Abstract
Processivity clamps that hold DNA polymerases to DNA for processivity were the first proteins known to encircle the DNA duplex. At the time, polymerase processivity was thought to be the only function of ring shaped processivity clamps. But studies from many laboratories have identified numerous proteins that bind and function with sliding clamps. Among these processes are mismatch repair and nucleosome assembly. Interestingly, there exist polymerases that are highly processive and do not require clamps. Hence, DNA polymerase processivity does not intrinsically require that sliding clamps evolved for this purpose. We propose that polymerases evolved to require clamps as a way of ensuring that clamps are deposited on newly replicated DNA. These clamps are then used on the newly replicated daughter strands, for processes important to genomic integrity, such as mismatch repair and the assembly of nucleosomes to maintain epigenetic states of replicating cells during development.
Keywords: DNA Replication, PCNA, β-clamp
1. Introduction
The simplicity and elegance of the DNA double helix structure suggested its replication might also be simple [1], but nothing could be further from the truth. The antiparallel orientation of the two strands of DNA makes the process much more complicated than initially anticipated. Watson and Crick correctly anticipated that one strand would template the other [2], and Arthur Kornberg discovered the DNA polymerase does exactly that, although template directed synthesis was unprecedented at the time [3]. Watson and Crick also realized a problem with their DNA model. The two strands were extensively wound about one another, and therefore the strands must be completely untwisted to be replicated, but they proposed the cell would somehow solve this problem [2]. This also proved correct, and we now know of many types of topoisomerases that twist, untwist, knot and unknot DNA [4]. Strand separation during replication also requires a helicase in all life forms. Indeed, the true complexity of the enzymatic machinery needed for DNA replication was completely unexpected. Take for example that the antiparallel geometry of the DNA duplex suggests DNA is made in two directions, 5’-3’ on one strand and 3’-5’ on the other. However, all nucleotide precursors are 5’ activated, and this constrains the direction of DNA synthesis to one chemical direction, 5’-3’ [4]. This implies that while both strands are chemically extended 5’-3’, they physically grow in opposite directions on the antiparallel strands of duplex DNA. This imposes a discontinuous mechanism of replication on one strand (i.e. the lagging strand), which is made as a series of fragments, while the other strand (i.e. leading strand) can be made continuously [4]. Semi-discontinuous replication requires a whole set of proteins just to accomplish replication of the discontinuous lagging strand, none of which were anticipated from the DNA structure. Thus RNA primase is needed to initiate each lagging strand fragment, single-strand (ss) DNA binding protein is needed to protect the single-strand (ss) DNA intermediate, RNase activity is required to remove the RNA primers, DNA polymerase is required to replace the RNA with DNA, and a ligase activity is required to seal the lagging strand fragments together [4]. Also unanticipated from the DNA structure is the requirement for DNA sliding clamps and their associated clamp loaders, essential to replication in all cell types [5, 6].
While most information transfer processes, such as transcription and translation, are performed by enzymes that share homology in cells from all domains of life, comparative genomics reveals the surprising finding that the core enzymes of DNA replication: polymerases, helicase, primase, and ssDNA binding proteins, share no homology between bacteria and eukaryotes/archaea, and thus are presumed to have evolved independently [7, 8]. This finding suggests that the first cell did not use these enzymes and perhaps had an RNA genome [8]. Given the lack of a common ancestor, the mechanics of replication in bacteria and eukaryotes may be quite distinct. Interestingly, the sliding clamp and clamp loader are homologous in all cell types, and thus were present in the primordial cell [7, 8]. Presumably the clamp served a different function than it does today. This review will briefly summarize the argument for independent evolution of replication enzymes, provide a current overview of the architecture of the bacterial and eukaryotic replisome machinery, and then will propose a new way of thinking about the role of sliding clamps in DNA replication.
2. Evolution of the core replication machinery
2.1 LUCA (Last Universal Common Ancestor)
DNA is the central repository of information in all modern day cells, bacteria, archaea and eukaryotes alike. However, it may not have always been that way. In the RNA World hypothesis, catalysis was performed by ribozymes [9] and the instructions for life were stored in the form of RNA [10]. The evolution of the protein synthesis machinery largely replaced ribozymes with proteins, although many functional RNAs still exist. For example, the process of translating RNA into protein is still performed by a ribozyme (i.e. the ribosome). Consistent with the existence of RNA before DNA is the fact that the metabolic path to deoxyribonucleotides in all cells occurs directly from the corresponding ribonucleotide, by removal of the 2’ OH from the ribose ring of the ribonucleotide, rather than assembling deoxyribonucleotides from small precursor molecules as the cell does to synthesize ribonucleotides. This reaction is performed by ribonucleotide reductase. This reductive reaction requires a free-radical mechanism and thus it is widely believed that a protein, and not a ribozyme have always performed this reaction, because a free radical intermediate would have destroyed an RNA enzyme. This suggests that the evolution of protein synthesis predated the existence of DNA. Interestingly, ribonucleotide reductase is homologous in cells from all three domains of life, indicating that LUCA, the Last Universal Common Ancestor cell, contained DNA [7, 8]. Indeed, LUCA must have had well-developed protein synthesis machinery because the genetic code, rRNAs, tRNAs and the twenty tRNA synthetases of all modern day cells are homologous.
Despite the near universal genetic code and many homologies among proteins in all cells, there are of course numerous biomolecules that do not have a common heritage. One of the most perplexing of these are the core proteins of DNA replication, suggesting LUCA used RNA as a genome and had not yet evolved a mechanism to replicate duplex DNA [7, 8] (Figure 1). Take for example the DNA polymerases. The replicative DNA polymerases of bacteria are in the C-family and are unrelated in sequence to the replicative polymerases of eukaryotes, which are in the B-family (Pols α, δ and ε) [11]. Although all DNA polymerases have a right hand shape, the sequence and chain folding topology of the B and C family polymerases are completely different [11, 12, 13, 14]. The differences between bacteria and eukaryotes go much further than DNA polymerase. The primase, helicase and ssDNA binding proteins lack homology and thus also lack a common ancestor [7, 8]. The RNA primase of bacteria is a single subunit sculpted from a Toprim fold (Topoisomerase and Primase fold), related to topoisomerase [15-17], while the primase of eukaryotes and archaea is a two subunit enzyme with a catalytic subunit related to X-family DNA polymerases [18-21]. Replicative helicases are hexameric rings; the bacterial helicase is based on the RecA fold, encircles the lagging strand and travels 5’-3’ [8, 22], while the eukaryotic/archaeal Mcm helicases (Mini-Chromosome Maintenance) are based on the AAA+ fold (ATPases Associated with a variety of cellular Activities), encircle the leading strand and travel in the 3’-5’ direction [23, 24]. Although the ssDNA binding proteins of all cells contain OB folds (Oligonucleotide/oligosaccharide Binding fold) [25], the architecture of the OB folds of the highly ordered bacterial homotetrameric SSB (Single-Strand DNA Binding protein) [26, 27], appears significantly distinct from the OB folds used in the largely disordered eukaryotic heterotrimer RPA (Replication Protein A) [28-30]. For these reasons, it is widely believed that the DNA replication machineries of bacteria and eukaryotes evolved independently from one another, after the major cellular domains of life split from LUCA (see Fig. 1) [7, 8].
Figure. 1. Proposed use of DNA in LUCA.
The ribosome, genetic code, tRNAs, and the DNA dependent RNA polymerase of RNA transcription are homologous in all three cellular domains of life, bacteria, archaea and eukaryotes. Thus, in this proposal, the translational and transcriptional pathways we re well developed in LUCA. The presence of Ribonucleotide Reductase, Recombinase and Thymidylate Synthase indicate that DNA was present, but the lack of homologous replication proteins suggest the DNA was not the genomic material. Proposed here, the DNA se rved the purpose of substrate for DNA recombination and transcription of mRNA and genomic RNA. Clamps and clamp loaders were present in LUCA, but their function is uncertain.
The DNA replication enzymes of the archaeal domain of life are related to those of eukaryotes, and thus the split of these cellular domains is believed to have occurred on the same branch of life from the primordial LUCA.
2.2. DNA usage in LUCA
It is interesting to note that LUCA almost certainly contained DNA, regardless of whether it used RNA as a genome. Not only is ribonucleotide reductase conserved in all cells, but thymidylate synthase is also broadly conserved, indicating that the letter “T” was already invented in LUCA [8]. The DNA recombinase (RecA, Rad51) is also conserved [8]. If LUCA did not use DNA as the genetic material, how did it produce DNA and what did it use DNA for? A simple proposal to answer these questions can be suggested from modern day retroviruses. Retroviruses utilize an RNA genome, yet produce duplex DNA as an intermediate in their life cycle [4]. The DNA intermediate is used for transcription of viral genes and to produce new copies of the RNA genome (i.e. replication). The DNA is synthesized from RNA in a very simple fashion, and does not require primase, helicase or single-strand (ss) DNA binding protein. Instead, a reverse transcriptase converts the RNA genome into a ssDNA (as a DNA/RNA hybrid), and synthesis is primed by a tRNA [4]. This simple replication process avoids the difficulties associated with replicating the antiparallel strands of duplex DNA. The DNA in LUCA could have been used to produce mRNA for protein synthesis, and for transcription of new RNA genomes, much as modern day retroviruses. Indeed DNA dependent RNA polymerase is conserved and thus presumed present in LUCA [7, 8]. The homologies between bacterial and eukaryotic DNA recombinases (i.e. RecA/Rad52) suggest that DNA was the substrate for recombination, a requirement for horizontal gene transfer. Before the genomes of organisms were known, the process of horizontal gene transfer was thought to be a relatively minor event. But with the sequence of numerous genomes in hand, it is now apparent that horizontal gene transfer was the major driving force of rapid evolution, allowing genetic material to be freely exchanged among viruses and pro-cells and fodder for the pressures of natural selection to sculpt free living organisms [7, 8]. Without recombination, evolution of cellular life as we know it would probably not have been possible.
With time, it is only natural that DNA became the genetic material for all cells because removal of the 2’ OH of the ribose results in a much more stable molecule than RNA. Hence, after the split of bacteria and eukaryotes/archaea from LUCA, two independent processes eventually solved the many problems inherent in duplication of antiparallel DNA. It may seem improbable that the same activities of helicase, primase, polymerase, and ssDNA binding protein are utilized for bacterial and eukaryotic replication, but these fundamental activities are inherent in any process that duplicates DNA. The use of sliding clamps for replication, and the fact that they were present in LUCA in the apparent absence of other replication enzymes, is not so obvious.
3. The Replication Fork
3.1. Asymmetry at the replication fork
Although the DNA duplex is a wonderfully symmetric molecule, the enzymology at the replication fork is highly asymmetric, based on the fact that synthesis can only occur in the 5’-3’ direction [4] which drives the positioning of different proteins at the replication fork.
