Abstract
Autism is a heritable disorder, with over 250 associated genes identified to date, yet no single gene accounts for more than 1–2% of cases. The clinical presentation, behavioural symptoms, imaging, and histopathology findings are strikingly heterogeneous. A more complete understanding of autism can be obtained by examining multiple genetic or behavioural mouse models of autism using MRI based neuroanatomical phenotyping. Twenty-six different mouse models were examined and the consistently found abnormal brain regions across models were the parieto-temporal lobe, cerebellar cortex, frontal lobe, hypothalamus, and the striatum. These models separated into three distinct clusters, two of which can be linked to the under and over-connectivity found in autism. These clusters also identified previously unknown connections between Nrxn1α, En2, and Fmr1; Nlgn3, BTBR, and Slc6A4; and also between X monosomy and Mecp2. With no single treatment for autism found, clustering autism using neuroanatomy and identifying these strong connections may prove to be a crucial step in predicting treatment response.
Keywords: Autism, Magnetic Resonance Imaging, Neuroanatomy, Heterogeneity, Mouse
Introduction
Autism is one of the most heritable psychiatric disorders, associated with a large number of rare genetic lesions with high penetrance1, 2. It is unified by clinical presentations involving deficits in social communication and language, alongside repetitive behaviours and restricted interests. Despite being diagnosed reliably3, autism’s clinical presentation, cognitive and behavioural symptoms, and imaging or histopathology findings show marked variability4. In fact, in a recent review, heterogeneity was reported as a hallmark characteristic in virtually all biological parameters related to autism5. The consequence of this heterogeneity is that no single treatment or diagnostic biomarker is likely for autism as a whole, and while few treatments exist at the moment, clinical trials suggest that only a subset of patients will respond to any given treatment6. Drs. Veenstra-VanderWeele and Blakely have recently discussed current autism treatment7 and indicated the need to examine subgroups of patients. Example subgroups suggested were patients with mTOR and 5-HT signaling abnormalities. Stessman and colleagues have also recently suggested a genotype first approach to subtyping autism8. This subgrouping of autism will enhance our understanding of which pathways need to be targeted for future therapies and, importantly, predict which subset of patients will respond to any given treatment.
Given this heterogeneity the question becomes, what is the appropriate way to group or subdivide autism? While grouping patients based on the behavioural symptoms or genetic underpinnings may seem appropriate, the specificity of behavioural findings and the 250+ implicated genes make these groupings less appealing. Moreover, the genetic causes of the majority of autistic patients remain unknown1. Neuroantomical similarities across groups, however, are specific and can transcend the genetic findings. Previous attempts have been made to cluster the neuroanatomical findings in human autism9, and that study highlights the importance and benefit to subtyping autism. Using a model system such as the mouse can build on this to eliminate environmental factors that may affect the behaviour and genetics.
There are currently over 70 mouse models related to autism10, either created through genetic manipulations based on human genetic findings11–13 or inbred strains defined as behaviourally autistic-like14. Additionally, the International Mouse Knockout Consortium has been working to mutate all protein-encoding genes in the mouse, which will allow for the inclusion of novel autism-related genes as they are discovered15. Several genetic mouse models related to autism have been created and examined and are often used to assess autistic behaviours. However, the behavioural phenotype in the mouse is very subtle and may not be reproducible as it can be dependent on the specific lab or method. One example of this are the controversial behaviour findings in the Neuroligin3 R451C mouse model13, 16, 17. The neuroanatomical phenotype, on the other hand, as defined by high-resolution magnetic resonance imaging (MRI) combined with advanced image processing techniques, is quite robust18, 19.
There are several additional advantages with using MRI for neuroanatomical phenotyping mouse models related to autism: 1) it provides whole brain coverage, thus not requiring prior hypotheses of implicated brain regions; 2) it is high throughput, with the MRI at the Mouse Imaging Centre (MICe) in Toronto designed to simultaneously image 16 mouse brains in parallel in a ~12 hour scan20, 21; and 3) the MRI methods used here to examine anatomy at a mesoscopic level of resolution directly translate to human imaging studies of patients with autism. Importantly, differences in neuroanatomy are tightly linked with behaviour, as evidenced by directly correlating local anatomy with behavioural measures22, by the retrospective finding that 90% of mouse models with a behavioural phenotype had imaging findings23, and by the realization that behavioural manipulations themselves alter local brain shape24.
The purpose of this study is to investigate how disparate etiologies create the autistic spectrum by examining the neuroanatomical phenotype of 26 mouse models related to the disorder (Supplementary Table 1). This provides information not only about the individual models, but also allows us to investigate the heterogeneity in structure, behavior, and genetics of human autism. This will lead us to a better understanding of autism and possibly lead to increased diagnostic specificity, thus allowing for more targeted treatment.
Materials and Methods
Animal Models
For each individual models there were ≥ 8 mice per group (required for statistical purposes18) and often there were 2 or 3 models for a given genotype depending on heterozygosity (Supplementary Table 1). The per-group numbers used throughout this work are acceptable for the recovery of differences on the order of 5% in volume18. In total, this work utilized scans of 432 animals. To qualify as an animal model of autism in this study, the mouse model would have to either 1) genetically modified to resemble or be related to a genetic lesion found in human autism (i.e. the Neuroligin3 R451C mouse) and listed on the SFARI gene database10, or 2) behaviourally tested such that all the core behavioural phenotypes that describe autism are found in the mouse (i.e. the BTBR mouse)25, 26.
Perfusion Protocol
Mice were sacrificed on approximately p60 (exact dates are listed in Supplementary Table 1). Therefore, the majority of the mice were adults and fully developed. Perfusions were performed at either the Mouse Imaging Centre (MICe) in Toronto or in the collaborator’s labs. All perfusion protocols were consistent across labs and followed a previously described and thoroughly tested perfusion protocol21, 27, 28. Specific details on the perfusion can be found in the supplementary methods.
Magnetic Resonance Imaging
Images were acquired on a 7 Tesla MRI scanner (Agilent, Palo Alto, CA) using a protocol that has been thoroughly tested and previously described29, 30. An in-house custom-built solenoid array was used to acquire anatomical images in parallel, allowing acquisition of the MRI images for up to 16 samples in one overnight session (MRI images acquired prior to 2010 were scanned 3 at a time prior to a hardware upgrade, see supplemental methods), which greatly increases the throughput and is essential for a project such as this. The sequence used was a T2 weighted Fast Spin Echo (FSE): parameters for this sequence have been optimized for high contrast between the gray and white matter required for the image registration process. Total imaging time for the MRI sequences was ~ 12 hours. Further sequence details and parameters are listed in the supplemental methods.
