Abstract
Triple negative (TN) (estrogen receptor [ER], progesterone receptor [PR] and Her2 negative) are highly aggressive, rapidly growing, hormone unresponsive tumors diagnosed at later stage that affect younger women with shorter overall survival. The majority of TN tumors are of the basal type. For the remainder identification of target markers for effective treatment strategies remains a challenge. Transgelin (TGLN) is a 22 kDa actin-binding protein of the calponin family. It is one of the earliest markers of smooth muscle differentiation. TGLN has been shown to have important biologic activities including regulating muscle fiber contractility, cell migration and tumor suppression. We examined TGLN expression in the different molecular subtypes of breast cancer.
TGLN expression was examined as a function of tumor size, grade, histologic type, lymph node (LN) status, patients’ age and overall survival, ER, PR, Her-2, Ki-67 in 101 tumors that included 35 luminal A, 28 luminal B, 4 Her2, and 34 TN types.
TGLN positivity (defined as 2+ or 3+) was associated with more aggressive tumors (10% of grade I/II tumors were TGLN+ vs. 53% of grade III tumors, P<0.001), high Ki-67 count and low ER and PR expression (p<0.001), but not with tumor size, age or LN metastasis. TN (n=34) tumors were 7.7 times more likely to be TGLN-positive than non-TN (n=67) tumors (77% vs. 10%, respectively, P<0.001).
TGLN may be an excellent diagnostic marker of TN tumors and could be useful in stratification of patients. TGLN may also prove a potential target for future treatment strategies.
Keywords: Transgelin, invasive breast cancer, molecular subtypes
1. Introduction
Breast cancer is a heterogeneous disease encompassing various entities with distinct morphological features and clinical behaviors. This diversity is the result of distinct genetic, epigenetic, and transcriptomic alterations [1–3]. Recently proposed classification schemes employ gene expression microarray analysis, to categorize breast cancer phenotypes based on their molecular features. The purpose of these classification systems is to facilitate identification of tumor markers that may serve as indicators of prognosis and potentially as therapeutic targets. Breast cancers are therefore categorized within five major molecular subtypes: luminal A, luminal B, normal breast-like, HER2, and basal-like [4–8]. However, the utility of such assignments of molecular subtyping, especially the basal-like subgroup, has generated much interest and has been called into question by scientists, pathologists and oncologists alike [9–11]. Triple-negative (TN) tumors (estrogen receptor [ER]-progesterone receptor [PR]-Her-2 negative) are highly aggressive, rapidly-growing, hormone-unresponsive tumors that tend to be diagnosed at a later stage, affect younger women, and are associated with shorter overall survival [4, 12, 13]. TN tumors have recently been shown to be molecularly, pathologically and clinically a heterogeneous subgroup, although the majority are basal-like. TGLN, also known as smooth muscle protein 22 a (SM22a) is a 22 kDa actin-binding protein of the calponin family that has been shown to stabilize loose actin gels, leading to actin filament gelation [14–16] It is one of the earliest markers of smooth muscle differentiation [14–16]. Although the precise function of TGLN remains unknown, it has been implicated to have a role in many biologic activities, including regulating muscle fiber contractility, cell differentiation, tissue invasion and tumor suppression [15, 17–19].
TGLN has recently been studied on a wide variety of tumors, including breast, colorectal, gastric, gall bladder, pancreatic, prostate and lung adenocarcinomas, with conflicting results. While some studies have demonstrated decreased TGLN expression in breast [15, 18, 20], colorectal [15, 18], gallbladder [21] and prostate cancers [22, 23], others have demonstrated increased expression in colorectal, lung, gastric and pancreatic cancers [24–26].
The goal of this study was to systematically study TGLN expression across molecular subtypes of breast cancer, with emphasis on comparing TGLN expression in TN and Non-TN tumors and correlating its expression with clinicopathologic parameters.
2. Patients and methods
2.1. Patient Cohort
This retrospective study was approved by the institutional review committee at the University of Kansas Medical Center. A total of 101 primary breast carcinomas diagnosed between 1997 (when Her2 testing became available) and 2010, and for which ER, PR and Her-2 status and follow-up information was available, were examined. The cohort included 94 invasive ductal, 7 invasive mixed ductal and lobular carcinomas. Based on the molecular subclassification, our dataset was composed of 35 luminal A (defined as ER+ and/or PR+, Her2 negative, Ki67 <14%), 28 luminal B (defined as ER+ and/or PR+ and Her2+ [Luminal-Her2+] or ER and/or PR+, Her2-, Ki67 ≥14%), 4 Her-2 positive (defined as ER negative, PR negative, Her2+), and 34 TN carcinomas (defined as ER and PR <1% and Her-2 negative by fluorescent in-situ hybridization [FISH] technology and automated immunohistochemistry [IHC]). The samples were taken from 52 mastectomy, 39 lumpectomy, 8 excisional biopsies and 2 core biopsy specimens. Histopathologic parameters, including histologic grade and type at the time of histopathologic diagnosis were extracted from patient pathology records. All tumors were graded using the modified “Nottingham” criteria of Bloom and Richardson. Additional parameters of patients’ age, tumor size and lymph node (LN) metastasis were also recorded for mastectomy, lumpectomy and excisional biopsy specimens.