3.1.1. Bacterial replisome
The bacterial homohexameric helicase, encircles only one of the two strands at a replication fork, and therefore is one factor that imparts asymmetric structure to the “replisome” machinery. Bacterial helicase encircles the lagging strand and travels 5’-3’ along the ssDNA using rNTPs as fuel [22]. It separates the strands of duplex DNA by excluding the opposite strand (leading strand) from the central channel of the ring as it steps forward along the strand that it encircles [31]. Also asymmetric, the bacterial replisome contains three copies of a C-family DNA polymerase (eg E. coli Pol III) [32, 33]. While three polymerases may seem like one too many polymerases for duplex DNA, cellular and in vitro studies have shown that two of the polymerases function on the lagging strand [34, 35]. Bacterial Okazaki fragments are 1-2 kb and the use of two polymerases for this strand ensures that lagging strand fragments are extended to completion. The lagging strand is primed by DnaG primase, a single subunit enzyme that is related to topoisomerase in sequence and structure; it generates short (<12 ntd) RNA primers [16, 17, 18]. The enzymatic activity of DnaG primase requires it to transiently interact with the helicase, thereby localizing RNA primers to replication fork junctions [4]. Both leading and lagging strand polymerase action require the sliding beta clamp. Without beta, Pol III is nearly inactive. But with the beta clamp, Pol III becomes rapid (>500bp/s) and highly processive (>5kb) during synthesis [36]. This rapid rate of synthesis is in keeping with the observed 650 ntd/s rate of synthesis of the E. coli chromosome [37]. Sliding clamps are assembled onto DNA at primed sites by a clamp loader apparatus that couples ATP hydrolysis to open and close beta clamps around primed sites [38]. Clamps and clamp loaders are the subject of the next section, but deserve some description here for the scaffolding role they play in the bacterial replisome. The subunits required for clamp loading function consist of a homotrimeric tau, and one each of delta and delta prime (Fig 2a) [32, 39].
Figure. 2. Replisomes of bacteria and eukaryotes.
A) The bacterial replisome is organized by the clamp loader, which contains a tau subunit homotrimer with extensions that bind three C-family DNA polymerases and connect to the helicase. The ho mohexameric helicase encircles the lagging strand. Primase is a single subunit based on the Toprim fold and acts to prime synthesis. DNA loops form during Okazaki fragment synthesis as a consequence of the connection between the leading and lagging strand polymerases via the clamp loader. B) The eukaryotic replisome is organized by the 11-subunit CMG helicase (composed of Cdc45, the Mcm2-7 hexamer that encircles the leading strand and the 4-subunit GINS heterotetramer. GINS binds to Pol ε, a B-family polymerase dedicated to the leading strand. The 4-subunit Pol α – Primase interacts with CMG through the Ctf4 homotrimer which also binds a GINS subunit of CMG. Pol α also contains a B-family polymerase that extends RNA primers to form hybrid RNA-DNA primers. Primers are further extended into Okazaki fragments by Pol δ (B-family polymerase) that functions with the PCNA clamp. Direct connections of Pol δ and the RFC clamp loader to other replisome components are currently unknown, unknown, and thus the lagging strand DNA may not form loops. The bacterial SSB tetramer and eukaryotic RPA ssDNA-binding proteins protect lagging strand ssDNA from nucleases and are not shown for clarity.
These subunits are members the AAA+ family, and each subunit contains three domains, two of which encompass the AAA+ region. The three tau subunits contain two additional C-terminal domains that bind directly to Pol III and connect to the helicase [39]. Hence, the clamp loader is the central organizer of the bacterial replisome, holding three polymerases together and interacting with the helicase [32, 34]. The single clamp loader places beta clamps onto both the leading and lagging strands [40]. During fork progression, ssDNA is generated on the lagging strand. SSB binds to the ssDNA, protecting it from nucleases and melting regions of secondary structure, greatly increasing the catalytic efficiency of Pol III-beta. It is interesting to note that the dnaX gene encoding the tau subunit also encodes a second protein in many bacteria, including E. coli [41]. This second protein is about 2/3 the N-terminal sequence of tau and referred to as gamma. In E. coli, gamma is generated by a translational frameshift that encounters a stop codon within two amino acids. Some bacteria utilize other methods to generate gamma, such as transcriptional slippage.
The gamma subunit can also assemble with delta and delta prime to form a clamp loader with similar catalytic activity to the tau-containing clamp loader [32]. Beta clamps are used by several other proteins in addition to the replicative Pol III polymerase, including several enzymes in DNA repair (MutS, MutL, ligase, translesion DNA polymerases) [42, 43]. Hence, it has been proposed that the gamma-containing clamp loader exists to assemble beta clamps onto DNA for repair. The most frequent repair process is the maturation of Okazaki fragments, which require removal of the RNA primer, fill-in with DNA, and ligation [44]. Pol I contains a 5’-3’ flap endonuclease that excises the RNA primer while the polymerase simultaneously fills-in DNA [4]. Ligase then seals the nick. Both Pol I and ligase interact with the beta clamp, and although their activity does not absolutely require its presence, interaction with the clamp increase their efficiency in locating the proper site of action [43].
3.1.2. Eukaryotic replisome
Eukaryotes handle the distinct jobs of leading and lagging strand replication quite differently from bacteria. The helicase consists of 11 distinct subunits, six of which comprise the Mcm2-7 heterohexamer that encircles ssDNA and act as a helicase that has the opposite polarity compared to the bacterial helicase [45-49]. Thus the Mcm2-7 motor of the eukaryotic helicase is on the leading strand instead of the lagging strand. Each of the Mcm subunits is a member of the AAA+ family, unrelated to the RecA-based structure of the bacterial helicase [50]. As their name implies AAA+ proteins are typically oligomers involved in numerous cellular processes including membrane fusion, proteolysis, replication, gene expression and many other functions [51, 52]. Five additional proteins are required for Mcm2-7 activity; these are Cdc45 and the heterotetramer GINS (Sld5, Psf1, Psf2 and Psf3) [45, 46, 49]. Thus the 11-subunit assembly is referred to as CMG (i.e. Cdc45-Mcm2-7-GINS) [49]. Interestingly, EM 3D reconstruction of CMG shows that the CMG helicase has two holes [45, 53]. The Mcm2-7 subunits form the largest cavity for encircling the leading strand. The other hole in CMG is smaller, and is formed by the outside portion of the Mcm ring bound to the GINS tetramer and Cdc45 accessory subunits. It is not yet known whether DNA enters this second cavity. The functions of GINS and Cdc45 are relatively unknown, but two of the GINS subunits bind other proteins, and the other two subunits may have partners yet to be discovered.
In eukaryotic cells the leading and lagging strands are duplicated by two different B-family DNA polymerases, Pol ε and Pol δ, and elegant genetic studies have demonstrated that Pol ε functions on the leading strand while Pol δ replicates the lagging strand [54, 55]. Biochemical studies reveal these two polymerases have distinctive properties suited for the asymmetric jobs of leading and lagging strand synthesis [56-58]. Pol ε consists of 4-subunits; the largest subunit contains the DNA polymerase and 3’-5’ exonuclease activities. The second largest subunit of Pol ε, Dpb2, is essential and has been demonstrated to bind the Psf1 subunit of the GINS complex [59, 60]. Pol ε is known to bind GINS during the activation of origins [61], and it is tempting to speculate that this Dpb2-Psf1 connection may also be utilized to attach Pol ε to CMG at the replication fork. Indeed, recent biochemical studies that reconstitute leading strand replication using pure proteins demonstrate that Pol ε is stabilized for function with CMG on the leading strand, while Pol δ is not stabilized by CMG [62].
The lagging strand Pol δ consists of four subunits (3 in yeast), the largest of which contains both polymerase and proofreading exonuclease activities [63, 64]. The function of the accessory subunits of Pol δ are not yet clear, although the second largest subunit shares homology to the Dpb2 subunit of Pol ε [65]. The C-terminal region of the polymerases has recently been shown to contain a Zn finger and a FeS cluster [65-67]. Like bacterial Pol III, Pol δ is essentially dependent on the sliding clamp for appreciable activity, and is capable of extending a primed site over 5 kb without dissociating from the DNA substrate [68]. Also specific to the lagging strand is the heterotrimeric RPA ssDNA binding protein [30]. Although apparently unrelated to bacterial SSB, RPA serves a similar function by protecting ssDNA against nuclease attack and removing secondary structures in ssDNA that act as blocks to forward progression by Pol δ-PCNA. Consistent with asymmetric functions on the leading and lagging strands, PCNA selects Pol δ over Pol ε, even when Pol ε is in 20-fold molar excess over Pol δ, for synthesis on PCNA primed ssDNA [62].
The eukaryotic primase is a four-subunit enzyme, Pol α that contains both an RNA primase and a B-family DNA polymerase [69, 70]. The primase and polymerase are connected by a B subunit (Pol12), with homology to the B subunits of Pols ε and δ. Pol α synthesizes a hybrid RNA/DNA primer of 25-35 nucleotides [69, 70]. The primase activity is performed by a Pri1/2 heterodimer; the catalytic site is located in the smaller subunit. The catalytic primase subunit is related to the X-family of DNA polymerases, unrelated to bacterial DnaG primase [71]. RNA primers of 7-12 nucleotides are transferred to the DNA polymerase subunit for further extension. The hybrid RNA/DNA primer is then recruited by Pol δ in a polymerase switch first discovered in the SV40 system [63, 64]. The RFC clamp loader facilitates this polymerase switch. It is interesting to note that eukaryotic Okazaki fragments are only about 160 bp on average, yet yeast Pol δ is highly processive with PCNA [62]. This seeming paradox will be discussed later in this review.
Okazaki fragment maturation is performed in eukaryotes quite differently than in bacteria. Eukaryotes have no polymerase like bacterial Pol I that contains a 5’-3’ exonuclease for RNA primer removal. Eukaryotic Okazaki fragment maturation is initiated by Pol δ, which is capable of limited strand displacement synthesis [63]. As the RNA is displaced, an exonuclease, usually Fen1, removes the displaced RNA [63]. Only after the RNA is removed can ligase seal Okazaki fragments together [72]. The DNA product of Pol α has low fidelity because Pol α lacks a 3’-5’ exonuclease. In fact Pol δ has been shown to proofread the DNA product of Pol α [73, 74]. It is also thought that during strand displacement, some of the DNA produced by Pol α may be removed by the same strand displacement/exonuclease reaction that eliminates the RNA [75].
