodont forms emerged during the middle Eocene at ~42 mya, and during the Oligocene, hypsodont forms represented more than 10% of the taxa (for details of emergence of different groups and their dynamics, see Janis et al. 2008 and Jardine et al. 2012). During the Miocene, hypselodont groups emerged, and by the Pliocene, brachydont phenotypes represented less than 20% of all rodents, while tall-crowned (hypsodont and hypselodont forms) became dominant, representing together ~75% of all North American species (Figure 2A). While mesodont and hypsodont phenotypes emerged 48 and 42 mya, respectively, hypselodonty did not arise until the Miocene at ~18 mya. The hypselodont expansion appears to be trimodal, with radiations at ~14, 8, and 5 mya. In general, the fossil record revealed a steady, progressive increase in the proportion of higher-crowned rodents, with punctuated changes and corresponding oscillations in species richness and environmental perturbations (Figure 2B,C and Table S2).
Figure 2. Summary of molar phenotype and taxonomic dynamics of North American rodents and climate over the past 50 million years.
(A) Molar phenotype prevalence of North American rodents. (B) Taxonomic richness as inferred by number of genera over time. (C) Global temperature changes as inferred by deep-sea benthic oxygen isotope curve. Temperature varied inversely with δ18O levels. 1. Paleocene-Eocene Thermal Maximum; 2. Eocene Climatic Optimum; 3. Oligocene Glaciation; 4. Late Oligocene Warming; 5. Miocene Glaciation; 6. Mid-Miocene Hyperthermal; 7. Northern Hemisphere Glaciation. Eo-Eocene; Oligo-Oligocene; Mio-Miocene; Plio-Pliocene. (D) Phylogeny and divergence times of extant North American rodent clades, depicting the convergent evolution of hypselodonty within Rodentia.
At the first order level, the evolutionary trends towards taller crowns are similar to the general cooling trend during the Cenozoic (Figure 2C). This observation agrees with recent paleontological patterns in large mammals, in which hypsodonty was previously reported to be mainly driven by aridity or water-stress (Eronen et al., 2010a, b) and not directly by temperature (Fortelius et al., 2002). While comparisons of continental climate data utilizing deep ocean oxygen isotope measurements can be problematic in establishing causality (Barnosky and Carrasco, 2002), our results do not refute the potential role of global climatic oscillations in influencing rodent tooth morphology and species richness dynamics and suggest a potential role in the subsequent evolution of hypselodont molar stem cell niches (Figure 2A,B, and C) (see e.g. discussion in Barnosky & Carrasco 2002).
To explore how molar height changed in individual rodent groups, we performed a molecular clock analysis to infer the divergence times of major extant rodent clades utilizing the complete Cytochrome B (CytB), Cytochrome Oxidase I (COI), and ATPase I gene sequences (Figure 2D). Twenty-nine species were chosen, representing all of the North American genera for which sequence data were available. The majority of extant clades originated during the Oligocene and Miocene, consistent with other reports (Fabre et al., 2012). Moreover, the divergence times of the extant hypselodont clades, with end-member genera Geomys, Thomomys, Lemmus, and Microtus, coincided with the radiation of hypselodont fossil taxa (Figure 2). As the hypselodont genus Aplodontia and hypsodont genus Castor are the only extant representatives of their clades, with divergence times (from fossil record data) dating to the Eocene, we further examined the tooth morphology dynamics of the Aplodontidae and Castoridae. In both cases, long-crowned subclades did not originate until late Oligocene or early Miocene (Figure S1). We also examined the intrafamily dynamics of Cricetdae, Dipododidae, Geomyidae, Heteromyidae, and Sciuridae, and found that each of those clades followed the trend towards higher crowns (Figure S1). These data are consistent with previously reported trends in biodiversity of North America during the Miocene (Badgley, 2010; Finarelli and Badgley, 2010) and indicate that multiple rodent groups acquired hypselodont molars independently.
Mathematical modeling yields patterns similar to the fossil record and indicates no delay in the evolution of hypselodonty from hypsodonty
Because the rodent pattern resembled a progressive brachydont-mesodont-hypsodont-hypselodont turnover (Figure 2), we next developed a four state continuous Markov chain stochastic model to examine whether any of the steps in tooth height evolution was delayed (Figure S2). The model is a stochastic process whose state space is the set where the values in each state (molar phenotype) at a given time correspond to the proportion of taxa that occur in that state. The transition matrix determines the probability of change in the proportion of taxa between states, which may be the same or different for each transition. Thus, each transition is characterized by a parameter (probability), and the model altogether is characterized by the number of unique probabilities of transition among states (parameters). We optimized the parameters for one-, two-, three- and six-parameter Markov models, and we used Akaike information criterion (AIC) to determine the best model fit to the fossil data (Figure S3 and Table S3) (see Materials and Methods for details).