Clinical parameters, including type of therapy, tumor recurrence and overall survival status, were obtained from the electronic medical records. Overall (as opposed to disease-specific) survival status was reported because discrepancies and/or incomplete data on cause of death are common.
2.2. Immunohistochemical Data
At diagnosis, tissue blocks containing the most representative and well-preserved tumor areas were selected for IHC. Immunohistochemical analysis was performed on tissue fixed with 10% neutral buffered formalin. IHC analyses for ER, PR, Her-2, and Ki-67, were performed at the time of diagnosis on all specimens. IHC analysis for TGLN was performed on tissue microarrays obtained from the same samples. Briefly, after review of the H and E slides and marking of tumor areas, 2 mm tissue cores of representative tumor areas were extracted and inserted in recipient blocks. Two cores from each tumor were analyzed in an attempt to account for the impact of tumor heterogeneity on TGLN expression. Her-2 antibody was detected using the HercepTest (DAKO, Carpinteria, CA). The individual antibodies, vendor, titration titer, time of titration, epitope retrieval method and method of detection are shown in Table 1.
Table 1.
Protocols for immunohistochemistry*
| Antibody | Vendor | Titer | Time | Epitope Retrieval | Method of Detection |
|---|---|---|---|---|---|
| ER | BioCare | 1:1000 | 30 min | BioCare Nuclear Decloaker | Envision + LP, mouse (Dako) |
| PR | Dako | 1:5000 | 30 min | Citrate, pH6 | Envision + LP, mouse (Dako) |
| Ki-67 | Dako | 1:1000 | 30 min | BioCare Reveal | Envision + LP, mouse (Dako) |
| HercepTest | Dako | P.D. Her2 | 30 min | Per Kit Instructions | Kit Components (K5204, Dako) |
| Transgelin (Anti SM22 alpha antibody) (ab14106) | Abcam | 1:3000 | 30 min | Biocare | Envision + LP, mouse (Dako) |
All stains were performed using the Dako Autostainer per manufacturer procedures. Buffer used was Tris-buffered saline with Tween; visualization with Dab+ (Dako)
Positive IHC reactions were defined as dark brown reaction cytoplasmic staining for TGLN, positive nuclear staining for ER, PR, and Ki-67 and dark brown reaction on the cell membrane for Her-2 with areas of high-density immunostaining selected for image analysis or manual scoring. For proliferation index (PI) of Ki-67, the percentage of nuclei with immunopositivity was determined using the PI program of first, the CAS (Cell Analysis System) 200 image analyzer (Bacus Laboratory, Chicago, IL) prior to 2001 and the Automated Cellular Imaging System (ACISTM) (San Juan Capistrano, CA) thereafter. For ER and PR both the CAS-200 and the ACISTM systems were used for scoring. Her-2 staining was quantified using a score of 0 or 1+ to indicate negative, and 2+ or 3+ to denote positive staining, per the scoring instructions included in the HercepTest kit. Results were validated using the Her-2 scoring system of the ACIS system and by FISH. TGLN staining was quantified manually using a score of 0 or 1+ as negative and 2+ or 3+ as positive. Staining of <1% of tumor cells with antibodies to ER and PR was considered ER-negative and PR-negative respectively.
2.3. Statistical Analysis
