Abstract
Membrane proteins may be influenced by the environment, and they may be unstable in detergents or fail to crystallize. As a result, approaches to characterize structures in a native environment are highly desirable. Here, we report a novel general strategy for precise distance measurements on outer membrane proteins in whole Escherichia coli cells and isolated outer membranes. The cobalamin transporter BtuB was overexpressed and spin labeled in whole cells and outer membranes and interspin distances were measured to a spin labeled cobalamin using pulse EPR. A comparative analysis of the data reveals a similar interspin distance between whole cells, outer membranes and synthetic vesicles. This approach provides an elegant way to study conformational changes or protein-protein/ligand interactions at surface-exposed sites of membrane protein complexes in whole cells and native membranes, and provides a method to validate outer membrane protein structures in their native environment.
Keywords: in-cell, PELDOR/DEER, vitamin B12 transporter, spin labelling, membrane protein
Membrane protein function often depends upon local and global motions that occur over a wide range of time scales (from ps to ms). These motions may be modulated by the surrounding environment and tools to study membrane proteins under native conditions are of great value.[1] However, determining membrane protein structure or dynamics with high resolution in whole cells is challenging and yet to be demonstrated. Like NMR, pulsed electron-electron double resonance (PELDOR), also known as double electron-electron resonance (DEER) is a tool with the potential to examine conformational changes in biomolecules in the cellular environment.[2] PELDOR enables distance measurements in the 1.5–8 nm range between two paramagnetic centers with high precision and reliability.[3, 4] It is more sensitive than NMR and there is no size limit to the protein of interest, both important features for in-cell spectroscopy.
Most biomolecules are not paramagnetic, and for EPR they must be modified with an appropriate spin label. Generally, labels are attached by covalently linking a methanethiosulfonate spin label (MTSSL) to cysteines generated by site-directed mutagenesis.[5] Alternatively, spin probes may be incorporated using genetic encoding in response to a nonsense codon.[6, 7] Both whole cell labeling of endogenously expressed proteins using MTSSL and genetic encoding have been demonstrated in E. coli.[7, 8] Distance measurements using PELDOR have also been performed by microinjection of spin labeled RNA, DNA, peptide and ubiquitin into eukaryotic cells.[9–13] However, the application of PELDOR on an endogenously expressed biomolecule (RNA, DNA or protein) has not been performed, primarily due to difficulties in obtaining high expression levels and specific and efficient labeling in a complex environment.
Here we introduce a general strategy to make accurate distance measurements on an arbitrary outer membrane protein in intact E. coli using PELDOR. Gram-negative bacteria are surrounded by a cell envelop (CE) composed of an outer membrane (OM) and an inner cytoplasmic membrane (CM) separated by a periplasmic space containing a thin peptidoglycan layer. The OM is intrinsically asymmetric and is composed of phospholipids (PL) lipopolysaccharides (LPS) and numerous β-barrel proteins that function in transport, signaling, motility, resistance to toxic compounds and membrane biogenesis.[14] The function of many of these proteins requires the presence of a proton motive force (pmf) and an interaction with proteins in the periplasm or the CM. For cysteine labeling with MTSSL, the target protein should lack reactive cysteines or have all the natural reactive cysteines replaced. Proteins in the OM of bacteria are generally free of reactive cysteines, thereby minimizing potential sources of non-specific signals.[8, 15]
Here, we chose the high affinity (KD < 1 nM)[16] vitamin B12 (or cyanocobalamin, CNCbl) outer membrane transporter BtuB from E. coli as a model system to determine whether in-cell PELDOR can be performed on a membrane protein. BtuB belongs to a family of proteins termed TonB-dependent transporters (TBDTs) that require the inner membrane ExbB-ExbD-TonB complex and a pmf for function. A conserved segment near the N-terminus of BtuB termed the Ton box acts as an energy-coupling segment and is believed to interact with the C-terminal region of TonB to release substrate into the periplasm.[17] BtuB has been crystalized in apo-, vitamin B12-bound[18] and TonB-bound[19] states. In these crystal structures the barrel is occluded with a central hatch (plug) domain and the mechanism by which substrate is released is presently not known.
