Abstract
Periodontitis is a complex immune-inflammatory disease that results from a preestablished infection in gingiva, mainly due to Gram-negative bacteria that colonize deeper in gingival sulcus and latter periodontal pocket. Host inflammatory and immune responses have both protective and destructive roles. Although cytokines, prostaglandins, and proteases struggle against microbial burden, these molecules promote connective tissue loss and alveolar bone resorption, leading to several histopathological changes, namely destruction of periodontal ligament, deepening of periodontal pocket, and bone loss, which can converge to attain tooth loss. Despite the efforts of genomics, transcriptomics, proteomics/peptidomics, and metabolomics, there is no available biomarker for periodontitis diagnosis, prognosis, and treatment evaluation, which could assist on the established clinical evaluation. Nevertheless, some genes, transcripts, proteins and metabolites have already shown a different expression in healthy subjects and in patients. Though, so far, ‘omics approaches only disclosed the host inflammatory response as a consequence of microbial invasion in periodontitis and the diagnosis in periodontitis still relies on clinical parameters, thus a molecular tool for assessing periodontitis lacks in current dental medicine paradigm. Saliva and gingival crevicular fluid have been attracting researchers due to their diagnostic potential, ease, and noninvasive nature of collection. Each one of these fluids has some advantages and disadvantages that are discussed in this review.
Keywords: Diagnosis, Gingival crevicular fluid, ‘Omics, Periodontitis, Saliva
1 Introduction
Periodontitis is a multifactorial infectious and immuno-inflammatory disease that, together with gingivitis, belongs to a more broad group of pathologies termed “periodontal diseases” [1]. It results from a complex interaction between colonizing microorganisms and host immune-inflammatory response, being characterized by irreversible histopathological changes, such as destruction of the periodontal ligament, bone destruction, and deepening of periodontal pockets, which can converge to tooth loss (Fig. 1) [2, 3]. Its complexity arises from the interplay between microbial pathogens and the host’s inflammatory and immune response as well as environmental and genetic factors [1-3]. However, not all inflammatory conditions of the gingival sulcus seem to progress to periodontitis [3]. Indeed, some potential risk factors have been established, which can be subcategorized in local plaque accumulation/oral hygiene, tobacco use, malocclusion, dental restorative procedures, iatrogenic factors, and systemic factors, such as age, race, gender, socioeconomic environment, genetic influences, and other systemic conditions, such as psychosomatic, nutritional, endocrine, metabolic and immunodeficient-related disorders [2-4].
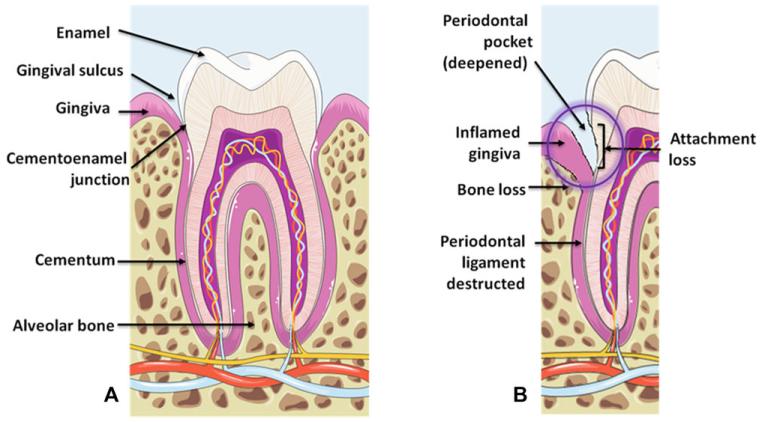
Figure 1.
Overview of the major histopathological changes found in periodontitis. (A) A periodontally healthy site with the most important histological structures depicted. (B) A periodontally affected site showing an inflamed gingiva due to microbial colonization, a deepened periodontal pocket, attachment loss and loss of periodontium structures (bone and periodontal ligament). The purple circle illustrates a critical area of inflammation and disease progression. Images were adapted from Servier Medical Art (http://www.servier.com).
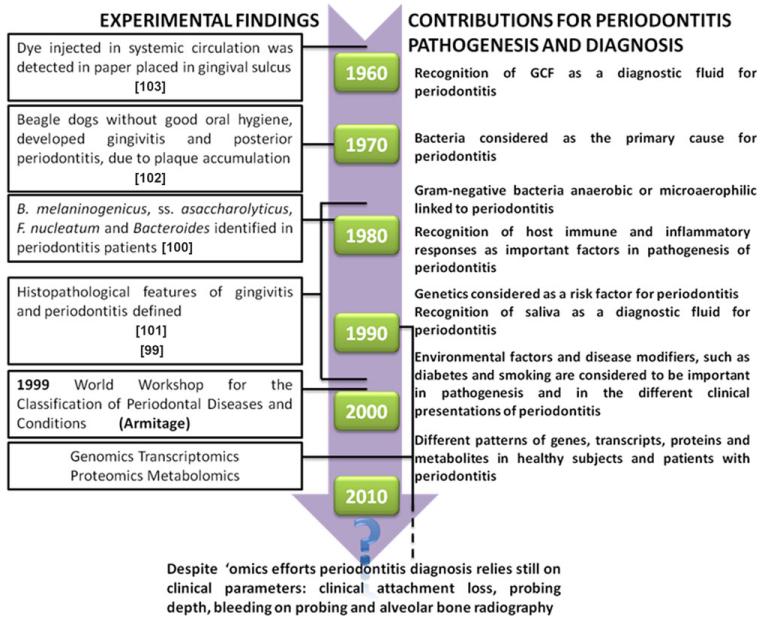
Since 1958, the knowledge of periodontitis pathogenesis has been growing, as depicted in Fig. 2. Nonetheless, periodontitis diagnosis is still performed with clinical tests and tools of low sensitivity and specificity. In addition, most of these clinical methods are fraught with a certain degree of subjectivity since they are examiner-dependent. Clinical evaluation relies on the assessment of oral hygiene, gingival status, clinical attachment loss (CAL), probing depth (PD), bleeding on probing (BOP), alveolar bone status, and other more involved procedures, such as periodontal microbiology testing, blood analysis for systemic health profiling, as well as histological studies [1,5]. Hence, the profession of periodontics lacks a reliable and objective arsenal to correctly perform diagnosis and prognosis of periodontitis-afflicted patients, which would allow an earlier diagnosis, and which could minimize interventions, such as periodontal surgery, in order to reduce periodontitis-related complications, such as tooth loss and subsequent rehabilitative therapeutics [1].
Figure 2.
Evolution on the view of periodontitis pathogenesis and diagnosis since 1958. The time frame was built based on published reports [5, 10, 11, 20, 99-103].
Following the ‘omics boom, researchers have made efforts to unravel molecular markers for periodontitis, resulting in the identification of several genes, transcripts, proteins, and metabolites related to periodontitis. While of interest, none of these parameters showed, so far, a highly selective and specific relationship to the disease [6]. In this pursuit, saliva and gingival crevicular fluid (GCF) have been the two main biofluids used to screen molecular profiles of periodontal disease, since they can reflect both the local oral microenvironment and the systemic environment related to health status [7, 8]. In this review, a brief account on the pathogenesis of periodontitis will be provided, highlighting saliva and GCF as diagnostic fluids, taking into account the contributions of several ‘omics perspectives focusing on the diagnosis of periodontitis. In an earlier review, Grant [6] provided an analysis of the major ‘omics contributions to the clinical field of periodontics. The current review aims at a further and in-depth analysis of the ‘omics approaches and, in addition, examine the relevance of the microbiome findings with respect to periodontitis. Also discussed are data on the overall contribution of genomics, transcriptomics, and proteomics/peptidomics, using the bioinformatics tool ClueGO [9] to gain insights into the complex molecular interactions and to address future directions in periodontitis diagnosis and research.
2 Material and methods
In order to analyze the “omics” contribution to the molecular diagnosis of periodontitis, database searching was first carried out on Pubmed, Google Scholar, and Web of Knowledge using “periodontitis,” “omics,” “genomics,” “transcriptomics,” “proteomics,” “peptidomics,” “metabolomics,” “microbioma” as keywords to retrieve recent publications of omics studies. Those publications were limited to the ones in which samples collected were entirely from human subjects and included saliva, GCF, blood, polymorphonuclear leukocyte, epithelial cells, gingival tissues samples, and bacterial biofilm.
This was followed by a new research on Pubmed using “periodontitis” as the first keyword and “genomics,” “transcriptomics,” “proteomics,” “peptidomics,” “metabolomics”/“metabonomics,” or “microbioma” as second keywords. Publication dates were limited to 2011 until present, retrieving close to 200 publications. Exclusion criteria included reviews and original articles regarding periodontitis treatment and antibiotic therapy, oral diseases, rather than periodontitis itself, such as endodontic infections, acute apical abcesses, dental root canal infections, dental periradicular lesions, dental implants, necrotizing periodontal diseases, or related to animal studies. The major goal was to analyze novel data concerning ‘omics contributions to periodontitis since Grants’ review [6] was submitted in 2011.
3 Periodontal diseases
3.1 Gingivitis and periodontitis
Since 1960, the model of periodontitis’ pathogenesis has been adapted to new findings, as reviewed by Kornman [10]. It was first assumed that periodontitis was a bacteria-induced disease, with bacterial dental plaque being responsible for gingivitis onset and, later on, to its development into periodontitis [10]. Currently, it is accepted that despite the pivotal role played by microorganisms, a complex set of factors seems to balance the initiation and the progression of the initial lesion to periodontitis. Those include genetic and environmental factors, summarized in Table 1, and may explain the range of host responses and clinical presentations of periodontitis [2,3,10].
