Summary
The biogenesis of autophagosomes commences at the phagophore assembly site (PAS), a protein-vesicle ultrastructure that is organized by the Atg1 complex. The Atg1 complex consists of the Atg1 protein kinase, the intrinsically disordered region-rich Atg13, and the dimeric double crescent-shaped Atg17-Atg31-Atg29 subcomplex. We show that the PAS contains a relatively uniform ~28 copies of Atg17, and upon autophagy induction, similar numbers of Atg1 and Atg13 molecules. We then apply ensemble refinement of small angle x-ray scattering (SAXS) to determine the solution structures of the Atg1-Atg13 and Atg17-Atg31-Atg29 subcomplexes and the Atg1 complex, using a trimmed “mini-pentamer” tractable to biophysical studies. We observe tetramers of Atg1 pentamers that assemble via Atg17-Atg31-Atg29. This leads to a model for the higher organization of the Atg1 complex in PAS scaffolding.
Keywords: autophagy, EAT domain, Atg13, Atg17, small angle x-ray scattering, protein structure, membrane remodeling, coarse-grained simulation, intrinsic disorder
Introduction
When eukaryotic cells are starved, macroautophagy (henceforward, “autophagy”) replenishes the pool of biosynthetic precursors(Reggiori and Klionsky, 2013). When intracellular pathogens, damaged mitochondria, or toxic inclusions threaten the cell, autophagy clears them out(Mizushima et al., 2008; Nixon, 2013). Autophagosome biogenesis commences as follows. A double-membrane structure known as the phagophore is initiated at unique loci in the cell (Ge et al., 2014; Lamb et al., 2013; Rubinsztein et al., 2012). In yeast, the phagophore originates from a single phagophore assembly site (PAS). The phagophore grows as it engulfs cellular material and eventually closes and is sealed, at which point it becomes the autophagosome. Many autophagy-related (Atg) proteins have been identified as factors in the pathway(Mizushima et al., 2011). Biochemical activities have been assigned to most of the Atg proteins, and crystal structures have been determined for an increasing number of them(Hurley and Schulman, 2014). There is almost no information, however, on how various protein structures assemble with one another to organize the PAS and thereby initiate phagophore biogenesis.
EM imaging of the yeast PAS reveals that it contains a small number of high curvature vesicles that are positive for the integral membrane protein Atg9(Mari et al., 2010; Yamamoto et al., 2012). Atg9 is one of several Atg proteins that are present at the PAS very early in autophagy initiation. Setting aside the Atg11 protein of the autophagy-like cytosol to vacuole transport (Cvt) pathway, the first proteins of canonical bulk autophagy to arrive at the PAS are Atg17, Atg29, and Atg31(Kabeya et al., 2009; Suzuki et al., 2007). These three proteins form a subcomplex that is denoted Atg17-Atg31-Atg29 because Atg31 bridges between Atg17 and Atg29, with the latter two making no direct contact (Ragusa et al., 2012). The Atg17 monomer has the shape of a crescent whose dimensions are similar to those of the Atg9 vesicles at the PAS (Ragusa et al., 2012). The Atg17-Atg31-Atg29 subcomplex dimerizes via the C-terminal helix of Atg17 such that it forms a double crescent (Chew et al., 2013; Ragusa et al., 2012). ATG17 alleles deficient in dimer formation do not assemble into a PAS and do not support autophagy(Ragusa et al., 2012). One goal of this study is to determine how many Atg17 dimers comprise the PAS.
The Atg1 protein kinase is a central regulator of autophagy initiation (Mizushima, 2010). Atg1 assembles with Atg17-Atg31-Atg29 subcomplex via the bridging protein Atg13 (Kabeya et al., 2005). Atg1 binds to Atg13 via an interaction between its C-terminal Early Autophagy Targeting/Tethering (EAT) domain and a short motif on the C-terminal intrinsically disordered region of Atg13 (Fujioka et al., 2014; Stjepanovic et al., 2014). The Atg1-Atg13 association is very tight, on the order of 100 nM (Fujioka et al., 2014; Stjepanovic et al., 2014), although the affinity decreases when Atg13 is phosphorylated (Fujioka et al., 2014). The Atg1-Atg13 subcomplex binds to Atg17-Atg31-Atg29 with a lower affinity (Fujioka et al., 2014; Stjepanovic et al., 2014). Structures are now known for the Atg1-Atg13 (Fujioka et al., 2014) and Atg17-Atg31-Atg29 (Ragusa et al., 2012) subcomplexes and for a fragment of Atg13 bound to Atg17-Atg31-Atg29 (Fujioka et al., 2014). The Atg1-Atg13 dimer is 90 Å across, while the Atg17-Atg31-Atg29 spans 340 Å. The Atg1-Atg13 binds near the distal tips of the Atg17 double crescent. This presents a structural puzzle in that it is not possible to align the two dimers about a common axis such that a closed 2:2:2:2:2 complex is formed. Thus, it is not straightforward to simply model the full Atg1 complex on the basis of the solved substructures, and an experimental analysis of the structure is required.
