Abstract
Obscurins, encoded by the single OBSCN gene, are giant cytoskeletal proteins with structural and regulatory roles. The OBSCN gene is highly mutated in different types of cancers. Loss of giant obscurins from breast epithelial cells confers them with a survival and growth advantage, following exposure to DNA-damaging agents. Here we demonstrate that the expression levels and subcellular distribution of giant obscurins are altered in human breast cancer biopsies compared with matched normal samples. Stable clones of non-tumorigenic MCF10A cells lacking giant obscurins fail to form adhesion junctions, undergo epithelial-to-mesenchymal transition and generate >100-μm mammospheres bearing markers of cancer-initiating cells. Obscurin-knockdown MCF10A cells display markedly increased motility as a sheet in 2-dimensional (2D) substrata and individually in confined spaces and invasion in 3D matrices. In line with these observations, actin filaments redistribute to extending filopodia where they exhibit increased dynamics. MCF10A cells that stably express the K-Ras oncogene and obscurin short hairpin RNA (shRNA), but not scramble control shRNA, exhibit increased primary tumor formation and lung colonization after subcutaneous and tail vein injections, respectively. Collectively, our findings reveal that loss of giant obscurins from breast epithelium results in disruption of the cell–cell contacts and acquisition of a mesenchymal phenotype that leads to enhanced tumorigenesis, migration and invasiveness in vitro and in vivo.
INTRODUCTION
Obscurins comprise a family of giant, multidomain, cytoskeletal proteins originally identified in striated muscles where they have key roles in their structural organization and contractile activity.1–3 The human OBSCN gene spans 150 kb on chromosome 1q42 and undergoes extensive splicing to give rise to at least four isoforms.4,5 The prototypical form of obscurin, obscurin A, is ~ 720 kDa and contains multiple signaling and adhesion domains arranged in tandem.1 The NH2-terminus of the molecule contains repetitive immunoglobulin (Ig) and fibronectin-III (Fn-III) domains, while the COOH-terminus includes several signaling domains, including an IQ motif, a src homology 3 domain, a Rho-guanine nucleotide exchange factor and a pleckstrin homology (PH) domain, interspersed by non-modular sequences. In addition to obscurin A, the OBSCN gene gives rise to another large isoform, obscurin B or giant (g) MLCK (Figure 1a), which has a molecular mass of ~ 870 kDa.4,5 Obscurin B contains two serine/threonine kinase domains, which replace the non-modular COOH-terminus of obscurin A.6 The two serine/threonine kinases may also be expressed independently as smaller isoforms, containing one (~55 kDa) or both (~145 kDa) kinase domains.7
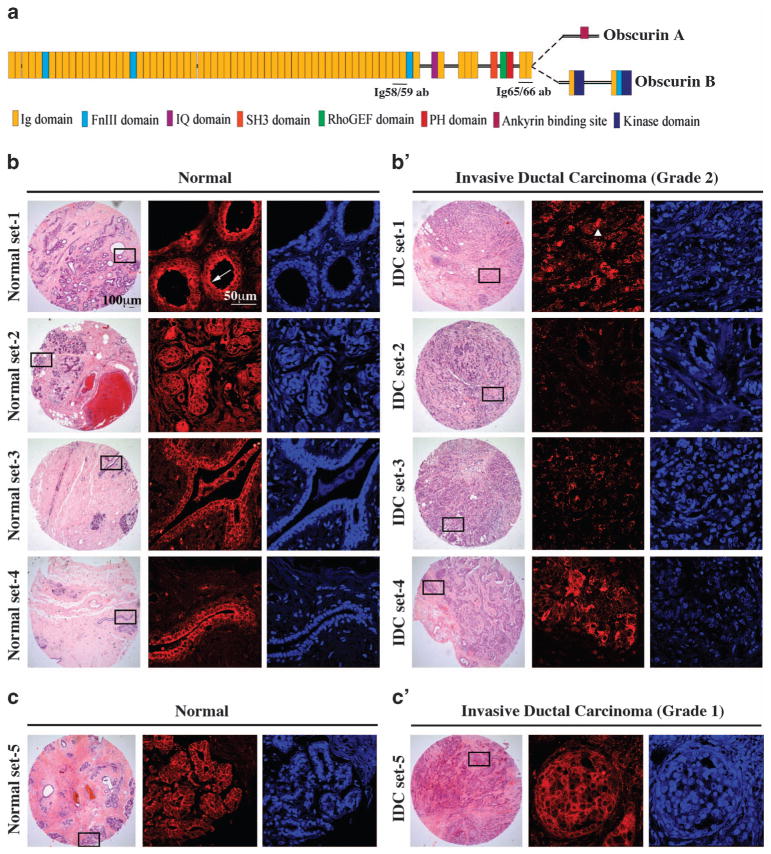
Figure 1.
