Cutaneous T-cell lymphomas (CTCL) are malignancies of skin-homing T lymphocytes. The etiology of CTCL remains uncertain, though epidemiologic features suggest an infectious origin, including a predilection for elderly individuals and a higher than expected incidence in immune-suppressed patients.1 CTCL specimens have been evaluated for evidence of infection, including Staphylococcus aureus and members of the retrovirus, herpesvirus, and polyomavirus families. However, studies have largely failed to reveal associations between infectious agents and CTCL.2-4
We utilized Digital Transcriptome Subtraction (DTS) to systematically search for oncogenic viruses in two types of CTCL, Sézary syndrome (SS) and mycosis fungoides (MF). DTS subtracts human nucleotide sequences from reference databases in silico, leaving a pool of candidate reads to be scrutinized for pathogenic origin.5-7
Six patients (MF, tumor stage=3, SS=3) diagnosed according to WHO-EORTC criteria8 were enrolled according to the University of Pittsburgh Institutional Review Board. MF tumors were biopsied, and SS patients' whole blood and healthy blood donor Leukopaks (Pittsburgh Central Blood Bank, n=3) underwent Ficoll-Paque (GE Healthcare Bio-Sciences Corp., Piscataway, NJ) centrifugation and CD4+ T cell isolation using the MACS CD4+ T Cell Isolation Kit II (Miltenyi Biotec Inc., Auburn CA). Total RNA was isolated from samples using TRIzol® Reagent (Invitrogen Corporation Carlsbad, CA) and assessed using the NanoVue Plus spectrophotometer (GE Healthcare, Piscataway, NJ), and the Agilent 2100 bioanalyzer (Quantum Analytics, Foster City, CA).
cDNA libraries were generated using a modified version of a described protocol (Supplementary Methods),7 and were pyrosequenced at the Duke University Genome Sequencing & Analysis Core Facility using the 454 GS-FLX Titanium platform (Roche Diagnostics Corporation, Branford, CT). A sequencing saturation analysis was done, and DTS was carried out as previously described (Supplementary Methods).6
Transcriptome sequencing yielded 631,142 Control reads, 605,318 MF reads, and 1,262,476 SS reads. After quality filtering, 88.2%, 85.2%, and 96.7% of Control, MF, and SS reads remained. Twenty percent of Reference Sequence (RefSeq)-aligning reads in each library were barcoded; a similar number of barcoded reads were derived from each sample, suggesting that no sample contributed disproportionately, and sequence saturation analysis suggested that the number of undetected unique transcripts was fractionally small (Supplementary Figure 1).
Of the remaining high fidelity reads, 99.4% of MF and 99.7% of HiFi SS reads aligned against human RefSeq RNA and DNA. From the reads that did not align against curated human DNA or RNA, MF and SS libraries yielded 18 and 24 candidates, respectively, that aligned with high specificity against sequences in curated viral databases (Figure 1). None of these candidates corresponded to established tumor viruses. When aligned against the NCBI's complete Non-redundant (nr) database, these reads had low complexity and aligned with nonviral sequences, suggesting artifactual alignment against viral sequences or correspondence to unannotated human transcript.
Figure 1. Digital Transcriptome Subtraction.