The architecture and function of the eukaryotic replisome apparatus is not yet as well defined as that of the bacterial replisome, but several fascinating aspects of its architecture have been established. As illustrated in Fig. 2b, the CMG and Pol ε function together on the leading strand [62]. Unlike Pol δ (and bacterial Pol III), biochemical studies show that the eukaryotic Pol δ is stabilized by CMG and does not absolutely require PCNA [62]. Eukaryotes also contain a Ctf4 protein that moves with replication forks, but has no homologue in bacteria [76, 77]. Ctf4 has recently been shown to be a homotrimer [78], and has long been known to bind Pol α [79]. The interaction site for Ctf4 within Pol α has been localized to a peptide motif in the N-terminal region of the large catalytic subunit [78]. Ctf4 also binds the Sld5 subunit of the GINS complex (within CMG) [78]. Hence, Ctf4 appears to fulfill a scaffolding role. Given the lack of homology between bacterial and eukaryotic replication proteins, it is somewhat surprising that the eukaryotic replisome contains a homotrimer at its core that binds helicase and the lagging strand Pol α. While this sounds similar to the role of C-terminal domains of the bacterial tau trimer, there is no detectable sequence similarity between the trimeric bacterial tau subunit and Ctf4, and in fact Ctf4 is not essential in yeast (although it is essential in other eukaryotes). The ligand for the third protomer of Ctf4, if any, is currently unknown. Unlike the bacterial replisome, there are no known connections of the RFC clamp loader or the lagging strand Pol δ to other subunits of the replisome. However, eukaryotic replication forks contain many other proteins that connect to CMG, identified by mass spectrometry from cell extract pullouts of CMG [76, 77]. This large assemblage, referred to as RPC (replisome progression complex) contains Mcm10, Ctf4, Csm3, Tof1, Mec1, FACT, Topo I, Pol α and probably other proteins as well [76, 77].
4. Processivity factors
4. 1. Sliding clamps and clamp loaders share a common ancestry.
The structure and function of clamps and clamp loaders were first identified in the E. coli system [80]. However, the existence of accessory factors for DNA polymerases had long been known in the E. coli, T4 phage and eukaryotic systems [81, 82]. The exact function of these accessory factors was unknown, although they were important to polymerase processivity and the T4 proteins were collectively referred to as a sliding clamp, while the idea that one, or any of them actually encircled DNA was not proposed. The fact that E. coli beta subunit topologically encircles DNA was first recognized from biochemical experiments [83], and then proven by crystal structure analysis [84]. This was the first demonstration of a protein that encircles DNA, previously unprecedented, and we now know that many proteins encircle DNA for their function. Clamps do not get onto DNA by themselves and require an ATP driven clamp loader complex [38]. The DNA sliding clamp and its cognate clamp loader are required for replication in all cell types and unlike the rest of the replication machinery, these components, are homologous and share a common ancestry [7, 8]. This was not immediately apparent from the sequence of the bacterial beta and eukaryotic PCNA, but the crystal structures of these proteins revealed that they are nearly superimposable (Fig. 3) [85].
Figure. 3. Architecture of sliding clamps.
A) Front view of the E. coli beta [pdb id:2POL] and yeast PCNA [pdb id:1PLQ] sliding clamps. Both rings are composed of six domains with the same chain folding topology. The six domains are arranged on a dimer of E. coli beta, and a trimer of eukaryotic PCNA. B) Side view of the clamps, showing the inherent asymmetry of the C- and N-terminal faces. Most clamp interactive proteins bind the C-face.
Their very different sequences with hard to detect homology is common for proteins that do not serve catalytic functions since their sequences are not constrained by the precise geometry required for catalytic active site residues. But advanced sequence algorithms recognize sequence homology between bacterial and eukaryotic clamps. Both beta and PCNA consist of six domains that form a ring (Fig. 3a). Bacterial beta is a homodimer and each subunit consists of 3 domains that have the same chain folding topology. Eukaryotic PCNA is a homotrimer and each subunit contains two domains, each of which share the same chain folding topology as the bacterial beta clamp. A continuous layer of antiparallel sheet structure, which includes the subunit interfaces, forms the outside perimeter of the clamps. The inside perimeter of the clamp is lined by 12 alpha helices. Side views of the clamps demonstrate that each face is structurally distinct, and are often referred to as the “C-face” (from which the C-termini protrude) and the N-face (Fig. 3b). The C-face is the surface that DNA polymerases and other proteins bind to [86, 87]. The clamp loaders of bacteria and eukaryotes also have similar structures, and their homology is obvious from the primary sequence analysis alone [88, 89]. Clamp loaders consist of five different proteins, arranged in a circle, that together form a central chamber that binds duplex DNA (Fig. 4) [90, 91]. There is a gap between two of the five subunits that provide access for DNA to enter the central chamber. Each of the five clamp loader subunits is a member of the AAA+ family [88, 89]. The first crystal structure of an AAA+ protein was the delta prime subunit of the E. coli clamp loader [92].
Figure. 4. Overview of clamp loader mechanism.
Clamp loaders are circular heteropentamers with ATP sites situated at subunit interfaces. Three domains in each subunit include the N-terminal AAA+ domains and a C-terminal oligomerization domain referred to as a collar. ATP binding enables binding to the C-face of the clamp, opening it at one interface. Primed DNA fits into a central chamber, accessed through a gap between two clamp loader subunits and the open clamp interface. The DNA brings the subunits and clamp into a right-hand spiral, triggering ATP hydrolysis that ejects the clamp loader, enabling the clamp to close around DNA. Polymerase, or other proteins, can then interact with the clamp.
The AAA+ region of homology folds into two domains, one domain contains the Walker A and B motifs for nucleotide binding, while the other domain contains conserved “sensor” residues thought to regulate hydrolysis in response to substrate binding. Clamp loaders are held into circular pentamers by a third domain, C-terminal to the AAA+ domains, which forms a tightly associated circular “collar” from which the AAA+ regions are suspended. Not all the clamp loader subunits bind ATP, as some have been mutated during evolution [93]. The ATP sites are located at subunit interfaces between the AAA+ domains, and catalysis requires an “arginine finger” provided by the subunit adjacent to the nucleotide binding subunit [94]. This bipartite catalytic site construction enables subunit communication during the catalytic reaction.
Many structural and biochemical experiments have been performed on clamp loaders of all types, and the crystal structure of the phage T4 clamp loader (gp44/62) in complex with the gp45 clamp and DNA has provided important details about how the clamp loading reaction works [95]. The overview of clamp loader function is illustrated in Fig. 4. Upon binding ATP, the clamp loader binds the clamp via motifs in the AAA+ domains and the bound clamp opens at one interface, which aligns with the gap in the side of the clamp loader. DNA then enters the central chamber and acts as a scaffold to bring the clamp loading subunits into a spiral pitch that aligns the active site residues at the subunit interfaces into the correct geometry to initiate ATP hydrolysis. Crystal structures of the T4 clamp loader with ADP-BeF in all three sites versus ADP in one site indicate that ATP is hydrolyzed in a particular order [95]. However, once started, all three ATP are destined to hydrolyze and the resulting conformation of the clamp loader breaks its connections to the clamp and DNA, ejecting the clamp loader and allowing the clamp to close. Polymerase, or other proteins, can then interact with the clamp.
4.2. The role of the sliding clamp in LUCA
We do not currently know the role of the clamp in LUCA, but we can make some reasonable proposals. In modern day cells, sliding clamps bind the replicative polymerase and enhance its processivity for DNA replication. Even if LUCA had an RNA genome it is possible that the sliding clamp in LUCA evolved to facilitate the reverse transcriptase in converting RNA to an RNA-DNA hybrid, and again to form the duplex DNA. Although modern day retroviruses do not require a clamp for reverse transcriptase activity, they have rather small genomes while LUCA must have had a much larger genome with genes for the rRNAs, the twenty tRNAs, the twenty tRNA synthetases, in addition to recombinase, RNA polymerase, clamp, clamp loader subunits and a few hundred other universal genes [96, 97]. Thus, the genome of LUCA may have been sufficiently large to require assistance of a sliding clamp, which may have helped hold the reverse transcriptase to DNA for processivity as it does for DNA polymerases in modern day cells. However, clamps function with many other proteins, as described earlier (Sections 3.1.1, 3.1.2), and in more detail below. Thus LUCA may have used clamps for some function other than reverse transcription.
Bacterial beta and eukaryotic PCNA interact with a wide variety of proteins, not just replicative DNA polymerases [43, 98]. E. coli beta interacts with the MutS and MutL proteins of mismatch repair, DNA ligase and all five DNA polymerases, including the replicative and translesion synthesis (TLS) polymerases of repair and mutagenesis [43, 99]. A very large number of proteins interact with eukaryotic PCNA, including those mentioned for bacterial beta, but also a plethora of proteins involved in DNA repair, cell cycle control and other DNA metabolic processes (Table I) [98]. The interaction is often, though not always, mediated by a peptide with a PIP (PCNA Interacting Peptide) motif [100]. The function of the interaction between PCNA and many of its partners is not clear, but one can expect that interaction of a protein with a ring shaped sliding clamp increases the rate of locating lesions, proteins or particular sequences on DNA, simply by converting the search from a 3D diffusion process to a 2D linear diffusion along DNA. This should effectively increase the local concentration of an enzyme that functions on DNA. In this fashion, LUCA could have effectively increased the concentration of proteins that function on DNA, without actually producing a high concentration of these proteins. In essence, this could amount to a cost saving measure in both time and energy for the cell. For LUCA, or at least the version of LUCA proposed here, one reaction of this type would be to increase the efficiency of transcription of the DNA by bringing the RNA polymerase in proximity to DNA via binding the clamp. In fact, the T4 phage offers excellent precedent for just this type of clamp function, in which the T4 gp45 clamp recruits the RNA polymerase to DNA for gene expression [101, 102].
Table 1. PCNA BINDING PROTEINS.
The table contains many proteins known to interact with PCNA. Many other proteins contain PIP motifs and may also bind the PCNA clamp (not listed here). Many entries are from the review by Hubscher & Maga, ref [69], although others have been added to this list.
| PCNA binding protein | Function | References |
|---|---|---|
| Polδ | DNA replication and repair | [117-119] |
| Polε | DNA replication and repair | [120, 121] |
| RF-C | DNA replication and repair | [122] |
| DNA ligase I | DNA replication and repair | [123] |
| Fen1 | DNA replication and repair | [124] |
| Topo I | DNA replication and repair | [125] |
| Topo IIα | DNA replication and repair | [126] |
| MLH1, MSH 2/3/6 | Mismatch DNA repair | [127, 128] |
| XP-G endonuclease | Nucleotide excision repair | [129] |
| WRN helicase | Double strand breaks DNA repair; linked to the Werner Syndrome disease | [130] |
| UBC9 | SUMOylation | [131] |
| Rad18 | Ubiquitin Ligase | [132] |
| AP-endonucleases APN1, APN2 | Base excision repair | [133, 134] |
| Uracil-DNA glycosylase | Base excision repair | [135] |
| Pol β | Base excision repair | [136] |
| Pol η | Translesion synthesis; linked to the XP-V disease | [137] |
| Pol ι | Translesion synthesis | [138] |
| Pol κ | Translesion synthesis | [139] |
| Pol λ | Translesion synthesis/BER/NHEJ | [140] |
| Pol ζ | Translesion synthesis | [141] |
| Rev1 | Translesion synthesis | [142] |
| Cyclin/CDKs | Cell cycle control | [143, 144] |
| p21 | Cell cycle control | [145] |
| Ctf18 | alternative clamp loader in sister chromatid cohesin | [114] |
| Elgl | alternative clamp loader in DNA repair | [146] |
| Pif1 helicase | break induced repair | [147] |
| Mgs1 | Replication stress | [148] |
| CAF-1 | Topological marker for CAF-1 | [100] |
| CAF-1 | Epigenetic inheritance | [149] |
| CAF-1 | Recruitment to DNA damages | [150] |
| P300 | Facilitation of PCNA function in DNA repair | [151] |
| MeCTr | Maintenance of methylation pattern | [152] |
| Ctf7p | Connection of sister chromatin cohesion to DNA Replication | [153] |
| CHL12 | Alternative clamp loader in sister chromatin cohesion | [154] |
| Gadd45 | Negative growth control, prevention of apoptosis | [155] |
| MyD118 | Negative growth control, prevention of apoptosis | [155] |
| Inglp33ING1 | Protection from UV-induced apoptosis | [156] |
4.3. PCNA as a “marker” of newly replicated DNA
One could make the case that clamps are not “fundamentally” required for processive replication, even though clamps are in fact processivity factors that are essential to replication in all cells. Specifically, there is ample modern day precedent for highly processive polymerases that do not utilize a clamp at all. Take for example phi29 DNA polymerase, a single subunit enzyme with high processivity in the absence of any other factors [103]. The DNA polymerases of the T7 phages are another example of highly processive enzymes that do not require a circular clamp [104]. In fact both bacterial and eukaryotic RNA polymerases are fantastically processive, staying on DNA for very long times, yet they do not use separate ring shaped sliding clamp proteins. Furthermore, it is not a “given” that an enzyme that replicates a genome must be highly processive, a distributive enzyme would seem sufficient. Primase is a case in point. E. coli primase is thought to be distributive during replication of the genome [105]. Indeed, many Okazaki fragments are thought to be left incomplete [106], and filled by soluble polymerases after the fork has passed, and thus even processive DNA synthesis during genome replication may not be required.