The expression profile of giant obscurins is altered in human breast cancer biopsies. (a) Schematic representation of giant obscurins A and B depicting their adhesion and signaling motifs. The regions used for the generation of the obscurin Ig58/59 and Ig65/66 antibodies are also denoted. (b, c′) Representative images of paired normal (b and c) and IDC biopsies of grade 2 (b′) and grade 1 (c′); hematoxylin and eosin (H&E)-stained tissue sections (left columns) with boxed areas corresponding to regions examined under confocal optics after immunolabeling with the obscurin Ig58/59 antibody (middle columns, shown in red) and 4,6-diamidino-2-phenylindole (DAPI; right columns, shown in blue). Obscurins are present at the cell membrane in normal samples, where they exhibit prominent luminal distribution (b, arrow). The expression of obscurins is significantly reduced in IDC grade 2 biopsies with residual proteins accumulating in cytoplasmic puncta (b′, arrowhead) but not in IDC grade 1 biopsies, where they are readily expressed at the plasma membrane (c′).
Early sequencing analysis of 13 023 genes in breast and colorectal cancers identified 189 candidate genes that were highly mutated.8 Of the 189 candidate genes, TP53 and OBSCN were the only commonly mutated genes in both tumor types.8 Additional analysis of OBSCN revealed a germline mutation in glioblastoma and novel somatic mutations in melanoma tumors.9 Moreover, whole genome array analysis of gastrointestinal stromal and leiomyosarcoma tumors indicated that the differential expression of OBSCN and PRUNE2 is a reliable two-gene expression classifier that can distinguish the two tumor types.10
We recently showed that obscurins are abundantly expressed in normal breast epithelial cells, where they localize at cell–cell junctions, the nucleus and in cytoplasmic puncta coinciding with the Golgi membrane, but their expression is markedly diminished in breast cancer cells.11 Downregulation of giant obscurins in non-tumorigenic MCF10A breast epithelial cells via shRNA technology conferred them with a survival advantage following exposure to DNA stress, due to reduced apoptosis, indicating that obscurins may have key roles in breast tumor suppression.11 Moreover, obscurin-KD MCF10A cells acquired a mesenchymal appearance and exhibited increased cell scattering compared with control cells, which formed epithelial clusters.11 Given that such phenotypic alterations are associated with major changes in the formation and stability of adherens junctions (AJs), we herein examine the role of giant obscurins in intercellular adhesion.12 Our studies document for the first time that the expression profile of giant obscurins is dramatically altered in advanced stage human breast cancer biopsies and that loss of giant obscurins from breast epithelial cells leads to disruption of AJs, induction of epithelial-to-mesenchymal transition (EMT) and acquisition of stem-like characteristics resulting in increased cell motility and invasion in vitro and tumorigenicity and metastasis in vivo.