The goal of this study is to gain insight into the structural organization of the PAS by determining the number of ATG17 molecules present, and by characterizing the structure of the Atg1-Atg13-Atg17-Atg31-Atg29 assembly. This supercomplex is the unit that actually functions in autophagy initiation. Our structural studies concentrate on a trimmed Atg1 complex consisting of the full Atg17-Atg31-Atg29 subcomplex, the Atg1 EAT domain (hereafter, “Atg1EAT”), and the Atg1- and Atg17-binding fragment of Atg13. This complex can be generated in quantities needed for biophysical studies. However, the flexibility of the Atg13 region between the Atg17 and Atg1 binding sites makes it inherently dynamic and thus unsuitable for crystallography. A number of methods have been developed to characterize structural ensembles of flexible protein systems in solution with the help of small angle x-ray scattering (SAXS) measurements using a variety of regularization strategies (Bernado et al., 2007; Boura et al., 2011; Foerster et al., 2008; Pelikan et al., 2009; Rozycki et al., 2011; Yang et al., 2010). We turned to SAXS to determine the structure of the trimmed Atg1 complex in solution. Together with the analysis of Atg1 complex subunit stoichiometry at the PAS, this makes it possible to advance a model for the higher order organization of the Atg1 complex at the PAS.
Results
Stoichiometry of Atg1 complex subunits at the PAS
We used quantitative fluorescence microscopy to determine how many copies of Atg1, Atg13, and Atg17 are present at the PAS. The yeast strain JBY404, which carries 128 LacO repeats and expresses the dimeric LacO binding protein LacI-GFP(Brickner and Walter, 2004), was used to standardize the measurement(Teis et al., 2008)(Fig. S1). The PAS was visualized in strains in which ATG1-GFP, ATG13-GFP, and ATG17-GFP were chromosomally integrated at their endogenous loci(Huh et al., 2003)(Fig. S1). Atg1-GFP and Atg17-GFP were previously shown to support autophagy (Cheong et al., 2005), and Atg13-GFP has been shown to incorporate normally into the PAS in an Atg17-dependent manner (Kawamata et al., 2008; Suzuki et al., 2007). When autophagy was induced with rapamycin, 28 ± 7 copies of Atg17-GFP were visualized at the PAS, along with 36–40 copies of Atg1-GFP and Atg13-GFP. In the absence of autophagy induction, the numbers of copies were similar, with 24 ± 4 copies of Atg17-GFP and 18–35 copies of Atg1-GTP and Atg13-GFP (Fig. 1A). The differences in Atg17 copy number are insignificant, consistent with previous observations that Atg17 is constitutively present at the PAS (Kawamata et al., 2008). The presence of Atg1 and Atg13-containing puncta in the absence of autophagy induction is attributed to Atg11(Suzuki et al., 2007). Autophagy induction had little effect on the frequency with which Atg17-positive puncta were seen in cells (Fig. 1B). Rapamycin treatment increased the frequency with which we observed Atg1- and Atg13-positive puncta from 10–20% to ~50% of cells examined (Fig. 1B). The effect of autophagy induction on the number of puncta is thus more pronounced than the effect on the number of molecules per punctum. These data suggested to us that the PAS under autophagy-inducing conditions contains a discrete, well-defined number of Atg1 complexes with an approximately equal number of subunits. Presumably, these Atg1 complexes must therefore be organized into some type of well-defined higher-order assembly at the PAS. In the rest of this study, we apply SAXS to probe the three-dimensional assembly of the Atg1 complex in order to obtain insights into the types of higher-order structures that might occur at the PAS.
Figure 1. Quantitation of Atg17 copy number at the PAS.
A. Estimated copy number of Atg17 from cells with a visible PAS. Values shown are based on the integrated intensity of the punctum and are an average of three independent experiments. The error bars represent the standard deviation of three replicates. B. Fraction of cells containing visible puncta in rich media or following rapamycin treatment. See also Figure S1.
Solution studies of Atg1EAT and Atg13
The crystal structure of the Kluveromyces marxianus Atg1EAT:Atg13441-500 subcomplex (Fujioka et al., 2014) shows that Atg1EAT consists of two tandem MIT domains, which dimerize via the first MIT domain. Atg13 binds to both of the MIT domains via two short motifs. To compare the crystal and solution structures, SAXS data were collected for the K. lactisAtg1EAT:Atg13400-475 complex. SAXS data were recorded on the Atg1EAT -Atg13400-475 at 0.8, 1.0, 2.4 and 5.2 mg/ml (Fig. 2A). Using the Guinier approximation, a radius of gyration Rg=33.1 ± 0.9 Å was obtained for Atg1EAT -Atg13400-475 with PRIMUS (Konarev et al., 2003). This is only in fair agreement with the crystal structure dimer, which has a radius of gyration of 27.1 Å; however, ~8% of the residues, including Atg1 598–653, are missing in the crystal structure, but would increase the Rg.
Figure 2. SAXS and solution structure of the Atg1-Atg13 subcomplex.
A. Experimental scattering intensity as a function of q obtained at three different protein concentrations (1mg/ml-blue, 2.4mg/ml-green, 5.2mg/ml-red). Inset: The region qRG < 1.3 where the Guinier approximation is valid is for a globular protein is shaded grey and the dashed line indicates the best fit to the Guinier approximation. B. Experimental (red) and computed scattering (green-crystal structure, blue-EROS ensemble) shown as I(q) vs q (top) and qI(q) vs. q (bottom) for Atg1-Atg13. C. One representative conformation from the EROS ensemble of Atg1-Atg13 subcomplex in solution.