Given that clamps are not an intrinsically fundamental requirement for highly processive polymerase action, one may turn the argument around and propose that replicative DNA polymerases evolved to piggyback on the use of clamps that are loaded onto DNA for other uses. This may have negated the need for replicative DNA polymerases to become highly processive on their own, since they would have access to clamps that easily provide the extra grip to DNA for high processivity. Interestingly, a dependence on clamps by replicative polymerases has the consequence of ensuring that clamps are used during replication and thus are faithfully deposited on newly replicated DNA, as illustrated in Fig. 5. The cell could then use clamps that are deposited onto newly replicated DNA as reporters, or markers, to distinguish new DNA from old DNA. There are at least two processes that depend on PCNA placement on newly replicated DNA. One of these processes is mismatch repair (MMR). Another process is nucleosome assembly, discussed in more detail below. MMR must not only recognize new DNA, but must also distinguish the new strand from the parental strand, and PCNA is required by MMR for this distinction [107, 108]. As described earlier, PCNA has two distinct faces, a C-face, and an N-face (Fig. 2). The C-face contains the hydrophobic pocket to which the PIP motif of most PCNA interactive proteins binds. At a primed template junction, PCNA is loaded onto DNA such that the C-face points in the direction of synthesis, enabling polymerases to bind the C-face for function. When the eukaryotic MutL homologue binds PCNA, its endonuclease is directed by the asymmetry inherent in the two faces of PCNA, and cleaves only the newly replicated strand [107, 108]. After nicking the new strand, subsequent excision of this new strand back to the mismatch (and a little beyond) ensures removal of the incorrect nucleotide of the mismatched base pair. This same clamp mediated strand discrimination process is believed to occur in bacterial cells that utilize MutL with endonuclease activity [109]. Hence, clamp directed MMR occurs in bacteria and eukaryotes alone. However, E. coli is an example of a bacterium that has evolved a different strategy of strand discrimination. In E coli, MutL lacks endonuclease activity, and a completely different process performs strand discrimination by MMR. E. coli contain MutH, an endonuclease that cleaves at hemimethylated GATC sites, and contain Dam methylase that methylates the A residue in the GATC site [4]. During replication, the new strand is transiently unmethylated in the wake of the fork, giving MutH time to cleave the new (unmethylated) strand in the event that MutS/L detects a mismatch. Why E. coli evolved methyl-directed strand discrimination for MMR is not clear.
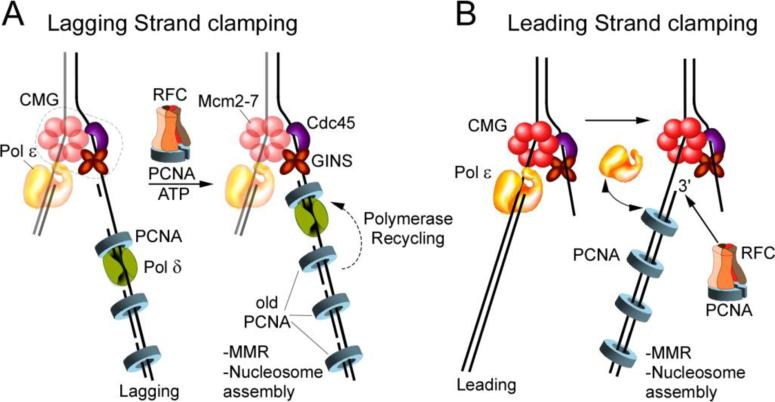
Figure. 5. PCNA marks newly replicated daughter strands for MMR and nucleosome assembly.
Eukaryotic factors are illustrated. A) PCNA clamps are deposited on each Okazaki fragment by virtue of Pol δ dependency on PCNA for function. Upon completing an Okazaki fragment, Pol δ ejects from its PCNA clamp and binds a new clamp at the next RNA primer, leaving PCNA on the newly replicated DNA. B) Proposed process of populating the leading strand with PCNA. Pol ε-CMG does not absolutely require PCNA, but Pol ε has weak affinity for DNA and likely comes on and off the 3’ terminus during leading strand extension, staying bound to the fork through connection to CMG. This provides RFC access to the leading strand for PCNA clamp loading. PCNA marks newly replicated daughter strands for MMR and nucleosome assembly.
Another process that uses the PCNA clamp is nucleosome assembly. Nucleosome assembly is thought to occur nearly coincident with DNA synthesis, directly in back of the fork [75]. New nucleosomes are assembled onto the daughter strands by the Caf1 and Hir nucleosome assembly factors, both of which require PCNA to initiate nucleosome assembly [100, 110]. Hence PCNA deposition onto DNA is important to signal nucleosome assembly onto daughter DNA duplexes.
These considerations imply that PCNA clamps must be left as markers on newly replicated daughter duplexes, to direct MMR and nucleosome assembly. It is therefore reasonable to speculate that replicative polymerases evolved to utilize clamps for processivity, even though polymerases exist that demonstrate intrinsic high processivity without need for a clamp. Specifically, DNA polymerases have evolved to “piggyback” on the existing tendency of clamps to be loaded onto newly replicated DNA in order to increase their processivity. We propose that polymerases that utilize clamps for processivity evolved this property (i.e. clamp usage) in order to ensure that newly replicated DNA has markers (i.e. clamps) to direct downstream processes line MMR. If replicative DNA polymerases did not required clamps, newly replicated DNA would not be able to be marked with clamps.
A well studied process has defined how the lagging strand is marked by sliding clamps. This process was initially identified in the E. coli system [80, 111], and considering the dependence of eukaryotic Pol δ on PCNA, it likely generalizes to eukaryotes [68]. Pol III requires a clamp for synthesis, but to depart from an Okazaki fragment it sacrifices the connection and leaves the clamp on the replicated DNA. In order for Pol III to extend the next Okazaki fragment, its dependence on clamps requires that a new clamp be pre-assembled on the RNA primer at the fork. Hence, polymerase dissociates from its clamp upon finishing an Okazaki fragments, and hops to a new clamp assembled at the next upstream primed site. Thus, clamps accumulate on the lagging strand during this polymerase recycling process during multiple rounds of Okazaki fragment synthesis. This process of polymerase recycling on the lagging strand leaves clamps on DNA where they can interact with other proteins; indeed, their accumulation on the lagging strand has been experimentally demonstrated [112].
Although clamps are abundant in both bacteria and eukaryotes, there are still more Okazaki fragments than clamps, and thus clamps are likely recycled off DNA. Both the E. coli and eukaryotic RFC clamp loaders can also remove clamps from DNA, and eukaryotes contain alternative clamp loaders in which one subunit is replaced by another protein [113]. One of these alternative clamp loaders is a superior clamp unloader and may fulfill this clamp recycling function [114]. Interestingly, the E. coli Pol III and eukaryotic Pol δ have evolved to be highly dependent on function with a clamp. Without a clamp they are nearly non-functional. Perhaps the selection pressure for a polymerase that is highly dependent on a clamp ensures that clamps are in fact used, and thus deposited on the lagging strand for downstream events, like MMR and nucleosome assembly.
The method, by which the leading strand is populated by clamps, if at all, has remained mysterious. The E. coli model system may not yield this answer since MMR in E. coli is directed by methylation. Nor are clamps needed for nucleosomes in bacteria, considering they lack nucleosomes altogether. But this is not the case in eukaryotes, which require PCNA for both MMR and nucleosome assembly, and thus PCNA clamps are expected to mark both the leading and lagging strands. Recent development of a eukaryotic replication fork using CMG helicase and several other enzymes in vitro provides a new clue to how this process might be solved in eukaryotes [62, 115]. Interestingly, the proposed solution has to do with the asymmetric functionality of the leading and lagging strand polymerases. Pol ε, unlike Pol δ, has a subunit (Dpb2) that forms a connection to the Psf1 subunit [60] (Psf1 is present in CMG). This may underlie the recently demonstrated stability of Pol ε-PCNA in leading strand synthesis with CMG compared to Pol δ-PCNA, which is not stabilized by CMG [62]. Unexpectedly, leading strand synthesis by Pol ε-CMG is not absolutely dependent on PCNA, unlike the case of Pol δ, which requires PCNA. Hence, Pol ε appears to gain processivity by connection to the CMG ring in front of it, rather than from the PCNA ring in back of it. Yet, model primed ssDNA systems show that yeast and human Pol ε are stimulated by PCNA under certain conditions, and therefore even though Pol ε lacks a PIP motif, it must have some affinity for PCNA. Considering that Pol ε does not bind PCNA tightly, as demonstrated for Pol δ [116], one would expect that Pol ε often dissociates from the 3’ terminus of a primed site during leading strand synthesis, but remains bound to CMG for re-association to DNA (eg. illustrated in Fig. 5b). This action could enable RFC access to the 3’ terminus of the leading strand for loading and populating the leading strand with PCNA clamps, as proposed [62, 115], and illustrated in Fig. 5.