RESULTS
Expression profile of giant obscurins in human breast cancer biopsies
We used fluorescent immunohistochemical methods and confocal imaging to investigate the expression profile of giant obscurins in human biopsies of invasive ductal carcinoma (IDC), which accounts for >80% of metastatic breast cancer, compared with normal matched samples, using an antibody that recognizes epitopes present in Ig domains 58 and 59 of obscurins A and B (Figure 1a). Obscurins are expressed in the ductal and lobular epithelium of normal breast tissue, where they exhibit membrane localization, with prominent concentration on the luminal side of epithelial cells in larger ducts and lobules (Figure 1b, arrow). In contrast, they are nearly absent from matched invasive ductal breast cancer tissue of grade ≥2, with residual obscurins accumulating in large cytoplasmic puncta throughout the tumor mass (Figure 1b′, arrowhead). Interestingly, invasive ductal breast cancer samples of grade 1 maintain membrane localization of obscurins similar to paired normal breast tissue samples (Figures 1c and c′).
To further elucidate changes in the expression levels of giant obscurins in human biopsies, we employed western blotting using an antibody that detects epitopes present in Ig domains 65 and 66 (Figure 1a). Analysis of protein homogenates prepared from age-, sex- and stage-matched normal and invasive ductal or infiltrating lobular carcinomas revealed a dramatic reduction in the amounts of giant obscurins A and B in the carcinoma specimens (Supplementary Figure S1a, top panel). Moreover, we consistently observed downregulation of an additional immunoreactive band with a calculated molecular mass of ~ 600 kDa that potentially corresponds to a novel giant obscurin isoform (Supplementary Figure S1a, middle panel). In agreement with this, the complete sequence of a novel obscurin isoform of human origin was recently deposited in the Ensembl database (accession number: ENSP00000355668) with a predicted molecular mass of ~ 611 kDa that starts in Ig31. Interestingly, homogenates prepared from cancer biopsies of either ductal or lobular origin contained smaller immunoreactive bands ranging in size between 300 and 90 kDa, which were absent from homogenates prepared from paired normal samples (Supplementary Figure S1b). These smaller obscurin forms may be novel, still uncharacterized, obscurin proteins or degradation products of giant obscurins due to the presence of missense or nonsense mutations in the OBSCN gene.8–10 Collectively, our findings indicate that the expression levels of giant obscurins are markedly diminished in breast cancer biopsies of both ductal and lobular origin, whereas residual obscurins, potentially representing mutant and/or truncated forms of giant obscurins, accumulate in large cytoplasmic puncta.
Obscurin-knockdown (KD) breast epithelial cells form mammospheres with stem-like characteristics
To study the role of giant obscurins in breast epithelium, we generated stable MCF10A obscurin-KD cell lines using shRNA constructs (shRNA 1–4) targeting different sequences within the common NH2-terminus and middle portion of giant obscurins A and B.11 In the current study, we utilized stable MCF10A clones transfected with obscurin shRNA-1 or shRNA-2 plasmids, which exhibited robust downregulation of giant obscurins (Figure 2a, upper part, arrows), while the expression of smaller isoforms remained unaltered (Figure 2a, lower part). We previously found that KD of giant obscurins from MCF10A cells bestows them with a survival advantage following exposure to DNA-damaging agents.11 We thus examined whether loss of obscurins also affects the survival of MCF10A cells in anchorage-independent conditions. Stable clones of MCF10A breast epithelial cells depleted of giant obscurins using shRNA-1 or shRNA-2 were allowed to form mammospheres in serum-free media and ultra-low attachment plates. Obscurin-KD MCF10A cells formed robust mammospheres >100 μm, while control cells stably transduced with scramble shRNA failed to do so (Figures 2b and c). To further investigate their self-renewing capabilities, primary mammospheres were dissociated and plated as single cells in 96-well culture dishes under similar culture conditions. Approximately 80% and 40% of obscurin shRNA-1 and obscurin shRNA-2 MCF10A cells formed secondary mammospheres ranging in size between 50–100 μm and 25–50 μm, respectively (Figure 2d).