Table 1.
Possible risk factors for periodontitis
| Nonmodifiable | Modifiable |
|---|---|
| Age | Oral hygiene |
| Gender | Smoking |
| Ethnicity | Diabetes control |
| Socioeconomic status | Obesity control |
| Genetic predisposition | Microbial flora |
| Some systemic diseases (Down’s syndrome, neutropenia, Papillon–Lefèvre syndrome, Chédiak–Higashi syndrome, AIDS, osteoporosis) |
Low dietary intake of calcium and vitamin D |
| Host immune factors | Local risk factors (restoration overhangs or deficiencies) |
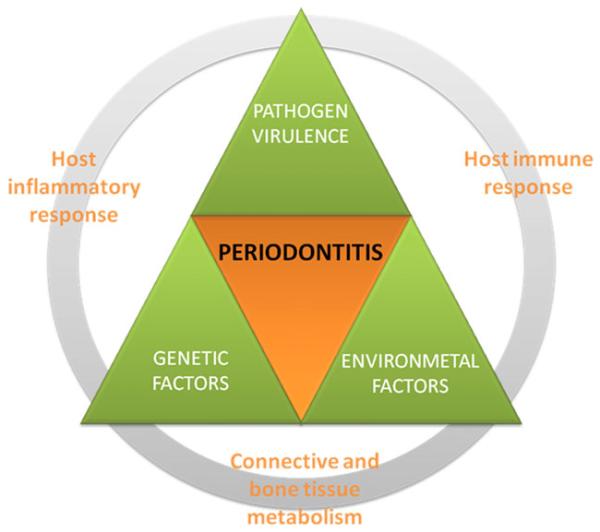
As such, the first hypothesis for periodontitis pathogenesis was replaced by a nonlinear model (1997; Fig. 3), where the interplay between pathogens, host inflammatory and immune response, connective and bone tissue metabolism, and the systemic environment were considered [10]. Additional risk factors, whether local or systemic, genetic, environmental, or acquired, were not excluded, and credited to modulate the clinical expression of the disease.
Figure 3.
Current view of periodontitis pathogenesis—a nonlinear model (1997) [10]. In the present model, microbial, genetic, and environmental factors play important roles in periodontitis pathogenesis. Besides, it recognizes the importance of host inflammatory and immune response and connective and bone tissue metabolism to the progression of periodontitis [2,3,10].
Depending on the clinical manifestation of periodontitis, seven classes of periodontal diseases were defined at the 1999 World Workshop for the Classification of Periodontal Diseases and Conditions [11]: chronic periodontitis (CP), localized aggressive periodontitis (LAP), generalized aggressive periodontitis (GAP), periodontitis as a manifestation of systemic disease associated with hematologic or genetic disorders, Necrotizing Ulcerative Periodontitis, abscesses of the periodontium, and combined periodontic-endodontic lesions. Although some of these conditions result from local manifestation of the disease, others result from systemic conditions. From those periodontitis subtypes, only CP and aggressive periodontitis (AP) will be discussed due to their epidemiologic relevance. CP is commonly found in the adult population, and its rate of progression is slow to moderate. Bacterial biofilms are consistent with the degree of tissue destruction depicted, even though there is some variability regarding the distribution of the lesions, which can be localized (LCP) or generalized chronic periodontitis (GCP). AP has a distinctly higher rate of progression and shows a remarkable familiar association. This particular form of periodontitis is less usual and can be further divided into LAP and GAP. The localized form is usually found in the adolescent population, while the latter generally affects people under 30 years of age. In LAP, the microbial biofilms are not consistent with the degree of tissue destruction, the defects are mainly localized in the region of the permanent first molars and incisors, whereas in GAP tissue defects are more largely distributed and microbial plaque accumulation is sometimes consistent with the severity of the periodontal tissue destruction [5].
3.2 Pathogenesis of periodontitis
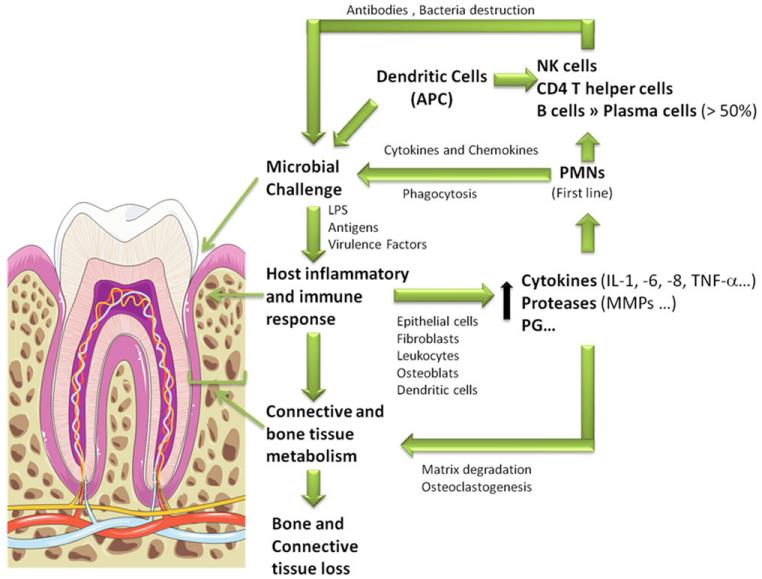
In spite of their distinct clinical presentation, CP and AP share some common features in terms of their pathogenesis, such as the activation of a pathogen-mediated inflammatory host response (Fig. 4). Several Gram-negative anaerobic and microaerophilic bacterial species have already been associated with periodontitis, noticeably, bacteria belonging to “red complex,” comprising Porphyromonas gingivalis, Treponema denticola and Tannerella forsythia. These species represent a typical group of pathogens active in periodontitis, which are able to adhere to host surfaces and coaggregate with other microorganisms, yielding biofilms [2, 3, 12]. That ability depends essentially on adhesins that recognize and interact with host elements, such as extracellular matrix components, or proteins expressed on host cell surfaces [12]. When the barrier to bacterial colonization and invasion is overcome, these pathogens induce a local inflammatory response through antigen stimulation and release of toxic products [13]. The defense response includes activation of both innate and acquired immunity with infiltration of the gingival tissues bordering the sulcular space with neutrophils and expression of antibodies by B cells. In an attempt to overcome the microbial burden, epithelial cells, periodontal ligament fibroblasts, leukocytes, osteoblasts, and dendritic cells release cytokines and chemokines, including interleukin 1 (IL-1), IL-6, chemokine (C-X-C motif) ligand 8, tumor necrosis factor α (TNF-α), and others as well as proteases, including matrix metalloproteinases (MMPs), prostaglandins, and other inflammatory mediators [3, 10, 13]. Despite the initial protection, these inflammatory molecules and proteases lead to the breakdown of the major tooth supporting structures affecting connective tissue and bone [13]. Furthermore, bacterial products can directly destruct supporting tissues and lead to further infiltration. For instance, P. gingivalis produces gingipains, extracellular cysteine proteinase–adhesin complexes that are able to adhere and digest fibronectin, collagen type V, and laminin [12, 14] as well as immune-related molecules, such as β-defensin 3 [15], IgG1, and IgG3 [16], and host protease inhibitors such as secretory leukocyte protease inhibitor [17] and even osteoprotegerin, which prevents osteoclastogenesis [18]. Thus, through bacterial and host inflammatory/immune factors, the periodontal tissue appears swollen, infected, and inflamed, leading to severe histological changes, namely, apical migration of the junctional epithelium, deepening of the periodontal pockets, destruction of connective tissue and bone, and, eventually, tooth loss [3].
Figure 4.
Cellular and molecular hallmarks of periodontitis—a summary scheme. In the presence of a microbial challenge, epithelial cells, fibroblasts, dendritic cells, leukocytes, and osteoclasts release inflammatory cytokines and chemokines that attract other leukocytes. MMPs are also released to allow leukocytes infiltration in periodontium. PMNs and dendritic cells activate both innate and acquired immune systems, which contributes to infection control through phagocytosis and destruction of bacteria [3, 10, 13]. The extensive release of cytokines, prostaglandins (PG), MMPs, and other proteases result in bone and connective tissue loss [13]. The tooth image was retrieved from Servier Medical Art (http://www.servier.com).
4 Saliva and GCF as diagnostic fluids
So far, periodontitis diagnosis has been accomplished predominantly with the periodontal probe measuring specific clinical parameters. These include the CAL. This measurement defines the distance from the cementoenamel junction—a fixed anatomical location that does not change throughout life—to the base of the probable base of the periodontal pocket. Other parameters include PD (a measure of the distance from the gingival margin to the base of the probable base of the periodontal pocket), BOP, and radiographically determined alveolar bone level [1, 5]. Hence, a more sensitive and specific molecular diagnostic tool is still missing, which could facilitate an earlier diagnosis, even before clinical manifestation of periodontitis, in generating a better and more accurate prognosis, and providing guidance in the decision for periodontal therapeutic interventions. In addition, the detection of biomarkers for active disease could be of tremendous benefit if they could be used for the assessing current disease activity and determining the risk of developing the disease.