Overall frequencies and percentages were summarized for tumor grade, histology, LN status, and evidence of expression by IHC for ER, PR, Her-2, Ki-67 and TGLN. Distributions of continuous variables were characterized by medians: age at diagnosis, tumor size, and percent positive cells for IHC (ER, PR, Her-2, TGLN and Ki-67). Categorical variables were compared across groups by Fisher’s exact test. Continuous variables were compared using non-parametric Mann-Whitney test. Logistic regression analysis was used to examine the effect of multiple variables on TGLN positivity and tumor recurrence.
3. Results
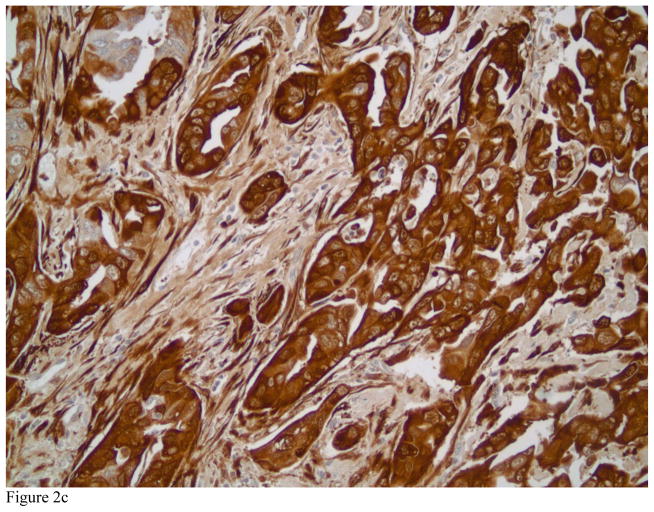
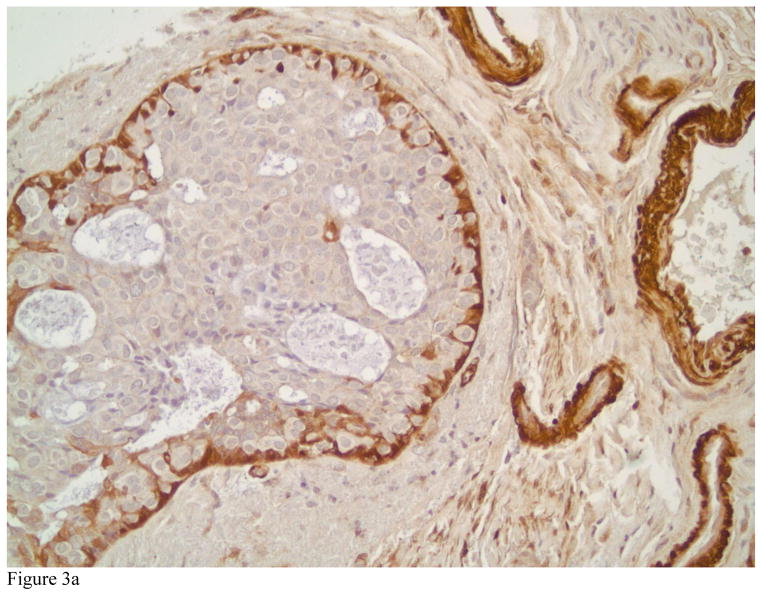
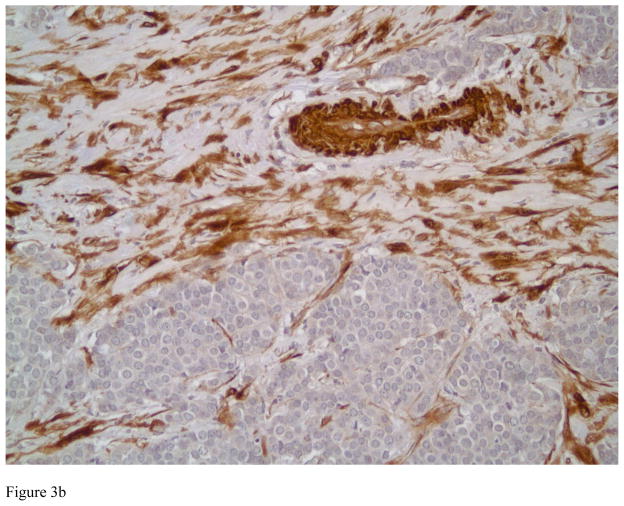
TGLN exclusively localized in the cytoplasm of benign and malignant tumor cells in breast tissue with no nuclear localization. Strong cytoplasmic staining could be observed in myoepithelial cells and fibroblastic cells of benign breast tissue. Figures 1A and B highlight the myoepithelial cells in a normal breast (A) and in a representative case with sclerosing adenosis (B). Normal luminal cells are predominately negative or display very weak cytoplasmic staining. TGLN was expressed in 33% of the invasive carcinomas selected for study (33 of 101 cases) (Figure 2). Examples of tumors with absent, 1+, 2+ and 3+ positively stained invasive carcinoma cells are shown in Figures 3A, B and C, respectively. Although limited by a small number of cases containing a component of ductal carcinoma in situ (DCIS), we also observed strong staining of myoepithelial cells demarcating the lesion (Figure 3A). In the overwhelming majority of samples with invasive tumors, TGLN was highly expressed in the surrounding stroma irrespective of its staining intensity within tumor cells (90%) (Figures 2C and 3B). Clinicopathological parameters of TGLN + and TGLN negative tumors are summarized in Table 2. Although 100% (N=7) of mixed ductal-lobular tumors were negative for TGLN expression, compared to 35% of pure ductal tumors, this difference was not statistically significant. On the other hand, the variation in TGLN expression by SBR grade was statistically significant, with 53% of grade III tumors being TGLN positive compared to only 10% (5/48) of grade I and II tumors. LN status did not vary by TGLN expression. TGLN expression was inversely associated with both ER and PR positivity, with two-thirds of hormone receptor negative tumors expressing TGLN, compared to only 10% of hormone receptor positive tumors (p<0.001). With the exception of 3 PR negative, TGLN negative tumors that were ER positive, there was concordance between the two hormone receptors. While Her-2 expression did not correlate with TGLN expression, there was a strong relationship between TN negative status and TGLN expression (p<0.001). High Ki-67 expression was similarly associated with TGLN expression. When treated as continuous variables, ER, PR, and Ki-67 expression remained different between TGLN + and TGLN -negative tumors. For ER, median values were 95% and 0% for TGLN negative and positive tumors, respectively; for PR median values were 32% and 0%; and for Ki-67, 10% and 50% (p<0.001 for all comparisons).