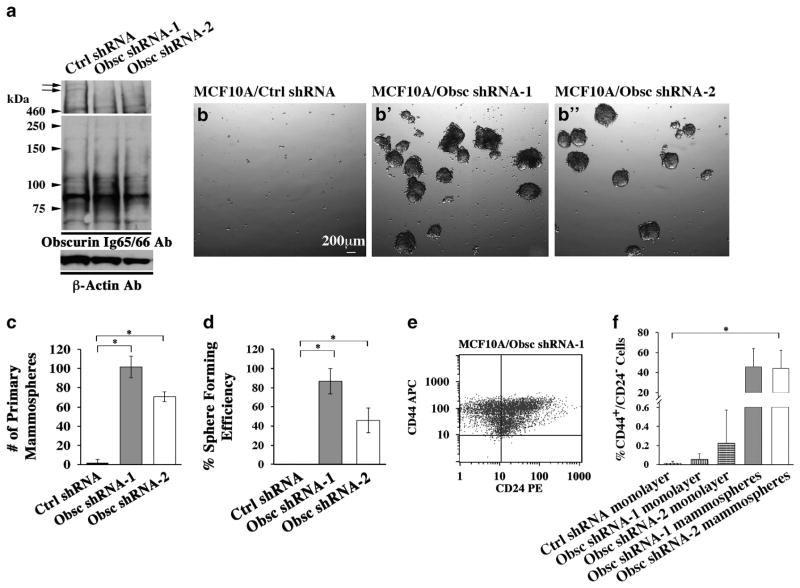
Figure 2.
Obscurin-KD MCF10A breast epithelial cells form mammospheres with enriched markers for cancer-initiating cells. (a) Giant obscurins (arrows) are downregulated in MCF10A cells stably transduced with obscurin shRNA-1 or obscurin shRNA-2 compared with control cells, as indicated by immunoblotting using the Ig65/66 antibody. Notably, the expression of small obscurins remains unchanged in MCF10A cells expressing obscurin shRNA-1 or obscurin shRNA-2 compared with control cells. Equal loading of protein homogenates was ensured by measuring protein concentration and probing for β-actin. (b, b′) MCF10A cells stably transduced with obscurin shRNA-1 (b′) or obscurin shRNA-2 (b″) are able to form primary mammospheres, while control cells are not (b). (c, d) Quantification of primary (c) and secondary (d) mammospheres formed by MCF10A cells stably expressing obscurin shRNA-1 or obscurin shRNA-2, compared with MCF10A cells expressing scramble control shRNA; n =3, error bars =s.d., *P<0.03; t-test. (e) Representative plot obtained from fluorescence activated cell sorting (FACS) analysis of primary mammospheres generated from MCF10A obscurin shRNA-1 expressing cells, indicating that they are highly enriched in a cell population with the surface marker signature CD44+/CD24−, which is associated with stem-like properties. (f) Approximately 40% of cells within the MCF10A obscurin shRNA-1 and obscurin shRNA-2 mammospheres are CD44+/CD24−, compared with <1% of adherent MCF10A control shRNA, obscurin shRNA-1 and obscurin shRNA-2 cell monolayers, as measured by FACS; n =3, error bars =s.d., *P<0.03; t-test.
Given that cultured mammospheres are often enriched in breast cancer-initiating cells containing the cell surface signature CD44+/CD24−,13 we examined the presence and relative abundance of CD44+/CD24− cells in MCF10A obscurin-KD primary mammospheres using flow cytometry. Approximately 40% of obscurin shRNA-1 or obscurin shRNA-2 MCF10A cells stained positive for CD44 and negative for CD24 compared with <1% of adherent control shRNA and obscurin shRNA-1 or obscurin shRNA-2 monolayers (Figures 2e and f). Thus, loss of giant obscurins from breast epithelial cells provides a survival and growth advantage in anchorage-independent conditions and leads to acquisition of stemness.