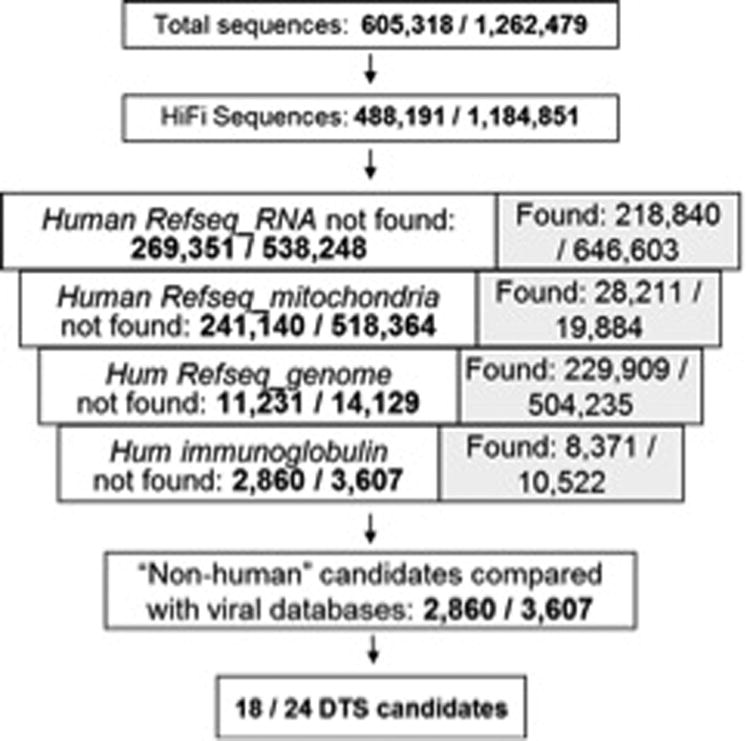
DTS of Mycosis Fungoides (MF) and Sézary Syndrome (SS) libraries yielded 18 and 24 potentially “non-human” reads, respectively, containing high viral homology. However, when analyzed manually and aligned against the NCBI's complete Non-redundant (nr) database, candidate reads were found to be of low complexity or align most highly against nonviral sequences, suggesting either artifactual alignment against viral sequences or correspondence to unannotated human transcript.
Because directly carcinogenic viruses actively express oncogenes in tumor cells, sequencing a tumor transcriptome to a sufficient depth should detect viral transcript if present. Together with other SS studies,3,4 we can conclude the possibility of MF or SS being caused by a directly oncogenic virus is exceedingly low, but not zero due to some limitations of DTS, such as failure to detect a novel virus lacking homology to known species. Additionally, viral sequences erroneously characterized as human, such as the endogenous retroviruses which comprise ∼10% of the human genome, could be falsely subtracted during DTS. Recently, increased expression of retrovirus transcripts as well as retrovirus-associated protein was identified in MF lesional skin, representing an important area of future study.9 Moreover, though the data presented here is derived from cells representative of the malignant populations of tumor-stage MF and SS, exhaustive studies including patch and plaque MF samples as well as SS skin samples would be advisable in the future to further confirm our conclusion.
Indirect pathogens could escape detection by DTS, such as a “hit-and-run” virus no longer present or not actively transcribing or bacteria that induce inflammation but are not integrated into the host genome. Staphylococcus aureus has been hypothesized to be related to CTCL pathogenesis, with studies showing an association between S. aureus carriage and CTCL, disease improvement with bacterial eradication, and disease-specific changes in T-cell receptor Vβ (TCR-Vβ) consistent with superantigen stimulation.2,10,11 To explore additional roles for pathogens in the etiology of CTCL, we conducted gene expression pathway analysis of SS samples compared with healthy controls. Gene expression data from the MF tumor samples was excluded due to a large amount of contamination from surrounding skin. Relative expression levels were determined (Supplementary Methods), and the resulting gene expression data was analyzed with Ingenuity Pathways Analysis (IPA, http://www.ingenuity.com) and Metacore (Thomson Reuters, http://thomsonreuters.com/metacore/) to identify those genes upregulated in SS with known roles in infectious disease processes.
Gene expression analysis of sequence-based transcriptome data did reveal upregulation of 216 genes involved in infectious disease pathways associated with viral, bacterial, and parasitic infections (Table 1). The pathway most closely associated with the CTCL transcriptome is the response to viral infection (Supplementary Figure 2). Activation of this pathway produces an antiviral and antistress response to a wide range of pathogens including viral dsRNA and bacterial lipopolysaccharides.12 These findings may indirectly support an early hypothesis of persistent antigen stimulation. Alternatively, highlighted pathways may be important to CTCL pathogenesis secondarily and contribute to immune response dysregulation occurring after pathogenesis. Further analysis of the relationship of bacterial and viral exposures in CTCL is warranted to delineate the relationship of these pathway alterations and disease onset and pathogenesis.