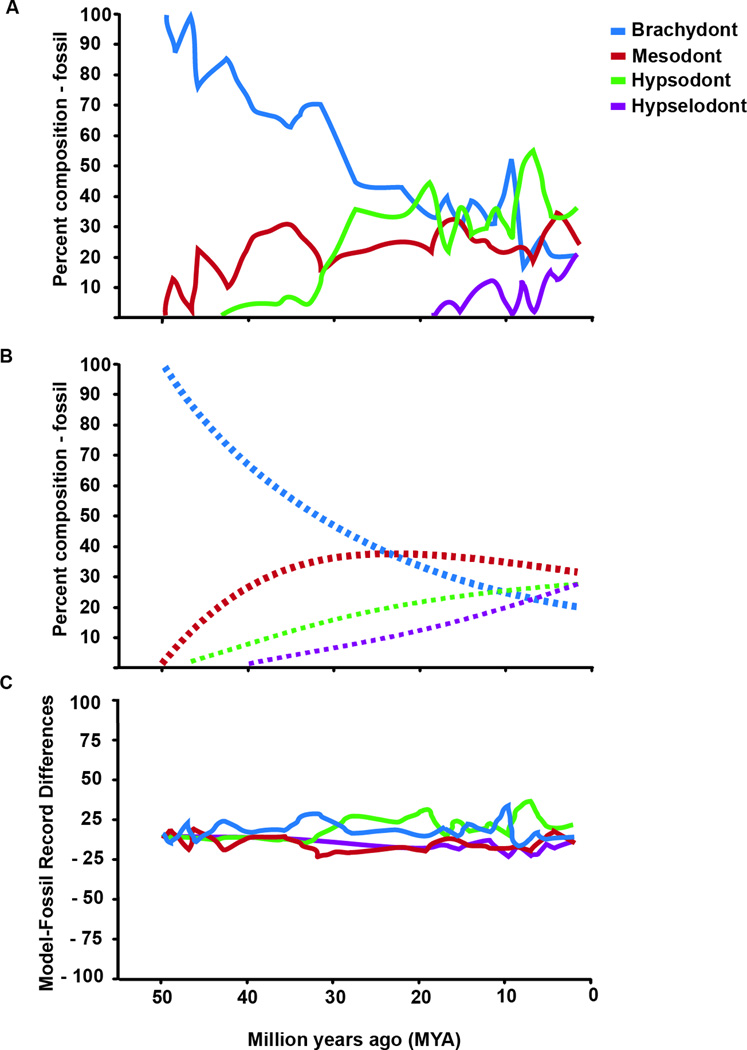
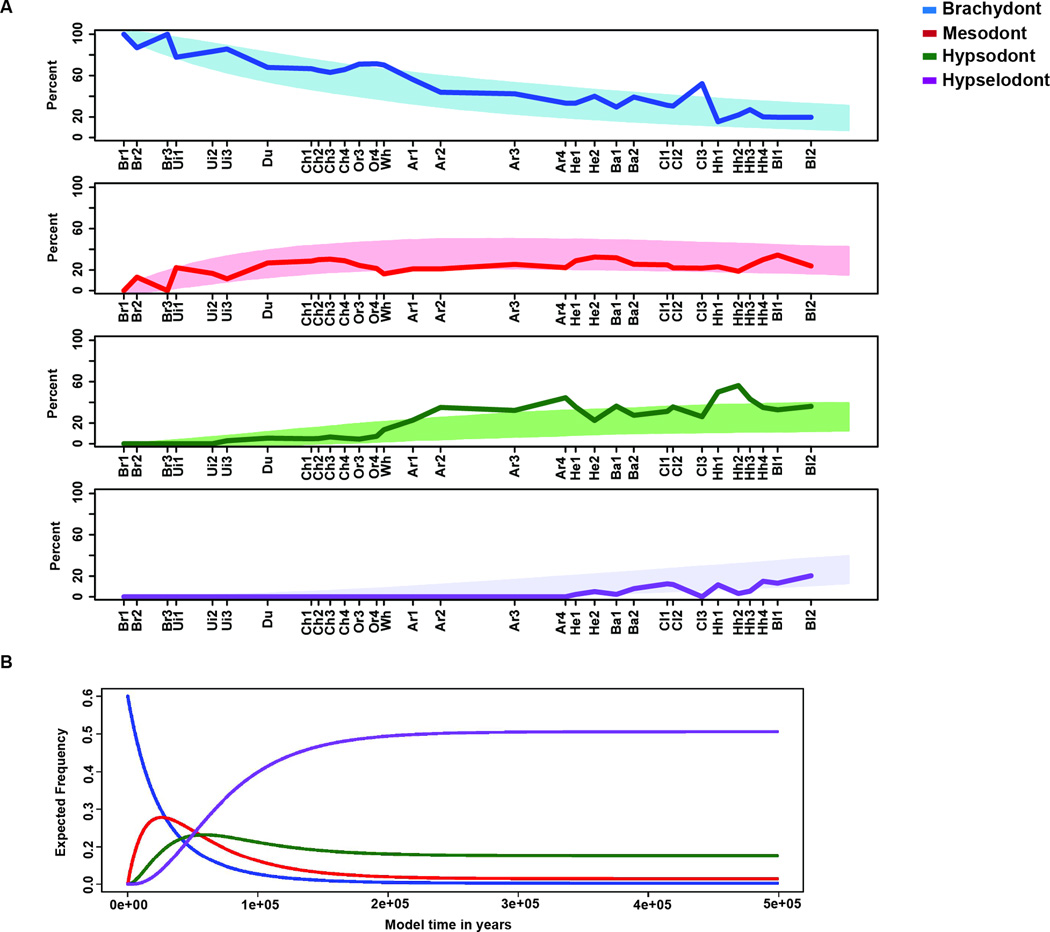
Our analyses revealed that the two-parameter Markov model was the best fit for the observed data, which sets equal all increases (i1 = i2 = i3) and decreases (d1 = d2 = d3) between states (Figure 3A,B and Figure S3). The optimized increase parameter was larger than the optimized decrease parameter by an order of magnitude (i=0.03981 and d=0.0075). Variation in the residuals between the model and the observed data was small (<0.3) and approximately normally distributed (Figure 3C). While most of the observed data fit within the 99% confidence interval of the simulated data (Figure 4A), there were a few cases where the observed frequencies were outside that range. This occurred in 2 of the 31 time bins for the brachydont and mesodont states and in 5 of the 31 time bins for the hypsodont and hypselodont states. The largest discrepancy was due to hypsodont taxa exhibiting greater prevalence than expected during time bins of the Arikareean (Ar2–Ar4: 28.1-18.8 mya) and hypselodont taxa exhibiting less prevalence than expected during the Clarendonian (Cl3: 10.1-9 mya) and Hemphillian (Hh2–Hh3: 7.6-5.9 mya) (Figure 4A and Table S4).
Figure 3. Observed data and two-parameter Markov model of rodent molar evolution.
Comparison of molar phenotype prevalence of North American rodents as inferred by fossil data (A) to prevalence calculated by the two-parameter Markov computer modeling (B). (C) Percentage difference between model and fossil data molar phenotype prevalence.
Figure 4. Markov model simulation confidence interval overlay and future predictions.
(A) Percent of the 100 simulated taxa that occur in each state. Blue represents brachydont (top), red represents mesodont (second), green represents hypsodont (third), and purple represents hypselodont (bottom). Shaded area represents the 99% Confidence Interval (CI) calculated from model simulation and dark solid lines are the observed frequencies from the fossil record converted to percent for comparison. The corresponding North American Land Mammal Ages (NALMA) are described in Table S4. (B) Expected frequency of states given the Markov two-parameter model. Model time is extended to show steady state near 250 million years of model time. The blue represents the brachydont state, the red represents the mesodont state, the green represents the hypsodont state, and the purple represents the hypselodont state.