Downregulation of giant obscurins in breast epithelial cells disrupts AJs and results in EMT
Loss of giant obscurins from MCF10A cells results in acquisition of a mesenchymal phenotype and increased cell scattering,11 both of which are associated with altered cell junctions.14 We thus analyzed the expression profile of various proteins within the cell–cell junctions of MCF10A cells stably expressing obscurin shRNA-1. We observed a dramatic decrease in the amounts of E-cadherin (98%), β-catenin (82%) and α-catenin (38%), whereas the levels of p120 catenin were significantly increased (114%) (Figures 3a and g). Similar results were obtained with MCF10A cells stably transduced with obscurin shRNA-2 (Supplementary Figure S2a). Consistent with the immunoblot analysis, immuno-staining of obscurin-KD MCF10A cells revealed a dramatic reduction of E-cadherin, β-catenin and α-catenin from the cell–cell contacts (Figures 3b and e′). Although residual E-cadherin assumed a homogeneous cytosolic distribution, residual β- and α-catenin exhibited discontinuous distributions at the cell–cell contacts. Notably, remaining β-catenin also showed nuclear accumulation (Figure 3c′, arrow). In contrast, p120 catenin localized at the cell–cell junctions similarly to control cells. Collectively, these results indicate that loss of giant obscurins markedly alters both the expression levels and subcellular distribution of proteins within AJs.
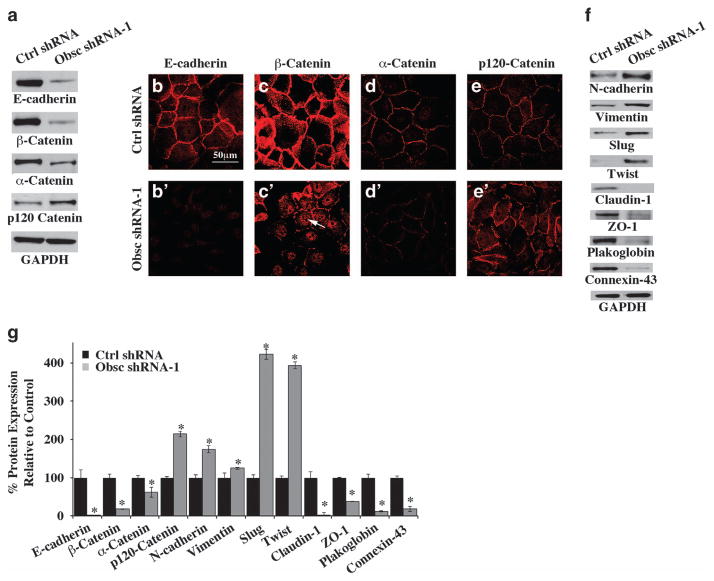
Figure 3.
Loss of giant obscurins results in disruption of AJs and induction of EMT. (a) Representative immunoblots of protein lysates prepared from MCF10A cells stably transduced with obscurin shRNA-1 or scramble control shRNA using a panel of antibodies to AJ proteins. (b–e′) Subcellular distribution of AJ proteins in obscurin-KD and control MCF10A cells. (f) Representative immunoblots of protein lysates prepared from obscurin-KD and control MCF10A cells probed with antibodies to proteins associated with EMT. Equal loading of protein lysates was ensured by measuring protein concentration and probing for glyceraldehyde 3-phosphate dehydrogenase (GAPDH). (g) Quantification of differences in the expression levels of AJ- and EMT-associated proteins in MCF10A cells stably expressing obscurin shRNA-1 compared with MCF10A cells transduced with scramble control shRNA, using densitometry and the Image J software; n =3, error bars =s.d., *P<0.05; t-test.