The missing residues were incorporated into a coarse-grained molecular model and subjected to EROS refinement. The resulting computed data are in nearly perfect agreement with the measured SAXS data over the entire q range (Fig. 2B). Fig. 2C shows a representative structure from the ensemble, and Fig. 3 shows the properties of the ensemble as a whole. The fitted ensemble reproduces the experimental data with a with χ2 = 1, and remains close to the unrefined simulation ensemble (S=−0.5; see Methods).
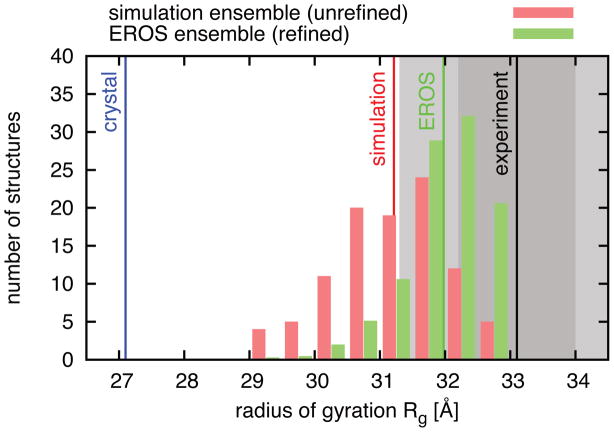
Figure 3. Properties of the Atg1-Atg13 ensemble.
The distribution of the radius of gyration of the simulation ensemble before refinement (red) and after refinement (green). Vertical lines indicate the radius of gyration of the crystal structure (blue), the average values of the radius of gyration of the unrefined ensemble (red) and of the refined ensemble (green), and the experimental value (black). The dark/light gray areas indicate values less than one/two standard deviations from experiment.
In Fig. 3, we illustrate the gain in structural information due to modeling, simulation, and ensemble refinement for the Atg1-Atg13 dimer by showing the distribution of the radius of gyration of the 100 structures drawn from the simulation ensemble before and after EROS refinement. The average value of the unrefined ensemble (31.1 Å) already shows a significantly improved agreement with experiment (33.1 ± 0.9 Å) compared to the value for the crystal structure (27.1 Å). Applying EROS to the simulation ensemble further improves this agreement by gently shifting the peak position of the distribution of the radius of gyration to larger values. The unrefined distribution and the refined distributions overlap nicely and the value of the radius of gyration of the refined ensemble (32 Å) deviates from the experimental value by only ~1.2 times the standard deviation. Consequently, the application of EROS to the scattering data over the whole q-range (q<0.28 Å−1) leads to a significant improvement of the estimate for the radius of gyration, which is determined by a just a small fraction of the available data (q < to ~0.046 Å−1). At the same time, the refined ensemble stays close to the simulation ensemble.
Solution structure of Atg17-Atg31-Atg29
To benchmark the refinement procedure against the best characterized component of the Atg complex, SAXS data were collected from samples of K. lactis Atg17-Atg31-Atg29 at three concentrations clow=1 mg/ml (~12.5 μM), cmed =2.5 mg/ml (~30 μM), and chigh =5.3 mg/ml (~64 μM) over q = 0.009 Å−1 to 0.26 Å−1 (Fig. 4A). The crystal structure of the closely related L. thermotolerans Atg17-Atg31-Atg29 (Ragusa et al., 2012) (Fig. 4B) was used as the basis for modeling the data (Fig. 4C). Comparison of the experimental P(r) distribution with those obtained from various crystal lattice-related dimers had previously led us to a model for the solution state of Atg17-Atg31-Atg29 (Ragusa et al., 2012). This model consisted of a double crescent shape with the monomers joined by zippering of their C-terminal helix, and is the starting point for the present analysis. Careful inspection of the qI(q) vs. q plot computed for the Atg17-Atg31-Atg29 double crescent revealed that the computed scattering from the dimer systematically underestimated the experimental scattering (Fig. 4C).
Figure 4. SAXS and solution ensemble model of Atg17-Atg31-Atg29.
A. Experimental scattering intensity as a function of q obtained at three different protein concentrations (1mg/ml-blue, 2.5mg/ml-green, 5.3mg/ml-red). Inset: The region qRG < 1.0 where the Guinier approximation is valid is for an elongated protein is shaded grey and the solid line indicates the scattering curve computed from the ensemble model. B. Crystallographic dimer. C. I(q) and qI(q) vs. q plots for a mixture of crystal dimers and crystal tetramers fitted to the highest concentration data. D. Crystallographic assembly of the Atg17-Atg31-Atg29 into tetramers. E. I(q) and q I(q) vs q plots of the data shown in (A) vs. computed scattering intensities of a mixture of dimers and tetramers at ratios consistent with analytical ultracentrifugation data (Stjepanovic et al., 2014).