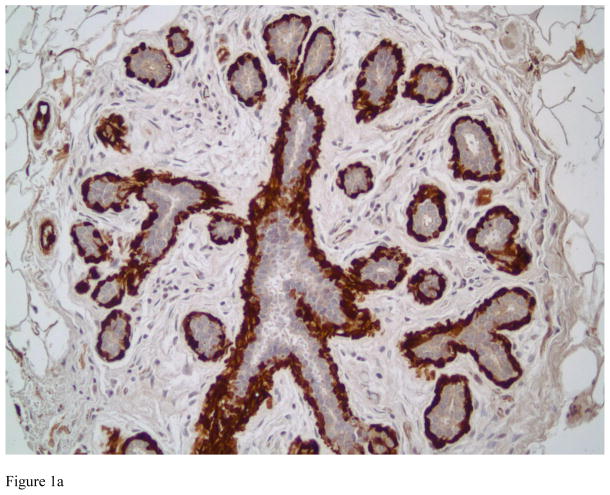
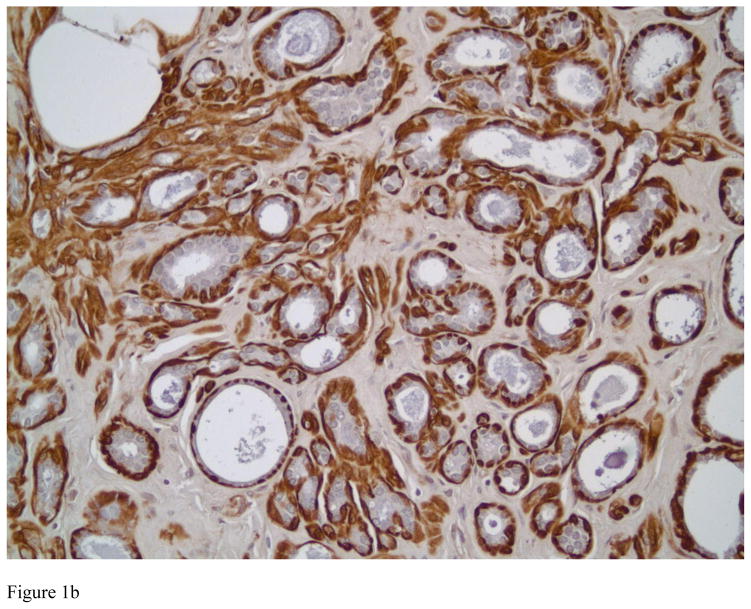
Figure 1.
Transgelin expression in benign breast tissue. Strong cytoplasmic staining is observed in myoepithelial cells in ducts and acini of normal breast (Figure 1A, immunostain, magnification × 200). Figure 1B shows Transgelin expression in sclerosing adenosis, a benign entity that can be confused with invasive carcinoma. Transgelin highlights the myoepithelial cells, confirming the benign nature of the lesion (immunostain, magnification × 200).
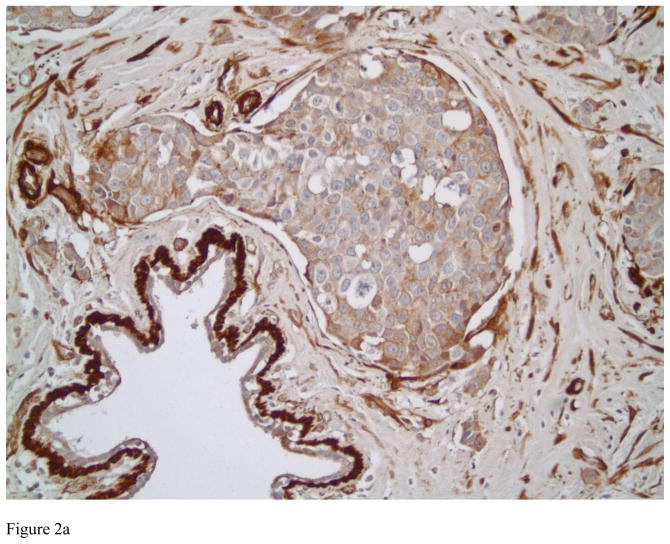
Figure 2.
Representative examples of high grade invasive ductal carcinomas with various staining patterns for Transgelin ranging from 1+ in Figure 2A, 2+ in Figure 2B and 3+ Figure 2C (immunostains, magnification × 200).
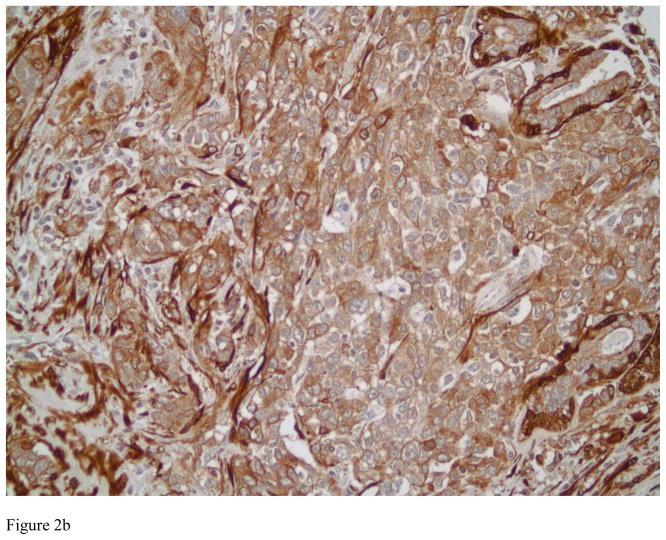
Figure 3.
Photomicrograph showing an example of ductal carcinoma in situ with negative staining by Transgelin in Figure 3A. Note the presence of myoepithelial cells positively staining for Transgelin, lending support to the in situ, noninvasive nature of the lesion. Also note several blood vessels with positive Transgelin staining in their smooth muscle walls (immunostain, magnification × 200). Figure 3B depicts an intermediate grade invasive ductal carcinoma with negative staining patterns for Transgelin and high Transgelin expression in the surrounding stroma (immunostains, magnification × 200).
Table 2.