We subsequently investigated whether MCF10A cells stably transduced with obscurin shRNA-1 undergo EMT. We observed a significant increase in the amounts of major mesenchymal proteins intimately associated with EMT, such as N-cadherin (175%) and vimentin (125%) (Figures 3f and g). Consistent with this, the expression levels of transcriptional regulators of EMT, including Slug (435%) and Twist (394%), were elevated (Figures 3f and g). In contrast, the amounts of junctional epithelial proteins, such as claudin-1 (100%), zona occludens-1 (ZO-1; 62%), connexin-43 (82%) and plakoglobin (88%), were drastically reduced (Figures 3f and g). Analysis of the expression levels of the same battery of proteins in MCF10A cells stably transduced with obscurin shRNA-2 yielded similar results (Supplementary Figure S2a). The concurrent increase in the expression levels of mesenchymal proteins and decrease in the expression levels of epithelial junctional proteins indicate that loss of giant obscurins induces EMT in breast epithelial cells. Interestingly, the use of siRNA technology to downregulate the expression of the EMT transcriptional regulators Twist and Slug in obscurin-KD MCF10A cells failed to or partially reversed their EMT phenotype, respectively. In particular, the expression levels of E-cadherin, β-catenin, N-cadherin and vimentin were unaltered following downregulation of Twist (data not shown). On the contrary, KD of Slug resulted in decreased expression of N-cadherin and increased expression of β-catenin, while the E-cadherin and vimentin levels remained unaltered (Supplementary Figure S2b). These findings indicate that the sole downregulation of either Twist of Slug is not sufficient to reverse EMT in MCF10A cells lacking giant obscurins.
To further validate that loss of giant obscurins from breast epithelial cells results in EMT, we used siRNA technology to transiently downregulate their expression in 184A1 human mammary epithelial cells (HMEC). At 72 h posttransfection, we observed >70% downregulation of giant obscurins in cells treated with obscurin siRNA targeting domain Ig60, compared with cells treated with scramble control siRNA (Supplementary Figure S2c). Similar to MCF10A obscurin-KD cells, 184A1 HMEC obscurin-KD cells exhibited reduced expression of β-catenin and E-cadherin (Supplementary Figure S2c). Thus 184A1 HMEC obscurin-KD cells also undergo EMT due to downregulation of giant obscurins.
Loss of giant obscurins increases F-actin dynamics at the cell–cell contacts
Given the disruption of the AJs in conjunction with the mesenchymal phenotype that obscurin-KD MCF10A cells acquire, we next assessed whether there are changes in the distribution and dynamics of actin filaments. Use of phalloidin staining to visualize actin localization in obscurin-KD cells revealed the presence of filopodia-like protrusions, enriched in actin filaments, emanating from the edges of the cells; such membrane protrusions were nearly absent in control cells, which contained actin bundles surrounding the circumference of the cells (Figures 4a and a′). In light of these alterations, we quantified actin dynamics at the cell–cell junctions using fluorescence recovery after photobleaching (FRAP). LifeAct-GFP (LifeAct-green fluorescent protein) was expressed in ~ 80% confluent monolayers of MCF10A cells stably transduced with control or obscurin shRNA-1 (Supplementary Figure S3) in order to visualize actin in live cells. Fluorescence intensity due to LifeAct-GFP expression was very intense at the cell–cell borders of MCF10A control and obscurin-KD cells, prior to photobleaching (Figures 4b and c, ‘prebleach’ panel). However, it was markedly attenuated in the exposed region, following photobleaching (Figures 4b and c, ‘bleach’ panel). Approximately 2.5 min later, the fluorescence intensity recovered at the cell–cell border, though not completely (Figures 4b and c; last two panels). Analysis of FRAP data (Figure 4d) revealed that the mobile fraction of LifeAct-GFP molecules was considerably larger in obscurin-KD MCF10A cells relative to control cells (Figure 4d), whereas no significant difference was noted on the half-life of LifeAct-GFP recovery (Figure 4e). Collectively, these data indicate that loss of giant obscurins from MCF10A cells results in the formation of membrane protrusions highly enriched in actin filaments, which exhibit increased dynamics compared with control cells.