Researchers have become increasingly interested in saliva and GCF as diagnostic fluids, since these biological fluids have shown the potential as diagnostic predictors of several conditions, ranging from systemic conditions, such as Sjögren syndrome, systemic sclerosis, diabetes mellitus, cerebrovascular/cardiovascular diseases, and acquired immunodeficiencies [7, 19, 20], to local manifestations, such as gingivitis, periodontitis, and caries [19], and even neoplasic disorders, such as salivary gland tumor, head and neck carcinoma, tumors of oral cavity, larynx carcinoma, and breast cancer [7,19].
Saliva is a complex body fluid composed by the secretions from major salivary glands comprising the parotid, sublingual, and submandibular glands and from about 600 minor salivary glands that are dispersed throughout the oral mucosa [7]. With regard to the molecular composition, saliva contains water (accounting for 99%), proteins, peptides, small organic molecules, electrolytes, nucleic acids, and hormones [7, 16]. Over 3000 proteins have been identified in saliva [21] with proline-rich proteins, mucins, cystatins, amylases, histatins, and statherin, representing the major families present in saliva [22, 23]. GCF exists as a serum transudate, changing into an inflammatory exudate as the inflammatory events progress. This fluid can be collected from the gingival sulcus or periodontal pocket. Like saliva, GCF is a complex fluid composed of molecules from different sources. The predominant components originate from serum or interstitial fluid, but other constituents reflect connective tissue and bone-derived molecules, inflammatory mediators, antibodies, and breakdown products from the periodontium [8,20].
Both saliva and GCF have advantages and disadvantages as diagnostic fluids, which are summarized in Table 2. Saliva collection is highly advantageous due to its noninvasive and painless nature, ease of collection that eliminates the need for technically trained professionals, and its minimal biological risks. Additional advantages of saliva collection relate to its higher safety when compared to other body fluids, lower cost of technical procedures and materials, applicability for large-scale populations, and higher collaboration from children, elderly people, handicapped, or anxious patients. In addition, saliva has a lower salt concentration when compared to urine, and the total protein concentration does not mirror the concentration of the most abundant proteins in blood and blood-derived products, such as serum albumin and globulins [7,24]. Nevertheless, the use of saliva as a diagnostic tool has some disadvantages, including the variation of molecular composition due to circadian rhythm, age, gender, dietary habits, method of collection, and the possible use of a stimulation technique. Moreover, special attention to sample collection and storage is needed, since saliva contains microorganisms, proteases, and proteins that can be enzymatically or physically destroyed [7,19,24]. In spite of these disadvantages, saliva remains as a resourceful diagnostic biofluid, as proteome coverage is not significantly affected by different collection methods and stimulation techniques [25].
Table 2.
Advantages and disadvantages of saliva and GCF as diagnostic fluids
| Fluid | Saliva | GCF |
|---|---|---|
| Parameter | ||
| Collection difficulty | Easy | Easy–medium |
| Yielded volume | High (>1.0 mL) | Very low (around 1 μL) |
| Invasiveness | Noninvasive | Can be minimally invasive |
| Speed of collection | Fast | Slow |
| Sensitivity to contamination |
Insensitive | Sensitive |
| Specificity to periodontitis |
Less specific than GCF |
Specific |
While saliva is produced in large volumes, ranging from 500 to 1500 mL per day in healthy adults and is easier to collect [7], GCF is produced in only microliter volumes that makes its collection technically more challenging. Furthermore, sampling times are longer, especially in healthy subjects, to achieve appreciable quantities of material. GCF collection is also prone to contamination during the commonly used paper strip collection method. This contamination includes components of saliva, blood, or bacterial plaque. This is not the case with saliva, since all nonexocrine contributions to saliva are part of its composition. Finally, GCF sampling can induce some discomfort, particularly if using “deep” intracrevicular method with paper strips [8, 20, 26]. Regardless of these disadvantages, GCF keeps attracting researchers because this fluid is more “periodontal specific” since it derives from local site involved in the actual periodontal disease manifestation. It is therefore considered to represent a mirror of the periodontal health state of an individual [8]. GCF can also be collected with a minimally invasive methods and in this case GCF is easier to handle than saliva due to its reduced viscosity when compared to whole saliva samples [8,24].
5 “Omics” approaches to periodontitis
Efforts have been made in several ‘omics disciplines in an attempt to achieve molecular tools for the diagnosis of periodontitis. Genomics, transcriptomics, proteomics/peptidomics, and metabolomics studies have a common goal that is the identification and characterization of a molecular signature of periodontitis, either on the level of DNA, RNA, proteins, or metabolites that hopefully could predict the development of periodontal disease, its prognosis, and assess the follow-up of periodontal treatment. Supporting Information Table 1 illustrates the published literature on the human periodontitis ‘omics studies.
As shown, some insight into potential diagnostic targets, such as single nucleotide polymorphisms (SNPs), genes, transcripts, proteins and metabolites, that could contribute to earlier diagnosis, prognosis, and follow-up of periodontitis have already been reported.. Nevertheless, none of the addressed markers has been validated as a clinical diagnostic tool of periodontitis. It is therefore not surprising that the classical clinical determinants, CAL, PD, BOP, and alveolar bone status, remain the gold standards of the clinical diagnostic armamentarium [1,5].
5.1 Genomics and transcriptomics approaches to periodontitis
Genomic and transcriptomic approaches aim to identify potential genes and SNPs associated with an increased risk to develop periodontitis, as well as to detect differences in gene expression in periodontitis-afflicted individuals. Genomic and transcriptomic studies have been focusing on one or a few candidate genes/transcripts [27-38]. There are also studies where genome- or transcriptome-wide approaches have been conducted, omitting an a priori positive discrimination of certain gene(s) [39-47]. Regardless of the approach, all studies require the extraction of DNA or RNA from blood, epithelial cells collected from an oral swab, gingival tissue, among other sources. While DNA can be directly amplified through PCR, purified RNA is first reverse-transcribed yielding cDNA. cDNA is then amplified in a quantitative real-time PCR device and is detected through fluorescence-based assays [48]. Depending upon the goal, the PCR method can be modified to increase sensitivity using sequence-specific primers to detect specific alleles in real time or to amplify several DNA templates with multiple primers (multiplex PCR) [49]. With regard to SNP or gene detection, different methods, including PCR-RFLP, TaqMan® Allelic Discrimination, Tetra-Primer Amplification Refractory Mutation System PCR, DNA arrays, among others, have been used in periodontitis biomarker studies (Supporting Information Table 1) [49,50].
Since periodontitis is characterized by an inflammatory burden, it is not surprising that several molecular markers related to inflammation are found differently expressed in periodontitis in omics studies. Indeed, classical proinflammatory cytokines, such as IL-1α, IL-1β, and TNF-α; key mediators of periodontal inflammation; and inducers of bone resorption by osteoclasts [13,40] have been linked to periodontitis. Moreover, RNA sequences of other transcripts of inflammatory mediators, such as IL-6, chemokine (C-X-C motif) ligand 8, regulated on activation normal t-cell expressed and secreted and macrophage chemotactic protein 1, were found to be expressed to higher degree in periodontitis-afflicted gingiva compared to healthy sites [40].
Also, several genomic studies showed an association of specific gene polymorphisms to different clinical presentations of periodontitis. For instance, the combination of IL-1A (−889, allele 2) with IL-1B (+3953, allele 2) genotypes are correlated with severe periodontitis [37]. Moreover, IL-1B (+3953TT) [35] and tumor necrosis factor α gene (−1031CC) genotypes are associated with CP, and tumor necrosis factor α gene (−308AA) with AP [34]. However, these genomic studies display some limitations, such as the use of highly homogeneous populations, which make conclusions valid only for the specific population studied. The limitations of these data relate to the focus on only a few genes, excluding the important possibility of gene–gene and gene–environment interactions. This aspect is important in an etiologically complex disease such as periodontitis. An additional confounder is the low number of evaluated subjects used in contrast to genome-wide association studies (GWAS) allowing the discrimination of intrapopulation genetic variability. Nevertheless, in large-scale studies [43-46], where these limitations are at least in part overcome, results are not very promising. For instance, in a report by Suzuki et al. [43], five promising genomic markers for periodontitis were detected. These are gonadotropin-releasing hormone 1 gene, phosphatidylinositol 3-kinase regulatory 1 gene, dipeptidylpeptidase 4 gene, fibrinogen-like 2 gene, and calcitonin receptor gene. These results, however, lack large-scale validation, due to the relative low number of participants (n = 41) and due to the homogeneity of the population consisting exclusively of Japanese subjects. DNA arrays, such as Affymetrix® Genome-Wide Human SNP Array 6.0 used by Teumer et al. [46], in a GWAS, allowed for the screening of millions of SNPs in a single assay, in a fast, accurate, and low-cost manner [50]. Despite the high number of evaluated subjects, GWAS yielded only one marker representing the glycosyltransferase 6 domain containing 1, GTL6D1 gene [45] and no genetic markers for periodontitis [46] after adjustment of the data for gender, age, diabetes, and smoking.