We find that the quality of the fits improves significantly if we use a mixture of Atg17-Atg31-Atg29 dimers and Atg17-Atg31-Atg29 tetramers. A likely candidate for a closed tetramer is suggested by crystal-lattice packing observed between two L. thermotolerans Atg17-Atg31-Atg29 dimers in the crystal (Fig. 4D). We refer to this structure as the “crystal tetramer”. These two dimers are related by non-crystallographic symmetry. Including the crystal tetramer, we can fit the highest concentration data using a mixture of ~70 % dimers and 30 % crystal tetramers with a χ2≈2.2 compared to χ2≈28 for a dimer only fit.
Ensemble refinement of SAXS (EROS) accounts fully for the Atg-17-31-29 data. In coarse-grained simulations, we find that two Atg17-Atg31-Atg29 dimers (Fig. 5A) can form open tetramers (Fig. 5B, C) or closed tetramers (Fig. 5D). In the open tetramers, Atg17-31-29 dimers are connected only at one of their ends. In the refined simulation ensemble, we can distinguish two cases: Atg31-Atg29 of one dimer binds either to the tip of Atg17 of the other dimer, or to the Atg31-Atg29 unit of the other dimer (Fig. 5B, C). The open states are very flexible, with motions between V-shaped and elongated structures.
Figure 5. Schematic of arrangements present in the Atg17-Atg31-Atg29 ensemble.
A. Schematic of the crystallographic dimer. B and C. Two examples of open tetramers. D. Closed tetramer. E. An alternative closed tetramer model that is also consistent with the SAXS data.
The scattering data can be explained very well by an ensemble consisting of dimers, closed tetramers, and open tetramers (Fig. 4E). From sedimentation velocity analytical ultracentrifugation (SV-AUC) data (see Methods), we estimated fractions clow~0.7, cmed~0.6, and chigh~0.5 of free dimers at the three concentrations of the SAXS samples. Using the crystal tetramer as model for the closed tetramer, we obtained excellent fits at each concentration with a χ2<1 and a relative entropy S>−0.1, indicating that the refined ensembles are close to the initial ensemble. The resulting fractions of [free dimers, closed tetramers, open tetramers] are given by [0.70, 0.15, 0.15], [0.61, 0.18, 0.21], and [0.50, 0.20, 0.30], for clow, cmed, and chigh, respectively. Approximately half of the open tetramers are structures with one Atg31-29 bound to the tip of Atg17 of the other dimer. The other half consists of structures with one Atg31-29 bound to one Atg31-29 of the other dimer.
A number of alternative closed models of the tetramer produce fits of comparable quality, including a low-energy structure spontaneously formed in coarse-grained Monte Carlo simulations (Fig. 5E). Different types of closed tetramers thus cannot be distinguished on the basis of the coarse-grained simulations and the scattering data alone.
Solution structure of the Atg1 mini-pentamer complex
A truncated version of the pentameric Atg1 complex was engineered such that the yields needed for biophysical studies could be expressed and purified. This complex consists of Atg1EAT, Atg13350-525, and full-length Atg17, Atg31and Atg291-85. We refer to this complex as the Atg1 minipentamer. It includes all of the elements known to interact in the assembly of the full complex. We carried out a SAXS solution structural analysis of the complex to understand how these components are arranged with respect to each other in three dimensions. SAXS data on the Atg1-Atg13-Atg17-Atg31-Atg29 mini-pentamer were collected over the range of q = 0.009–0.26 Å−1 at 0.7, 1.2 and 2.6 mg ml−1 (Fig. 6A).
Figure 6. SAXS and solution structure of Atg1-Atg13-Atg17-Atg31-Atg29.
A. Experimental scattering intensity as a function of q (0.7mg/ml-blue, 1.2mg/ml-green, 2.6mg/ml-red). Inset: The region qRG < 1.0 where the Guinier approximation is valid is for an elongated protein is shaded grey and the solid line indicates the scattering curve computed from the ensemble model. B. Poor fits are obtained at low q for all models except for cis-tetramers. C. Experimental and computed I(q) vs. q and qI(q) vs. q scattering from EROS ensemble for the four different trans-tetramers (green) and the two different cis-tetramers (blue). Arrows indicate systematic deviations from experiment for the trans-tetramer structures.
The crystal structures of the Atg1EAT:Atg13 and Atg13-Atg17-Atg31-Atg29 were used as the basis for refining the solution structure of the minipentamer against the SAXS data using the EROS procedure. After sequence alignment using BLAST (Altschul et al., 1997; Altschul et al., 2005), we added the Atg13 residues that were absent from these crystal structures, treating them as flexible linkers. Dimers of minipentamers were constructed on the basis of the crystallographic Atg17-Atg31-Atg29 dimer. Tetramers of minipentamers were constructed on the basis of the major non-crystallographic lattice contact in Atg17-Atg31-Atg29 crystals (Fig. 4D). The following models were analyzed for their fits to the SAXS data, alone and in combinations: Atg17-31-29 dimer (Fig. 7A); Atg1-13 dimer (Fig. 7B); Atg17-31-29 dimer connected to one Atg1-13 dimer (Fig. 7C); Atg17-31-29 dimer connected to two Atg1-13 dimers (Fig. 7D); Atg17-31-29 tetramer with one Atg1-13 dimer bound to one of its ends (Fig. 7E); Atg17-31-29 tetramer with two Atg1-13 dimers bound “in cis” to nearby dimer tips (Fig. 7F); Atg17-31-29 tetramer with two Atg1-13 dimers bound “in trans” to distant dimer tips (Fig. 7G); and two freely moving Atg17-31-29 dimers connected via a single Atg1-13 dimer (Fig. 7H).