Registration and Analysis
To visualize and compare the mouse brains, the images from the anatomical MRI scans were linearly (6 parameter followed by a 12 parameter) and nonlinearly registered together for each of the individual models using a combination of the mni_autoreg tools31 and ANTS32. This registration process uses deformation based morphometry to calculate the volume of 62 segmented structures encompassing cortical lobes, large white matter structures (i.e. corpus callosum), ventricles, cerebellum, brain stem, and olfactory bulbs33. The volumes of each of the 62 regions were calculated as percentages of total brain volume for each of the 26 different models. The brains can also be assessed on a voxelwise basis to examine the localized changes within regions and throughout the brain. Further detail on the registration process can be found in the supplemental methods.
Clustering of Models
All statistical analysis and clustering were performed using the R statistical package (www.r-project.com). For each of the 62 brain regions in each of the 26 different mouse models effect sizes were calculated, measured as Cohen’s d. Effect size was used in order to not just examine significant findings but also to compare trends between groups. Effect sizes were then compared across models using hierarchical clustering, with correlation as the distance function. Effect sizes were computed for each anatomical structure for every mouse line, and dendrograms linking either the brain regions (X-axis) or models together (Y-axis) are displayed.
Defining the Groups
To determine the consistency of the clustering we bootstrapped each of the 26 models. This consisted of random sampling from mice within each group per model (i.e. genetic mutant and control) with repeats such that the ‘n’ was consistent per model but a new random sampling of the group was used each time. This was repeated 1000 times to determine the consistency of the clustering. How often regions were clustered together (assuming 8 different groups) was determined for each of the 1000 repeats. Regions were termed “connected” when they were grouped with each other over 50% of the time. Clustering of the models was performed in a similar fashion. How often models were clustered together (assuming 5 different groups) was determined for each of the 1000 repeats, and a 26 × 26 matrix was created in which the proportion of time (of the 1000 repeats) a model was grouped with another model. The assumption of groups (8 for the regions and 5 for the models) was based on the original dendrogram for the real data prior to the bootstrapping. Different group assumptions (±1) and connection thresholds (±5%) were tested and the results were consistent. Clusters were determined using the R statistical library pvclust, which assesses clustering uncertainty via multiscale bootstrap resampling34. The hierarchical clustering method used was “complete,” and the distance metric was “correlation” which used the Pearson method. An alpha <0.10 was used to determine the clusters.
Verification of Clusters
In order to confirm that the clustering was indicative of actual relationships between the models several verifications were performed. Two random simulated data sets were created in which the volumes for each of the 62 structures were randomized based on the mean and standard deviation of the given structure across all models. The first data set was consistent with the real data, in that the groups had overlapping controls (i.e. 16p11 (df/+) and 16p11(dp/+) were compared to the same control). The second data set compared each of the 26 models to its own randomized control (i.e. 16p11 (df/+) and 16p11 (dp/+) both had a separate controls). These new randomized data sets were remade 100 times and run through the same analysis as the real data to determine the likelihood of the connections. That data was then thresholded at 20, 25, 30, and 50% in order to determine the connection strength of the models (i.e. how often in the 100 samples that models were grouped together). This was a bootstrapping based threshold, where chance for 5 groups would be 20%. At a connection threshold of 25% no connections existed in the data set without overlapping controls, therefore no model was connected to another in >25% of the 100 bootstrapped samples (Supplementary Table 2). Lastly, while we attempted to minimize the variation in our study by using a specific age and sex, this was not always possible. Turner syndrome (XO), for example, made using male subjects impossible. To verify our interpretations of the data and groups, we reanalyzed the data with only male subjects and a restricted age range (8–15 weeks). With this restriction, 19 different models were still included and the clusters remained consistent.
Results
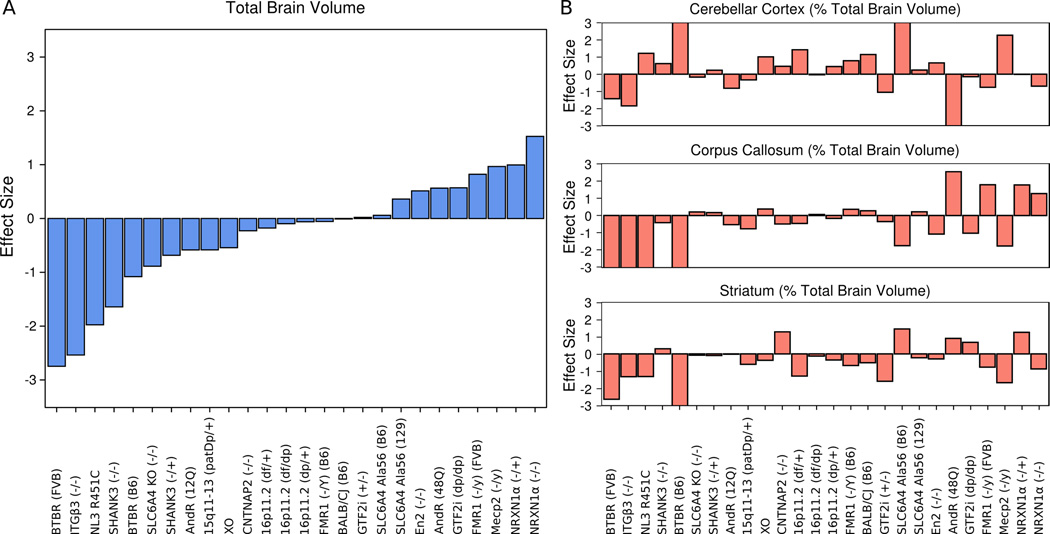
Head circumference and brain size differences are commonly reported findings in human autism, though somewhat controversial5, 35, 36, and the heterogeneity in these measurements was recapitulated with the 26 mouse models examined. Effect size differences in total brain volume between the model and corresponding control ranged from −2.7 to 1.5 (Figure 1A). Of the 26 models examined 8 models were smaller (effect sizes < −0.5) and 5 models were larger (effect size > 0.5). In order to account for the differences in total brain volume, relative volume (% total brain volume) was used for the regional comparisons of the 62 different regions33. There regional differences were still quite heterogeneous with relative volume (Figure 1B). Supplementary Table 3 lists the number of significant regional differences for each of the 26 models measured in both absolute (mm3) and relative volume. Seventeen of the 26 models examined had significant regional relative volume differences at a false discovery rate of less than 10%.