Distribution of clinical and histopathologic parameters versus Transgelin expression in breast cancer patients
| Clinical feature or frequency of biomarker expression | Total N = 101 | Transgelin Negative N = 68 | Transgelin Positive N = 33 | Frequency positive within category | P=Value |
|---|---|---|---|---|---|
|
| |||||
| Age at diagnosis, years; median (range) | 53 (34–94) | 56 (36–94) | 51 (34–90) | 0.22 | |
|
| |||||
| Tumor size, cm (median) | 2.2 (0.6–12.2) | 2.2 {0.6–12.2) | 2.5 (1.0–5.4) | 0.81 | |
|
| |||||
| Surgical Procedure | 0.83 | ||||
| Biopsy | 2 (2%) | 1 (2%) | 1 (3%) | 50% | |
| Excisional Biopsy | 8 (8%) | 5 (7%) | 3 (9%) | 38% | |
| Lumpectomy | 39 (39%) | 25 (37%) | 14 (42%) | 36% | |
| Mastectomy | 52 (51%) | 37 (54%) | 15 (46%) | 29% | |
|
| |||||
| Histology | 0.092 | ||||
| Ductal | 94 (93%) | 61 (90%) | 33 (100%) | 35% | |
| Mixed Lobular | 7 (7%) | 7 (10%) | 0 (0%) | 0% | |
|
| |||||
| Grade (SBR) | 3.1 × 10−5 | ||||
| I | 14 (14%) | 12 (18%) | 2 (6%) | 14% | |
| II | 34 (34%) | 31 (46%) | 3 (9%) | 9% | |
| III | 53 (52%) | 25 (36%) | 28 (85%) | 53% | |
|
| |||||
| Lymph Node Procedure | 0.12 | ||||
| Axillary | 57 (56%) | 35 (52%) | 22 (67%) | 39% | |
| Sentinel | 23 (23%) | 19 (28%) | 4 (12%) | 17% | |
| Both | 11 (11%) | 9 (13%) | 2 (6%) | 18% | |
| None | 10 (10%) | 5 (7%) | 5 (15%) | 50% | |
|
| |||||
| Lymph node | 0.65 | ||||
| Negative | 45 (50%) | 30 (48%) | 15 (54%) | 33% | |
| Positive | 46 (50%) | 33 (52%) | 13 (46%) | 28% | |
|
| |||||
| ER Positivity, ≥1% | 1 × 10−6 | ||||
| No | 38 (38%) | 11 (16%) | 27 (82%) | 71% | |
| Yes | 63 (62%) | 57 (84%) | 6 (18%) | 10% | |
|
| |||||
| PR Positivity, ≥1% | 1 × 10−6 | ||||
| No | 41 (41%) | 14 (21%) | 27 (82%) | 66% | |
| Yes | 60 (59%) | 54 (79%) | 6 (18%) | 10% | |
|
| |||||
| Her-2 Positivity, ACIS ≥2.0 | 1.0 | ||||
| No | 90 (89%) | 60 (88%) | 30 (91%) | 33% | |
| Yes | 11 (11%) | 8 (12%) | 3 (9%) | 27% | |
|
| |||||
| Triple Negative | 1 × 10−6 | ||||
| No | 67 (66%) | 60 (88%) | 7 (21%) | 10% | |
| Yes | 34 (34%) | 8 (12%) | 26 (79%) | 77% | |
|
| |||||
| High Ki-67 expression, ≥ 10% | 0.0025 | ||||
| No | 44 (44%) | 37 (54%) | 7 (21%) | 16% | |
| Yes | 57 (56%) | 31 (46%) | 26 (79%) | 46% | |
A significantly higher percentage of TGLN positive tumors were noted in the TN subtype (77%) than other subtypes (10%, (p <0.001) (Table 3). TGLN expression was 11%, 7% and 25% in luminal A, luminal B and Her-2 subtypes, respectively.
Table 3.
Distributions of molecular subtype for invasive breast carcinomas in relationship to Transgelin expression
| Tumor Subtype | Total N=101 | Transgelin Negative | Transgelin Positive | Frequency positive cases |
|---|---|---|---|---|
| Luminal A | 35 (35%) | 31 (46%) | 4 (12%) | 11% |
| Luminal B | 28 (28%) | 26 (38%) | 2 (6%) | 7% |
| Her-2 | 4 (4%) | 3 (4%) | 1 (3%) | 25% |
| TN | 34 (34%) | 8 (12%) | 26 (79%) | 77% |
| Total | 101 | 68 | 33 | 33% |
Although there were strong and statistically significant associations between TGLN expression and a variety of tumor characteristics (grade, ER expression, PR expression, Ki-67 proliferative status, and TN subtype), many of these are themselves correlated with each other. Thus, a multivariable logistic regression analysis was conducted to determine which factors best and independently predicted for TGLN positivity. On multivariate logistic regression, TN subtype remained positively associated with TGLN positivity (P<0.001). With lack of Her-2 expression exhibited in 4 of the remaining 7 tumors (p=0.067). Thus, TGLN expression is a very robust marker for the TN TGLN expression showed 76% sensitivity and 90% specificity as a predictor of TN subtype.