Figure 7. Possible arrangements of subunits Atg1-Atg-13-Atg17-Atg31-Atg29 considered in fitting the solution ensemble.
Subunit color codes are indicated at the lower right corner.
Performing EROS on these ensembles, the only acceptable models were stoichiometric tetramers, i.e., those containing two Atg1-13 dimers. At q < 0.03 Å−1, only the cis-tetramer models were able to fit the data (Fig. 6B, C). Two representative cis-tetramer models are shown in Fig. 8. At medium and low experimental protein concentrations, no systematic deviation from the experimental data can be observed for the ensemble represented by the two cis-tetramer models shown in Fig. 8. Residual systematic deviations at very low q-values observed at the highest protein concentration can probably be attributed to concentration-dependent effects. The cis-tetramer thus exhibits the most consistent fit to the scattering data and thus emerges as the most likely model.
Figure 8. Structural model for the minipentameric Atg1 complex in solution.
Two representative structures of cis-tetramers from the minipentamer ensemble are shown. Subunits are colored as in Fig. 7.
Discussion
The goal of this study was to obtain a deeper understanding of the structural organization of the PAS. We found that the yeast PAS contains, on average, just 28 molecules of Atg17. This number is reasonably consistent with the suggestion that as few as ~80 molecules of Atg9 are present at the PAS (Yamamoto et al., 2012). It is also consistent with our observation of ~20–40 copies of Atg1 and Atg13 at the PAS. Autophagy induction does not greatly increase the number of copies of Atg1 subunits at the PAS, if the PAS can be observed in a given cell. However, autophagy induction does markedly increase the proportion of yeast cells that contain an Atg1- and Atg13-positive PAS. The observation that the PAS seems to contain a relatively uniform number of copies of Atg1 complex subunits suggests that the PAS is likely to have a consistent and regular structure.
Previously, we found that the Atg17-Atg31-Atg29 dimer is the minimal building block of the PAS, and that dimerization-deficient Atg17 proteins did not form a PAS and did not support autophagy (Ragusa et al., 2012). The copy number analysis shows that ~14 Atg17 dimers are likely to be involved in scaffolding the PAS. This raises the crucial question as to how these dimers are organized with respect to each other into a higher-order structure. Here we found that both Atg17-Atg31-Atg29 and the mini-pentamer Atg1-Atg13-Atg17-Atg31-Atg29 are capable of forming tetramers. The tetramers are organized in a similar manner, with the long axes of the Atg17 double crescent aligned roughly parallel to one another. The SAXS data do not have the resolution to independently identify the tetramer interface between the two Atg17 molecules. However, models based on the crystal lattice dimer:dimer contact are fully consistent with the scattering data, both for Atg17-Atg31-Atg29 and for the minipentamer Atg1-Atg13-Atg17-Atg31-Atg29.
Taking these in vivo and in vitro observations together, we propose that the early yeast PAS consists, at a minimum, of approximately 7 Atg17-Atg31-Atg29 tetramers. Atg17 is present at the PAS even in fed cells, whilst Atg1 and Atg13 are probably recruited to the PAS following starvation and TORC1 inactivation(Kamada et al., 2000; Kamada et al., 2010; Reggiori et al., 2004). In one mode, Atg1 and Atg13 associate with one another following starvation (Fujioka et al., 2014). This model implies that a stoichiometric ratio of Atg1-Atg13 dimers joins the Atg17-Atg31-Atg29 complex when autophagy is triggered. In an alternative model, fully assembled yeast Atg1 complexes have been observed even when isolated from fed cells (Kraft et al., 2012). In either event, the solution structure model suggests that the Atg1 complex assembly pathway culminates in the presence of seven cis-tetramers of 4:4:4:4:4 stoichiometry at the PAS.
At one end, the cis-tetramers bind two free Atg13 molecules that are not partnered with an Atg17 molecule. At the other end, they offer two unfilled Atg13 binding sites at the distal tips of the Atg17 crescents. The Atg17 double crescent is 350 Å long, while the Atg1EAT-Atg13 dimer is less than 100 Å across its maximum dimension. Thus it is not possible for the open Atg13 and Atg17 contacts to be satisfied within a single tetramer. The observation that the Atg13:Atg17 contacts are not saturated at the concentrations used in this study is consistent with the relatively high Kd value, ~10 μM for the constructs used (Stjepanovic et al., 2014). It is interesting that the cis-tetramer fits the SAXS data much better than Atg17 tetramers containing a single Atg1-Atg13 dimer. This suggests that there is some cooperativity in the binding of Atg1-Atg13 dimers to the same end of the tetramer. The portions of Atg1 and Atg13 crystallized by (Fujioka et al., 2014) do not contact each other directly in most models of the cis-tetramer. The cooperativity therefore seems most likely to be explained by direct interactions between the intrinsically disordered portions of Atg1 and Atg13 with one another.