Table 1. Gene expression analysis identifies infectious disease pathways upregulated in SS.
Analysis of gene expression data using Ingenuity revealed that pathways of viral, bacterial, and parasitic infection response were all upregulated in SS samples compared with normal controls. The above table shows the p-value for each functional pathway, the total number of molecules dysregulated in each pathway, and a selection of key molecules.
| Functional Pathway | p-Value | # Molecules | Seclected Molecules |
|---|---|---|---|
| Viral Infection | 5.24E-12 | 187 | APOBEC3G,CD48, CD63, CXCR4, DEFA4, DNAJA3, DUSP1, HCP5, IRF1, ISG20, ITGB1, LCN2, MYC, MYD88, NFKBIA, OAS1, PDCD1, PGLYRP1, PSME2, RELA, S100A9, TOX, VAV1, ZAP70 |
| Bacterial Infection | 6.50E-04 | 43 | BPI, C1QBP, CD27, CEBPB, CHIT1, CSK, DEFA4, DUSP1, E2F4, IL6ST, LY86, MAP2K3, MIF, MYD88, PILRB, REL, RELA, S100A9, SLAMF6, |
| Parasitic Infection | 4.69E-03 | 18 | AHNAK, ATP2B4, CXCR4, IL4R, ITGB7, MYD88, REL, SLAMF6 |
Importantly, DTS failed to detect human T-cell leukemia/lymphotropic virus type 1 (HTLV-1), a virus identified in several studies of CTCL samples in mid 1990's, with findings coming largely from a single laboratory.13 Since that time, technological progress has allowed for evaluation of the presence of HTLV-1 in CTCL using modern, powerful methodology. Presently, none of the recent studies looking for evidence of HTLV-1 in CTCL were able to identify this sequence.14,15 Because studies utilizing digital transcriptome subtraction techniques did not detect clonally integrated HTLV-1 into the host genome,3,4 we can conclude with a high degree of certainty that HTLV-1 is not a directly oncogenic virus in CTCL pathogenesis. However, it does not exclude a possibility that concurrent infection with HTLV-1 could contribute to a chronic antigen stimulation model which has long been proffered.16
This study excludes an entire class of important human pathogens—namely, directly oncogenic viruses—in CTCL pathoetiology while providing supportive evidence for a potential role of indirectly oncogenic infectious agents in CTCL pathogenesis.
Supplementary Material
Acknowledgments
DTS experiments were performed in the laboratory of Yuan Chang and Patrick S. Moore, Cancer Virology Program, University of Pittsburgh Cancer Institute, and were supported through NIH CA136363, CA136806 and Research Professorships from the American Cancer Society.
This publication was made possible in part by NIH SPORE Program Grant# 5P50CA121973-05 to LG and LF, Grant Number UL1 RR024153 from the National Center for Research Resources (NCRR), and University of Pittsburgh Clinical and Translational Research Center.
BOD and EM were supported by the Howard Hughes Medical Fellows Program.
Funding: DTS experiments were performed in the laboratory of Yuan Chang and Patrick S. Moore, Cancer Virology Program, University of Pittsburgh Cancer Institute, and were supported through NIH CA136363, CA136806 and Research Professorships from the American Cancer Society. This publication was made possible in part by NIH SPORE Program Grant# 5P50CA121973-05 to LG and LF, Grant Number UL1 RR024153 from the National Center for Research Resources (NCRR), and University of Pittsburgh Clinical and Translational Research Center. BOD and EM were supported by the Howard Hughes Medical Fellows Program.
Footnotes
Disclosures: The authors report no disclosures, financial interest, or conflicts of interest.