Other evidence of inflammation in periodontitis were derived from microarrays studies, a higher throughput technology for transcriptome analysis, which were employed to assess differences in gene expression between healthy and periodontitis patients [39, 41, 42]. The high-throughput nature of the transcriptomic and genomic techniques requires an equally high-throughput bioinformatic tools to analyze long lists of genes and validate its statistical significance using annotation databases, such as gene ontology [51]. It was found, for example, that the leukocyte transendothelial migration pathway was upregulated in periodontitis, which can be explained by the dependency on leukocyte migration through endothelial cells being part of the first line of defense against bacterial invasion [39]. Moreover, genes related to apoptosis, antimicrobial humoral response, antigen presentation, regulation of metabolism, signal transduction and angiogenesis were found to be differently expressed in periodontitis and healthy subjects [41]. With the aid of microarray technologies, it could be shown that cell communication pathways were downregulated in periodontitis-affected tissues, specifically connexin, desmogrein 1, desmocollin 1, and nestin. This finding could result from attachment loss of communicating structures, either in cell-to-cell communications at the soft tissue level, or in cell-to-tooth signaling as a consequence of the inflammatory status of the periodontium [39]. Despite enabling the detection of distinctive patterns of biological pathways in health and periodontitis, the gene expression data obtained were gathered from heterogeneous cell populations that comprised epithelial cells, connective tissue fibroblasts, and several infiltrating cell types. While these data are important, additional approaches using proteomic/peptidomic tools are required for further confirmation and validation of the reported data [41].
5.2 Proteomics and peptidomics approaches to periodontitis
Evaluation of the complete protein and peptide profile in health and periodontitis has been the aim of proteomics/peptidomics approaches. Concerning the methodological strategy, both bottom-up approaches, where proteins are digested to peptides, and top-down approaches, where proteins are kept intact, have been employed [52]. The majority of the studies depicted in Supporting Information Table 1 follow a bottom-up approach [53-61], while just two studies [62, 63] follow a top-down approach. In both methods sample collection and processing occur prior to proteomic analysis. Whole saliva, stimulated or unstimulated, and GCF are widely used as starting materials. To remove cell debris, food remnants, and to prevent protein digestion, whole saliva is centrifuged at low temperatures with a cocktail of protease inhibitors or kept on ice, and the recovered supernatant is immediately frozen at −80°C [53,55,56,59,61-64]. GCF is recovered from paper strips with different organic or aqueous elution methods [54,57,58,60]. In bottom-up proteomics, peptides are fractionated with multidimensional resolving techniques before MS analysis in order to decrease the complexity of the mixture [24,65]. Electrophoresis, either 1DE or 2DE, is one of the most widely used techniques to separate the proteins in the sample. In 2DE, proteins are separated according to their pI in the first dimension, and in the second dimension according to their molecular weight, using denaturing conditions, in the presence of SDS-PAGE. In the bottom-up approach, resolved proteins present in bands (SDS-PAGE, 1DE) or in spots (2D-PAGE) are digested with trypsin for identification by MS [65, 66].There are, however, also gel-free-based approaches to decrease the complexity of protein mixtures, such as chromatographic separation methods. RP chromatography and ion-exchange chromatography (IEC) represent the most common chromatographic procedures used in peptide fractionation. RP chromatography separates proteins based on their hydrophobicity, and IEC protein separation is related to the differences in net charge of each protein or peptide at the prevailing pH. IEC resins can be classified into strong cation or anion exchangers or into weak cation or anion exchangers. The salient difference of these resins relates to the charge of the functional groups covalently bound to the matrix and the degree of retaining this charge at the employed pH [65]. In the majority of bottom-up proteomic studies summarized in Supporting Information Table 1, a combination of both electrophoretic and chromatographic separation methods is used for achieving multidimensional separation of proteins. In top-down proteomic/peptidomic studies, as carried out by Taiyeb-Ali and his colleagues [63] and Zhang and his colleagues [62], a combination of electrophoretic and chromatographic separation techniques was employed for protein fractionation without any protein digestion. After these steps, the resolved proteins/peptides are analyzed by MS. First, proteins are ionized by ESI or by MALDI. Once ionized, proteins are resolved in mass analyzers accordingly to m/z stability, using instrumentation parameters such as quadrupoles, TOF, or m/z resonance frequency (ion trap, orbitrap and ion cyclotron resonance). Some experiments use hybrid technologies with more than one analyzer to further resolve proteins [52]. Finally, detected proteins are identified using algorithms like SEQUEST or MASCOT [66]. In proteomic studies, the technique of zymography also plays an important role for the analysis of proteases. In this electrophoretic technique, a substrate for a specific protease is copolymerized within the polyacrylamide gel matrix to assess enzymatic activity within specific zones of the electrophoretogram. After the run, SDS is washed out and the gel is stained to distinguish between digested and nondigested zones [67].
Some studies have been aimed at the characterization of the GCF proteome, identifying between 199 [57] and 327 [54] proteins. These included proteins related to early inflammation, immune response, protease activity, modulators of cytokines, response to stimulus, biological regulation, metabolic processes, and extracellular matrix constituents [54,57]. There are studies that have provided some insight into the proteomic GCF patterns in healthy and in periodontitis-related conditions. For example, Nagata et al. [60] identified 64 proteins specific to periodontally healthy sites and 63 proteins specific to periodontally affected sites. These proteins were related to blood, cytoskeleton, immunity, inflammation, lipids, and some enzymes [60]. Another study used label-free quantitative proteomics (LC/MSE) of the GCF exudatome to compare healthy with periodontally diseased sites. The results showed that the sites from the latter group displayed higher amounts of microbial proteins and l-plastin, and lower amounts of annexin-1, neutrophil defensins, cystatin B, and IgG [58]. Similarly to these GCF data, plastin-2, profilin-1, neutrophil collagenase, α-2-macroglobulin have been found to be differently expressed in health and periodontitis with the aid of salivary proteomics. Other salivary proteome studies demonstrated complement C3, lactotransferrin, MMP-9, serum albumin, Ig γ2 and α2 chain C region, vitamin D binding protein, α-amylase, zinc-α2 glycoprotein, S100A8, –A9, –A6, Ig heavy chain V-III region, and hemoglobin to be overexpressed [53, 55, 56, 59] and lactotransferrin, elongation factor 2, 14–3–3 sigma, short palate lung and nasal epithelium carcinoma-associated protein 2 precursor, carbonic anhydrase 6 and cystatin SN precursor to be underexpressed in periodontitis [55,59].
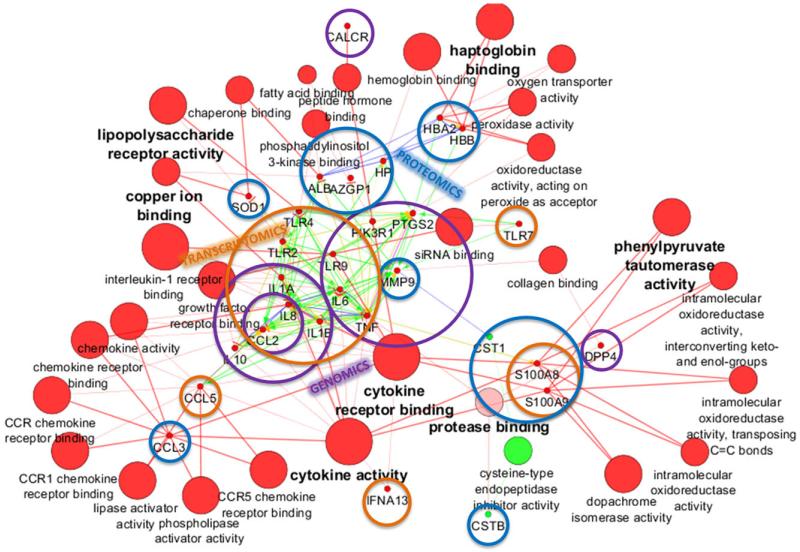
The inflammatory picture of periodontitis can clearly be shown with a network of genes, transcripts, and proteins, constructed according to molecular function with the bioinformatic tool ClueGO [9] depicted in Cytoscape [68] (Fig. 5). This network was built based on the molecular findings of the studies described in Supporting Information Table 1, with genomics, transcriptomics, or proteomics/peptidomics methodology. The scheme aims to translate the reported data from saliva or GCF obtained with the various omics approaches into a perspective view of their potential use for specific marker application in the field of periodontology. As can be seen in Fig. 5, several genes and proteins are up-regulated in periodontitis, remarkably those that belong to cytokine receptor binding, cytokine activity, lipopolysaccharide (LPS) receptor activity, haptoglobin binding, copper ion binding, and phenylpyruvate tautomerase activity. Only genes with cysteine-type endopeptidase inhibitory activity were downregulated, which include cystatin SN and cystatin B. Low levels of protease inhibitors are consistent with high proteolytic activity, which can explain matrix degradation and connective tissue and bone loss in periodontitis. On the other hand, the upregulation of cytokine receptor binding and activity genes as well as genes of LPS receptor activity and phenylpyruvate tautomerase activity could be a consequence of the infectious and inflammatory process underlying the pathogenesis of periodontitis. Moreover, upregulation of haptoglobin binding genes could be indicative of increased level of ruptured erythrocytes and the liberation of free hemoglobin in inflammation sites.
Figure 5.
Network of genes, transcripts, and proteins differentially expressed in periodontitis built with the ClueGO tool. Red nodes represent upregulated molecular markers in periodontitis, while green nodes represent downregulated molecular markers. Enrichment analysis was performed with CluePedia: activation, green arrows; binding, blue arrows; expression, yellow arrows; and PTMs, pink arrows. Contributions of genomics, transcriptomics, and proteomics are shown with transparent purple, orange, and blue circles, respectively.