Prognostic Utility of Transgelin Expression
Although a comprehensive review and analysis of patient outcome was not possible, we found 18 of 92 patients (20%) have died (all causes) and that 19 of 80 (24%) have experienced a recurrence of their breast cancer. While there was no correlation between TGLN expression and overall survival, there was a trend (p=0.096) toward higher recurrence rate (9/25, 36%) among patients with TGLN expression versus those without TGLN expression (10/55, 18%). SBR grade III, ER percent and positivity, molecular subtype (specifically TN), Ki-67 percent, and tumor diameter were also associated with recurrence. On multivariable logistic regression analysis, only proliferation and tumor size were statistically significant (p=0.009) factors.
4. Discussion
We show there is a differential expression of TGLN among molecular breast cancer subtypes. TGLN was predominantly upregulated in higher-grade TN subtype breast cancers. TGLN expression positively correlated with high Ki-67 and low ER and PR status but was not associated with patients’ age, tumor size, or LN metastasis. Interestingly, even though the Her-2 subtype of non-TN tumors showed higher TGLN positivity in 1 of the 4 samples studied (25%), the number of tumors in this subgroups wasn’t high enough. Given the lack of previously documented literature and the smaller subset of those subtypes, the authors of this study suggest further confirmation through future studies in order to reconcile the noted findings are needed. Our cohort was selected based on availability of tissue and biomarkers profiles and with a special interest in the TN tumor subtypes, thus is imbalanced for the different molecular subtypes as compared to real life prevalence of each subtype. One would notice that the number of TN cases was very high about 34%, while the prevalence of TN tumors is usually within the 12–15% range.
Recent studies have shown the usefulness of TGLN in improving diagnostic accuracy of soft tissue tumors with smooth muscle differentiation [16]. The higher TGLN expression in a significant number of the TN tumors compared to other subtypes is consistent with the fact that the majority of them are of basal type with myoepithelial differentiation. It is not a surprise seeing TGLN localization in the basal cell layer of benign breast tissue. TGLN staining has potential utility in highlighting lesions such as sclerosing adenosis and in differentiating in situ from invasive lesions, which lends support to its addition to the battery of immunohistochemical markers for myoepithelial cells including SMM, p63, calponin, actin and S100. In addition to its expression in cells with smooth muscle differentiation, it is also expressed in the cytoplasm of fibroblasts and some epithelial cells. TGLN expression has been used to aid in histologic identification of tumors ranging from colorectal, gastric carcinoma, gall bladder, pancreatic cancer, prostate and lung adenocarcinoma and uterine tumors of smooth muscle differentiation [15, 16, 18, 20–26].
Our description of TGLN expression across molecular subtypes of breast cancer may help to clarify discrepant findings of TGLN expression in previous reports. For example, previous studies demonstrating decreased expression of TGLN in breast cancer have often used cell lines or tumors that are of the luminal type without accounting for its differential expression in the various molecular breast cancer types. In a report by Shields et al, all cell lines included for analysis are known to express estrogen receptors [18]. The authors reported lower or no expression of TGLN, which is consistent with our results. TGLN overexpression in aggressive breast cancer histologic types such as TN and poorly differentiated tumors is similar to findings reported by Zhang et al [25] and Lin et al [27], wherein TGLN was highly expressed in aggressive colorectal cancers associated with local and distant metastasis, advanced clinical stage and shorter overall survival. In another report, TGLN expression was 25fold higher in tumorigenic cells than nontumorigenic hepatocellular, colon and prostate cancer cells [28}. These studies suggest that TGLN may promote migration and invasion of malignant cells of different origins [28].
The recent finding of differential and compartment-specific expression of the homologs TGLN and TGLN2 in lung adenocarcinoma and its stroma by Rho et al [24] adds another level of complexity in evaluating TGLN data in the literature. The authors report TGLN was strictly localized to tumor-induced reactive myofibroblastic stromal tissue compartment, whereas over-expression of TGLN2 was exclusively localized to the neoplastic glandular compartment. Our results showed variability in TGLN localization both in tumor cells and in the surrounding stroma. Benign breast stroma usually lacked TGLN expression, whereas stromal tissue in 90% of specimens over-expressed TGLN irrespective of the molecular subtype. These findings suggest TGLN acts at the interface of the epithelium and surrounding mesenchyme in the setting of malignant invasion. Recent studies have shown TGLN involvement in many biologic activities including cell differentiation and tissue invasion and endometriosis and as a tumor suppressor [17–19]. Further study should aim to better describe the biologic activity of the TGLN protein in invasive breast carcinoma.
The characteristics of basal-like and TN breast cancer have been extensively reviewed and remain a subject of controversy [4, 9, 29]. With no internationally accepted definition of basal-like breast cancers there are numerous similarities between basal-like and TN breast cancers [4,30,31]. Both affect younger patients, are more prevalent in African-American women, often present as interval cancers and are highly aggressive with a very low 5-year survival compared to tumors of other molecular subtypes [4, 32]. A recent study by Lehmann et al [33] defined six unique TN sub-types by analyzing gene expression profiles from 21 breast cancer datasets. These included two basal-like subtypes (BL1 and BL2), a mesenchymal (M), a mesenchymal stem-like (MSL), a luminal androgen receptor (LAR) subtype and an immunomodulatory subtype wherein BL1 and BL2 showed higher DNA damage response genes expression. M and MSL demonstrated higher expression of epithelial mesenchymal transition genes, and androgen receptor signaling gene expression was noted in LAR sub-type. These subtypes showed varying responses to different targeted therapies [12].