5. Summary
Sliding clamps and clamp loaders are the only pieces of the replication machinery that are homologous in all domains of life, and thus are presumed to have been present in LUCA. LUCA may have used RNA as a genome, and DNA as a substrate for transcription and recombination. In this scenario, sliding clamps may have had a function(s) other than, or in addition to use in genome replication, possibly as a way of increasing efficiency of protein action on DNA. Evolution of the complicated process of semi-discontinuous DNA replication of antiparallel duplex DNA may have been a late development, and could have selected for DNA polymerases that depend on clamps for activity as a way of ensuring that clamps are used and deposited on daughter strands to mark newly synthesized DNA. In eukaryotes, populating the leading and lagging daughter duplex products with clamps may have been achieved by separate mechanisms, facilitated by use of distinct polymerases on these two strands. Populating clamps on the leading strand may be facilitated by a relatively distributive polymerase that binds the helicase in front of it to stay with the fork, and thus periodically leaves the 3’ terminus for access by RFC in clamp loading. Populating the lagging strand with clamps could be facilitated by a polymerase that evolved to be completely dependent on the clamp for activity, thus requiring a new clamp for every Okazaki fragment. In support of this proposal, there is ample precedent for highly processive DNA and RNA polymerases that do not utilize clamps. Thus clamps are not fundamentally required for high processivity. The use of clamps to mark new DNA may be essential to genome stability, by enabling strand discrimination in MMR. Nucleosome deposition is also facilitated by PCNA, and inheritance of epigenetic modifications during development of a multicellular organism may require rapid targeted assembly of epigenetic modified nucleosomes behind the replication fork. In this perspective, clamps did not evolve to be processivity factors, but as DNA markers instead, and polymerases evolved to utilize them in order to ensure that newly replicated DNA is properly marked by clamps that direct processes in back of the replication fork.
Highlights.
Sliding clamps and clamp loaders share a common ancestry.
Sliding clamps were present in LUCA (Last Universal Common Ancestor)
Helicase, primase, DNA polymerase may not have been present in LUCA
We propose PCNA marks new DNA strands for MMR and nucleosome assembly.
The use of clamps to mark newly replicated DNA may be essential to genome stability.
Acknowledgement
The authors are grateful for funding from US NIH GM38839 and HHMI. We also appreciate help from Nina Yao for artwork in the figure illustrations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. http://dx.doi.org/10.1038/171738a0. [DOI] [PubMed] [Google Scholar]
- 2.Watson JD, Crick FH. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953;171:964–967. doi: 10.1038/171964b0. http://dx.doi.org/10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- 3.Lehman IR, Bessman MJ, Simms ES, Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. I. Preparation of substrates and partial purification of an enzyme from Escherichia coli. J Biol Chem. 1958;233:163–170. [PubMed] [Google Scholar]
- 4.Kornberg A, Baker TA. DNA Replication. 2nd ed. University Science Books; New York: 2005. [Google Scholar]
- 5.Kelch BA, Makino DL, O'Donnell M, Kuriyan J. Clamp loader ATPases and the evolution of DNA replication machinery. BMC Biol. 2012;10:34. doi: 10.1186/1741-7007-10-34. http://dx.doi.org/10.1186/1741-7007-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Donnell M, Kuriyan J. Clamp loaders and replication initiation. Curr Opin Struct Biol. 2006;16:35–41. doi: 10.1016/j.sbi.2005.12.004. http://dx.doi.org/10.1016/j.sbi.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 7.Forterre P, Filee J, Myllykallio H. Origin and Evolution of DNA and DNA Replication Machineries. Landes Bioscience. 2004 Oct 4;:24. 2004. [Google Scholar]
- 8.Leipe DD, Aravind L, Koonin EV. Did DNA replication evolve twice independently? Nucleic Acids Res. 1999;27:3389–3401. doi: 10.1093/nar/27.17.3389. http://dx.doi.org/10.1093/nar/27.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. http://dx.doi.org/10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 10.Woese CR. Genetic Code. New Edition ed. Joanna Cotler Books; 1968. [Google Scholar]
- 11.Steitz TA. DNA polymerases: structural diversity and common mechanisms. J Biol Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. http://dx.doi.org/10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 12.Yang W. An overview of Y-Family DNA polymerases and a case study of human DNA polymerase eta. Biochemistry. 2014;53:2793–2803. doi: 10.1021/bi500019s. http://dx.doi.org/10.1021/bi500019s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beese LS, Steitz TA. Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steitz TA. Visualizing polynucleotide polymerase machines at work. EMBO J. 2006;25:3458–3468. doi: 10.1038/sj.emboj.7601211. http://dx.doi.org/10.1038/sj.emboj.7601211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aravind L, Leipe DD, Koonin EV. Toprim--a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 1998;26:4205–4213. doi: 10.1093/nar/26.18.4205. http://dx.doi.org/10.1093/nar/26.18.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keck JL, Roche DD, Lynch AS, Berger JM. Structure of the RNA polymerase domain of E. coli primase. Science. 2000;287:2482–2486. doi: 10.1126/science.287.5462.2482. http://dx.doi.org/10.1126/science.287.5462.2482. [DOI] [PubMed] [Google Scholar]
- 17.Podobnik M, McInerney P, O'Donnell M, Kuriyan J. A TOPRIM domain in the crystal structure of the catalytic core of Escherichia coli primase confirms a structural link to DNA topoisomerases. J Mol Biol. 2000;300:353–362. doi: 10.1006/jmbi.2000.3844. http://dx.doi.org/10.1006/jmbi.2000.3844. [DOI] [PubMed] [Google Scholar]
- 18.Frick DN, Richardson CC. DNA primases. Annu Rev Biochem. 2001;70:39–80. doi: 10.1146/annurev.biochem.70.1.39. http://dx.doi.org/10.1146/annurev.biochem.70.1.39. [DOI] [PubMed] [Google Scholar]
- 19.Kilkenny ML, Longo MA, Perera RL, Pellegrini L. Structures of human primase reveal design of nucleotide elongation site and mode of Pol alpha tethering. Proc Natl Acad Sci U S A. 2013;110:15961–15966. doi: 10.1073/pnas.1311185110. http://dx.doi.org/10.1073/pnas.1311185110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirk BW, Kuchta RD. Arg304 of human DNA primase is a key contributor to catalysis and NTP binding: primase and the family X polymerases share significant sequence homology. Biochemistry. 1999;38:7727–7736. doi: 10.1021/bi990247c. http://dx.doi.org/10.1021/bi990247c. [DOI] [PubMed] [Google Scholar]
- 21.Kuchta RD, Stengel G. Mechanism and evolution of DNA primases. Biochim Biophys Acta. 2010;1804:1180–1189. doi: 10.1016/j.bbapap.2009.06.011. http://dx.doi.org/10.1016/j.bbapap.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LeBowitz JH, McMacken R. The Escherichia coli dnaB replication protein is a DNA helicase. J Biol Chem. 1986;261:4738–4748. [PubMed] [Google Scholar]
- 23.Bochman ML, Bell SP, Schwacha A. Subunit organization of Mcm2-7 and the unequal role of active sites in ATP hydrolysis and viability. Mol Cell Biol. 2008;28:5865–5873. doi: 10.1128/MCB.00161-08. http://dx.doi.org/10.1128/MCB.00161-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bochman ML, Schwacha A. The Mcm2-7 complex has in vitro helicase activity. Mol Cell. 2008;31:287–293. doi: 10.1016/j.molcel.2008.05.020. http://dx.doi.org/10.1016/j.molcel.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 25.Murzin AG. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat Struct Biol. 2000;7:648–652. doi: 10.1038/77943. http://dx.doi.org/10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 27.Roy R, Kozlov AG, Lohman TM, Ha T. Dynamic structural rearrangements between DNA binding modes of E. coli SSB protein. J Mol Biol. 2007;369:1244–1257. doi: 10.1016/j.jmb.2007.03.079. http://dx.doi.org/10.1016/j.jmb.2007.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bochkarev A, Bochkareva E. From RPA to BRCA2: lessons from single-stranded DNA binding by the OB-fold. Curr Opin Struct Biol. 2004;14:36–42. doi: 10.1016/j.sbi.2004.01.001. http://dx.doi.org/10.1016/j.sbi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 29.Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. http://dx.doi.org/10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. http://dx.doi.org/10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 31.Kaplan DL. The 3′-tail of a forked-duplex sterically determines whether one or two DNA strands pass through the central channel of a replication-fork helicase. J Mol Biol. 2000;301:285–299. doi: 10.1006/jmbi.2000.3965. http://dx.doi.org/10.1006/jmbi.2000.3965. [DOI] [PubMed] [Google Scholar]
- 32.McInerney P, Johnson A, Katz F, O'Donnell M. Characterization of a triple DNA polymerase replisome. Mol Cell. 2007;27:527–538. doi: 10.1016/j.molcel.2007.06.019. http://dx.doi.org/10.1016/j.molcel.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 33.Reyes-Lamothe R, Sherratt DJ, Leake MC. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science. 2010;328:498–501. doi: 10.1126/science.1185757. http://dx.doi.org/10.1126/science.1185757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Georgescu RE, Kurth I, O'Donnell ME. Single-molecule studies reveal the function of a third polymerase in the replisome. Nat Struct Mol Biol. 2011;19:113–116. doi: 10.1038/nsmb.2179. http://dx.doi.org/10.1038/nsmb.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lia G, Michel B, Allemand JF. Polymerase exchange during Okazaki fragment synthesis observed in living cells. Science. 2012;335:328–331. doi: 10.1126/science.1210400. http://dx.doi.org/10.1126/science.1210400. [DOI] [PubMed] [Google Scholar]
- 36.Kuriyan J, O'Donnell M. Sliding clamps of DNA polymerases. J Mol Biol. 1993;234:915–925. doi: 10.1006/jmbi.1993.1644. http://dx.doi.org/10.1006/jmbi.1993.1644. [DOI] [PubMed] [Google Scholar]
- 37.Breier AM, Weier HU, Cozzarelli NR. Independence of replisomes in Escherichia coli chromosomal replication. Proc Natl Acad Sci U S A. 2005;102:3942–3947. doi: 10.1073/pnas.0500812102. http://dx.doi.org/10.1073/pnas.0500812102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeruzalmi D, O'Donnell M, Kuriyan J. Clamp loaders and sliding clamps. Curr Opin Struct Biol. 2002;12:217–224. doi: 10.1016/s0959-440x(02)00313-5. http://dx.doi.org/10.1016/S0959-440X(02)00313-5. [DOI] [PubMed] [Google Scholar]