There is only a single peptidomic study [62] showing none of the identified peptides to be clustered with the other markers. The promise and potential of peptidomics for diagnostics considering the high proteolytic activity associated with inflammatory lesions justify further investigation in the peptidomics field. A few genes/proteins have already been validated with more than one ‘omics methodology as indicated in the graphic representation with overlapping circles in Fig. 5. For instance, upregulation of S100A8 and S100A9 in periodontitis was corroborated by transcriptomics and proteomics studies. Overexpression of IL-1α, IL-1β, toll-like receptor 9, and TNF-α is another example of validated data by two ‘omics approaches (genomics and transcriptomics). MMP-9 upregulation in periodontal disease was shown by genomics and proteomics. Nevertheless, a similar trend has also been observed for other diseases such as Sjögren syndrome, diabetes mellitus, acquired immunodeficiency syndrome (AIDS) [20,24,34,69,70], or neoplasic conditions such as oral squamous cell carcinoma [71], which highlight a degree of nonspecificity of these targets.
5.3 Microbioma findings in periodontitis
To date, the majority of microbiome studies have relied on genomics approaches, such as 16S rRNA genes pyrosequencing, DNA–DNA hybridization, and microarrays [72-74]. They attempt to find possible associations among bacterial species identified from subgingival and supragingival biofilm samples with different periodontitis conditions [75-80], clinical parameters regarding periodontal health [81, 82], with systemic conditions, such as diabetes mellitus [83-86], and rheumatoid arthritis [87], Down syndrome [88], or with other factors, such as smoking, race, or presence of caries [89]. Overall, the results show that a community of microorganisms rather than a single identity should be associated to periodontitis pathogenesis. For instance, Griffen et al. [80] used a genomic methodology to identify CP-specific bacteria showing that Spirochaetes, Synergistetes, Bacteroidetes, Clostridia, Negativicutes, and Erysipelotrichia were disease-associated, while Proteobacteria and the class Bacilli were health-associated. Another research group was able to link Aggregatibacter actinomycetemcomitans, Filifactor alocis, Tannerella sp., Solobacterium moorei, Parvimonas micra, and Capnocytophaga sp. to LAP [79].
Proteomics and metabolomics represent an important complement/alternative to genomics and transcriptomics studies on microbiome, since it is possible to address bacterial phenotypic profiles in gingival sulci rather than their genotype. These approaches avoid problems related to cultivation-based methods, such as the cultivation of fastidious bacteria, which are difficult or impossible to culture and characterize in available culture media [90]. So far, proteomics and metabolomics studies regarding microbiome of periodontitis had distinct purposes. For instance, a proteomic study aimed to unravel potential bacterial biomarkers in GCF [91]. Another studies focused on the characterization of red complex biofilms proteome [92] and on the identification of immunoreactive antigens from A. actinomycetemcomitans infected subjects [93].
It is noticeable, though, the lack of global-approached proteomics/peptidomics and metabolomics studies regarding microbiome, particularly with the same goals as the ones included in Supporting Information Table 1, that is to say, unraveling a molecular signature of periodontitis. For instance, more ‘omics studies are needed to identify bacterial adhesins, their targets, and virulence factors as well as human proteases, which can be potential predictors of disease progression and a discriminatory tool between different clinical presentations of periodontitis. For example, a glycomic study was already performed, which revealed the importance of fucose in host glycan moieties to P. gingivalis’ fimbriae binding [94]. Moreover, P. gingivalis ATCC 33277 fimbriae have been shown to adhere to acidic proline-rich proteins, proline-rich glycoproteins, and statherins, proteins present in parotid-derived salivary secretion [95]. Another study found that LPS of the same bacteria can bind to α-amylase, cystatins, prolactin-inducible protein, lysozyme C, Ig components, serum albumin, lipocalin-1, and submaxillary gland androgen-regulated protein 3B, using P. gingivalis LPS-immobilized beads [96]. Indeed, the proteins referred to above are glycoproteins and present themselves differentially expressed in CP [53, 59]. AP has been associated with increased levels of several salivary glycoproteins as well. These include serum albumin, Ig γ2 chain C region, Ig α2 chain C region, vitamin D binding protein, α-amylase, and zinc-α2 glycoprotein [55]. Hence, glycoproteomics may be also a helpful tool to identify distinctive glycoproteomic profiles among the different clinical stages of periodontitis.
5.4 Metabolomics approaches to periodontitis
Among the omics studies, metabolomics approaches are of a more recent vintage, but could represent a very productive avenue regarding potential contributions with regard to the diagnosis of periodontitis. Metabolomics ultimate concern is to obtain a complete screen of all metabolites of a given biological sample and interpret how the metabolic profile changes with a given pathophysiological state. The key techniques used in this field are NMR, GC-MS LC-MS [97]. The results of an NMR-based study of periodontitis in which the metabolic profiles of GCP patients were compared to those of healthy subjects are provided in Supporting Information Table 1. NMR spectra from processed saliva samples were obtained by applying Fourier transform. An unsupervised principal component analysis was performed to determine the variance of the NMR profiles [98]. NMR patterns of whole saliva showed a distinct metabolic profile in GCP patients, which revealed increased levels of acetate, γ-aminobutyrate, n-butyrate, succinate, trimethylamine, propionate, phenylalanine, and valine, and decreased levels of pyruvate and N-acetyl groups, which could be explained by host tissue degradation and metabolic and fermentative activity of the pathogens [98].
5.5 “Omics” challenges for the next decade
The challenge for the field of omics in the next decade will be to unravel new molecular patterns of biomarkers useful for the field of periodontics. Particularly promising to achieve this goal are approaches in peptidomics and metabolomics. While few studies have been conducted in these fields, they appear promising tools to achieve the desired molecular information for the development of a diagnostic tool applicable for the diagnosis, monitoring, and prospective evaluation of this widespread oral disease. Peptide fingerprinting in addition to enzymatic studies, such as zymography, can be the solution to address which proteases and peptides are differentially expressed in the various clinical presentations of periodontitis, and may detect specific types or families of microorganisms being particularly virulent and destructive for the periodontium. Glycoproteomics will also be an important tool to detect molecular targets for pathogen adhesins, and metabolomics approaches may help to define characteristic patterns associated uniquely with CP, AP, and the healthy state of the periodontium.
6 Concluding remarks
In summary, periodontitis is a complex inflammatory disease that leads to the destruction of the periodontium and ultimately to loss of the dentition. It is clearly known that genetic, environmental, and microbiological factors seem to determine periodontitis predisposition, onset, and progression. In the molecular realm, the various omics studies conducted so far have pointed to several genes, SNPs, transcripts, and proteins/peptides, revealing a significant association with the different clinical presentations of periodontitis. The few studies comprising larger numbers of subjects have not confirmed these associations. Some proteomics studies of saliva and GCF have uncovered distinct protein profiles in health and periodontitis, but closer inspection of these protein markers reveals a lack of specificity. Proteolytic activity underlying periodontitis is at the outset of being explored showing collagenolytic and gelatinolytic activities being more intense in periodontitis patients. Metabolomics and peptidomics studies have begun to disclose the pattern of metabolites and the “fragmentome” signature of periodontitis. So far, ‘omics studies have described the host inflammatory response as a consequence of pathogen invasion. In the future, a comprehensive study of bacterial adhesins and virulence factors, their molecular targets, and host proteases through proteomic and peptidomic approaches could be valuable for finding molecular predictors of disease progression and to discriminate between the various manifestations of periodontitis.
From the molecular studies carried out so far, it is increasingly evident that adequate specificity and sensitivity for a diagnostic tool is required. The complexity of the disease makes it clear that this can only be achieved by a multiplex approach in which more than one biomarker is measured simultaneously. It also requires study cohorts of adequate size to maximize the chances to reach the goal of validity. The ultimate diagnostic tool will be acceptable by the clinicians only if the application of such a tool provides data superior or at least adding so far elusive and critically important information to what can be achieved by currently employed clinical assessment procedures.
Supplementary Material
Acknowledgments
This work was supported by Portuguese Foundation for Science and Technology (FCT), European Union, QREN, FEDER and COMPETE for funding the QOPNA research unit (project PEst-C/QUI/UI0062/2013), RNEM (Portuguese Mass Spectrometry Network), and the research project (FCT PTDC/EXPL/BBB-BEP/0317/2012); QREN (FCOMP-01-0124-FEDER-027554). NIH/NIDCR/NIAID grants DE07652 (FGO), DE05672 (FGO), AI087803 (EJH), and AI101067 (EJH).
Abbreviations
- AP
aggressive periodontitis
- BOP
bleeding on probing
- CAL
clinical attachment loss
- GAP
generalized aggressive periodontitis
- GCF
gingival crevicular fluid
- GCP
generalized chronic periodontitis
- GWAS
genome-wide association study
- IEC
ion exchange chromatography
- IFN-α1
interferon α 1
- IL
interleukin
- LAP
localized aggressive periodontitis
- LCP
localized chronic periodontitis
- LPS
lipopolysaccharide
- MIP-1α
macrophage inflammatory protein 1α
- MMP
matrix metalloproteinase
- NOS3
endothelial nitric oxide synthase
- PD
probing depth
- SNP
single nucleotide polymorphism
- TNF-α
tumor necrosis factor α
Footnotes
The authors have declared no conflict of interest.