To specifically test whether the evolution of hypselodonty was delayed, we set up a three-parameter Markov model, keeping constant the brachydont-mesodont-hypsodont transition probability, but decreasing the probability (increasing difficulty) of the hypsodont-hypselodont step. This resulted in AIC values higher than that of the two-parameter model (Figure S3 and Table S3), suggesting that the evolution of hypselodonty was not delayed with respect to the evolution of other phenotypes. Taken together, the two-parameter Markov model indicates that constant transition probabilities account for most of the trends in the increase of molar crown height throughout the Cenozoic, including the evolution of hypselodonty. Thus, the evolution of the adult hypselodont stem cell niches followed the same predictable quantitative trend as the overall gradual increase in rodent crown height.
Finally, because our simple model accounted for the changes in tooth crown height for 50 million years within a 99% confidence interval, we explored future trends of molar height proportions by extending the number of generations in the Markov model into the future to achieve a steady state of phenotype prevalence (Figure 4B). The steady state occurred at around 250 million years in the future, with a rodent taxa composition of 0.5% brachydont, 2.9% mesodont, 15.4% hypsodont, and 81.2% hypselodont. Moreover, extending the model (with all other settings being equal) only 25,000 time steps results in a transient composition of 9.7%, 19.1%, 25.5% and 45.7%, respectively. While obviously highly conjectural, the model thus predicts that the trend of an increasing proportion of hypselodont taxa will continue and ultimately result in a hypselodontdominated rodent community. Ecologically this scenario does not require grassland dominated future ecosystems, as might be intuitively assumed. Rather, hypsodonty and hypselodonty allow the utilization of broad range of diets, as is the case in living voles (Kays and Wilson, 2009; Zimmerman, 1965).