An important implication of the presence of two unpartnered Atg13 and Atg17 binding sites per cis-tetramer is that each tetramer is capable of forming up to two Atg13-mediated interactions with other cis-tetramers. A cis-tetramer could either form a branched interaction with two other cis-tetramers, each linked by a single Atg13-Atg17 connection. Such a branched connection would not be possible for the dimeric complex, and the possibility only emerges once the tetramer is formed. The Atg1 complex has been proposed to tether Atg9 vesicles at the PAS(Sekito et al., 2009). The possibility of branched chains of tetramers offers an appealing model for the scaffolding of a cluster of vesicles (Fig. 9). Clearly, if as few as seven tetramers scaffold the PAS, the chain, whether linear or branched, could not extend beyond ~250 nm. These dimensions are consistent with the size of the Atg9 vesicle cluster at the PAS (Mari et al., 2010).
Figure 9.
Speculative model for the organization of the PAS by Atg1 complexes
Methods
Quantitative Imaging of the PAS
The following yeast strains were grown in YPD media supplemented with 2% glucose: JBY404 (Mari et al., 2010), a kind gift from Dr. Jason Brickner (Northwestern University) which was used as the GFP intensity standard; and (MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ATG1-GFP, ATG13-GFP, and ATG17-GFP), a kind gift from Dr. William Prinz (NIH). Cells were grown at 30°C and harvested at an OD600 of 0.9–1.0. For rapamycin incubation, cells were further incubated with 0.2μg/ml rapamycin for three hours at room temperature. For cell fixation, 1ml of cells was centrifuged at 1000 × g for 5min, resuspended in 500 μl 1x PBS with 4% PFA and incubated for 15 minutes at room temperature. Cells were then centrifuged at 1000 × g for 4min and resuspended in 500 μl of 1x PBS with 1.6M sorbitol two times. Samples were kept in the dark at 4°C.
For imaging, Fisher microscope cover glasses (12-545-102) were ultrasonically cleaned for 30min in 1:1 water:isopropyl alcohol followed by rinsing with water and air-drying. 1 μl of fixed yeast cells was enclosed between two cover glasses. Samples were loaded into an Attofluor chamber (Invitrogen) for imaging. Wide field fluorescence imaging was performed with a Nikon Ti-E based microscope with an ASI automatic stage. The excitation laser source was a Lumencor sola coupled to the microscope by optical fiber. The Chroma GFP optical filter set of excitation dichroic, excitation wavelength filter, emission wavelength filters was used. A 100X TIRF NA1.49 oil objective (Nikon) was used with no further emission path magnification. An Andor Ixon Ultra EMCCD camera was used for image acquisition without using electron multiplication. For each sample, z-stacks were obtained from several different positions. 11 stepwise images of 500ms exposure time were acquired spanning 4.5μm in the z-direction. This condition was sufficient to cover the entire width of sample yeast cells. In subsequent analysis, fluorescent puncta of LacI-GFP and Atg17-GFP within the focal region were chosen for analysis from each cell of interest. Single z section analysis was carried out, as the estimated size of the PAS is less than the width of a single z-section.
Yeast cells with clearly visible fluorescent puncta were used for analysis. Cells with unusual morphology or evidence of diffuse fluorescence were not considered. For each sample, more than 30 individual cells were analyzed. For intensity integration, emission intensity was summed near the region of each punctum. The background was calculated from non-punctate regions of the same yeast cell and as subtracted. LacI-GFP puncta served as fixed copy number standard with N=256 and the Atg17-GFP copy number was determined by calculating the intensity ratio and multiplying by N. For calculation of the percentage of cells with puncta, more than 100 cells were analyzed for each sample. The number of cells with visible fluorescence puncta was calculated and divided by the total number of cells analyzed.
Protein Expression and Purification
The Atg17-31-291-85 complex from Kluyveromyces lactis was expressed and purified as described previously (Brickner and Walter, 2004). DNA coding for K. lactis Atg1 residues 562–831 with an N-terminal His6 tag and TEV cleavage site was subcloned into pST39 (cite). Subsequently, DNA coding for K. lactis Atg13 residues 400–475 was subcloned into pST39 containing Atg1 562–831. Atg1 562–831 and Atg13 350–525 were also subcloned into the pST39 containing Atg17-Atg31-Atg291-85. Atg1EAT-Atg13400-475, and Atg1EAT-Atg13350-525-Atg17-Atg31-Atg291-85proteins were expressed in E. coli BL21 DE3 cells. Cells were grown to log phase and then induced with 0.75 mM IPTG for 3 hours at 37°C. Cells were harvested and stored at −80°C until lysis. Cells were lysed by high pressure homogenization using an Emulsiflex C3 (Avestin) in 50 mM Tris pH 8.0, 500 mM NaCl and an EDTA-free complete protease inhibitor table (Roche). Lysate was clarified by centrifugation at ~40,000 × g for 1 hour at 4°C. The proteins were purified using Talon affinity resin (BD Biosciences) and eluted in buffer containing 20 mM Tris pH 8.0, 200 mM NaCl and 250 mM imidazole. The elution was diluted four fold in a buffer containing 50 mM Tris pH 8.0 and further purified by Hitrap-Q (GE Healthcare). Protein was eluted from Hitrap-Q using a gradient ranging from 0to 1M NaCl. Finally, protein was purified using size exclusion chromatography on a Superdex 200 16/60 (GE Healthcare) in a buffer containing 20mM Tris pH8.0, 200 mM NaCl. To purify the Atg1 minipentamer a three fold molar excess of Atg1EAT-Atg13350-525 was mixed with Atg17-31-29. After a 30 minute incubation at 4°C protein was purified using a Superose 6 column in 20 mM Tris pH 8.0 and 200 mM NaCl.