Additional supporting information may be found in the online version of this article at the publisher’s web-site
References
- [1].Slots J. Periodontology: past, present, perspectives. Periodontology 2000. 2013;62:7–19. doi: 10.1111/prd.12011. [DOI] [PubMed] [Google Scholar]
- [2].Page RC, Kornman KS. The pathogenesis of human periodontitis: an introduction. Periodontology 2000. 1997;14:9–11. doi: 10.1111/j.1600-0757.1997.tb00189.x. [DOI] [PubMed] [Google Scholar]
- [3].Kinane DF. Causation and pathogenesis of periodontal disease. Periodontology 2000. 2001;25:8–20. doi: 10.1034/j.1600-0757.2001.22250102.x. [DOI] [PubMed] [Google Scholar]
- [4].Albandar JM, Rams TE. Risk factors for periodontitis in children and young persons. Periodontology 2000. 2002;29:207–222. doi: 10.1034/j.1600-0757.2002.290110.x. [DOI] [PubMed] [Google Scholar]
- [5].Armitage GC. Periodontal diagnoses and classification of periodontal diseases. Periodontology 2000. 2004;34:9–21. doi: 10.1046/j.0906-6713.2002.003421.x. [DOI] [PubMed] [Google Scholar]
- [6].Grant MM. What do ‘omic technologies have to offer periodontal clinical practice in the future? J. Periodontal Res. 2012;47:2–14. doi: 10.1111/j.1600-0765.2011.01387.x. [DOI] [PubMed] [Google Scholar]
- [7].Pfaffe T, Cooper-White J, Beyerlein P, Kostner K, Punyadeera C. Diagnostic potential of saliva: current state and future applications. Clin. Chem. 2011;57:675–687. doi: 10.1373/clinchem.2010.153767. [DOI] [PubMed] [Google Scholar]
- [8].Gupta G. Gingival crevicular fluid as a periodontal diagnostic indicator-I: host derived enzymes and tissue breakdown products. J. Med. Life. 2012;5:390–397. [PMC free article] [PubMed] [Google Scholar]
- [9].Bindea G, Mlecnik B, Hackl H, Charoentong P, et al. ClueGO: a cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformation. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kornman KS. Mapping the pathogenesis of periodontitis: a new look. J. Periodontol. 2008;79:1560–1568. doi: 10.1902/jop.2008.080213. [DOI] [PubMed] [Google Scholar]
- [11].Armitage GC. Development of a classification system for periodontal diseases and conditions. Ann. Periodontol. 1999;4:1–6. doi: 10.1902/annals.1999.4.1.1. [DOI] [PubMed] [Google Scholar]
- [12].Amano A. Bacterial adhesins to host components in periodontitis. Periodontology 2000. 2010;52:12–37. doi: 10.1111/j.1600-0757.2009.00307.x. [DOI] [PubMed] [Google Scholar]
- [13].Di Benedetto A, Gigante I, Colucci S, Grano M. Periodontal disease: linking the primary inflammation to bone loss. Clin. Dev. Immunol. 2013;2013:503754. doi: 10.1155/2013/503754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ruggiero S, Cosgarea R, Potempa J, Potempa B, et al. Cleavage of extracellular matrix in periodontitis: gingipains differentially affect cell adhesion activities of fibronectin and tenascin-C. Biochim. Biophys. Acta. 2013;1832:517–526. doi: 10.1016/j.bbadis.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Maisetta G, Brancatisano FL, Esin S, Campa M, Batoni G. Gingipains produced by Porphyromonas gingivalis ATCC49417 degrade human-β-defensin 3 and affect peptide’s antibacterial activity in vitro. Peptides. 2011;32:1073–1077. doi: 10.1016/j.peptides.2011.02.003. [DOI] [PubMed] [Google Scholar]
- [16].Vincents B, Guentsch A, Kostolowska D, von Pawel-Rammingen U, et al. Cleavage of IgG1 and IgG3 by gingi-pain K from Porphyromonas gingivalis may compromise host defense in progressive periodontitis. FASEB J. 2011;25:3741–3750. doi: 10.1096/fj.11-187799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Into T, Inomata M, Kanno Y, Matsuyama T, et al. Arginine-specific gingipains from Porphyromonas gingivalis deprive protective functions of secretory leucocyte protease inhibitor in periodontal tissue. Clin. Exp. Immunol. 2006;145:545–554. doi: 10.1111/j.1365-2249.2006.03156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yasuhara R, Miyamoto Y, Takami M, Imamura T, et al. Lysine-specific gingipain promotes lipopolysaccharide- and active-vitamin D3-induced osteoclast differentiation by degrading osteoprotegerin. Biochem. J. 2009;419:159–166. doi: 10.1042/BJ20081469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Castagnola M, Picciotti PM, Messana I, Fanali C, et al. Potential applications of human saliva as diagnostic fluid. Acta Otorhinolaryngol. Ital. 2011;31:347–357. [PMC free article] [PubMed] [Google Scholar]
- [20].Lamster IB, Ahlo JK. Analysis of gingival crevicular fluid as applied to the diagnosis of oral and systemic diseases. Ann. N. Y. Acad. Sci. 2007;1098:216–229. doi: 10.1196/annals.1384.027. [DOI] [PubMed] [Google Scholar]
- [21].Rosa N, Correia MJ, Arrais JP, Lopes P, et al. From the salivary proteome to the OralOme: comprehensive molecular oral biology. Arch. Oral Biol. 2012;57:853–864. doi: 10.1016/j.archoralbio.2011.12.010. [DOI] [PubMed] [Google Scholar]
- [22].Carpenter GH. The secretion, components, and properties of saliva. Annu. Rev. Food Sci. Technol. 2013;4:267–276. doi: 10.1146/annurev-food-030212-182700. [DOI] [PubMed] [Google Scholar]
- [23].Amado F, Lobo MJC, Domingues P, Duarte JA, Vitorino R. Salivary peptidomics. Expert Rev. Proteomics. 2010;7:709–721. doi: 10.1586/epr.10.48. [DOI] [PubMed] [Google Scholar]
- [24].Amado FML, Ferreira RP, Vitorino R. One decade of salivary proteomics: current approaches and outstanding challenges. Clin. Biochem. 2013;46:506–517. doi: 10.1016/j.clinbiochem.2012.10.024. [DOI] [PubMed] [Google Scholar]
- [25].Golatowski C, Gesell Salazar M, Dhople VM, Hammer E, et al. Comparative evaluation of saliva collection methods for proteome analysis. Clin. Chim. Acta. 2013;419:42–46. doi: 10.1016/j.cca.2013.01.013. [DOI] [PubMed] [Google Scholar]
- [26].Griffiths GS. Formation, collection and significance of gingival crevice fluid. Periodontology 2000. 2003;31:32–42. doi: 10.1034/j.1600-0757.2003.03103.x. [DOI] [PubMed] [Google Scholar]
- [27].Holla LI, Vokurka J, Hrdlickova B, Augustin P, Fassmann A. Association of Toll-like receptor 9 haplotypes with chronic periodontitis in Czech population. J. Clin. Periodontology. 2010;37:152–159. doi: 10.1111/j.1600-051X.2009.01523.x. [DOI] [PubMed] [Google Scholar]
- [28].Kajita K, Honda T, Amanuma R, Domon H, et al. Quantitative messenger RNA expression of Toll-like receptors and interferon-α1 in gingivitis and periodontitis. Oral Microbiol. Immunol. 2007;22:398–402. doi: 10.1111/j.1399-302X.2007.00377.x. [DOI] [PubMed] [Google Scholar]
- [29].Daing A, Singh SV, Saimbi CS, Khan MA, Rath SK. Cyclooxygenase 2 gene polymorphisms and chronic periodontitis in a north Indian population: a pilot study. J. Periodontal Implant Sci. 2012;42:151–157. doi: 10.5051/jpis.2012.42.5.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Jaradat SM, Ababneh KT, Jaradat SA, Abbadi MS, et al. Association of interleukin-10 gene promoter polymorphisms with chronic and aggressive periodontitis. Oral Dis. 2012;18:271–279. doi: 10.1111/j.1601-0825.2011.01872.x. [DOI] [PubMed] [Google Scholar]
- [31].Sahingur SE, Xia X-J, Gunsolley J, Schenkein HA, et al. Single nucleotide polymorphisms of pattern recognition receptors and chronic periodontitis. J. Periodontal Res. 2011;46:184–192. doi: 10.1111/j.1600-0765.2010.01327.x. [DOI] [PubMed] [Google Scholar]
- [32].Erciyas K, Pehlivan S, Sever T, Igci M, et al. Endothelial nitric oxide synthase gene polymorphisms associated with periodontal diseases in Turkish adults. African J. Biotechnol. 2010;9:3042–3047. [Google Scholar]
- [33].Kdkhodazadeh M, Hajilooi M, Houshmand B, Khazaei S, et al. Evaluation of PECAM-1 gene polymorphism in patients with periodontal disease and healthy individuals. ISRN Dent. 2012;2012:751920. doi: 10.5402/2012/751920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Yang W, Jia Y, Wu H. Four tumor necrosis factor alpha genes polymorphisms and periodontitis risk in a Chinese population. Hum. Immunol. 2013;74:1684–1687. doi: 10.1016/j.humimm.2013.08.009. [DOI] [PubMed] [Google Scholar]