Discussion
The fossil record and the mathematical model indicated that molar height dynamics were remarkably orderly for most of the Cenozoic. Environmental factors, such as large scale cooling (Zachos et al., 2001) or aridification (Eronen et al., 2012), and the uplift of the Rocky Mountain range (Mix et al., 2010), appear to have constantly favored an increase in the proportion of taxa with higher molar crowns. These environmental changes resulted in a shift in North American landscape towards open grassland communities. Furthermore, the expansion of hypsodont forms in the late Oligocene and early Miocene occurred during the appearance of saltatory rodents and coincided with the appearance of cursorial ungulates (Janis et al., 2002), locomotor types associated with more open habitats (typical of more arid environmental conditions). However, the steady constant change observed in both the fossil record and the model suggest that no specific environmental events are needed to explain the increase in crown height, but rather that rodents with taller teeth had small but constant favor, perhaps following from the Red Queen hypothesis, while shorter-crowned species and entire family clades fell out of favor and disappeared (Quental and Marshall, 2013), due to increased range of abrasive dietary intake. Thus, the abiotic (aridification from climatic cooling and geological uplift resulting in more grass and grit) acted as an ecological driving pressure for higher crowns, while the mechanism for achieving this phenotype was continuous, quantitative, and biotic. Those factors, however, are not the only possible catalysts for evolution of hypselodonty, as South American rodents appear to have evolved this trait under the pressure of increased dietary grit from volcanic ash (Stromberg et al., 2013).
In the fossil data, hypselodonty shows a relatively steep rise in the late Miocene. While still largely within the model prediction that hypselodont rodents evolved from endemic hypsodont forms, one possible explanation for the relatively abrupt increase of hypselodont taxa in North America following the slow initial increase could be immigration. Whereas Microtus (Initial presence, I.P. 2.5 Ma) is an immigrant from Asia to North America, and Hydrochoerus (I.P. 2.2 Ma) and Neochoerus (I.P. 5.4 Ma) are immigrants from South America, the fossil data contain hypselodont Cratogeomys (I.P. 3.7 Ma), Cupidinimus (I.P. 18 Ma), Dipodomys (I.P. 8.3 Ma), Eodipodomys (I.P. 10.5 Ma), Geomys (I.P. 13.7 Ma), Guildayomys (I.P. 2.20 Ma), Microdipodops (I.P. 3.7 Ma), Orthogeomys (I.P. 2.20 Ma), Phelosaccomys (I.P. 12.5 Ma), and Thomomys (I.P. 11.1 Ma), which likely originated in North America. Therefore, as all of the mid-Miocene and late Miocene hypselodont taxa appear to have originated in North America, the increase is unlikely to have been the result of immigration. Thus, with migration not a significant factor, as all genera started out as brachydont, any change of the frequency of phenotypes would suggest evolution within those taxa.
The evolution of hypsodonty has been extensively studied in ungulates (Fortelius et al., 2002; Jernvall et al., 1996; Mihlbachler et al., 2011). However, an important constraining factor for the evolution of hypselodonty in ungulate molars is the presence of occlusal pockets of enamel, which are developmentally incompatible with continuous tooth growth (Janis and Fortelius, 1988). With regard to temporal differences with rodent molar evolution, hypsodont ungulate taxa were unknown (or at a minimum exceedingly rare) before the late Eocene, and these forms were relatively infrequent until the middle Miocene (Janis, 2000). Interestingly, in both Eurasia (Jernvall and Fortelius, 2002) and North America (Jardine et al., 2012), hypsodont ungulates did not dominate the ungulate communities until the late Miocene, the time when hypselodont rodents radiated.
Comparative developmental biology studies between molars in mouse (brachydont) and vole (hypselodont) have shown that many genes important for development of incisor cervical loops, such as members of the Fgf, Notch, and Bmp pathways, are also expressed in adult molar stem cell niches (Keranen et al., 1998; Setkova et al., 2006; Tummers and Thesleff, 2003). The maintenance of expression of these genes results in delayed closure of root apex, a shorter root, and prolonged deposition of enamel by the embryonic dental epithelial stem cells, leading to progressively taller crowns (Janis, 1988; Tummers and Thesleff, 2003; Yokohama-Tamaki et al., 2006). This suggests that hypsodont phenotypes can act as reservoirs for evolution of hypselodont phenotypes. Indeed, experimental activation of Fgf10 in developing mouse molars after embryonic day 17 (E17) in vitro resulted in taller crowns (Yokohama-Tamaki et al., 2006). These studies raise the possibility that the transition from hypsodont to hypselodont molars in rodents results from retaining signalling involved in stem-cell regulation and perhaps incorporation of aspects of stem-cell regulation already in place in rodent incisors.