Figure 1.
Heterogeneity of Volume Measurements Across Models - A) Total brain volume across all models. Individual models ordered from smallest to largest effect size compared to their corresponding control. As brain volume or head circumference is a widely used indicator of an autism-like phenotype it is noteworthy to see a range of total brain volume differences that are consistent with human findings in autism. B) Relative volumes of 3 example regions are shown (cerebellar cortex, corpus callosum, and striatum across all models. Models ordered identical to A). The variability in B) shows that the total brain volume differences are not the only factor driving the heterogeneity in these models.
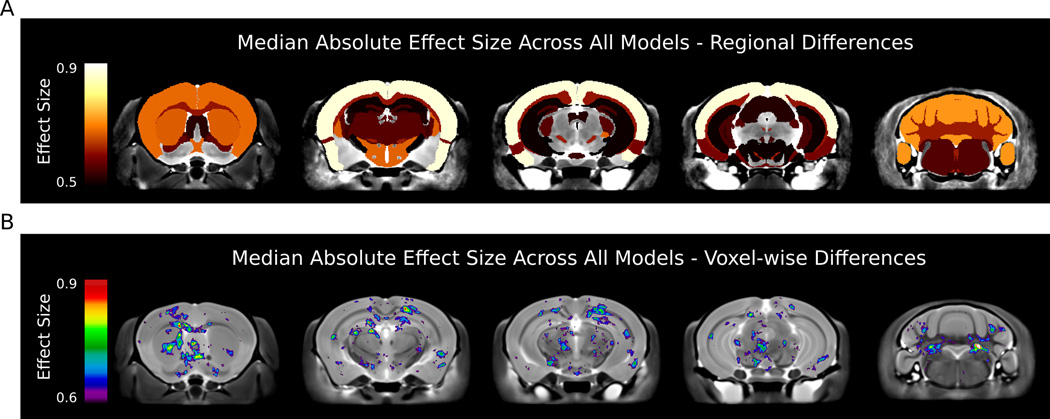
The most affected regions across all models, regardless of whether the region was increased or decreased, were the parieto-temporal lobe (median absolute effect size of 0.96), cerebellar cortex (0.77), frontal lobe (0.73), hypothalamus (0.72), and the striatum (0.72). Figure 2A highlights the significant regions with median effect sizes above 0.5 on 5 different coronal slices. When examining sub-regional (i.e. per voxel) median absolute effect sizes (Figure 2B) additional alterations were identified located in hippocampal CA1, deep cerebellar nuclei, and dorsal raphe nuclei.
Figure 2.
Median Absolute Effects Across all Models - Coronal slices indicating regions that were affected with a median absolute effect size greater than 0.5 for regional comparison and 0.6 for voxelwise comparisons. A) The most affected regions across all models were the parieto-temporal lobe, cerebellar cortex, frontal lobe, hypothalamus, and the striatum. B) voxelwise differences highlighted additional areas affected across all models. Decreases are seen in CA1 and the dentate of the hippocampus as well as increases in the dorsal raphe nuclei.
Median effect sizes were computed for each anatomical structure for every mouse line from the bootstrapped dataset (see methods), and dendrograms linking either the regions (X-axis) or models together (Y-axis) were computed using correlation as the distance metric (Figure 3). The network of the regions and models that resulted from this can be seen in Figure 4 and 5, respectively.
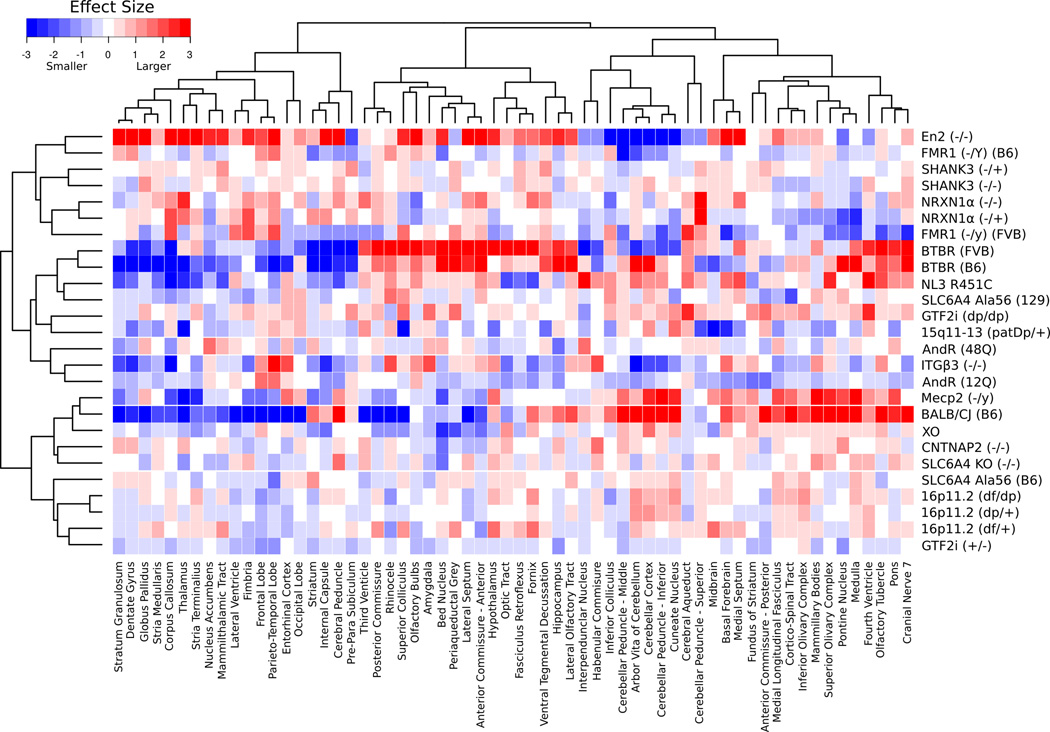
Figure 3.