- 39.Gao D, McHenry CS. tau binds and organizes Escherichia coli replication proteins through distinct domains. Domain IV, located within the unique C terminus of tau, binds the replication fork, helicase, DnaB. J Biol Chem. 2001;276:4441–4446. doi: 10.1074/jbc.M009830200. http://dx.doi.org/10.1074/jbc.M009830200. [DOI] [PubMed] [Google Scholar]
- 40.Turner J, Hingorani MM, Kelman Z, O'Donnell M. The internal workings of a DNA polymerase clamp-loading machine. EMBO J. 1999;18:771–783. doi: 10.1093/emboj/18.3.771. http://dx.doi.org/10.1093/emboj/18.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dallmann HG, McHenry CS. DnaX complex of Escherichia coli DNA polymerase III holoenzyme. Physical characterization of the DnaX subunits and complexes. J Biol Chem. 1995;270:29563–29569. [PubMed] [Google Scholar]
- 42.Lopez de Saro FJ, Marinus MG, Modrich P, O'Donnell M. The beta sliding clamp binds to multiple sites within MutL and MutS. J Biol Chem. 2006;281:14340–14349. doi: 10.1074/jbc.M601264200. http://dx.doi.org/10.1074/jbc.M601264200. [DOI] [PubMed] [Google Scholar]
- 43.Lopez de Saro FJ, O'Donnell M. Interaction of the beta sliding clamp with MutS, ligase, and DNA polymerase I. Proc Natl Acad Sci U S A. 2001;98:8376–8380. doi: 10.1073/pnas.121009498. http://dx.doi.org/10.1073/pnas.121009498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benkovic SJ, Valentine AM, Salinas F. Replisome-mediated DNA replication. Annu Rev Biochem. 2001;70:181–208. doi: 10.1146/annurev.biochem.70.1.181. 70/1/181 [pii] 10.1146/annurev.biochem.70.1.181. [DOI] [PubMed] [Google Scholar]
- 45.Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol. 2011;18:471–477. doi: 10.1038/nsmb.2004. http://dx.doi.org/10.1038/nsmb.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. http://dx.doi.org/10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kang YH, Farina A, Bermudez VP, Tappin I, Du F, Galal WC, Hurwitz J. Interaction between human Ctf4 and the Cdc45/Mcm2-7/GINS (CMG) replicative helicase. Proc Natl Acad Sci U S A. 2013;110:19760–19765. doi: 10.1073/pnas.1320202110. http://dx.doi.org/10.1073/pnas.1320202110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kang YH, Galal WC, Farina A, Tappin I, Hurwitz J. Properties of the human Cdc45/Mcm2-7/GINS helicase complex and its action with DNA polymerase epsilon in rolling circle DNA synthesis. Proc Natl Acad Sci U S A. 2012;109:6042–6047. doi: 10.1073/pnas.1203734109. http://dx.doi.org/10.1073/pnas.1203734109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci U S A. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. http://dx.doi.org/10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bochman ML, Schwacha A. The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol Mol Biol Rev. 2009;73:652–683. doi: 10.1128/MMBR.00019-09. http://dx.doi.org/10.1128/MMBR.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. http://dx.doi.org/10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 52.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. http://dx.doi.org/10.1101/gr.9.1.27. [PubMed] [Google Scholar]
- 53.Costa A, Renault L, Swuec P, Petojevic T, Pesavento J, Ilves I, MacLellan-Gibson K, Fleck RA, Botchan MR, Berger JM. DNA binding polarity, dimerization, and ATPase ring remodeling in the CMG helicase of the eukaryotic replisome. Elife. 2014:e03273. doi: 10.7554/eLife.03273. 10.7554/eLife.03273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kunkel TA, Burgers PM. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 2008;18:521–527. doi: 10.1016/j.tcb.2008.08.005. http://dx.doi.org/10.1016/j.tcb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. http://dx.doi.org/10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hogg M, Johansson E. DNA polymerase epsilon. Subcell Biochem. 2012;62:237–257. doi: 10.1007/978-94-007-4572-8_13. http://dx.doi.org/10.1007/978-94-007-4572-8_13. [DOI] [PubMed] [Google Scholar]
- 57.Hogg M, Osterman P, Bylund GO, Ganai RA, Lundstrom EB, Sauer-Eriksson AE, Johansson E. Structural basis for processive DNA synthesis by yeast DNA polymerase varepsilon. Nat Struct Mol Biol. 2014;21:49–55. doi: 10.1038/nsmb.2712. http://dx.doi.org/10.1038/nsmb.2712. [DOI] [PubMed] [Google Scholar]
- 58.Johansson E, Dixon N. Replicative DNA polymerases. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a012799. http://dx.doi.org/10.1101/cshperspect.a012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Isoz I, Persson U, Volkov K, Johansson E. The C-terminus of Dpb2 is required for interaction with Pol2 and for cell viability. Nucleic Acids Res. 2012;40:11545–11553. doi: 10.1093/nar/gks880. http://dx.doi.org/10.1093/nar/gks880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sengupta S, van Deursen F, de Piccoli G, Labib K. Dpb2 integrates the leading-strand DNA polymerase into the eukaryotic replisome. Curr Biol. 2013;23:543–552. doi: 10.1016/j.cub.2013.02.011. http://dx.doi.org/10.1016/j.cub.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 61.Muramatsu S, Hirai K, Tak YS, Kamimura Y, Araki H. CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol (epsilon}, and GINS in budding yeast. Genes Dev. 2010;24:602–612. doi: 10.1101/gad.1883410. http://dx.doi.org/10.1101/gad.1883410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Georgescu RE, Langston LD, Yao NY, Yurieva O, Zhang D, Finkelstein J, Agarwal T, O'Donnell ME. Mechanism of Asymmetric Polymerase Assembly at the Eukaryotic Replication Fork. Nat Struct Mol Biol. 2014 doi: 10.1038/nsmb.2851. http://dx.doi.org/10.1038/nsmb.2851. [DOI] [PMC free article] [PubMed]
- 63.Garg P, Burgers PM. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit Rev Biochem Mol Biol. 2005;40:115–128. doi: 10.1080/10409230590935433. http://dx.doi.org/10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- 64.Stillman B. DNA polymerases at the replication fork in eukaryotes. Mol Cell. 2008;30:259–260. doi: 10.1016/j.molcel.2008.04.011. http://dx.doi.org/10.1016/j.molcel.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johansson E, Macneill SA. The eukaryotic replicative DNA polymerases take shape. Trends Biochem Sci. 2010;35:339–347. doi: 10.1016/j.tibs.2010.01.004. http://dx.doi.org/10.1016/j.tibs.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 66.Netz DJ, Stith CM, Stumpfig M, Kopf G, Vogel D, Genau HM, Stodola JL, Lill R, Burgers PM, Pierik AJ. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat Chem Biol. 2011;8:125–132. doi: 10.1038/nchembio.721. http://dx.doi.org/10.1038/nchembio.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sanchez Garcia J, Ciufo LF, Yang X, Kearsey SE, MacNeill SA. The C-terminal zinc finger of the catalytic subunit of DNA polymerase delta is responsible for direct interaction with the B-subunit. Nucleic Acids Res. 2004;32:3005–3016. doi: 10.1093/nar/gkh623. http://dx.doi.org/10.1093/nar/gkh623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Langston LD, O'Donnell M. DNA polymerase delta is highly processive with proliferating cell nuclear antigen and undergoes collision release upon completing DNA. J Biol Chem. 2008;283:29522–29531. doi: 10.1074/jbc.M804488200. http://dx.doi.org/10.1074/jbc.M804488200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kaguni LS, Rossignol JM, Conaway RC, Banks GR, Lehman IR. Association of DNA primase with the beta/gamma subunits of DNA polymerase alpha from Drosophila melanogaster embryos. J Biol Chem. 1983;258:9037–9039. [PubMed] [Google Scholar]
- 70.Kaguni LS, Rossignol JM, Conaway RC, Lehman IR. Isolation of an intact DNA polymerase-primase from embryos of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1983;80:2221–2225. doi: 10.1073/pnas.80.8.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hubscher U, Maga G. DNA replication and repair bypass machines. Curr Opin Chem Biol. 2011;15:627–635. doi: 10.1016/j.cbpa.2011.08.009. http://dx.doi.org/10.1016/j.cbpa.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 72.Garg P, Stith CM, Sabouri N, Johansson E, Burgers PM. Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 2004;18:2764–2773. doi: 10.1101/gad.1252304. http://dx.doi.org/10.1101/gad.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Clausen AR, Zhang S, Burgers PM, Lee MY, Kunkel TA. Ribonucleotide incorporation, proofreading and bypass by human DNA polymerase delta. DNA Repair (Amst) 2013;12:121–127. doi: 10.1016/j.dnarep.2012.11.006. http://dx.doi.org/10.1016/j.dnarep.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pavlov YI, Frahm C, Nick McElhinny SA, Niimi A, Suzuki M, Kunkel TA. Evidence that errors made by DNA polymerase alpha are corrected by DNA polymerase delta. Curr Biol. 2006;16:202–207. doi: 10.1016/j.cub.2005.12.002. http://dx.doi.org/10.1016/j.cub.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 75.Smith DJ, Whitehouse I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 2012;483:434–438. doi: 10.1038/nature10895. http://dx.doi.org/10.1038/nature10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. http://dx.doi.org/10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 77.Gambus A, van Deursen F, Polychronopoulos D, Foltman M, Jones RC, Edmondson RD, Calzada A, Labib K. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J. 2009;28:2992–3004. doi: 10.1038/emboj.2009.226. http://dx.doi.org/10.1038/emboj.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Simon AC, Zhou JC, Perera RL, van Deursen F, Evrin C, Ivanova ME, Kilkenny ML, Renault L, Kjaer S, Matak-Vinkovic D, Labib K, Costa A, Pellegrini L. A Ctf4 trimer couples the CMG helicase to DNA polymerase alpha in the eukaryotic replisome. Nature. 2014 doi: 10.1038/nature13234. http://dx.doi.org/10.1038/nature13234. [DOI] [PMC free article] [PubMed]
- 79.Miles J, Formosa T. Protein affinity chromatography with purified yeast DNA polymerase alpha detects proteins that bind to DNA polymerase. Proc Natl Acad Sci U S A. 1992;89:1276–1280. doi: 10.1073/pnas.89.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.O'Donnell ME. Accessory proteins bind a primed template and mediate rapid cycling of DNA polymerase III holoenzyme from Escherichia coli. J Biol Chem. 1987;262:16558–16565. [PubMed] [Google Scholar]