The evolutionary analyses presented here indicate that increases in the proportion of high-crowned taxa, from brachydont to mesodont, then from mesodont to hypsodont, and finally from hypsodont to hypselodont, are largely quantitative changes (changes occurring on an quantifiable steady continuum). Thus, at least in the case of rodent teeth, the evolution of the stem cell niche appears to have been a quantitative rather than a qualitative step. Our results thus demonstrate that studies of the fossil record can provide important insight into the developmental potential to evolve stem cells.
Experimental Procedures
Computerized tomography of rodent molars
Mandible specimens of Mus musculus, Microtus pinetorum, Microtus epiroticus, and Allactaga elater were obtained from the University of Central Oklahoma and the California Academy of Sciences. Mandibular first molar samples were scanned using a micro-focused X-ray tomographic system (MicroXCT-200, Xradia, Pleasanton, CA) at 55 kV and 144 eA. 500 projection images at an exposure time of 10 s with a linear magnification of 2× were taken. The final pixel size was 20 em. The volume was reconstructed using a back projection filtered algorithm (XRadia, Pleasanton, CA). Following reconstruction, teeth were manually segmented and rendered as 3D surfaces using MicroView (Version 5.2.2, GE Healthcare, Pittsburgh, PA).
NOW database of North American rodent fossils
3,550 fossils from the New and Old World (NOW) database (http://helsinki.fi/science/now/) were examined. These data are based on Janis et al. 1998 and Janis et al. 2008 and range from 50 Mya to 2 Mya. Occurrence location, age, molar phenotype (brachydont, mesodont, hypsodont, or hypselodont) and crown height were documented. The fossil data were identified up to the genus level. To control for sampling bias of having multiple samples of the same genus per time point, we removed duplicate occurrences (more than one unique genus per time point per), unless more than one phenotype was shared within the same genus, at which point both would be kept. This resulted in resulting in 1169 rodent fossils (Table S1). Such sample control did not require further correction for phenotype prevalence calculation or phyletic reconstruction (Rosenberg and Kumar, 2001), as calculations of rodent dental phenotype prevalence were based on the proportion of genera with different tooth phenotype in each time bin (see also discussion in Dunhill et al 2014 about the preservation quality of the fossil record) (Dunhill et al., 2014). Tooth crown height was recorded in the database as brachydont (low), mesodont (medium high), hypsodont (high), and hypselodont (high with no root formation). Crown/root proportions for extant brachydont, mesodont, and hypsodont taxa are < 0.9, 0.9–1.5, and > 1.5, respectively. As time bins we used the North American Land Mammal Ages (NALMAs) based on Woodburne 2004 as used in the NOW database (Woodburne, 2004) and calculated the number of unique genus occurrences per bin and molar phenotype prevalence as a function of number of genera with each molar phenotype per bin over total number of genera in the bin (Table S2). All data were plotted in SigmaPlot.
Molecular clock and phylogeny
Complete cytochrome B (CytB), cytochrome oxidase I (COI), and ATPase I gene sequences data for 29 North American rodent species were obtained from Gen-Bank. Species were selected to include representatives of every North American genus with available genetic data. Sequences were analysed in BEAST 1.6.1 (Bayesian evolutionary analysis by sampling trees), which uses a Markov Chain Monte-Carlo (MCMC) algorithm to average over tree space and create posterior probability distribution for analysis parameters (Drummond and Rambaut, 2007). Set parameters were divergence time and time of most recent ancestor (TMRCA). The Relaxed Clock: Uncorrelated Log-normal clock model was chosen to account for lineage specific rate heterogeneity and Yule tree prior as a simple model of speciation (Ronquist et al., 2012). The MCMC chain length was set to 10 million generations, with sampling done every 10,000th generation. Clock calibration was obtained by Mus-Rattus, Muridae-Sciuridae, and Rodentia-Leporidae divergences (Benton et al., 2009). A Normal distribution prior was chosen for MRCA with 0.5 my standard deviation (Ho and Phillips, 2009; Huchon et al., 2002; Yang and Rannala, 2006). The output BEAST file was analysed in TRACER 1.5 and maximum clade credibility trees were produced in TreeAnnotator 1.6.1.