Tumor markers currently have important diagnostic and therapeutic implications for pathologists and oncologists alike. In the case of TN tumors, current treatment regimens are of limited efficacy. Of patients with TN tumors, 17–58% have complete pathological response after anthracycline- or anthracycline -taxane-based neoadjuvant chemotherapy, and <20% of TN tumors demonstrate complete pathological response after platinum-based neoadjuvant chemotherapy [34]. Patients who fail to achieve complete pathological response despite chemotherapy have a dismal prognosis. It is the authors’ hope that markers like TGLN might inform the research and development of potential treatment strategies. A report by Cai et al [35] highlights the potential role TGLN in the development of drug resistance in breast cancer. The authors identified a substance called Paeonol, derived from the root bark of Paeonia Suffruticosa, that successfully reversed paclitaxel resistance in human breast cancer cells by regulating the expression of TGLN2 [35]. Yang et al, have shown that TGLN blocks androgen stimulated cell growth by preventing binding of an androgen receptor co-activator with androgen receptor and thereby thwarting subsequent translocation of the androgen to the nucleus in prostate cancer [23]. We show that TGLN is differentially expressed among the molecular breast cancer subtypes. Limitations to our study include a paucity of lobular and HER2-positive tumors and our categorization of molecular subtypes by immunohistochemical parameters, as opposed to genomic parameters. Although the introduction of tissue microarray technology has been validated in many retrospective studies of many tissue types, uncertainty persists as to whether this technique is useful in accurately evaluating intratumor heterogeneity of expression. Two cores from each tumor were analyzed in an attempt to account for the impact of tumor heterogeneity on TGLN expression.
Even when tumors are compared based on histological grading, TGLN expression proves to be an excellent diagnostic marker. TGLN is a potentially valuable diagnostic marker for high-grade/TN breast cancer. TGLN may also provide a potential target for future treatment of TN tumors, which carry a notoriously poor prognosis despite current chemotherapeutic regimens.
Acknowledgments
Financial support for the study was partially provided by the NIH under grant NIHR01HD0690431-01A1.
Footnotes
Disclosure: All authors do NOT have potentially biasing relationship of a financial, professional or personal nature with any commercial interests that would constitute a conflict of interest related to the studies reported in this manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Correa Geyer F, Reis-Filho JS. Microarray-based gene expression profiling as a clinical tool for breast cancer management: are we there yet? Int J Surg Pathol. 2009;17:285–302. doi: 10.1177/1066896908328577. [DOI] [PubMed] [Google Scholar]
- 2.Geyer FC, Marchio C, Reis-Filho JS. The role of molecular analysis in breast cancer. Pathology. 2009;41:77–88. doi: 10.1080/00313020802563536. [DOI] [PubMed] [Google Scholar]
- 3.Reis-Filho JS, Lakhani SR. Breast cancer special types: why bother? J Pathol. 2008;216:394–8. doi: 10.1002/path.2419. [DOI] [PubMed] [Google Scholar]
- 4.Badve S, Dabbs DJ, Schnitt SJ, et al. Basal-like and triple-negative breast cancers: a critical review with an emphasis on the implications for pathologists and oncologists. Mod Pathol. 2011;24:157–67. doi: 10.1038/modpathol.2010.200. [DOI] [PubMed] [Google Scholar]
- 5.Hu Z, Fan C, Oh DS, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parker JS, Mullins M, Cheang MC, et al. Supervised Risk Predictor of Breast Cancer Based on Intrinsic Subtypes. J Clin Oncol. 2009;27:1160–7. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 8.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Pro Natl Acad Sci USA. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gusterson B. Do ‘basal-like’ breast cancers really exist? Nat Rev Cancer. 2009;9:128–34. doi: 10.1038/nrc2571. [DOI] [PubMed] [Google Scholar]
- 10.Pusztai L, Mazouni C, Anderson K, Wu Y, Symmans WF. Molecular classification of breast cancer: Limitations and potential. Oncologist. 2006;11:868–77. doi: 10.1634/theoncologist.11-8-868. [DOI] [PubMed] [Google Scholar]
- 11.Weigelt B, Baehner FL, Reis-Filho JS. The contribution of gene expression profiling to breast cancer classification, prognostication and prediction: a retrospective of the last decade. J Pathol. 2010;220:263–80. doi: 10.1002/path.2648. [DOI] [PubMed] [Google Scholar]
- 12.Criscitiello C, Azim HA, Jr, Schouten PC, Linn SC, Sotiriou C. Understanding the biology of triple-negative breast cancer. Ann Oncol. 2012;(Suppl 6):vi13–8. doi: 10.1093/annonc/mds188. [DOI] [PubMed] [Google Scholar]
- 13.Elias AD. Triple-negative breast cancer: a short review. Am J Clin Oncol. 2010;33:637–45. doi: 10.1097/COC.0b013e3181b8afcf. [DOI] [PubMed] [Google Scholar]
- 14.Lees-Miller JP, Heeley DH, Smillie LB, Kay CM. Isolation and characterization of an abundant and novel 22-kDa protein (SM22) from chicken gizzard smooth muscle. J Biol Chem. 1987;262:2988–93. [PubMed] [Google Scholar]