Volume Differences and Clustering of the Regions Examined – This heatmap displays the median effect size differences in relative volume between the 26 different mouse models and their specific controls for each of the 62 different regions across the 1000 bootstrapped samples. Red represents an increase in volume compared to control and blue represents a decrease. The dendrograms on the x and y-axes represent the correlation between regions (x-axis) and models (y-axis). For regions that are closely correlated, such as the stratum granulosum and dentate gyrus, the dendrogram joins close to the data, whereas regions such as the periaqueductal grey and corpus callosum are not as closely correlated so they join higher.
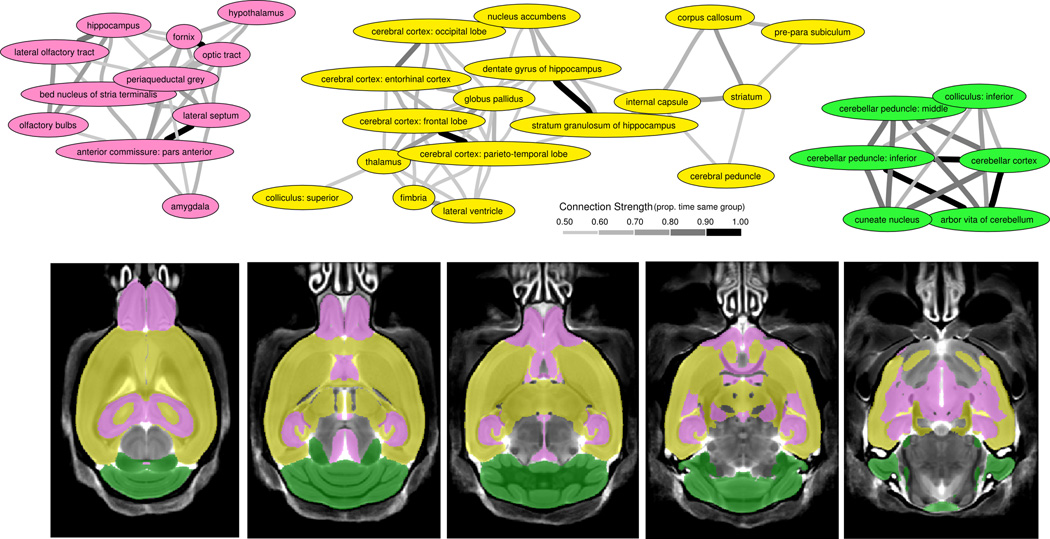
Figure 4.
Clustering of Regions - A) Bootstrapping the regions from Figure 3 revealed 3 large clusters. These clusters are connected based on the proportion of time within the same group over the 1000 bootstrapped samples. Anything above 50% was considered connected. To generalize these regions, the first (pink) cluster includes regions involved with social perception and autonomic regulation as well as some of the most sexually dimorphic regions in the brain, the second (yellow) cluster contains the majority of white matter regions, which could be representative of connectivity, and the third (green) cluster represents the cerebellar regions, which are commonly implicated in autism. B) Highlights the clusters on 5 axial slices throughout the brain and shows the interspersed nature of the pink and yellow clusters.
Figure 5.
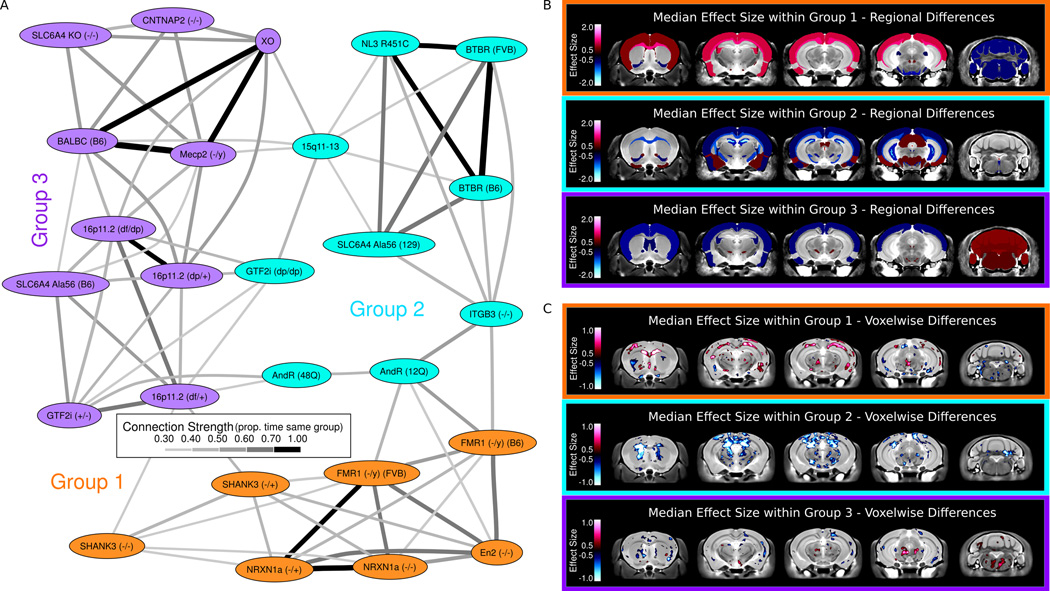
A) Clustering of the Autism Models - Clustering of the models, based on the bootstrapping within models shown in Figure 3, was created in a similar fashion to the regions shown in Figure 4. The hierarchical clustering segregated the models into three specific groups. These groups are connected based on the proportion of time within the same group over the 1000 bootstrapped samples. Anything above 30% was considered connected as random connections were only found below 25%. B) Regional Differences within Groups - The most affected regions in each of the three groups are highlighted. Group 1 is characterized by increases in many of the white matter structures, specifically the corpus callosum and fimbria, and the cortex, and decreases in the cerebellar cortex. Group 2 is characterized by decreases in many white matter structures, and again the corpus callosum is implicated, as well as the striatum and hippocampus. Group 3 is characterized by increases in the cerebellum and decreases in the thalamus and lateral septum. For a full listing of the differences in these groups see Supplementary Table 4. C) Voxelwise Differences within Groups – Similar to the regions Group 1 is characterized by increases in many of the white matter structures, specifically the corpus callosum and external capsule are outlined here. Group 2 is characterized by decreases in many white matter structures, and again the corpus callosum is drastically decreased in size. Group 3 is characterized by bilateral decreases in the striatum as well as an increase in the dorsal raphe nuclei.