Markov model for the evolution of crown height
To describe the evolution of molar crown height, we used a four state continuous Markov chain stochastic model similar to the M/M/1 queuing model (Figure S2). The four states were brachydont, mesodont, hypsodont, and hypselodont. When each state had a different probability to increase (i) and decrease (d), the Markov model had six-parameters (i1, i2, i3, d1, d2, and d3). When all increases were constant (i1 = i2 = i3), and all decreases were constant (d1 = d2 = d3), the Markov model reduces down to two-parameters (i and d). When constant increase equalled constant decrease (i1 = i2 = i3 = d1 = d2 = d3), the Markov model reduced down to a one-parameter model.
The transition matrix (P) for the Markov model represents the probability of transitions from one state to another. For example, the probability of increasing from brachydont to mesodont from one time step to the next was PBrach,Meso = i1 and the probability of staying in the brachydont state from one time step to the next was PBrach,Brach = 1 - i1.
The initial frequencies of the four states were given by
because the probability of being brachydont at the start of the model was 1. The probability of frequencies of the four states at the first generation (t = 1) was
The probability of frequencies of the four states at any time t was
We optimized the model parameters to obtain the best fit between the model and the observed frequencies of the four states in the fossil record by minimizing the sum of squares of the residuals for all time bins. We considered the fits of a six-parameter model, a two-parameter model and a one-parameter model. Figure S3 shows the two-parameter Markov model fitted to fossil data. We concluded that the six-parameter model was likely over-parameterized and we did not consider the one-parameter model to be realistic, as the reversals in fossil data are extremely rare. We used Nelder-Mead minimization algorithm to optimize parameters in the two-parameter and six-parameter model. We used Brent minimization algorithm for the one-parameter model.
To assess the overall fit of the Markov model with optimized parameters to the observed data from the fossil record, we used computer simulation. The two-parameter model fit the data better than the one-parameter model and was as good a fit as the six-parameter model (Table S3), so we continued the simulation with the two-parameter model. To simulate the evolution of molar crown height, we started with a pool of rodent taxa (N=100), where all taxa were in the brachydont state. The probability for any taxon to increase state was i and the probability for any taxon to decrease state was d. We used a random number generator to simulate the composition (frequency of crown type) of the rodent taxa on a geological time scale, ~49 million years. Computational times were prolonged for long time scales, so we set 1,000 generations to equal one model time step, assuming that one year equals one generation for rodent taxa. After repeating the simulation 1,000 times, we calculated the 99% confidence intervals based on the expected frequencies given the model for each modeled time step. All analyses were performed in the R Statistical Programing Language.
Supplementary Material
Acknowledgements
We thank Jan Schnitzler, Jan Prochazka, Jimmy Hu, Kenneth Locey, and Mikael Fortelius for comments and assistance.
Funding:
This work was funded by the National Institutes of Health through the NIH Director’s New Innovator Award Program, DP2-OD007191, and by R01-DE021420, both to O.D.K, by F30-DE022482 to V.M., F32-DE023702 to A.S., and a Marie Curie fellowship to J.T.E., and the Academy of Finland to J.J.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
The experimental approach was designed by V.M., J.E, J.J., and O.D.K. Fossil record analysis and climate correlation were carried out by V.M. and J.E. Micro-CT scans were carried out by A.S. Molecular clock analysis was carried out by V.M. C.J. analyzed the tooth evolution patterns based on the molecular clock and fossil record. Markov model was designed by J.E. and M.L. and analyzed by V.M. All authors contributed to writing the manuscript and analysis of data.
Competing Financial Interests: The authors declare no competing financial interests.