- [35].Masamatti SS, Kumar A, Baron TKA, Mehta DS, Bhat K. Evaluation of interleukin-1B (+3954) gene polymorphism in patients with chronic and aggressive periodontitis: a genetic association study. Contemp. Clin. Dent. 2012;3:144–149. doi: 10.4103/0976-237X.96815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Heo S-H, Choi Y-J, Lee J-H, Lee J-M, Cho J-Y. S100A2 level changes are related to human periodontitis. Mol. Cells. 2011;32:445–450. doi: 10.1007/s10059-011-0132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kornman KS, Crane A, Wang H-Y, Giovlne F. S. d., et al. The interleukin-1 genotype as a severity factor in adult periodontal disease. J. Clin. Periodontol. 1997;24:72–77. doi: 10.1111/j.1600-051x.1997.tb01187.x. [DOI] [PubMed] [Google Scholar]
- [38].Polk DE, Wang X, Feingold E, Shaffer JR, et al. Effects of smoking and genotype on the PSR index of periodontal disease in adults aged 18–49. Int. J. Environ. Res. Public Health. 2012;9:2839–2850. doi: 10.3390/ijerph9082839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Abe D, Kubota T, Morozumi T, Shimizu T, et al. Altered gene expression in leukocyte transendothelial migration and cell communication pathways in periodontitis-affected gingival tissues. J. Periodontal Res. 2011;46:345–353. doi: 10.1111/j.1600-0765.2011.01349.x. [DOI] [PubMed] [Google Scholar]
- [40].Davanian H, Stranneheim H, Båge T, Lagervall M, et al. Gene expression profiles in paired gingival biopsies from periodontitis-affected and healthy tissues revealed by massively parallel sequencing. PLoS One. 2012;7:e46440. doi: 10.1371/journal.pone.0046440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Demmer RT, Behle JH, Wolf DL, Handfield M, et al. Transcriptomes in healthy and diseased gingival tissues. J. Periodontol. 2008;79:2112–2124. doi: 10.1902/jop.2008.080139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lakschevitz FS, Aboodi GM, Glogauer M. Oral neutrophil transcriptome changes result in a pro-survival phenotype in periodontal diseases. PLoS One. 2013;8:e68983. doi: 10.1371/journal.pone.0068983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Suzuki A, Ji G, Numabe Y, Ishii K, et al. Large-scale investigation of genomic markers for severe periodontitis. Odontology. 2004;92:43–47. doi: 10.1007/s10266-004-0035-4. [DOI] [PubMed] [Google Scholar]
- [44].Hou T, Gao L, Zheng J, Liu Z, et al. Matrix metalloproteinase-1 gene polymorphisms and periodontitis susceptibility: a meta-analysis based on 11 case-control studies. Gene. 2013;521:111–115. doi: 10.1016/j.gene.2013.02.014. [DOI] [PubMed] [Google Scholar]
- [45].Schaefer AS, Richter GM, Nothnagel M, Manke T, et al. A genome-wide association study identifies GLT6D1 as a susceptibility locus for periodontitis. Hum. Mol. Genet. 2010;19:553–562. doi: 10.1093/hmg/ddp508. [DOI] [PubMed] [Google Scholar]
- [46].Teumer A, Holtfreter B, Völker U, Petersmann A, et al. Genome-wide association study of chronic periodontitis in a general German population. J. Clin. Periodontol. 2013;40:977–985. doi: 10.1111/jcpe.12154. [DOI] [PubMed] [Google Scholar]
- [47].Wright HJ, Chapple ILC, Matthews JB, Cooper PR. Fusobacterium nucleatum regulation of neutrophil transcription. J. Periodontal Res. 2011;46:1–12. doi: 10.1111/j.1600-0765.2010.01299.x. [DOI] [PubMed] [Google Scholar]
- [48].Bustin S, Mueller R. Real-time reverse transcription PCR (qRT-PCR) and its potential use in clinical diagnosis. Clin. Sci. 2005;109:365–379. doi: 10.1042/CS20050086. [DOI] [PubMed] [Google Scholar]
- [49].Kim S, Misra A. SNP genotyping: technologies and biomedical applications. Annu. Rev. Biomed. Eng. 2007;9:289–320. doi: 10.1146/annurev.bioeng.9.060906.152037. [DOI] [PubMed] [Google Scholar]
- [50].Beaudet AL, Belmont JW. Array-based DNA diagnostics: let the revolution begin. Annu. Rev. Med. 2008;59:113–129. doi: 10.1146/annurev.med.59.012907.101800. [DOI] [PubMed] [Google Scholar]
- [51].Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Yates JR, Ruse CI, Nakorchevsky A. Proteomics by mass spectrometry: approaches, advances, and applications. Annu. Rev. Biomed. Eng. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- [53].Salazar MG, Jehmlich N, Murr A, Dhople VM, et al. Identification of periodontitis associated changes in the proteome of whole human saliva by mass spectrometric analysis. J. Clin. Periodontol. 2013;40:825–32. doi: 10.1111/jcpe.12130. [DOI] [PubMed] [Google Scholar]
- [54].Carneiro LG, Venuleo C, Oppenheim FG, Salih E. Proteome data set of human gingival crevicular fluid from healthy periodontium sites by multidimensional protein separation and mass spectrometry. J. Periodontal Res. 2012;47:248–262. doi: 10.1111/j.1600-0765.2011.01429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Wu Y, Shu R, Luo L-J, Ge L-H, Xie Y-F. Initial comparison of proteomic profiles of whole unstimulated saliva obtained from generalized aggressive periodontitis patients and healthy control subjects. J. Periodontal Res. 2009;44:636–644. doi: 10.1111/j.1600-0765.2008.01172.x. [DOI] [PubMed] [Google Scholar]
- [56].Haigh BJ, Stewart KW, Whelan JRK, Barnett MPG, et al. Alterations in the salivary proteome associated with periodontitis. J. Clin. Periodontol. 2010;37:241–247. doi: 10.1111/j.1600-051X.2009.01525.x. [DOI] [PubMed] [Google Scholar]
- [57].Tsuchida S, Satoh M, Umemura H, Sogawa K, et al. Proteomic analysis of gingival crevicular fluid for discovery of novel periodontal disease markers. Proteomics. 2012;12:2190–2202. doi: 10.1002/pmic.201100655. [DOI] [PubMed] [Google Scholar]
- [58].Bostanci N, Heywood W, Mills K, Parkar M, et al. Application of label-free absolute quantitative proteomics in human gingival crevicular fluid by LC/MS E (gingival exudatome) J. Proteome Res. 2010;9:2191–2199. doi: 10.1021/pr900941z. [DOI] [PubMed] [Google Scholar]
- [59].Da L, Gonçalves R, Regina M, Nogueira FCS, et al. Comparative proteomic analysis of whole saliva from chronic periodontitis patients. J. Proteomics. 2010;73:1334–1341. doi: 10.1016/j.jprot.2010.02.018. [DOI] [PubMed] [Google Scholar]
- [60].Kido J, Bando M, Hiroshima Y, Iwasaka H, et al. Analysis of proteins in human gingival crevicular fluid by mass spectrometry. J. Periodontal Res. 2012;47:488–499. doi: 10.1111/j.1600-0765.2011.01458.x. [DOI] [PubMed] [Google Scholar]
- [61].Rangé H, Léger T, Huchon C, Ciangura C, et al. Salivary proteome modifications associated with periodontitis in obese patients. J. Clin. Periodontol. 2012;39:799–806. doi: 10.1111/j.1600-051X.2012.01913.x. [DOI] [PubMed] [Google Scholar]
- [62].Zhang J, Zhou S, Li R, Cao T, et al. Magnetic bead-based salivary peptidome profiling for periodontal-orthodontic treatment. Proteome Sci. 2012;10:1–8. doi: 10.1186/1477-5956-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Chan HH, Rahim ZHA, Jessie K, Hashim OH, Taiyeb-Ali TB. Salivary proteins associated with periodontitis in patients with type 2 diabetes mellitus. Int. J. Mol. Sci. 2012;13:4642–4654. doi: 10.3390/ijms13044642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Thomadaki K, Helmerhorst EJ, Tian N, Sun X, et al. Whole-saliva proteolysis and its impact on salivary diagnostics. J. Dent. Res. 2011;90:1325–1330. doi: 10.1177/0022034511420721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Manadas B, Mendes VM, English J, Dunn MJ. Peptide fractionation in proteomics approaches. Expert Rev. Proteomics. 2010;7:655–663. doi: 10.1586/epr.10.46. [DOI] [PubMed] [Google Scholar]
- [66].Penque D. Two-dimensional gel electrophoresis and mass spectrometry for biomarker discovery. Proteomics Clin. Appl. 2009;3:155–172. doi: 10.1002/prca.200800025. [DOI] [PubMed] [Google Scholar]
- [67].Wilkesman J, Kurz L. Protease analysis by zymography: a review on techniques and patents. Recent Pat. Biotechnol. 2009;3:175–184. doi: 10.2174/187220809789389162. [DOI] [PubMed] [Google Scholar]
- [68].Shannon P, Markiel A, Ozier O, Baliga NS, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Fleissig Y, Deutsch O, Reichenberg E, Redlich M, et al. Different proteomic protein patterns in saliva of Sjögren’s syndrome patients. Oral Dis. 2009;15:61–68. doi: 10.1111/j.1601-0825.2008.01465.x. [DOI] [PubMed] [Google Scholar]
- [70].Zheng L, Zhang Z, Yu C, Yang C. Expression of Toll-like receptors 7, 8, and 9 in primary Sjögren’s syndrome. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2010;109:844–850. doi: 10.1016/j.tripleo.2010.01.006. [DOI] [PubMed] [Google Scholar]
- [71].Rhodus NL, Ho V, Miller CS, Myers S, Ondrey F. NF-κB dependent cytokine levels in saliva of patients with oral preneoplastic lesions and oral squamous cell carcinoma. Cancer Detect. Prev. 2005;29:42–45. doi: 10.1016/j.cdp.2004.10.003. [DOI] [PubMed] [Google Scholar]