References
- 1.Pomerantz RG, Campbell LS, Jukic DM, Geskin LJ. Posttransplant cutaneous T-cell lymphoma: case reports and review of the association of calcineurin inhibitor use with posttransplant lymphoproliferative disease risk. Arch Dermatol. 2010;146(5):513–516. doi: 10.1001/archdermatol.2010.60. [DOI] [PubMed] [Google Scholar]
- 2.Mirvish ED, Pomerantz RG, Geskin LJ. Infectious agents in cutaneous T-cell lymphoma. J Am Acad Dermatol. 2011;64(2):423–431. doi: 10.1016/j.jaad.2009.11.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee CS, Ungewickell A, Bhaduri A, et al. Transcriptome sequencing in Sezary syndrome identifies Sezary cell and mycosis fungoides-associated LncRNAs and novel transcripts. Blood. 2012 doi: 10.1182/blood-2012-04-423061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dereure O, Cheval J, Du Thanh A, et al. No Evidence for Viral Sequences in Mycosis Fungoides and Szary Syndrome Skin Lesions: A High-Throughput Sequencing Approach. J Invest Dermatol. 2012 doi: 10.1038/jid.2012.371. [DOI] [PubMed] [Google Scholar]
- 5.Xu Y, Stange-Thomann N, Weber G, et al. Pathogen discovery from human tissue by sequence-based computational subtraction. Genomics. 2003;81(3):329–335. doi: 10.1016/s0888-7543(02)00043-5. [DOI] [PubMed] [Google Scholar]
- 6.Feng H, Taylor JL, Benos PV, et al. Human transcriptome subtraction by using short sequence tags to search for tumor viruses in conjunctival carcinoma. J Virol. 2007;81(20):11332–11340. doi: 10.1128/JVI.00875-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319(5866):1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willemze R, Jaffe ES, Burg G, et al. WHO-ERTC classification for cutaneous lymphomas. Blood. 2005;105:3768–3788. doi: 10.1182/blood-2004-09-3502. [DOI] [PubMed] [Google Scholar]
- 9.Maliniemi P, Vincendeau M, Mayer J, et al. Expression of human endogenous retrovirus-w including syncytin-1 in cutaneous T-cell lymphoma. PloS one. 2013;8(10):e76281. doi: 10.1371/journal.pone.0076281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackow CM, Cather JC, Hearne V, Asano AT, Musser JM, Duvic M. Association of erythrodermic cutaneous T-cell lymphoma, superantigen-positive Staphylococcus aureus, and oligoclonal T-cell receptor V beta gene expansion. Blood. 1997 Jan 1;89(1):32–40. [PubMed] [Google Scholar]
- 11.Talpur R, Bassett R, Duvic M. Prevalence and treatment of Staphylococcus aureus colonization in patients with mycosis fungoides and Sezary syndrome. The British journal of dermatology. 2008 Jul;159(1):105–112. doi: 10.1111/j.1365-2133.2008.08612.x. [DOI] [PubMed] [Google Scholar]
- 12.Sadler AJ, Williams BR. Structure and function of the protein kinase R. Current topics in microbiology and immunology. 2007;316:253–292. doi: 10.1007/978-3-540-71329-6_13. [DOI] [PubMed] [Google Scholar]
- 13.Pancake BA, Zucker-Franklin D, Coutavas EE. The cutaneous T cell lymphoma, mycosis fungoides, is a human T cell lymphotropic virus-associated disease. A study of 50 patients. The Journal of clinical investigation. 1995 Feb;95(2):547–554. doi: 10.1172/JCI117697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bazarbachi A, Soriano V, Pawson R, et al. Mycosis fungoides and Sezary syndrome are not associated with HTLV-I infection: an international study. British journal of haematology. 1997 Sep;98(4):927–933. doi: 10.1046/j.1365-2141.1997.3213138.x. [DOI] [PubMed] [Google Scholar]
- 15.Pawlaczyk M, Filas V, Sobieska M, Gozdzicka-Jozefiak A, Wiktorowicz K, Breborowicz J. No evidence of HTLV-I infection in patients with mycosis fungoides and Sezary syndrome. Neoplasma. 2005;52(1):52–55. [PubMed] [Google Scholar]
- 16.Tan RS, Butterworth CM, McLaughlin H, et al. Mycosis fungoides--a disease of antigen persistence. Br J Dermatol. 1974;91:607–616. doi: 10.1111/j.1365-2133.1974.tb12449.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