- 81.Kelman Z, O'Donnell M. DNA replication: enzymology and mechanisms. Curr Opin Genet Dev. 1994;4:185–195. doi: 10.1016/s0959-437x(05)80044-9. http://dx.doi.org/10.1016/S0959-437X(05)80044-9. [DOI] [PubMed] [Google Scholar]
- 82.Kelman Z, O'Donnell M. Structural and functional similarities of prokaryotic and eukaryotic DNA polymerase sliding clamps. Nucleic Acids Res. 1995;23:3613–3620. doi: 10.1093/nar/23.18.3613. http://dx.doi.org/10.1093/nar/23.18.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stukenberg PT, Studwell-Vaughan PS, O'Donnell M. Mechanism of the sliding beta-clamp of DNA polymerase III holoenzyme. J Biol Chem. 1991;266:11328–11334. [PubMed] [Google Scholar]
- 84.Kong XP, Onrust R, O'Donnell M, Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. http://dx.doi.org/10.1016/0092-8674(92)90445-I. [DOI] [PubMed] [Google Scholar]
- 85.Gulbis JM, Kelman Z, Hurwitz J, O'Donnell M, Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. http://dx.doi.org/10.1016/S0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 86.Mossi R, Jonsson ZO, Allen BL, Hardin SH, Hubscher U. Replication factor C interacts with the C-terminal side of proliferating cell nuclear antigen. J Biol Chem. 1997;272:1769–1776. doi: 10.1074/jbc.272.3.1769. http://dx.doi.org/10.1074/jbc.272.3.1769. [DOI] [PubMed] [Google Scholar]
- 87.Naktinis V, Turner J, O'Donnell M. A molecular switch in a replication machine defined by an internal competition for protein rings. Cell. 1996;84:137–145. doi: 10.1016/s0092-8674(00)81000-4. http://dx.doi.org/10.1016/S0092-8674(00)81000-4. [DOI] [PubMed] [Google Scholar]
- 88.Cullmann G, Fien K, Kobayashi R, Stillman B. Characterization of the five replication factor C genes of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:4661–4671. doi: 10.1128/mcb.15.9.4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.O'Donnell M, Onrust R, Dean FB, Chen M, Hurwitz J. Homology in accessory proteins of replicative polymerases--E. coli to humans. Nucleic Acids Res. 1993;21:1–3. doi: 10.1093/nar/21.1.1. http://dx.doi.org/10.1093/nar/21.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bowman GD, O'Donnell M, Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature. 2004;429:724–730. doi: 10.1038/nature02585. http://dx.doi.org/10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- 91.Simonetta KR, Kazmirski SL, Goedken ER, Cantor AJ, Kelch BA, McNally R, Seyedin SN, Makino DL, O'Donnell M, Kuriyan J. The mechanism of ATP-dependent primer-template recognition by a clamp loader complex. Cell. 2009;137:659–671. doi: 10.1016/j.cell.2009.03.044. http://dx.doi.org/10.1016/j.cell.2009.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guenther B, Onrust R, Sali A, O'Donnell M, Kuriyan J. Crystal structure of the delta' subunit of the clamp-loader complex of E. coli DNA polymerase III. Cell. 1997;91:335–345. doi: 10.1016/s0092-8674(00)80417-1. http://dx.doi.org/S0092-8674(00)80417. [DOI] [PubMed] [Google Scholar]
- 93.Hedglin M, Kumar R, Benkovic SJ. Replication clamps and clamp loaders. Cold Spring Harb Perspect Biol. 2013;5:a010165. doi: 10.1101/cshperspect.a010165. 5/4/a010165 [pii] 10.1101/cshperspect.a010165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Johnson A, Yao NY, Bowman GD, Kuriyan J, O'Donnell M. The replication factor C clamp loader requires arginine finger sensors to drive DNA binding and proliferating cell nuclear antigen loading. J Biol Chem. 2006;281:35531–35543. doi: 10.1074/jbc.M606090200. http://dx.doi.org/10.1074/jbc.M606090200. [DOI] [PubMed] [Google Scholar]
- 95.Kelch BA, Makino DL, O'Donnell M, Kuriyan J. How a DNA polymerase clamp loader opens a sliding clamp. Science. 2011;334:1675–1680. doi: 10.1126/science.1211884. http://dx.doi.org/10.1126/science.1211884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Koonin EV. Carl Woese's vision of cellular evolution and the domains of life. RNA Biol. 2014;11:197–204. doi: 10.4161/rna.27673. http://dx.doi.org/10.4161/rna.27673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Puigbo P, Wolf YI, Koonin EV. Genome-wide comparative analysis of phylogenetic trees: the prokaryotic forest of life. Methods Mol Biol. 2012;856:53–79. doi: 10.1007/978-1-61779-585-5_3. http://dx.doi.org/10.1007/978-1-61779-585-5_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Maga G, Hubscher U. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J Cell Sci. 2003;116:3051–3060. doi: 10.1242/jcs.00653. http://dx.doi.org/10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- 99.Indiani C, Langston LD, Yurieva O, Goodman MF, O'Donnell M. Translesion DNA polymerases remodel the replisome and alter the speed of the replicative helicase. Proc Natl Acad Sci U S A. 2009;106:6031–6038. doi: 10.1073/pnas.0901403106. http://dx.doi.org/10.1073/pnas.0901403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. http://dx.doi.org/S0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- 101.Kolesky SE, Ouhammouch M, Geiduschek EP. The mechanism of transcriptional activation by the topologically DNA-linked sliding clamp of bacteriophage T4. J Mol Biol. 2002;321:767–784. doi: 10.1016/s0022-2836(02)00732-5. http://dx.doi.org/S0022283602007325 [pii]. [DOI] [PubMed] [Google Scholar]
- 102.Tinker-Kulberg RL, Fu TJ, Geiduschek EP, Kassavetis GA. A direct interaction between a DNA-tracking protein and a promoter recognition protein: implications for searching DNA sequence. EMBO J. 1996;15:5032–5039. [PMC free article] [PubMed] [Google Scholar]
- 103.Blanco L, Salas M. Replication of phage phi 29 DNA with purified terminal protein and DNA polymerase: synthesis of full-length phi 29 DNA. Proc Natl Acad Sci U S A. 1985;82:6404–6408. doi: 10.1073/pnas.82.19.6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Andraos N, Tabor S, Richardson CC. The highly processive DNA polymerase of bacteriophage T5. Role of the unique N and C termini. J Biol Chem. 2004;279:50609–50618. doi: 10.1074/jbc.M408428200. http://dx.doi.org/10.1074/jbc.M408428200. [DOI] [PubMed] [Google Scholar]
- 105.Wu CA, Zechner EL, Marians KJ. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. I. Multiple effectors act to modulate Okazaki fragment size. J Biol Chem. 1992;267:4030–4044. [PubMed] [Google Scholar]
- 106.Yang J, Zhuang Z, Roccasecca RM, Trakselis MA, Benkovic SJ. The dynamic processivity of the T4 DNA polymerase during replication. Proc Natl Acad Sci U S A. 2004;101:8289–8294. doi: 10.1073/pnas.0402625101. http://dx.doi.org/10.1073/pnas.0402625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Goellner EM, Smith CE, Campbell CS, Hombauer H, Desai A, Putnam CD, Kolodner RD. PCNA and Msh2-Msh6 Activate an Mlh1-Pms1 Endonuclease Pathway Required for Exo1-Independent Mismatch Repair. Mol Cell. 2014;55:291–304. doi: 10.1016/j.molcel.2014.04.034. http://dx.doi.org/10.1016/j.molcel.2014.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pluciennik A, Dzantiev L, Iyer RR, Constantin N, Kadyrov FA, Modrich P. PCNA function in the activation and strand direction of MutLalpha endonuclease in mismatch repair. Proc Natl Acad Sci U S A. 2010;107:16066–16071. doi: 10.1073/pnas.1010662107. http://dx.doi.org/10.1073/pnas.1010662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bolz NJ, Lenhart JS, Weindorf SC, Simmons LA. Residues in the N-terminal domain of MutL required for mismatch repair in Bacillus subtilis. J Bacteriol. 2012;194:5361–5367. doi: 10.1128/JB.01142-12. http://doi.org/10.1128/JB.01142-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sharp JA, Fouts ET, Krawitz DC, Kaufman PD. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr Biol. 2001;11:463–473. doi: 10.1016/s0960-9822(01)00140-3. http://dx.doi.org/S0960-9822(01)00140-3. [DOI] [PubMed] [Google Scholar]
- 111.Stukenberg PT, Turner J, O'Donnell M. An explanation for lagging strand replication: polymerase hopping among DNA sliding clamps. Cell. 1994;78:877–887. doi: 10.1016/s0092-8674(94)90662-9. http://dx.doi.org/S0092-8674(94)90662-9. [DOI] [PubMed] [Google Scholar]
- 112.Yuzhakov A, Turner J, O'Donnell M. Replisome assembly reveals the basis for asymmetric function in leading and lagging strand replication. Cell. 1996;86:877–886. doi: 10.1016/s0092-8674(00)80163-4. http://dx.doi.org/S0092-8674(00)80163-4. [DOI] [PubMed] [Google Scholar]
- 113.Johnson A, O'Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. http://dx.doi.org/10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 114.Bylund GO, Burgers PM. Replication protein A-directed unloading of PCNA by the Ctf18 cohesion establishment complex. Mol Cell Biol. 2005;25:5445–5455. doi: 10.1128/MCB.25.13.5445-5455.2005. http://dx.doi.org/10.1128/MCB.25.13.5445-5455.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kunkel TA, Burgers PM. Delivering nonidentical twins. Nat Struct Mol Biol. 2014 doi: 10.1038/nsmb.2852. http://dx.doi.org/10.1038/nsmb.2852. [DOI] [PMC free article] [PubMed]
- 116.Chilkova O, Stenlund P, Isoz I, Stith CM, Grabowski P, Lundstrom EB, Burgers PM, Johansson E. The eukaryotic leading and lagging strand DNA polymerases are loaded onto primer-ends via separate mechanisms but have comparable processivity in the presence of PCNA. Nucleic Acids Res. 2007;35:6588–6597. doi: 10.1093/nar/gkm741. http://dx.doi.org/10.1093/nar/gkm741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ducoux M, Urbach S, Baldacci G, Hubscher U, Koundrioukoff S, Christensen J, Hughes P. Mediation of proliferating cell nuclear antigen (PCNA)-dependent DNA replication through a conserved p21(Cip1)-like PCNA-binding motif present in the third subunit of human DNA polymerase delta. J Biol Chem. 2001;276:49258–49266. doi: 10.1074/jbc.M106990200. http://dx.doi.org/10.1074/jbc.M106990200. [DOI] [PubMed] [Google Scholar]
- 118.Zhang P, Mo JY, Perez A, Leon A, Liu L, Mazloum N, Xu H, Lee MY. Direct interaction of proliferating cell nuclear antigen with the p125 catalytic subunit of mammalian DNA polymerase delta. J Biol Chem. 1999;274:26647–26653. doi: 10.1074/jbc.274.38.26647. http://dx.doi.org/10.1074/jbc.274.38.26647. [DOI] [PubMed] [Google Scholar]
- 119.Zhou JQ, He H, Tan CK, Downey KM, So AG. The small subunit is required for functional interaction of DNA polymerase delta with the proliferating cell nuclear antigen. Nucleic Acids Res. 1997;25:1094–1099. doi: 10.1093/nar/25.6.1094. http://dx.doi.org/10.1093/nar/25.6.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dua R, Levy DL, Li CM, Snow PM, Campbell JL. In vivo reconstitution of Saccharomyces cerevisiae DNA polymerase epsilon in insect cells. Purification and characterization. J Biol Chem. 2002;277:7889–7896. doi: 10.1074/jbc.M108546200. http://dx.doi.org/10.1074/jbc.M108546200. [DOI] [PubMed] [Google Scholar]