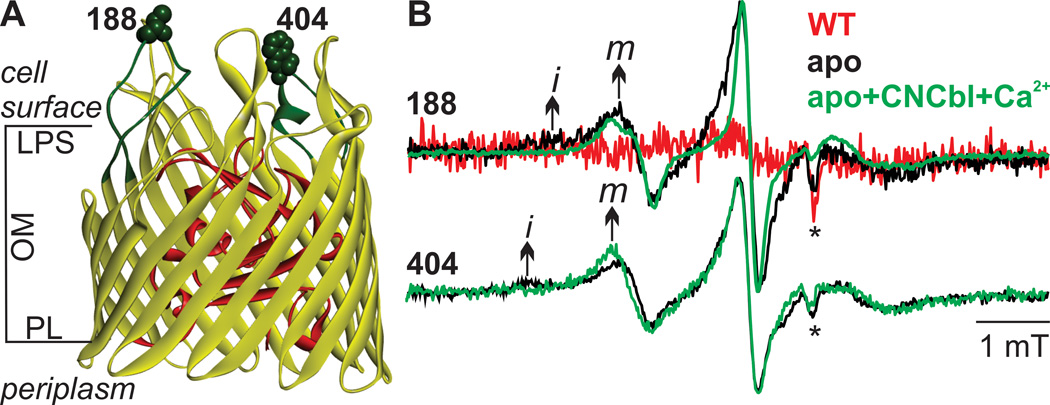
Typically, 200–300 copies of BtuB are expressed per E. coli cell when CNCbl is omitted from the medium,[20] which is far below the level required for a PELDOR experiment (10–50 µM). Therefore we over expressed wild-type (WT) BtuB and the cysteine mutants 188C and 404C located on extracellular loops 2 and 7, respectively (Figure. 1A), and spin labeled the E. coli whole cells (termed whole cells) with MTSSL. Reaction of the MTSSL with cysteine forms the spin-labeled side chain R1 (Figure S1A), to produce 188R1 and 404R1 derivatives of BtuB.
Figure 1.
MTSSL labeling of BtuB in live E. coli cells. A) Spin labeled positions (shown in space-filling model) and the loops carrying them highlighted in green using PDB 1NQH. The core domain inside the barrel is shown in red. B) X-band RT CW EPR spectra for MTSSL-labeled E. coli expressing WT, 188C or 404C BtuB using a 20 µL suspension containing 2×109 cells (see Supplementary Material). For 188R1, the WT control and the apo- samples were measured under identical conditions for a quantitative comparison. The spectra of 188 apo+CNCbl+Ca2+ (100 µM CNCbl + 1 mM CaCl2) is scaled to the intensity of 188-apo sample. For position 404, the spectra are scaled to the same intensity. The arrow indicates the mobile (m) and the immobile (i) components in the spectra and the asterisk indicates artifacts from sample tubes.
The X-band (9.4 GHz) room temperature continuous wave (RT CW) EPR of whole cells is shown in Figure 1B and yielded a two-component spectrum composed of a mobile and an immobile component (marked ‘m’ and ‘i’) for both 188R1 and 404R1. These two components are resolved due to their different correlation times and suggests the existence of two distinct modes of R1 motion at these positions in whole cells. When CNCbl and Ca2+ were added to E. coli, the immobile components vanish, and for 188R1 the spectra narrowed as a result of stronger averaging of the magnetic anisotropy of R1 reflecting an increased motion of the loop. This observation confirms that overexpression does not affect the ability of BtuB to bind the ligand and undergo conformational changes. An earlier study with 188R1 reconstituted in POPC vesicles revealed a different lineshape and the presence of the ligands reduced loop mobility,[21] showing that the dynamics and conformational changes are modulated by the surrounding environment. Unlike the short stability of nitroxide spin labels against reduction observed inside the cells,[7, 9, 22] the signal intensity for both 188R1 and 404R1 was stable for at least 7 hours when the cells were kept on ice. When WT BtuB was expressed at comparable levels to the mutants (Table S1), the EPR signal was very weak indicating that most of the signal from the cysteine mutants originated from specific reaction with the MTSSL. In contrast to residues 188 and 404 that face the cell surface, residue 10 in the Ton box that faces the periplasm could not be labeled in whole cells. Thus in whole cells, labeling is limited to those surface-exposed residues with direct access to MTSL from outside.