Three large clusters were identified from a subset of regions (Figure 4). Regions were only included for this if their volume was larger than 0.7 mm3 or had direct relevance to another structure (i.e. anterior commissure – pars posterior). The first two clusters contained regions that were interspersed throughout the rest of the brain. The first (pink) cluster includes regions commonly identified with the limbic system and involved with social perception and autonomic regulation, such as the bed nucleus of the stria terminalis, amygdala, hypothalamus, lateral septum, hippocampus, and olfactory regions. These are also among the most sexually dimorphic areas of the brain28, 37. The second (yellow) cluster includes many of the large white matter structures in the mouse brain such as the cerebral peduncle, corpus callosum, internal capsule, and fimbria, which together account for ~72% of the white matter tracts in the brain (outside the cerebellum). Some of the smaller white matter structures are not included within this grouping, the fornix and anterior commissure, but are likely influenced by their close association to the olfactory bulbs (anterior commissure) and hypothalamus (fornix). Regardless, these findings may indicate that the changes in white matter are a global difference in connectivity. In addition, the basal ganglia to cortex circuit (nucleus accumbens, globus pallidus, striatum, thalamus, and the cortex) is within this group. The third (green) cluster consisted of 6 structures linking cerebellar regions and the inferior colliculus.
The clustering of the 26 different mouse lines produced three large groups (Figure 5A), which differed in both direction of the volume difference as well as localization of anatomical phenotypes. The strongest connections (>60%) in the clustering of models were between Nrxn1α, En2, and Fmr1 in Group 1, Nlgn3 KI, BTBR, and Slc6A4 KI (129) in Group 2, and Mecp2, XO, and BALB/C, as well as Gtf2i (+/−) and 16p11. A full listing of the groups and their most affected regions can be found in Supplementary Table 4. A visual representation of the most affected regions for each group is shown in Figure 5B. Group 1 (consisting of En2, both Fmr1 mutations, Nrxn1α, and Shank3) has increases in large white matter structures including the corpus callosum, fimbria, and fornix, as well as increases in the frontal and parieto-temporal lobe and decreases in the cerebellar cortex (Figure 5B, Supplementary Table 4). Conversely, in group 2 (consisting of AndR, BTBR, Gtf2i (dp/dp), Itgβ3, 15q11–13, Slc6A4 KI (129), and Nlgn3 KI) the majority of structures were found to be smaller. Decreases in white matter structures such as the cerebral peduncle, corpus callosum, and internal capsule, as well as the globus pallidus, hippocampus and striatum were found in Group 2. Group 3 (consisting of 16p11, BALB/c, Cntnap2 (−/−), Gtf2i (+/−), Mecp2, Slc6A4 KI (B6), Slc6A4 KO, and XO) was a mixture of increases and decreases, where the frontal and parieto-temporal lobes were decreased in size and the cerebellum was increased in size. Median voxelwise differences in each of the 3 groups followed similar trends to the regional changes (Figure 5C) with Group 1 displaying increases in major white matter structures like the corpus callosum, Group 2 showing the opposite change, and Group 3 displaying no differences. Furthermore, all three groups had differences in the cerebellum, however none of those changes were consistent in direction or precise location across models.
Discussion
Expectedly, the mouse models used in this study recapitulate the heterogeneity seen with the neuroanatomical findings in autism patients38, 39. Furthermore, the parieto-temporal lobe, cerebellar cortex, frontal lobe, hypothalamus, and the striatum are some of the most implicated regions in human autism40, 41. While those regions were the most affected across all models, they were not consistently affected in all groups and the changes were not consistently in the same direction. The cerebellar cortex, for example, was oppositely affected in Group 1 compared to Group 3 and not affected in Group 2. This highlights the importance of the clustering performed here, because in spite of the cerebellar cortex being one of the most affected regions across all models it is not intrinsic to the autism phenotype, and it is imperative that this variability be taken into account.
Overall, the clustering of anatomical regions revealed three separate circuits: a cortex to basal ganglia loop likely implicated in repetitive behaviours, executive function, and communication, a more dispersed set of brains regions involved in social recognition and autonomic regulation22, and a third cluster localized in the cerebellum, which has repeatedly been shown to be affected in autism42. These groupings also appear to be consistent with the abnormal connectivity theory in autism43, where the neuroanatomy of long range connections in the models within Group 3 was the least affected by the genetic manipulation, Group 1 consisted of increases in white matter structures across models, and Group 2 consisted of decreases in similar white matter structures.
While behavioural phenotyping of these 26 different models was not performed in this study, it is worth examining the published behavioural findings of these models to determine any over-lapping behaviours that could explain the groups. The known behaviours of the 26 models relevant to autism are listed in Supplementary Table 5, and an attempt at clustering the behavioural findings has been previously published44, but their clustering is confounded by missing and mixed results. Autism is primarily a disorder of sociability, and in total across all 3 groups there are 10 models that have a reported social deficit and 7 that have either no difference noted or mixed results. Supplementary Figure 1 shows the regional median effect size differences for the poor social models as well as the normal/mixed social models. Interestingly, the hippocampus shows up in both groups, decreased in the normal social models and increased in the poor social models. The hypothalamus is also substantially larger in the poor social models compared to their corresponding controls. These findings from the social groups are not consistent with any of the neuroanatomical groupings, with the hypothalamus and hippocampus not affected in any of the three groups. Furthermore, the behavioural findings are confounded by many of relevant findings either not reported, not reproducible, or having mixed results. The mixed results are especially troubling, and as noted in a 2006 paper, differences in noise, light, home cage environment, handling and diet can dramatically alter behaviour45. This further illustrates the need for a highly specific reproducible technique for the large-scale high throughput phenotyping and clustering of the autistic mouse models.
Similar to the reported behavioural findings for these models there is only marginal genetic overlap. For example, the genes that are related to the synapse, Cntnap2, Fmr1, Mecp2, Nlgn3, Nrxn1α, and Shank3 are spread across the three groups, although 50% are found in Group 1. Furthermore, Nlgn3 and Nrxn1α, in spite of connecting across the synaptic cleft, are found in different groups and have remarkably divergent findings. This was also true when multiple models were clustered with known behavioural findings44. While genetics and related genetic pathways seem like a reasonable clustering metric, an inherent problem is that genetic clustering can be too specific as an unreasonable number of clusters can emerge. Furthermore, the genetics are only known in 20–50% of autism patients2, and therefore the neuroanatomical clustering performed here is an attractive alternative as it does not rely on genetic information.