- [72].Boyd SD. Diagnostic applications of high-throughput DNA sequencing. Annu. Rev. Pathol. Mech. Dis. 2013;8:381–410. doi: 10.1146/annurev-pathol-020712-164026. [DOI] [PubMed] [Google Scholar]
- [73].Pozhitkov AE, Beikler T, Flemmig T, Noble PA. High-throughput methods for analysis of the human oral microbiome. Periodontology 2000. 2011;55:70–86. doi: 10.1111/j.1600-0757.2010.00380.x. [DOI] [PubMed] [Google Scholar]
- [74].Wade WG. Has the use of molecular methods for the characterization of the human oral microbiome changed our understanding of the role of bacteria in the pathogenesis of periodontal disease? J. Clin. Periodontol. 2011;38:7–16. doi: 10.1111/j.1600-051X.2010.01679.x. [DOI] [PubMed] [Google Scholar]
- [75].You M, Mo S, Leung WK, Watt RM. Comparative analysis of oral treponemes associated with periodontal health and disease. BMC Infect. Dis. 2013;13:1–13. doi: 10.1186/1471-2334-13-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].López R, Dahlén G, Retamales C, Baelum V. Clustering of subgingival microbial species in adolescents with’ periodontitis. Eur. J. Oral Sci. 2011;119:141–150. doi: 10.1111/j.1600-0722.2011.00808.x. [DOI] [PubMed] [Google Scholar]
- [77].Da Silva-Boghossian CM, do Souto RM, Luiz RR, Colombo APV. Association of red complex, A. actinomycetemcomitans and non-oral bacteria with periodontal diseases. Arch. Oral Biol. 2011;56:899–906. doi: 10.1016/j.archoralbio.2011.02.009. [DOI] [PubMed] [Google Scholar]
- [78].Benrachadi L, Bouziane A, Azziman Z, Bouziane-Ouartini F, Ennibi O. Screening for periodontopathogenic bacteria in severe chronic periodontitis in a Moroccan population. Méd. Mal. Infect. 2012;42:599–602. doi: 10.1016/j.medmal.2012.10.003. [DOI] [PubMed] [Google Scholar]
- [79].Shaddox LM, Huang H, Lin T, Hou W, et al. Microbiological characterization in children with aggressive periodontitis. J. Dent. Res. 2012;91:927–933. doi: 10.1177/0022034512456039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Griffen AL, Beall CJ, Campbell JH, Firestone ND, et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012;6:1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Fujinaka H, Takeshita T, Sato H, Yamamoto T, et al. Relationship of periodontal clinical parameters with bacterial composition in human dental plaque. Arch. Microbiol. 2013;195:371–383. doi: 10.1007/s00203-013-0883-9. [DOI] [PubMed] [Google Scholar]
- [82].López R, Dahlén G, Baelum V. Subgingival microbial consortia and the clinical features of periodontitis in adolescents. Eur. J. Oral Sci. 2012;119:455–462. doi: 10.1111/j.1600-0722.2011.00875.x. [DOI] [PubMed] [Google Scholar]
- [83].Casarin RCV, Barbagallo A, Meulman T, Santos VR, et al. Subgingival biodiversity in subjects with uncontrolled type-2 diabetes and chronic periodontitis. J. Periodontal Res. 2013;48:30–36. doi: 10.1111/j.1600-0765.2012.01498.x. [DOI] [PubMed] [Google Scholar]
- [84].Field C. a, Gidley MD, Preshaw PM, Jakubovics N. Investigation and quantification of key periodontal pathogens in patients with type 2 diabetes. J. Periodontal Res. 2012;47:470–478. doi: 10.1111/j.1600-0765.2011.01455.x. [DOI] [PubMed] [Google Scholar]
- [85].Castrillon CA, Hincapie JP, Yepes FL, Roldan N, et al. Occurrence of red complex microorganisms and Aggregatibacter actinomycetemcomitans in patients with diabetes. J. Investig. Clin. Dent. 2013;4:1–7. doi: 10.1111/jicd.12051. [DOI] [PubMed] [Google Scholar]
- [86].Zhou M, Rong R, Munro D, Zhu C, et al. Investigation of the effect of type 2 diabetes mellitus on subgingival plaque microbiota by high-throughput 16S rDNA pyrosequencing. PLoS One. 2013;8:e61516. doi: 10.1371/journal.pone.0061516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Scher JU, Ubeda C, Equinda M, Khanin R, et al. Periodontal disease and the oral microbiota in new-onset rheumatoid arthritis. Arthritis Rheum. 2012;64:3083–3094. doi: 10.1002/art.34539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Khocht A, Yaskell T, Janal M, Turner BF, et al. Subgingival microbiota in adult down syndrome periodontitis. J. Periodontal Res. 2012;47:500–507. doi: 10.1111/j.1600-0765.2011.01459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Ge X, Rodriguez R, Gunsolley J, Xu P. Oral microbiome of deep and shallow dental pockets in chronic periodontitis. PLoS One. 2013;8:e65520. doi: 10.1371/journal.pone.0065520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Parahitiyawa NB, Scully C, Leung WK, Yam WC, et al. Exploring the oral bacterial flora: current status and future directions. Oral Dis. 2010;16:136–145. doi: 10.1111/j.1601-0825.2009.01607.x. [DOI] [PubMed] [Google Scholar]
- [91].Baliban RC, Sakellari D, Li Z, DiMaggio PA, et al. Novel protein identification methods for biomarker discovery via a proteomic analysis of periodontally healthy and diseased gingival crevicular fluid samples. J. Clin. Periodontol. 2012;39:203–212. doi: 10.1111/j.1600-051X.2011.01805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Zainal-Abidin Z, Veith PD, Dashper SG, Zhu Y, et al. Differential proteomic analysis of a polymicrobial biofilm. J. Proteome Res. 2012;11:4449–4464. doi: 10.1021/pr300201c. [DOI] [PubMed] [Google Scholar]
- [93].Rylev M, Abduljabar AB, Reinholdt J, Ennibi O-K, et al. Proteomic and immunoproteomic analysis of Aggregatibacter actinomycetemcomitans JP2 clone strain HK1651. J. Proteomics. 2011;74:2972–2985. doi: 10.1016/j.jprot.2011.07.022. [DOI] [PubMed] [Google Scholar]
- [94].Sojar HT, Smith DF. Porphyromonas gingivalis fimbriae carbohydrate specificity assessment by glycomics. FEMS Immunol. Med. Microbiol. 2012;66:83–87. doi: 10.1111/j.1574-695X.2012.00989.x. [DOI] [PubMed] [Google Scholar]
- [95].Amano A, Shizukuishi S, Horie H, Kimura S, et al. Binding of Porphyromonas gingivalis fimbriae to proline-rich glycoproteins in parotid saliva via a domain shared by major salivary components. Infect. Immun. 1998;66:2072–2077. doi: 10.1128/iai.66.5.2072-2077.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Choi S, Baik JE, Jeon JH, Cho K, et al. Identification of Porphyromonas gingivalis lipopolysaccharide-binding proteins in human saliva. Mol. Immunol. 2011;48:2207–2213. doi: 10.1016/j.molimm.2011.06.434. [DOI] [PubMed] [Google Scholar]
- [97].Smolinska A, Blanchet L, Buydens LMC, Wijmenga SS. NMR and pattern recognition methods in metabolomics: from data acquisition to biomarker discovery: a review. Anal. Chim. Acta. 2012;750:82–97. doi: 10.1016/j.aca.2012.05.049. [DOI] [PubMed] [Google Scholar]
- [98].Aimetti M, Cacciatore S, Graziano A, Tenori L. Metabonomic analysis of saliva reveals generalized chronic periodontitis signature. Metabolomics. 2012;8:465–474. [Google Scholar]
- [99].Kinane DF, Lindhe J. Pathogenesis of periodontal diseases. In: Lindhe J, editor. Textbook of Periodontology. Munksgaard; Copenhagen: 1997. [Google Scholar]
- [100].Slots J. Subgingival microflora and periodontal disease. J. Clin. Periodontol. 1979;6:351–382. doi: 10.1111/j.1600-051x.1979.tb01935.x. [DOI] [PubMed] [Google Scholar]
- [101].Page RC, Schroeder HE. Pathogenesis of inflammatory periodontal disease. Asummary of current work. Lab. Invest. 1976;34:235–249. [PubMed] [Google Scholar]
- [102].Lindhe J, Hamp S-E, Löe H. Experimental periodontitis in the Beagle dog. J. Periodontal Res. 1973;8:1–10. doi: 10.1111/j.1600-0765.1973.tb00735.x. [DOI] [PubMed] [Google Scholar]
- [103].Brill N, Krasse BO. The passage of tissue fluid into the clinically healthy gingival pocket. Acta Odontol. Scand. 1958;16:233–245. [Google Scholar]
- [104].Salvi GE, Lawrence HP, Offenbacher S, Beck JD. Influence of risk factors on the pathogenesis of periodontitis. Periodontology 2000. 1997;14:173–201. doi: 10.1111/j.1600-0757.1997.tb00197.x. [DOI] [PubMed] [Google Scholar]
- [105].Thomadaki K, Bosch JA, Oppenheim FG, Helmerhorst EJ. The diagnostic potential of salivary protease activities in periodontal health and disease. Oral Dis. 2013;19:781–788. doi: 10.1111/odi.12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Al-Sabbagh M, Alladah A, Lin Y, Kryscio RJ, et al. Bone remodeling-associated salivary biomarker MIP-1α distinguishes periodontal disease from health. J. Periodontal Res. 2012;47:389–395. doi: 10.1111/j.1600-0765.2011.01445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.