In addition to whole cells we also isolated CE membranes (OM+CM) following over expression of the protein. Our attempts to spin label BtuB in these CE membranes was not successful due to a high level of non-specific labeling. Positions 188 and 404 were tested and found to have identical spectra to the WT control samples (Figure S2A). Previously, it was shown that sites in the Ton box of BtuB (residues 6–12) could be labeled with the MTSSL in isolated OM preparations.[15] Therefore, we prepared isolated OM membranes by selective removal of the CM with sarkosyl solubilization.[15, 23] As expected, removal of CM reduced non-specific labeling (Figure S2B). However, the spectra for 188R1 and 404R1 still contained significant contributions from non-specific signals, making a direct comparison to the in-cell or POPC spectra difficult. We succeeded in labeling position 399 on loop7 (see Figure S9) and position 10 in the Ton box at the periplasmic face (see Figure 4) for which labeling failed in whole cells.
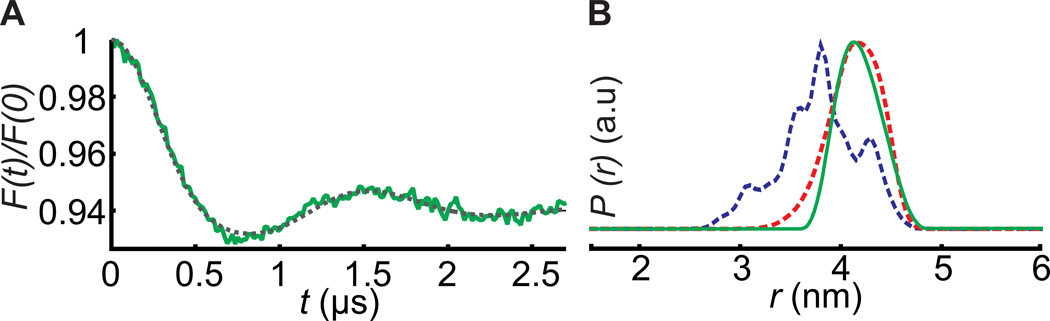
Figure 4.
PELDOR between BtuB10R1 and TEMPO-CNCbl spin pairs in OM A) Normalized form factor obtained after intermolecular background correction of the primary data (Figure S8B) using a 3D spin distribution with the fitting overlaid in dotted grey line. B) Distance distributions calculated using the DeerAnalysis2013[25] software with the Tikhonov regularization parameter α set to 10. The blue dotted line is the distance distribution obtained from the simulation for PELDOR between 10R1 and TEMPO-CNCbl using BtuB-CNCbl structure (PDB 1NQH). The red dotted line is an additional simulation using the “any rotamers” function in MMM software, in which the side chain packing is disregarded to match the experimental data.
Non-specific spin-labeling influences PELDOR distance distributions if the resulting dipolar interactions are within the measureable range. If the non-specifically attached labels are far apart or the two cysteines on BtuB are only partially spin labeled, the modulation amplitude and the S/N are reduced. Another complication could arise from overexpression if this promotes protein oligomerization or aggregation. In order to determine the feasibility of measuring PELDOR signals in native systems with intrinsic background signals, we synthesized a TEMPO-labeled cyanocobalamin (TEMPO-CNCbl, see Supplementary Material) (Figure 3 and S3 A-H), which should yield a specific distance to another spin label on BtuB. TEMPO-CNCbl binds to both BtuB (Figure 2) and the periplasmic binding protein BtuF (Figure S4). Moreover, TEMPO-CNCbl supported the growth of E. coli RK5016 cells on minimal media (Figure S3H), indicating that the modification did not significantly disturb structure and function. Using the binding of TEMPO-CNCbl, we estimated the amount of BtuB in whole cells and OM (see Supplementary Material). For whole cells, we could achieve up to 30 µM BtuB concentration with 97% specific labeling, whereas for the OM preparations we obtain only 20–25% specific labeling due to a large background signal (Table S1).