While this study attempted to minimize the variation between groups to the genetics alone, 16p11 model was included with mixed sex and XO included females only, and while many of models were ~p60 in age, there were some models where this was not the case. However, if the models are restricted to only males, and the age range is restricted to 8–15 weeks, 19 different models would still be included. Overall the differences across the restricted models remained consistent, the parieto-temporal lobe (median absolute effect size of 1.14), cerebellar cortex (0.76), frontal lobe (0.71), hypothalamus (0.75), and the striatum (0.86) all remained strongly affected across models. Furthermore, several white matter structures were highlighted in the restricted group, the corpus callosum (1.26), cerebral peduncle (1.02), and internal capsule (0.86) were all strongly affected across the 19 models. This white matter finding lends credence to the abnormal connectivity being an important factor in autism. When these 19 models were run through the same analysis pipeline they clustered into a similar three groups. Group 1 was the same group minus the Shank3 models and Fmr1 (B6), as they did not fit within the age range. Group 2 and 3 were slightly different but kept the core of their individual groups with several models lost due to the age and sex restriction, 16p11s and XO for example. These similarities in the smaller more restrictive group indicate that in spite of the variability of age and sex in with the larger 26 models, the overall story remains the same: That autism–like anatomical phenotypes in mouse models both preferentially affect key regions of the brain, but also divide into distinct clusters based on directionality and localization of anatomical changes in the brain.
Independent MRI studies of 4 of these models have been previously published. The NL3 KO has been examined by Radyushkin and colleagues46. However, they only examined 6 large regions, including the total brain volume. The only difference found was a total brain volume decrease of 5%, which is similar to the 8% decrease found with the NL3 R451C KI. Previous MRI examinations of the Mecp2 mouse have reported findings of decreased total brain volume and cerebellar a motor cortex decreases are consistent with the absolute volume differences in the Mecp2308 model used here47, 48. A recent study on the BTBR mouse model49 is in good agreement with the results BTBR (vs. B6) mice shown here22, including reductions in the fronto-cortical areas and thalamus, and increases in the hippocampus and cerebellum. Another recent study by Portmann and colleagues has reported several neuroanatomical differences in the 16p11.2 mouse model at a young age (p7)50. Despite the independent creation and age and sex differences, the 16p11.2 differences in the Portmann mouse are in remarkable agreement with the 16p11.2 differences seen here, with several subcortical midline structures like the hypothalamus significantly increased in both studies. Overall the strong agreement with independent studies on these four mouse lines increases our confidence in the generalizability of the results reported herein.
With the number of children being diagnosed with autism increasing, early diagnosis and appropriate treatments is key. The heterogeneity of autism makes this difficult. While genetics may link children of a certain group together, for example, 16p11.2 disorders or Phelan-McDermid Syndrome (22q13 deletion), classification of autism based on neuroanatomical similarities allow these groups to expand beyond a single specific gene or chromosomal region as well as include autistic patients that may have an unknown genetic link. These neuroanatomical similarities provide new links between models/genes. For example, Nrxn1α, Fmr1, and En2 are very strongly connected, and therefore, examining the effects of an mGlur5 inhibitor, like CTEP, which has been shown to useful in Fmr1 mice51, could have a similar effect for Nrxn1α and En2.
The work here has shown that, while there is not a single neuroanatomical pattern defining autism, there also is not, as seen from the overlapping groups in Figure 5, a distinct neuroanatomical pattern for each of the 26 models examined here as several of the models have overlapping findings. How our groupings overlap or differ from strategies using protein-protein interactions or using behaviour needs to be explored. Similarly, the developmental profiles of these groups need to be explored to determine is their developmental trajectory remains consistent. The ultimate test of these groupings, however, will be how they predict treatment, whether it is behavioural or pharmaceutical. With neuroanatomical differences driving the groupings, we hypothesize similar treatment responses for models within groups.
Supplementary Material
Supplementary Figure 1 – Regions Associated with Social Deficits - A) Highlighted are the most affected regions in the normal social models. B) Similar to A) the most affected regions are highlighted for the poor social models, which included models that had no social differences or mixed results (Supplementary Table 5). Interestingly the hippocampus was inversely related to the sociability of the mice. Furthermore, the size of the hypothalamus is substantially increased in the poor social models but was not found in any of the three groups in Figure 4.
Acknowledgements
This work was primarily funded by the Canadian Institute for Health Research (CIHR) and the Ontario Brain Institute (OBI). J.E received salary support from the Ontario Mental Health Foundation (OHMF) and R.M.H. holds a Canada Research Chair.
Footnotes
Competing Interests - The authors declare competing financial intrests: E.A. has received consultation fees from Novartis and Seaside therapeutics, and has an unrestricted grant from Sanofi Canada. J.V-VW. receives research funding from Seaside Therapeutics, Novartis, Roche Pharmaceuticals, Forest, Sunovion, and SynapDx and sits on the advisory board for Novartis and Roche Pharmaceuticals. The remaining authors declare no conflicts of interest.