SAXS data collection and processing
SAXS data were recorded on Atg1EAT-Atg13400-475, Atg17-Atg31-Atg291-85 and the Atg1EAT -Atg13350-525-Atg17-Atg31-Atg291-85minipentamer at the Advanced Light Source SIBYLS beamline 12.3.1 at Lawrence Berkeley National Laboratory. Samples were dialyzed overnight against 20 mM Tris pH 8.0, 200 mM NaCl and 2% Glycerol. 2% glycerol was used to decrease effects due to radiation damage during data collection. After dialysis samples were added to a microplate (Axygen 321-60-051) and the plate was sealed with an Axymat (Axygen 521-01-151). The plate was snap frozen in liquid nitrogen and stored at −80°C. Upon arrival at the beamline the plate was thawed and samples spun at 3700 rpm for 10 min at 4°C. Samples were held at 10°C until data were recorded, which took approximately 4 hours for the entire plate.
SAXS data were recorded on buffer before and after each different protein sample was recorded. SAXS data on Atg1EAT-Atg13400-475 were recorded at 1.0, 2.4 and 5.2 mg/ml. SAXS data on the Atg17-31-29 complex were recorded at 1.0, 2.5 and 5.3 mg/ml.
EROS analysis of SAXS data
To analyze the SAXS data, we applied the Ensemble Refinement of SAXS (EROS) method (Rozycki et al., 2011). EROS uses a maximum entropy approach to minimally refine an initial ensemble of structures by fitting to the experimental data (Boura et al., 2011). Monte-Carlo simulations of structural models coarse-grained at the amino-acid level generate the initial ensembles of structures (Kim and Hummer, 2008). Amino acids interact via Lennard-Jones-type effective pair potentials and Debye-Hückel-type electrostatic potentials. Structured domains are kept rigid, and disordered domains are modeled either as Gaussian chains or as polymer chains with stretching, bending, and torsion-angle potentials. Here, we used the Gaussian chain model for performance reasons.
We performed replica exchange simulations (REMC) using 20 replicas with temperatures ranging from room temperature to approximately three times room temperature. We simulated a single Atg17-Atg31-Atg29 dimer (5000 structures) with flexible hinges (residues 113 to 122 of Atg31) and a pair of dimers with flexible hinges (5000 structures). For Atg1-Atg13, we sampled 2000 different linker configurations. We ran simulations of each of the six minipentamer tetramer topologies, each producing 2000 structures. For each of the other complex arrangements tested here, we produced at least 2000 structures. For analysis in EROS, we randomly chose 100 structures from each of these ensembles.
To refine Atg17-31-29 scattering data, which exhibit a clear concentration dependence, we estimated the fractions of free dimers from SV-AUC experiments (Ragusa et al., 2012). At the 10 μM concentration of the SV-AUC experiments, we find ~80% free dimers. From the ratios of the peak areas of the dimer and tetramer peaks of the c(s) curve, we crudely estimated Kd≈ 20 μM. With this value of the tetramer:dimer dissociation constant, we calculated fractions of free dimers of ~0.7, ~0.6, and ~0.5 at concentrations clow, cmed, and chigh, respectively. These fractions of free dimers entered the EROS refinement as initial weights. For open tetramers and closed tetramers, we used equal initial weights.
Supplementary Material
Highlights.
The phagophore assembly site (PAS) contains a uniform number of Atg1 complexes
Solution structures of Atg1-Atg13 dimer and Atg17-Atg31-Atg29 tetramer defined
The Atg1-Atg13-Atg17-Atg31-Atg29 complex is a closed tetramer in solution
Model for vesicle scaffolding at the PAS by approximately seven Atg1 tetramers
Acknowledgments
We thank Greg Hura for help with SAXS data collection and interpretation, and Jason Brickner and Will Prinz for yeast strains. This work was supported by the National Institutes of Health GM111730 (J. H. H), Ruth Kirschtein NRSA Fellowship GM099319 (M. J. R.), and the Max Planck Society (J. K., G. H.). This work was conducted in part at the Advanced Light Source (ALS), a national user facility operated by Lawrence Berkeley National Laboratory on behalf of the Department of Energy, Office of Basic Energy Sciences, through the Integrated Diffraction Analysis Technologies (IDAT) program, supported by DOE Office of Biological and Environmental Research and NIH grant GM105404.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schaffer AA, Yu YK. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 2005;272:5101–5109. doi: 10.1111/j.1742-4658.2005.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernado P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Structural characterization of flexible proteins using small-angle X-ray scattering. J Am Chem Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- Boura E, Rozycki B, Herrick DZ, Chung HS, Vecer J, Eaton WA, Cafiso DS, Hummer G, Hurley JH. Solution structure of the ESCRT-I complex by small angle x-ray scattering, EPR, and FRET spectroscopy. Proc Natl Acad Sci U S A. 2011;108:9437–9442. doi: 10.1073/pnas.1101763108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner JH, Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2:1843–1853. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H, Yorimitsu T, Reggiori F, Legakis JE, Wang CW, Klionsky DJ. Atg17 regulates the magnitude of the autophagic response. Mol Biol Cell. 2005;16:3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew LH, Setiaputra D, Klionsky DJ, Yip CK. Structural characterization of the Saccharomyces cerevisae autophagy regulatory complex Atg17-Atg31-Atg29. Autophagy. 2013;9:1467–1474. doi: 10.4161/auto.25687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foerster F, Webb B, Krukenberg KA, Tsuruta H, Agard DA, Sali A. Integration of small-angle X-ray scattering data into structural modeling of proteins and their assemblies. J Mol Biol. 2008;382:1089–1106. doi: 10.1016/j.jmb.2008.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka Y, Suzuki SW, Yamamoto H, Kondo-Kakuta C, Kimura Y, Hirano H, Akada R, Inagaki F, Ohsumi Y, Noda NN. Structural basis of starvation-induced assembly of the autophagy initiation complex. Nat Struct Mol Biol. 2014;21:513–521. doi: 10.1038/nsmb.2822. [DOI] [PubMed] [Google Scholar]
- Ge L, Baskaran S, Schekman R, Hurley JH. The Protein-Vesicle Network of Autophagy. Curr Opin Cell Biol. 2014;29:18–24. doi: 10.1016/j.ceb.2014.02.005. [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Schulman BA. Atomistic Autophagy: The Structures of Cellular Self-Digestion. Cell. 2014;157:300–311. doi: 10.1016/j.cell.2014.01.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Kamada Y, Baba M, Takikawa H, Sasaki M, Ohsumi Y. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell. 2005;16:2544–2553. doi: 10.1091/mbc.E04-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Noda NN, Fujioka Y, Suzuki K, Inagaki F, Ohsumi Y. Characterization of the Atg17-Atg29-Atg31 complex specifically required for starvation-induced autophagy in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2009;389:612–615. doi: 10.1016/j.bbrc.2009.09.034. [DOI] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, Ohsumi Y. Tor Directly Controls the Atg1 Kinase Complex To Regulate Autophagy. Mol Cell Biol. 2010;30:1049–1058. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell. 2008;19:2039–2050. doi: 10.1091/mbc.E07-10-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Hummer G. Coarse-grained models for simulations of multiprotein complexes: application to ubiquitin binding. J Mol Biol. 2008;375:1416–1433. doi: 10.1016/j.jmb.2007.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J Appl Crystallogr. 2003;36:1277–1282. [Google Scholar]
- Kraft C, Kijanska M, Kalie E, Siergiejuk E, Lee SS, Semplicio G, Stoffel I, Brezovich A, Verma M, Hansmann I, et al. Binding of the Atg1/ULK1 kinase to the ubiquitin-like protein Atg8 regulates autophagy. EMBO J. 2012;31:3691–3703. doi: 10.1038/emboj.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14:759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010;190:1005–1022. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. The Role of Atg Proteins in Autophagosome Formation. Annu Rev Cell Devel Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19:983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- Pelikan M, Hura GL, Hammel M. Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen Physiol Biophys. 2009;28:174–189. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragusa MJ, Stanley RE, Hurley JH. Architecture of the Atg17 Complex as a Scaffold for Autophagosome Biogenesis. Cell. 2012;151:1501–1512. doi: 10.1016/j.cell.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Klionsky DJ. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics. 2013;194:341–361. doi: 10.1534/genetics.112.149013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- Rozycki B, Kim YC, Hummer G. SAXS ensemble refinement of ESCRT-III CHMP3 conformational transitions. Structure. 2011;19:109–116. doi: 10.1016/j.str.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Shpilka T, Elazar Z. Mechanisms of Autophagosome Biogenesis. Curr Biol. 2012;22:R29–R34. doi: 10.1016/j.cub.2011.11.034. [DOI] [PubMed] [Google Scholar]
- Sekito T, Kawamata T, Ichikawa R, Suzuki K, Ohsumi Y. Atg17 recruits Atg9 to organize the pre-autophogsomal structure. Genes Cells. 2009;14:525–538. doi: 10.1111/j.1365-2443.2009.01299.x. [DOI] [PubMed] [Google Scholar]
- Stjepanovic G, Davies CW, Stanley RE, Ragusa MJ, Kim DJ, Hurley JH. Assembly and dynamics of the autophagy initiating Atg1 complex. Proc Natl Acad Sci U S A. 2014;111:12793–12798. doi: 10.1073/pnas.1407214111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- Teis D, Saksena S, Emr SD. Ordered Assembly of the ESCRT-III Complex on Endosomes Is Required to Sequester Cargo during MVB Formation. Dev Cell. 2008;15:578–589. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, Ichikawa R, Kinjo M, Ohsumi Y. Atg9 Vesicles are an Important Membrane Source During Early Steps of Autophagosome Formation. J Cell Biol. 2012;198:219–233. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Blachowicz L, Makowski L, Roux B. Multidomain assembled states of Hck tyrosine kinase in solution. Proc Natl Acad Sci U S A. 2010;107:15757–15762. doi: 10.1073/pnas.1004569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.