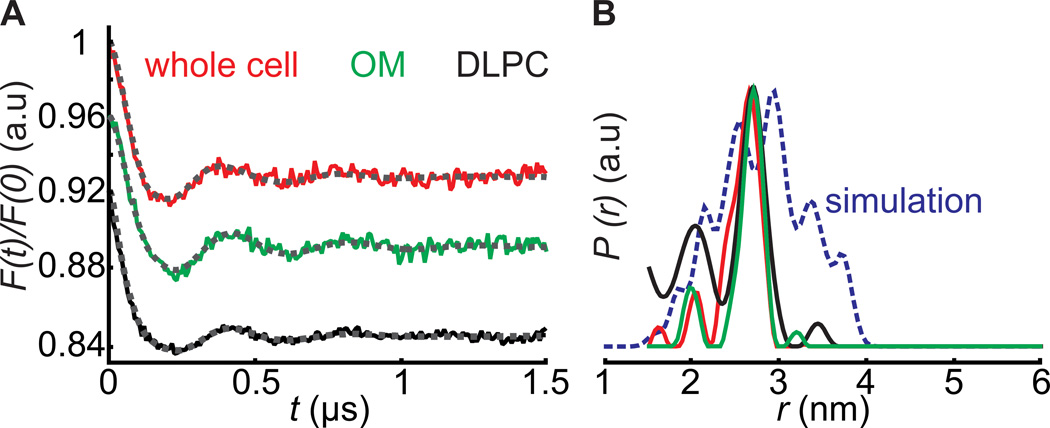
Figure 3.
PELDOR between BtuB188R1 and TEMPO-CNCbl spin pairs in whole cells, OM and DLPC vesicles. The whole cells or outer membrane preparations contained 25–30 µM BtuB after MTSSL labeling, and PELDOR was performed after adding an equal amount of TEMPO-CNCbl. A) Normalized form factors obtained after intermolecular background correction of the primary data (Figure S6 C-E) using a 3D spin distribution with the fittings overlaid in dotted grey lines. B) Distance distributions calculated with the DeerAnalysis2013[25] software with the Tikhonov regularization parameter α set to 10. In DLPC, the modulation amplitude obtained is not directly comparable to the other samples due to the bidirectional orientation and unknown concentration of BtuB. Even with a large fraction of non-specific labeling (Table S1) the OM sample gave ~8% modulation amplitude. Although devoid of non-specific signals, whole cell samples as well gave similar modulation amplitudes, which might be due to the transport of a fraction of TEMPO-CNCbl into the periplasm. The blue line shows the MMM simulation[24] (www.epr.ethz.ch/software/index) for PELDOR between 188R1 and TEMPO-CNCbl on PDB 1NQH.