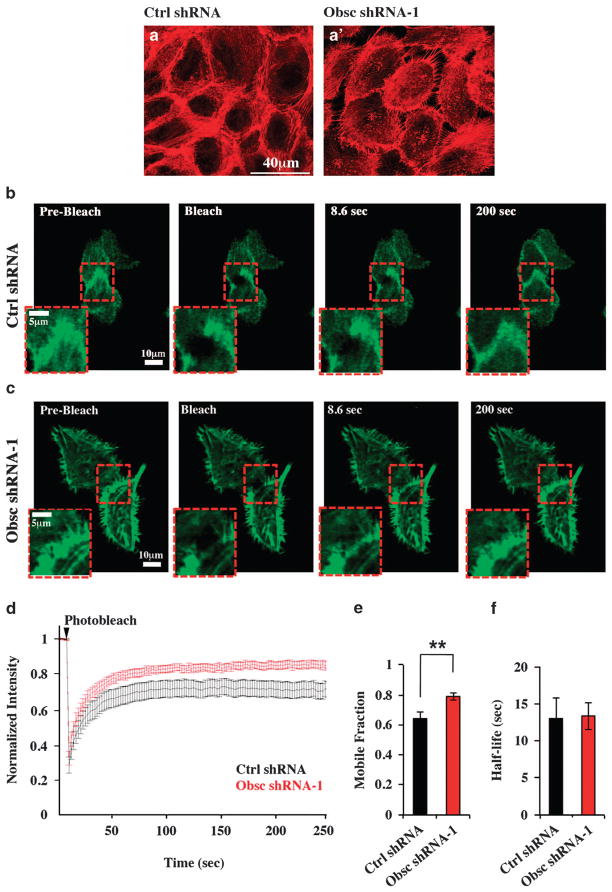
Figure 4.
Obscurin-KD MCF10A cells show increased actin dynamics at the cell–cell borders, as measured by FRAP. (a) Actin localization visualized using phalloidin staining of scramble control and obscurin shRNA-1 expressing MCF10A cells. Control cells (a) exhibit actin bundles around regions of the cell–cell contact, while obscurin shRNA-1 cells (a′) exhibit filopodia-like actin protrusions, emanating from the cell periphery. (b, c) LifeAct-GFP was expressed in ~80% confluent MCF10A cell monolayers stably transduced with control shRNA (b) or obscurin shRNA-1 (c). A circular region of 4 μm in diameter at a cell–cell junction was photobleached, and LifeAct-GFP expression was imaged during fluorescence recovery at different time points. The red dotted square regions correspond to the zoomed images shown in insets. (d) FRAP curves indicate the fraction of initial fluorescence intensity as a function of time for MCF10A cells expressing control shRNA (black) or obscurin shRNA-1 (red); the time of photobleaching is indicated by the arrowhead. (e) Mobile fraction of LifeAct-GFP molecules for cells expressing control shRNA (n =19) or obscurin shRNA-1 (n =21); error bars =s.e., **P<0.01, t-test. (f) Half-life of LifeAct-GFP recovery for cells expressing control shRNA or obscurin shRNA-1.
KD of giant obscurins in breast epithelial cells promotes migration and invasion
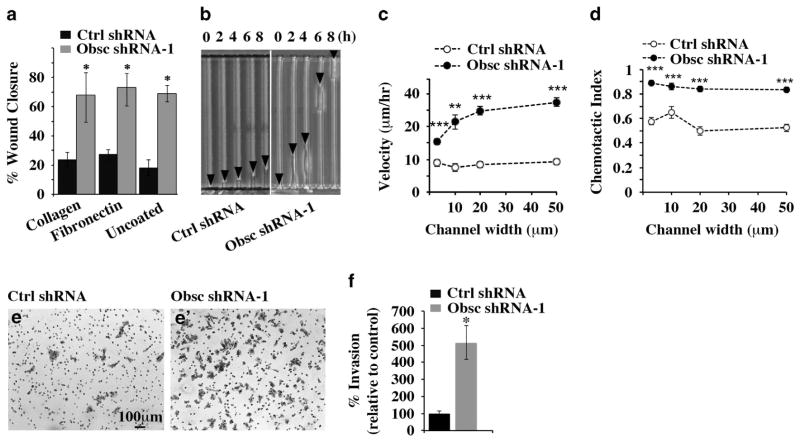
We next investigated whether the alterations in actin dynamics in obscurin-KD MCF10A cells are associated with an increase in cell migration and invasion. Fluorescence imaging of LifeAct-GFP indicated that obscurin-KD, but not control, cells displayed increased migration and persistence along 2-dimensional substrates (Supplementary Video S1, Supplementary Still Image S1). To quantitatively study the migratory potential of obscurin-KD MCF10A cells, we first used a wound-healing assay in which stable clones of control or obscurin shRNA-1 cells were plated on collagen, fibronectin or uncoated wells. MCF10A cells lacking giant obscurins exhibited significantly increased directional migration as a sheet compared with control cells, after a 12-h incubation period either in the presence of collagen or fibronectin (Figure 5a). A marked difference between obscurin-KD and scramble control cells was also detected in the absence of a substrate, indicating that obscurin-KD cells are capable of migrating in a substrate-independent manner, presumably due to their inability to form stable adhesion contacts.