References
- Badgley C. Tectonics, topography, and mammalian diversity. Ecogeography. 2010;33:220–231. [Google Scholar]
- Barnosky AD, Carrasco MA. Effects of Oligo-Miocene global climate changes on mammalian species richness in the northwestern quarter of the USA. Evolutionary Ecology Research. 2002;4:811–841. [Google Scholar]
- Benton MJ, Donoghue PCJ, Asher RJ. Calibrating and constraining molecular clocks. In: Hedges S, Kumar S, editors. The Timetree of Life. Oxford University Press; 2009. pp. 35–86. [Google Scholar]
- Damuth J, Janis CM. On the relationship between hypsodonty and feeding ecology in ungulate mammals, and its utility in palaeoecology. Biological reviews of the Cambridge Philosophical Society. 2011;86:733–758. doi: 10.1111/j.1469-185X.2011.00176.x. [DOI] [PubMed] [Google Scholar]
- Davies VA, Kemble ED. Social play behaviours and insect predation in northern grasshopper mice (Onychomys leucogaster) Behavioural processes. 1983;8:197–204. doi: 10.1016/0376-6357(83)90008-6. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC evolutionary biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunhill AM, Hannisdal B, Benton MJ. Disentangling rock record bias and common-cause from redundancy in the British fossil record. Nature communications. 2014;5:4818. doi: 10.1038/ncomms5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eronen J, Puolamaki K, Liu L, Lintulaakso KJD, Janis C, Fortelius M. Precipitation and large herbivorous mammals, part II: Application to fossil data. Evolutionary Ecology Research. 2010a;12:235–248. [Google Scholar]
- Eronen J, Puolamaki K, Liu L, Lintulaakso KJD, Janis C, Fortelius M. Precipitation and large herbivorous mammals, part I: Estimates from present-day communities. Evolutionary Ecology Research. 2010b;12:217–233. [Google Scholar]
- Eronen JT, Fortelius M, Micheels A, Portmann FT, Puolamäki K, Janis CM. Neogene Aridification of the Northern Hemisphere. Geology. 2012;40:823–826. [Google Scholar]
- Esselstyn J, Achmadi A, Rowe K. Evolutionary novelty in a rat wth no molars. Biology Letters. 2012;8:990–993. doi: 10.1098/rsbl.2012.0574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre PH, Hautier L, Dimitrov D, Douzery EJ. A glimpse on the pattern of rodent diversification: a phylogenetic approach. BMC evolutionary biology. 2012;12:88. doi: 10.1186/1471-2148-12-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finarelli JA, Badgley C. Diversity dynamics of Miocene mammals in relation to the history of tectonism and climate. Proceedings Biological sciences / The Royal Society. 2010;277:2721–2726. doi: 10.1098/rspb.2010.0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortelius M, Eronen J, Jernval J, Liu L, Pushkina D, Tesakov J, Vislobokova I, Zhang Z, Zhou L. Fossil Mammals Resolve Regional Patterns of Eurasian Climate Change During 20 Million Years. Evolutionary Ecology Research. 2002;4:1005–1016. [Google Scholar]
- Harada H, Kettunen P, Jung HS, Mustonen T, Wang YA, Thesleff I. Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling. J Cell Biol. 1999;147:105–120. doi: 10.1083/jcb.147.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard S, Hauser D. Key evolutionary innovations and their ecological mechanisms. Historical Biology. 1995;10:151–173. [Google Scholar]
- Ho SY, Phillips MJ. Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. Syst Biol. 2009;58:367–380. doi: 10.1093/sysbio/syp035. [DOI] [PubMed] [Google Scholar]
- Huchon D, Madsen O, Sibbald MJ, Ament K, Stanhope MJ, Catzeflis F, de Jong WW, Douzery EJ. Rodent phylogeny and a timescale for the evolution of Glires: evidence from an extensive taxon sampling using three nuclear genes. Mol Biol Evol. 2002;19:1053–1065. doi: 10.1093/oxfordjournals.molbev.a004164. [DOI] [PubMed] [Google Scholar]
- Janis C. An estimation if tooth volume and hypsodonty indices in ungulate mammals, and the correlation of these factors with dietary preferences. Teeth Revisited: Proceedings of the VIIth Internaitonal Symposium on Dental Morphology. 1988:367–387. [Google Scholar]
- Janis C. Origin and Evolution of Herbivory in Terrestrial Vertebrates. Cambridge, UK: Cambridge University Press; 2000. Patterns in the evolution of herbivory in large terrestrial mammals: the Paleogene of North America. [Google Scholar]
- Janis C, Damuth J, Theodor J. The origins and evolution of the North American grassland biome: the story from the hoofed mammals. Palaeogeography, Palaeoclimatology, Palaeoecology. 2002;117:183–198. [Google Scholar]
- Janis CM, Fortelius M. On the means whereby mammals achieve increased functional durability of their dentitions, with special reference to limiting factors. Biological reviews of the Cambridge Philosophical Society. 1988;63:197–230. doi: 10.1111/j.1469-185x.1988.tb00630.x. [DOI] [PubMed] [Google Scholar]
- Jardine P, Janis C, Sahney S, Bentoc M. Grit not grass: Concordant patterns of early origin of hypsodonty in Great Plains ungulates and glires. Palaeogeography, Palaeoclimatology, Palaeoecology. 2012;366:1–10. [Google Scholar]