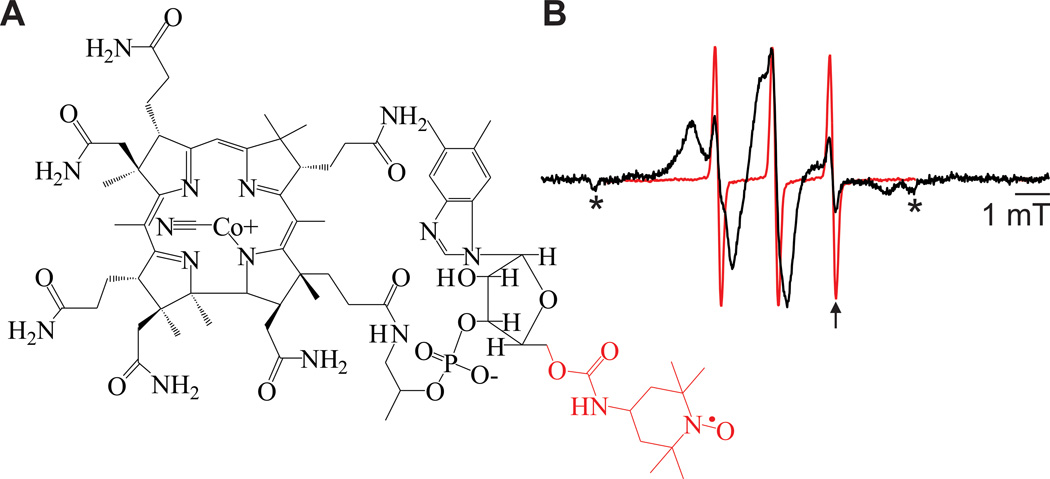
Figure 2.
TEMPO-CNCbl structure and binding. A) Structure of TEMPO-CNCbl with the TEMPO and the bonds to the 5′ carbon of ribose highlighted in red. B) RT CW EPR spectra of 10 µM TEMPO-CNCbl (red) or 10 µM TEMPO-CNCbl + 1 mM CaCl2 + BtuB reconstituted in POPC vesicles (black). The arrow indicates the characteristic resonance position for the third hyperfine line of free TEMPO-CNCbl due to its smaller correlation time and the asterisks indicate artifacts from sample tubes.
In a second step, we performed PELDOR measurements using 188R1 in whole cells, OM and reconstituted DLPC vesicles after addition of TEMPO-CNCbl (in 1:1 molar ratio to BtuB). The presence of 15% glycerol-d8 significantly improved the spin echo decay both for whole cells and OM samples (Figure S5). In the absence of TEMPO-CNCbl, both whole cells and OM gave only an exponentially decaying signal (Figure S6 A-B), demonstrating that overexpression of BtuB does not cause oligomerization or aggregation and that the non-specific labeling particularly in the OM results in spins spatially separated by distances > 7 nm. In presence of TEMPO-CNCbl, whole cells gave a PELDOR trace yielding a bimodal distance distribution with mean values at 2.02 ± 0.07 and 2.60 ± 0.15 nm respectively (Figure 3). Despite the dramatic differences in the surrounding environments, the measured distance distributions in whole cells, OM and DLPC vesicles were similar. Remarkably, even with a large amount of non-modulated signals from nonspecific labeling, OM preparations also gave clean PELDOR traces, confirming that there is specific binding of TEMPO-CNCbl to BtuB. To compare the experimental data with the crystal structure, we computed 188R1 - TEMPO-CNCbl distance distributions in BtuB-CNCbl structure (PDB 1NQH) using rotamer libraries calculated for R1[24] and TEMPO-CNCbl (see Supplementary Material). In addition to distances seen experimentally, simulations revealed the presence of longer distances (Figure 3), which arose primarily from rotamers of TEMPO-CNCbl seen in the crystal-structure based simulation that were not populated in the membrane environment (Figure S7).
To determine whether distances could be measured to sites that were not labeled in whole cells, we attempted PELDOR across the OM between 10R1 located in the Ton box and TEMPO-CNCbl (Figure S8A). Interestingly, PELDOR gave a single distance with mean value at 4.17 ± 0.21 nm (Figure 4). Simulations on BtuB-CNCbl structure (1NQH) revealed a distance distribution between 2.6–4.75 nm with mean distance at 3.78 ± 0.40 nm (Figure 4). This discrepancy with experimental data may be accounted for by the differences in side chain packing predicted in the crystal structure when compared to the OM (red line in Figure 4). Additionally, PELDOR between spin labels attached to loop2 and loop7 (Figure S9) in OM indicates that this approach can be used to follow conformational changes of the loops and other flexible regions in native environments.