- 121.Eissenberg JC, Ayyagari R, Gomes XV, Burgers PM. Mutations in yeast proliferating cell nuclear antigen define distinct sites for interaction with DNA polymerase delta and DNA polymerase epsilon. Mol Cell Biol. 1997;17:6367–6378. doi: 10.1128/mcb.17.11.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fotedar R, Mossi R, Fitzgerald P, Rousselle T, Maga G, Brickner H, Messier H, Kasibhatla S, Hubscher U, Fotedar A. A conserved domain of the large subunit of replication factor C binds PCNA and acts like a dominant negative inhibitor of DNA replication in mammalian cells. EMBO J. 1996;15:4423–4433. [PMC free article] [PubMed] [Google Scholar]
- 123.Levin DS, Bai W, Yao N, O'Donnell M, Tomkinson AE. An interaction between DNA ligase I and proliferating cell nuclear antigen: implications for Okazaki fragment synthesis and joining. Proc Natl Acad Sci U S A. 1997;94:12863–12868. doi: 10.1073/pnas.94.24.12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Warbrick E, Lane DP, Glover DM, Cox LS. Homologous regions of Fen1 and p21Cip1 compete for binding to the same site on PCNA: a potential mechanism to co-ordinate DNA replication and repair. Oncogene. 1997;14:2313–2321. doi: 10.1038/sj.onc.1201072. http://dx.doi.org/10.1038/sj.onc.1201072. [DOI] [PubMed] [Google Scholar]
- 125.Loor G, Zhang SJ, Zhang P, Toomey NL, Lee MY. Identification of DNA replication and cell cycle proteins that interact with PCNA. Nucleic Acids Res. 1997;25:5041–5046. doi: 10.1093/nar/25.24.5041. http://dx.doi.org/10.1093/nar/25.24.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Niimi A, Suka N, Harata M, Kikuchi A, Mizuno S. Co-localization of chicken DNA topoisomerase IIalpha, but not beta, with sites of DNA replication and possible involvement of a C-terminal region of alpha through its binding to PCNA. Chromosoma. 2001;110:102–114. doi: 10.1007/s004120100140. http://dx.doi.org/10.1007/s004120100140. [DOI] [PubMed] [Google Scholar]
- 127.Clark AB, Valle F, Drotschmann K, Gary RK, Kunkel TA. Functional interaction of proliferating cell nuclear antigen with MSH2-MSH6 and MSH2-MSH3 complexes. J Biol Chem. 2000;275:36498–36501. doi: 10.1074/jbc.C000513200. http://dx.doi.org/10.1074/jbc.C000513200. [DOI] [PubMed] [Google Scholar]
- 128.Umar A, Buermeyer AB, Simon JA, Thomas DC, Clark AB, Liskay RM, Kunkel TA. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. http://dx.doi.org/10.1016/S0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- 129.Gary R, Ludwig DL, Cornelius HL, MacInnes MA, Park MS. The DNA repair endonuclease XPG binds to proliferating cell nuclear antigen (PCNA) and shares sequence elements with the PCNA-binding regions of FEN-1 and cyclin-dependent kinase inhibitor p21. J Biol Chem. 1997;272:24522–24529. doi: 10.1074/jbc.272.39.24522. http://dx.doi.org/10.1074/jbc.272.39.24522. [DOI] [PubMed] [Google Scholar]
- 130.Lebel M, Spillare EA, Harris CC, Leder P. The Werner syndrome gene product co-purifies with the DNA replication complex and interacts with PCNA and topoisomerase I. J Biol Chem. 1999;274:37795–37799. doi: 10.1074/jbc.274.53.37795. http://dx.doi.org/10.1074/jbc.274.53.37795. [DOI] [PubMed] [Google Scholar]
- 131.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. http://dx.doi.org/10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 132.Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. http://dx.doi.org/10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dianova II, Bohr VA, Dianov GL. Interaction of human AP endonuclease 1 with flap endonuclease 1 and proliferating cell nuclear antigen involved in long-patch base excision repair. Biochemistry. 2001;40:12639–12644. doi: 10.1021/bi011117i. http://dx.doi.org/10.1021/bi011117i. [DOI] [PubMed] [Google Scholar]
- 134.Unk I, Haracska L, Gomes XV, Burgers PM, Prakash L, Prakash S. Stimulation of 3′-->5′ exonuclease and 3′-phosphodiesterase activities of yeast apn2 by proliferating cell nuclear antigen. Mol Cell Biol. 2002;22:6480–6486. doi: 10.1128/MCB.22.18.6480-6486.2002. http://dx.doi.org/10.1128/MCB.22.18.6480-6486.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Krokan HE, Otterlei M, Nilsen H, Kavli B, Skorpen F, Andersen S, Skjelbred C, Akbari M, Aas PA, Slupphaug G. Properties and functions of human uracil-DNA glycosylase from the UNG gene. Prog Nucleic Acid Res Mol Biol. 2001;68:365–386. doi: 10.1016/s0079-6603(01)68112-1. [DOI] [PubMed] [Google Scholar]
- 136.Kedar PS, Kim SJ, Robertson A, Hou E, Prasad R, Horton JK, Wilson SH. Direct interaction between mammalian DNA polymerase beta and proliferating cell nuclear antigen. J Biol Chem. 2002;277:31115–31123. doi: 10.1074/jbc.M201497200. http://dx.doi.org/10.1074/jbc.M201497200. [DOI] [PubMed] [Google Scholar]
- 137.Haracska L, Johnson RE, Unk I, Phillips B, Hurwitz J, Prakash L, Prakash S. Physical and functional interactions of human DNA polymerase eta with PCNA. Mol Cell Biol. 2001;21:7199–7206. doi: 10.1128/MCB.21.21.7199-7206.2001. http://dx.doi.org/10.1128/MCB.21.21.7199-7206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Haracska L, Johnson RE, Unk I, Phillips BB, Hurwitz J, Prakash L, Prakash S. Targeting of human DNA polymerase iota to the replication machinery via interaction with PCNA. Proc Natl Acad Sci U S A. 2001;98:14256–14261. doi: 10.1073/pnas.261560798. http://dx.doi.org/10.1073/pnas.261560798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Haracska L, Unk I, Johnson RE, Phillips BB, Hurwitz J, Prakash L, Prakash S. Stimulation of DNA synthesis activity of human DNA polymerase kappa by PCNA. Mol Cell Biol. 2002;22:784–791. doi: 10.1128/MCB.22.3.784-791.2002. http://dx.doi.org/10.1128/MCB.22.3.784-791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Maga G, Villani G, Ramadan K, Shevelev I, Tanguy Le Gac N, Blanco L, Blanca G, Spadari S, Hubscher U. Human DNA polymerase lambda functionally and physically interacts with proliferating cell nuclear antigen in normal and translesion DNA synthesis. J Biol Chem. 2002;277:48434–48440. doi: 10.1074/jbc.M206889200. http://dx.doi.org/10.1074/jbc.M206889200. [DOI] [PubMed] [Google Scholar]
- 141.Garg P, Stith CM, Majka J, Burgers PM. Proliferating cell nuclear antigen promotes translesion synthesis by DNA polymerase zeta. J Biol Chem. 2005;280:23446–23450. doi: 10.1074/jbc.C500173200. http://dx.doi.org/10.1074/jbc.C500173200. [DOI] [PubMed] [Google Scholar]
- 142.Wood A, Garg P, Burgers PM. A ubiquitin-binding motif in the translesion DNA polymerase Rev1 mediates its essential functional interaction with ubiquitinated proliferating cell nuclear antigen in response to DNA damage. J Biol Chem. 2007;282:20256–20263. doi: 10.1074/jbc.M702366200. http://dx.doi.org/10.1074/jbc.M702366200. [DOI] [PubMed] [Google Scholar]
- 143.Xiong Y, Zhang H, Beach D. D type cyclins associate with multiple protein kinases and the DNA replication and repair factor PCNA. Cell. 1992;71:505–514. doi: 10.1016/0092-8674(92)90518-h. http://dx.doi.org/0092-8674(92)90518-H. [DOI] [PubMed] [Google Scholar]
- 144.Zhang H, Xiong Y, Beach D. Proliferating cell nuclear antigen and p21 are components of multiple cell cycle kinase complexes. Mol Biol Cell. 1993;4:897–906. doi: 10.1091/mbc.4.9.897. http://dx.doi.org/10.1091/mbc.4.9.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Waga S, Hannon GJ, Beach D, Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature. 1994;369:574–578. doi: 10.1038/369574a0. http://dx.doi.org/10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- 146.Ulrich HD. New insights into replication clamp unloading. J Mol Biol. 2013;425:4727–4732. doi: 10.1016/j.jmb.2013.05.003. http://dx.doi.org/10.1016/j.jmb.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 147.Schmidt KH, Derry KL, Kolodner RD. Saccharomyces cerevisiae RRM3, a 5′ to 3′ DNA helicase, physically interacts with proliferating cell nuclear antigen. J Biol Chem. 2002;277:45331–45337. doi: 10.1074/jbc.M207263200. http://dx.doi.org/10.1074/jbc.M207263200. [DOI] [PubMed] [Google Scholar]
- 148.Hishida T, Ohya T, Kubota Y, Kamada Y, Shinagawa H. Functional and physical interaction of yeast Mgs1 with PCNA: impact on RAD6-dependent DNA damage tolerance. Mol Cell Biol. 2006;26:5509–5517. doi: 10.1128/MCB.00307-06. http://dx.doi.org/10.1128/MCB.00307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhang Z, Shibahara K, Stillman B. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature. 2000;408:221–225. doi: 10.1038/35041601. http://dx.doi.org/10.1038/35041601. [DOI] [PubMed] [Google Scholar]
- 150.Moggs JG, Grandi P, Quivy JP, Jonsson ZO, Hubscher U, Becker PB, Almouzni G. A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol Cell Biol. 2000;20:1206–1218. doi: 10.1128/mcb.20.4.1206-1218.2000. http://dx.doi.org/10.1128/MCB.20.4.1206-1218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hasan S, Hassa PO, Imhof R, Hottiger MO. Transcription coactivator p300 binds PCNA and may have a role in DNA repair synthesis. Nature. 2001;410:387–391. doi: 10.1038/35066610. http://dx.doi.org/10.1038/35066610. [DOI] [PubMed] [Google Scholar]
- 152.Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. http://dx.doi.org/10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 153.Skibbens RV, Corson LB, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 1999;13:307–319. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ohta S, Shiomi Y, Sugimoto K, Obuse C, Tsurimoto T. A proteomics approach to identify proliferating cell nuclear antigen (PCNA)-binding proteins in human cell lysates. Identification of the human CHL12/RFCs2-5 complex as a novel PCNA-binding protein. J Biol Chem. 2002;277:40362–40367. doi: 10.1074/jbc.M206194200. http://dx.doi.org/10.1074/jbc.M206194200. [DOI] [PubMed] [Google Scholar]
- 155.Vairapandi M, Azam N, Balliet AG, Hoffman B, Liebermann DA. Characterization of MyD118, Gadd45, and proliferating cell nuclear antigen (PCNA) interacting domains. PCNA impedes MyD118 AND Gadd45-mediated negative growth control. J Biol Chem. 2000;275:16810–16819. doi: 10.1074/jbc.275.22.16810. http://dx.doi.org/275/22/16810 [pii]. [DOI] [PubMed] [Google Scholar]
- 156.Scott M, Bonnefin P, Vieyra D, Boisvert FM, Young D, Bazett-Jones DP, Riabowol K. UV-induced binding of ING1 to PCNA regulates the induction of apoptosis. J Cell Sci. 2001;114:3455–3462. doi: 10.1242/jcs.114.19.3455. [DOI] [PubMed] [Google Scholar]