References
- 1.Abrahams BS, Geschwind DH. Connecting genes to brain in the autism spectrum disorders. Arch Neurol. 2010;67:395–399. doi: 10.1001/archneurol.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geschwind DH. Genetics of autism spectrum disorders. Trends Cogn Sci (Regul Ed) 2011;15:409–416. doi: 10.1016/j.tics.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lord C, Risi S, DiLavore PS, Shulman C, Thurm A, Pickles A. Autism from 2 to 9 years of age. Arch Gen Psychiatry. 2006;63:694–701. doi: 10.1001/archpsyc.63.6.694. [DOI] [PubMed] [Google Scholar]
- 4.Huerta M, Lord C. Diagnostic evaluation of autism spectrum disorders. Pediatr Clin North Am. 2012;59 doi: 10.1016/j.pcl.2011.10.018. 103–11–xi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amaral DG. The promise and the pitfalls of autism research: an introductory note for new autism researchers. Brain Res. 2011;1380:3–9. doi: 10.1016/j.brainres.2010.11.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canitano R. Novel treatments in autism spectrum disorders: From synaptic dysfunction to experimental therapeutics. Behavioural Brain Research. 2012 doi: 10.1016/j.bbr.2012.11.024. [DOI] [PubMed] [Google Scholar]
- 7.Veenstra-VanderWeele J, Blakely RD. Networking in autism: leveraging genetic, biomarker and model system findings in the search for new treatments. Neuropsychopharmacology. 2012;37:196–212. doi: 10.1038/npp.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stessman HA, Bernier R, Eichler EE. A genotype-first approach to defining the subtypes of a complex disease. Cell. 2014;156:872–877. doi: 10.1016/j.cell.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hrdlicka M, Dudova I, Beranova I, Lisy J, Belsan T, Neuwirth J, et al. Subtypes of autism by cluster analysis based on structural MRI data. Eur Child Adolesc Psychiatry. 2005;14:138–144. doi: 10.1007/s00787-005-0453-z. [DOI] [PubMed] [Google Scholar]
- 10.Banerjee-Basu S, Packer A. SFARI Gene: an evolving database for the autism research community. Dis Model Mech. 2010;3:133–135. doi: 10.1242/dmm.005439. [DOI] [PubMed] [Google Scholar]
- 11.Nakatani J, Tamada K, Hatanaka F, Ise S, Ohta H, Inoue K, et al. Abnormal behavior in a chromosome-engineered mouse model for human 15q11–13 duplication seen in autism. Cell. 2009;137:1235–1246. doi: 10.1016/j.cell.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horev G, Ellegood J, Lerch JP, Son Y-EE, Muthuswamy L, Vogel H, et al. Dosage-dependent phenotypes in models of 16p11.2 lesions found in autism. Proc Natl Acad Sci USA. 2011;108:17076–17081. doi: 10.1073/pnas.1114042108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tabuchi K, Blundell J, Etherton MR, Hammer RE, Liu X, Powell CM, et al. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 2007;318:71–76. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moy SS, Nadler JJ, Young NB, Perez A, Holloway LP, Barbaro RP, et al. Mouse behavioral tasks relevant to autism: phenotypes of 10 inbred strains. Behavioural Brain Research. 2007;176:4–20. doi: 10.1016/j.bbr.2006.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.International Mouse Knockout Consortium. Collins FS, Rossant J, Wurst W. A mouse for all reasons. Cell. 2007;128:9–13. doi: 10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 16.Chadman KK, Gong S, Scattoni ML, Boltuck SE, Gandhy SU, Heintz N, et al. Minimal aberrant behavioral phenotypes of neuroligin-3 R451C knockin mice. Autism Res. 2008;1:147–158. doi: 10.1002/aur.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Etherton M, Földy C, Sharma M, Tabuchi K, Liu X, Shamloo M, et al. Autism-linked neuroligin-3 R451C mutation differentially alters hippocampal and cortical synaptic function. Proc Natl Acad Sci USA. 2011;108:13764–13769. doi: 10.1073/pnas.1111093108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lerch JP, Gazdzinski L, Germann J, Sled JG, Henkelman RM, Nieman BJ. Wanted dead or alive? The tradeoff between in-vivo versus ex-vivo MR brain imaging in the mouse. Front Neuroinform. 2012;6:6. doi: 10.3389/fninf.2012.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Eede MC, Scholz J, Chakravarty MM, Henkelman RM, Lerch JP. Mapping registration sensitivity in MR mouse brain images. Neuroimage. 2013;82:226–236. doi: 10.1016/j.neuroimage.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 20.Bock NA, Konyer NB, Henkelman RM. Multiple-mouse MRI. Magn Reson Med. 2003;49:158–167. doi: 10.1002/mrm.10326. [DOI] [PubMed] [Google Scholar]
- 21.Lerch JP, Sled JG, Henkelman RM. MRI phenotyping of genetically altered mice. Methods Mol Biol. 2011;711:349–361. doi: 10.1007/978-1-61737-992-5_17. [DOI] [PubMed] [Google Scholar]
- 22.Ellegood J, Babineau BA, Henkelman RM, Lerch JP, Crawley JN. Neuroanatomical analysis of the BTBR mouse model of autism using magnetic resonance imaging and diffusion tensor imaging. Neuroimage. 2013;70:288–300. doi: 10.1016/j.neuroimage.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nieman BJ, Lerch JP, Bock NA, Chen XJ, Sled JG, Henkelman RM. Mouse behavioral mutants have neuroimaging abnormalities. Hum Brain Mapp. 2007;28:567–575. doi: 10.1002/hbm.20408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lerch JP, Yiu AP, Martinez-Canabal A, Pekar T, Bohbot VD, Frankland PW, et al. Maze training in mice induces MRI-detectable brain shape changes specific to the type of learning. Neuroimage. 2011;54:2086–2095. doi: 10.1016/j.neuroimage.2010.09.086. [DOI] [PubMed] [Google Scholar]
- 25.Brodkin ES. BALB/c mice: low sociability and other phenotypes that may be relevant to autism. Behavioural Brain Research. 2007;176:53–65. doi: 10.1016/j.bbr.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 26.McFarlane HG, Kusek GK, Yang M, Phoenix JL, Bolivar VJ, Crawley JN. Autism-like behavioral phenotypes in BTBR T+tf/J mice. Genes, Brain and Behavior. 2008;7:152–163. doi: 10.1111/j.1601-183X.2007.00330.x. [DOI] [PubMed] [Google Scholar]
- 27.Cahill LS, Laliberté CL, Ellegood J, Spring S, Gleave JA, Eede MCV, et al. Preparation of fixed mouse brains for MRI. Neuroimage. 2012;60:933–939. doi: 10.1016/j.neuroimage.2012.01.100. [DOI] [PubMed] [Google Scholar]