In summary, we have demonstrated for the first time the application of PELDOR in whole cell and native membrane environments to extract precise distance constraints for an endogenously expressed membrane protein. In this study, the concentration of BtuB used (30 µM) corresponds to about 105 copies per cell, which is comparable to the expression level of some endogenous outer membrane proteins. A comparison of 188R1 and TEMPO-CNCbl PELDOR data between three different membrane environments indicates that lipid composition does not significantly influence loop conformation. Further measurements with other positions on BtuB will tell us whether this observation holds in general. OM preparations provide a versatile tool to study membrane proteins and unlike the alternate approaches such as nanodiscs,[26] it does not require the tedious processes of solubilization, purification and membrane reconstitution of the target protein. The approach presented here provides an opportunity to validate outer membrane protein structures and to make new structural and functional investigations of several outer membrane macromolecular complexes critical for gram-negative bacterial physiology under native conditions.
Supplementary Material
Footnotes
We would like to thank Prof. Dr. Enrico Schleiff for providing the facilities for protein preparation, Katja Barth for support with cobalamin spin labelling and Prof. Robert K. Nakamoto for helpful discussions. This work was financially supported by Deutsche Forschungsgemeinschaft (SFB 807 to T.F.P.), Marie-Curie GO-IN Fellowship (to B.J.) and the National Institutes of Health, GM035215 (to D.S.C)
Supporting information for this article is available on the WWW
Contributor Information
Dr. Benesh Joseph, Institut für physikalische und theoretische Chemie, und Biomolekulares Magnetresonanz Zentrum, Universität Frankfurt, Max-von-Laue-Strasse 7, 60438 Frankfurt am, Main (Germany)
Arthur Sikora, Department of Chemistry, University of Virginia, McCormick Road, Charlottesville VA 22904-4319, USA.
Dr. Enrica Bordignon, Department of Physics, Free University of Berlin, Arnimallee 14, 14195 Berlin, Germany
Dr. Gunnar Jeschke, Laboratory for Physical Chemistry, ETH Zurich, Vladimir-Prelog-Weg 2, 8093 Zürich, Switzerland
Dr. David S Cafiso, Email: cafiso@virginia.edu, Department of Chemistry, University of Virginia, McCormick Road, Charlottesville VA 22904-4319, USA.
Dr. Thomas F Prisner, Email: prisner@prisner.de, Institut für physikalische und theoretische Chemie, und Biomolekulares Magnetresonanz Zentrum, Universität Frankfurt, Max-von-Laue-Strasse 7, 60438 Frankfurt am, Main (Germany).
References
- 1.Mchaourab HS, Steed PR, Kazmier K. Structure. 2011;19:1549–1561. doi: 10.1016/j.str.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hansel R, Luh LM, Corbeski I, Trantirek L, Dotsch V. Angew. Chem. Int. Ed. 2014;53:10300–10314. doi: 10.1002/anie.201311320. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2014;126:10466–10480. [Google Scholar]
- 3.Jeschke G. Annu. Rev. Phys. Chem. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 4.Schiemann O, Prisner TF. Q. Rev. Biophys. 2007;40:1–53. doi: 10.1017/S003358350700460X. [DOI] [PubMed] [Google Scholar]
- 5.Hubbell WL, Altenbach C. Curr. Opin. Struct. Biol. 1994;4:566–573. [Google Scholar]