- Jernvall J, Fortelius M. Common mammals drive the evolutionary increase of hypsodonty in the Neogene. Nature. 2002;417:538–540. doi: 10.1038/417538a. [DOI] [PubMed] [Google Scholar]
- Jernvall J, Hunter JP, Fortelius M. Molar tooth diversity, disparity, and ecology in Cenozoic ungulate radiations. Science. 1996;274:1489–1492. doi: 10.1126/science.274.5292.1489. [DOI] [PubMed] [Google Scholar]
- Kays R, Wilson D. Mammals of North America. 2 edn. Princeton Press; 2009. [Google Scholar]
- Keranen SV, Aberg T, Kettunen P, Thesleff I, Jernvall J. Association of developmental regulatory genes with the development of different molar tooth shapes in two species of rodents. Development genes and evolution. 1998;208:477–486. doi: 10.1007/s004270050206. [DOI] [PubMed] [Google Scholar]
- Klein OD, Lyons DB, Balooch G, Marshall GW, Basson MA, Peterka M, Boran T, Peterkova R, Martin GR. An FGF signaling loop sustains the generation of differentiated progeny from stem cells in mouse incisors. Development. 2008;135:377–385. doi: 10.1242/dev.015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihlbachler MC, Rivals F, Solounias N, Semprebon GM. Dietary change and evolution of horses in North America. Science. 2011;331:1178–1181. doi: 10.1126/science.1196166. [DOI] [PubMed] [Google Scholar]
- Mix HT, Mulch A, Kent-Corson ML, Chamberlain CP. Cenozoic migration of topography in the North American Cordillera. Geology. 2010;39:87–90. [Google Scholar]
- Quental TB, Marshall CR. How the Red Queen drives terrestrial mammals to extinction. Science. 2013;341:290–292. doi: 10.1126/science.1239431. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Klopfstein S, Vilhelmsen L, Schulmeister S, Murray DL, Rasnitsyn AP. A total-evidence approach to dating with fossils, applied to the early radiation of the hymenoptera. Syst Biol. 2012;61:973–999. doi: 10.1093/sysbio/sys058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg MS, Kumar S. Incomplete taxon sampling is not a problem for phylogenetic inference. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10751–10756. doi: 10.1073/pnas.191248498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setkova J, Lesot H, Matalova E, Witter K, Matulova P, Misek I. Proliferation and apoptosis in early molar morphogenesis-- voles as models in odontogenesis. The International journal of developmental biology. 2006;50:481–489. doi: 10.1387/ijdb.052067js. [DOI] [PubMed] [Google Scholar]
- Soholt LF. Water balance of Merriam's kangaroo rat, Dipodomys merriami, during cold exposure. Comparative biochemistry and physiology A, Comparative physiology. 1975;51:369–372. doi: 10.1016/0300-9629(75)90382-5. [DOI] [PubMed] [Google Scholar]
- Stromberg CA, Dunn RE, Madden RH, Kohn MJ, Carlini AA. Decoupling the spread of grasslands from the evolution of grazer-type herbivores in South America. Nature communications. 2013;4:1478. doi: 10.1038/ncomms2508. [DOI] [PubMed] [Google Scholar]
- Tummers M, Thesleff I. Root or crown: a developmental choice orchestrated by the differential regulation of the epithelial stem cell niche in the tooth of two rodent species. Development. 2003;130:1049–1057. doi: 10.1242/dev.00332. [DOI] [PubMed] [Google Scholar]
- Vermeij GJ. Historical contingency and the purported uniqueness of evolutionary innovations. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1804–1809. doi: 10.1073/pnas.0508724103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XP, Suomalainen M, Felszeghy S, Zelarayan LC, Alonso MT, Plikus MV, Maas RL, Chuong CM, Schimmang T, Thesleff I. An integrated gene regulatory network controls stem cell proliferation in teeth. PLoS Biol. 2007;5:e159. doi: 10.1371/journal.pbio.0050159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodburne MO. Biostratigraphy and geochronology. New York: Columbia University Press; 2004. Late Cretaceous and Cenozoic mammals of North America. [Google Scholar]
- Yang Z, Rannala B. Bayesian estimation of species divergence times under a molecular clock using multiple fossil calibrations with soft bounds. Mol Biol Evol. 2006;23:212–226. doi: 10.1093/molbev/msj024. [DOI] [PubMed] [Google Scholar]
- Yokohama-Tamaki T, Ohshima H, Fujiwara N, Takada Y, Ichimori Y, Wakisaka S, Ohuchi H, Harada H. Cessation of Fgf10 signaling, resulting in a defective dental epithelial stem cell compartment, leads to the transition from crown to root formation. Development. 2006;133:1359–1366. doi: 10.1242/dev.02307. [DOI] [PubMed] [Google Scholar]
- Zachos J, Pagani M, Sloan L, Thomas E, Billups K. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science. 2001;292:686–693. doi: 10.1126/science.1059412. [DOI] [PubMed] [Google Scholar]
- Zimmerman EG. A comparison of habitat and food of two species of Microtus. Journal of Mammalogy. 1965;46:605–612. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.