- 28.Spring S, Lerch JP, Henkelman RM. Sexual dimorphism revealed in the structure of the mouse brain using three-dimensional magnetic resonance imaging. Neuroimage. 2007;35:1424–1433. doi: 10.1016/j.neuroimage.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 29.Nieman BJ, Flenniken AM, Adamson SL, Henkelman RM, Sled JG. Anatomical phenotyping in the brain and skull of a mutant mouse by magnetic resonance imaging and computed tomography. Physiol Genomics. 2006;24:154–162. doi: 10.1152/physiolgenomics.00217.2005. [DOI] [PubMed] [Google Scholar]
- 30.Nieman BJ, Bock NA, Bishop J, Chen XJ, Sled JG, Rossant J, et al. Magnetic resonance imaging for detection and analysis of mouse phenotypes. NMR Biomed. 2005;18:447–468. doi: 10.1002/nbm.981. [DOI] [PubMed] [Google Scholar]
- 31.Collins DL, Neelin P, Peters TM, Evans AC. Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J Comput Assist Tomogr. 1994;18:192–205. [PubMed] [Google Scholar]
- 32.Avants BB, Yushkevich P, Pluta J, Minkoff D, Korczykowski M, Detre J, et al. The optimal template effect in hippocampus studies of diseased populations. Neuroimage. 2010;49:2457–2466. doi: 10.1016/j.neuroimage.2009.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dorr AE, Lerch JP, Spring S, Kabani N, Henkelman RM. High resolution three-dimensional brain atlas using an average magnetic resonance image of 40 adult C57Bl/6J mice. Neuroimage. 2008;42:60–69. doi: 10.1016/j.neuroimage.2008.03.037. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 35.Courchesne E, Campbell K, Solso S. Brain growth across the life span in autism: age-specific changes in anatomical pathology. Brain Res. 2011;1380:138–145. doi: 10.1016/j.brainres.2010.09.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raznahan A, Wallace GL, Antezana L, Greenstein D, Lenroot R, Thurm A, et al. Compared to what? Early brain overgrowth in autism and the perils of population norms. Biological Psychiatry. 2013;74:563–575. doi: 10.1016/j.biopsych.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gleave JA, Wong MD, Dazai J, Altaf M, Henkelman RM, Lerch JP, et al. Neuroanatomical phenotyping of the mouse brain with three-dimensional autofluorescence imaging. Physiol Genomics. 2012;44:778–785. doi: 10.1152/physiolgenomics.00055.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toal F, Daly EM, Page L, Deeley Q, Hallahan B, Bloemen O, et al. Clinical and anatomical heterogeneity in autistic spectrum disorder: a structural MRI study. Psychol Med. 2010;40:1171–1181. doi: 10.1017/S0033291709991541. [DOI] [PubMed] [Google Scholar]
- 39.Amaral DG, Schumann CM, Nordahl CW. Neuroanatomy of autism. Trends Neurosci. 2008;31:137–145. doi: 10.1016/j.tins.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 40.Stanfield AC, McIntosh AM, Spencer MD, Philip R, Gaur S, Lawrie SM. Towards a neuroanatomy of autism: A systematic review and meta-analysis of structural magnetic resonance imaging studies. European Psychiatry. 2008;23:289–299. doi: 10.1016/j.eurpsy.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 41.Kurth F, Narr KL, Woods RP, O'Neill J, Alger JR, Caplan R, et al. Diminished gray matter within the hypothalamus in autism disorder: a potential link to hormonal effects? Biological Psychiatry. 2011;70:278–282. doi: 10.1016/j.biopsych.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fatemi SH, Aldinger KA, Ashwood P, Bauman ML, Blaha CD, Blatt GJ, et al. Consensus paper: pathological role of the cerebellum in autism. Cerebellum. 2012;11:777–807. doi: 10.1007/s12311-012-0355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kana RK, Libero LE, Moore MS. Disrupted cortical connectivity theory as an explanatory model for autism spectrum disorders. Phys Life Rev. 2011;8:410–437. doi: 10.1016/j.plrev.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 44.Ey E, Leblond CS, Bourgeron T. Behavioral profiles of mouse models for autism spectrum disorders. Autism Res. 2011;4:5–16. doi: 10.1002/aur.175. [DOI] [PubMed] [Google Scholar]
- 45.Bailey KR, Rustay NR, Crawley JN. Behavioral phenotyping of transgenic and knockout mice: practical concerns and potential pitfalls. ILAR J. 2006;47:124–131. doi: 10.1093/ilar.47.2.124. [DOI] [PubMed] [Google Scholar]
- 46.Radyushkin K, Hammerschmidt K, Boretius S, Varoqueaux F, El-Kordi A, Ronnenberg A, et al. Neuroligin-3-deficient mice: model of a monogenic heritable form of autism with an olfactory deficit. Genes, Brain and Behavior. 2009;8:416–425. doi: 10.1111/j.1601-183X.2009.00487.x. [DOI] [PubMed] [Google Scholar]
- 47.Saywell V, Viola A, Confort-Gouny S, Le Fur Y, Villard L, Cozzone PJ. Brain magnetic resonance study of Mecp2 deletion effects on anatomy and metabolism. Biochemical and Biophysical Research Communications. 2006;340:776–783. doi: 10.1016/j.bbrc.2005.12.080. [DOI] [PubMed] [Google Scholar]
- 48.Ward BC, Agarwal S, Wang K, Berger-Sweeney J, Kolodny NH. Longitudinal brain MRI study in a mouse model of Rett Syndrome and the effects of choline. Neurobiology of Disease. 2008;31:110–119. doi: 10.1016/j.nbd.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 49.Dodero L, Damiano M, Galbusera A, Bifone A, Tsaftsaris SA, Scattoni ML, et al. Neuroimaging evidence of major morpho-anatomical and functional abnormalities in the BTBR T+TF/J mouse model of autism. PLoS ONE. 2013;8:e76655. doi: 10.1371/journal.pone.0076655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Portmann T, Yang M, Mao R, Panagiotakos G, Ellegood J, Dolen G, et al. Behavioral Abnormalities and Circuit Defects in the Basal Ganglia of a Mouse Model of 16p11.2 Deletion Syndrome. Cell Rep. 2014 doi: 10.1016/j.celrep.2014.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Michalon A, Sidorov M, Ballard TM, Ozmen L, Spooren W, Wettstein JG, et al. Chronic pharmacological mGlu5 inhibition corrects fragile X in adult mice. Neuron. 2012;74:49–56. doi: 10.1016/j.neuron.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 – Regions Associated with Social Deficits - A) Highlighted are the most affected regions in the normal social models. B) Similar to A) the most affected regions are highlighted for the poor social models, which included models that had no social differences or mixed results (Supplementary Table 5). Interestingly the hippocampus was inversely related to the sociability of the mice. Furthermore, the size of the hypothalamus is substantially increased in the poor social models but was not found in any of the three groups in Figure 4.