- 6.Fleissner MR, Brustad EM, Kalai T, Altenbach C, Cascio D, Peters FB, Hideg K, Peuker S, Schultz PG, Hubbell WL. Proc. Natl. Acad. Sci. USA. 2009;106:21637–21642. doi: 10.1073/pnas.0912009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt MJ, Borbas J, Drescher M, Summerer D. J. Am. Chem. Soc. 2014;136:1238–1241. doi: 10.1021/ja411535q. [DOI] [PubMed] [Google Scholar]
- 8.Jiang X, Payne MA, Cao Z, Foster SB, Feix JB, Newton SM, Klebba PE. Science. 1997;276:1261–1264. doi: 10.1126/science.276.5316.1261. [DOI] [PubMed] [Google Scholar]
- 9.Krstic I, Hansel R, Romainczyk O, Engels JW, Dotsch V, Prisner TF. Angew. Chem. Int. Ed. 2011;50:5070–5074. doi: 10.1002/anie.201100886. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2011;123:5176–5180. [Google Scholar]
- 10.Azarkh M, Singh V, Okle O, Seemann IT, Dietrich DR, Hartig JS, Drescher M. Nat. Protoc. 2013;8:131–147. doi: 10.1038/nprot.2012.136. [DOI] [PubMed] [Google Scholar]
- 11.Qi M, Gross A, Jeschke G, Godt A, Drescher M. J. Am. Chem. Soc. 2014;136:15366–15378. doi: 10.1021/ja508274d. [DOI] [PubMed] [Google Scholar]
- 12.Igarashi R, Sakai T, Hara H, Tenno T, Tanaka T, Tochio H, Shirakawa M. J. Am. Chem. Soc. 2010;132:8228–8229. doi: 10.1021/ja906104e. [DOI] [PubMed] [Google Scholar]
- 13.Martorana A, Bellapadrona G, Feintuch A, Di Gregorio E, Aime S, Goldfarb D. J. Am. Chem. Soc. 2014;136:13458–13465. doi: 10.1021/ja5079392. [DOI] [PubMed] [Google Scholar]
- 14.Fairman JW, Noinaj N, Buchanan SK. Curr. Opin. Struct. Biol. 2011;21:523–531. doi: 10.1016/j.sbi.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merianos HJ, Cadieux N, Lin CH, Kadner RJ, Cafiso DS. Nat. Struct. Biol. 2000;7:205–209. doi: 10.1038/73309. [DOI] [PubMed] [Google Scholar]
- 16.Bradbeer C, Reynolds PR, Bauler GM, Fernandez MT. J. Biol. Chem. 1986;261:2520–2523. [PubMed] [Google Scholar]
- 17.Heller KJ, Kadner RJ, Gunther K. Gene. 1988;64:147–153. doi: 10.1016/0378-1119(88)90488-x. [DOI] [PubMed] [Google Scholar]
- 18.Chimento DP, Mohanty AK, Kadner RJ, Wiener MC. Nat. Struct. Biol. 2003;10:394–401. doi: 10.1038/nsb914. [DOI] [PubMed] [Google Scholar]
- 19.Shultis DD, Purdy MD, Banchs CN, Wiener MC. Science. 2006;312:1396–1399. doi: 10.1126/science.1127694. [DOI] [PubMed] [Google Scholar]
- 20.Di Masi DR, White JC, Schnaitman CA, Bradbeer C. J. Bacteriol. 1973;115:506–513. doi: 10.1128/jb.115.2.506-513.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim M, Xu Q, Murray D, Cafiso DS. Biochemistry. 2008;47:670–679. doi: 10.1021/bi7016415. [DOI] [PubMed] [Google Scholar]
- 22.Jagtap AP, Krstic I, Kunjir NC, Hansel R, Prisner TF, Sigurdsson ST. Free. Radic. Res. 2015;49:78–85. doi: 10.3109/10715762.2014.979409. [DOI] [PubMed] [Google Scholar]
- 23.Filip C, Fletcher G, Wulff JL, Earhart CF. J. Bacteriol. 1973;115:717–722. doi: 10.1128/jb.115.3.717-722.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polyhach Y, Bordignon E, Jeschke G. Phys. Chem. Chem. Phys. 2011;13:2356–2366. doi: 10.1039/c0cp01865a. [DOI] [PubMed] [Google Scholar]
- 25.Jeschke G, Chechik V, Ionita P, Godt A, Zimmermann H, Banham J, Timmel CR, Hilger D, Jung H. Appl. Magn. Reson. 2006;30:473–498. [Google Scholar]
- 26.Bayburt TH, Sligar SG. FEBS Lett. 2010;584:1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.