- 15.Assinder SJ, Stanton JA, Prasad PD. Transgelin: an actin-binding protein and tumour suppressor. Int J Biochem Cell Biol. 2009;41:482–6. doi: 10.1016/j.biocel.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 16.Robin YM, Penel N, Pérot G, et al. Transgelin is a novel marker of smooth muscle differentiation that improves diagnostic accuracy of leiomyosarcomas: a comparative immunohistochemical reappraisal of myogenic markers in 900 soft tissue tumors. Mod Pathol. 2013;26:502–10. doi: 10.1038/modpathol.2012.192. [DOI] [PubMed] [Google Scholar]
- 17.Lawson D, Harrison M, Shapland C. Fibroblast transgelin and smooth muscle SM22alpha are the same protein, the expression of which is down-regulated in many cell lines. Cell Motil Cytoskeleton. 1997;38:250–7. doi: 10.1002/(SICI)1097-0169(1997)38:3<250::AID-CM3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 18.Shields JM, Rogers-Graham K, Der CJ. Loss of transgelin in breast and colon tumors and in RIE-1 cells by Ras deregulation of gene expression through Raf-independent pathways. J Biol Chem. 2002;277:9790–9. doi: 10.1074/jbc.M110086200. [DOI] [PubMed] [Google Scholar]
- 19.Albiges-Rizo C, Destaing O, Fourcade B, Planus E, Block MR. Actin machinery and mechanosensitivity in invadopodia, podosomes and focal adhesions. J Cell Sci. 2009;122(Pt 17):3037–49. doi: 10.1242/jcs.052704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wulfkuhle JD, Sgroi DC, Krutzsch H, et al. Proteomics of human breast ductal carcinoma in situ. Cancer Res. 2002;62:6740–9. [PubMed] [Google Scholar]
- 21.Sahasrabuddhe NA, Barbhuiya MA, Bhunia S, et al. Identification of prosaposin and transgelin as potential biomarkers for gallbladder cancer using quantitative proteomics. Biochem Biophys Res Commun. 2014;446:863–9. doi: 10.1016/j.bbrc.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prasad PD, Stanton JA, Assinder SJ. Expression of the actin-associated protein transgelin (SM22) is decreased in prostate cancer. Cell Tissue Res. 2010;339:337–47. doi: 10.1007/s00441-009-0902-y. [DOI] [PubMed] [Google Scholar]
- 23.Yang Z, Chang YJ, Miyamoto H, et al. Transgelin functions as a suppressor via inhibition of ARA54-enhanced androgen receptor transactivation and prostate cancer cell growth. Mol Endocrinol. 2007;21:343–58. doi: 10.1210/me.2006-0104. [DOI] [PubMed] [Google Scholar]
- 24.Rho JH, Roehrl MH, Wang JY. Tissue proteomics reveals differential and compartment-specific expression of the homologs transgelin and transgelin-2 in lung adenocarcinoma and its stroma. J Proteome Res. 2009;8:5610–8. doi: 10.1021/pr900705r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Ye Y, Shen D, et al. Identification of transgelin-2 as a biomarker of colorectal cancer by laser capture microdissection and quantitative proteome analysis. Cancer Sci. 2010;101:523–9. doi: 10.1111/j.1349-7006.2009.01424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou L, Zhang R, Zhang L, et al. Upregulation of transgelin is an independent factor predictive of poor prognosis in patients with advanced pancreatic cancer. Cancer Sci. 2013;104:423–30. doi: 10.1111/cas.12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin Y, Buckhaults PJ, Lee JR, et al. Association of the actin-binding protein transgelin with lymph node metastasis in human colorectal cancer. Neoplasia. 2009;11:864–73. doi: 10.1593/neo.09542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee EK, Han GY, Park HW, Song YJ, Kim CW. Transgelin promotes migration and invasion of cancer stem cells. J Proteome Res. 2010;9:5108–17. doi: 10.1021/pr100378z. [DOI] [PubMed] [Google Scholar]
- 29.Rakha E, Ellis I, Reis-Filho J. Are triple-negative and basal-like breast cancer synonymous? Clin Cancer Res. 2008;14:618. doi: 10.1158/1078-0432.CCR-07-1943. author reply 618–9. [DOI] [PubMed] [Google Scholar]
- 30.Livasy CA, Karaca G, Nanda R, et al. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol. 2006;19:264–71. doi: 10.1038/modpathol.3800528. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen TO, Hsu FD, Jensen K, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–74. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 32.Morris GJ, Naidu S, Topham AK, et al. Differences in breast carcinoma characteristics in newly diagnosed African-American and Caucasian patients: a single-institution compilation compared with the National Cancer Institute’s Surveillance, Epidemiology, and End Results Database. Cancer. 2007;110:876–84. doi: 10.1002/cncr.22836. [DOI] [PubMed] [Google Scholar]
- 33.Lehmann BD, Bauer JA, Chen X, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–67. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanchez-Munoz A, García-Tapiador AM, Martínez-Ortega E, et al. Tumour molecular subtyping according to hormone receptors and HER2 status defines different pathological complete response to neoadjuvant chemotherapy in patients with locally advanced breast cancer. Clin Transl Oncol. 2008;10:646–53. doi: 10.1007/s12094-008-0265-y. [DOI] [PubMed] [Google Scholar]
- 35.Cai J, Chen S, Zhang W, et al. Paeonol reverses paclitaxel resistance in human breast cancer cells by regulating the expression of transgelin 2. Phytomedicine. 2014;21:984–91. doi: 10.1016/j.phymed.2014.02.012. [DOI] [PubMed] [Google Scholar]