Figure 5.
Obscurin-KD MCF10A cells exhibit increased motility and invasiveness in vitro. (a) Quantification of percentage (%) of wound closure of MCF10A cells stably expressing scramble shRNA or obscurin shRNA-1 between 0 and 12 h after plating on dishes coated with collagen, fibronectin or no substrate, growing to confluency and being wounded; n =3, error bars =s.d., *P<0.03; t-test. (b) Image of control and obscurin shRNA-1-expressing cells migrating through 3-μm wide microchannels. Arrowheads point to the cell’s leading edge at the indicated time points. (c, d) Cell velocity (c) and chemotactic index (d) as a function of channel width for MCF10A cells stably expressing scramble shRNA or obscurin shRNA-1; n =3, error bars =s.e. of at least 30 cells analyzed per condition, **P<0.01 and ***P<0.001; t-test. (e, e′) Obscurin-KD and control MCF10A cells were added to a Matrigel-coated chamber and allowed to invade for 16 h. Staining of invaded cells with crystal violet was followed by quantification of the percentage of invasion of each cell population. (f) The percentage of invasion of obscurin-KD cells was compared with that of control cells, which was arbitrarily set to 100%; n =3, error bars =s.d., *P<0.03; t-test.
We then evaluated the effects of obscurin-KD on single cell motility through microchannels of varying widths (that is, 3, 10, 20 and 50 μm) using a microfluidic-based migration chamber combined with live-cell phase-contrast imaging (Figure 5b).15–18 Obscurin-KD MCF10A cells migrated faster (Figure 5c, Supplementary Video S2, Supplementary Still Image S2) and more persistently (Figure 5d, Supplementary Video S2) than scramble control MCF10A cells, as evidenced by their higher migration velocity and chemotactic index, which represents the ratio of the net cell displacement to the total distance traveled by the cell. MCF10A cells stably transduced with obscurin shRNA-2 showed similarly increased migration in both wound-healing and microchannel assays compared with scramble control cells (Supplementary Figures S4a–c).
We also examined the invasive potential of obscurin-KD MCF10A cells through a Matrigel-coated chamber. Similar to the wound-healing and microchannel assays, loss of giant obscurins dramatically increased the invasiveness of MCF10A breast epithelial cells (>400%) compared with scramble control cells (Figures 5e and f). Additionally, MCF10A cells expressing obscurin shRNA-2 showed increased invasive capabilities (>700%) compared with control cells (Supplementary Figures S4d and e). Notably, at 16 h, which is the time point that percentage of invasion was measured (Figures 5e and f and Supplementary Figures S4d and e), scramble control and obscurin shRNA-1 or shRNA-2 transduced cells did not exhibit statistically significant differences in their proliferative capacities (Supplementary Figure S4f).
Taken altogether, our data demonstrate that loss of giant obscurins from breast epithelial cells significantly increases their migratory and invasive potentials.
Downregulation of giant obscurins promotes tumorigenicity and distant colonization in vivo
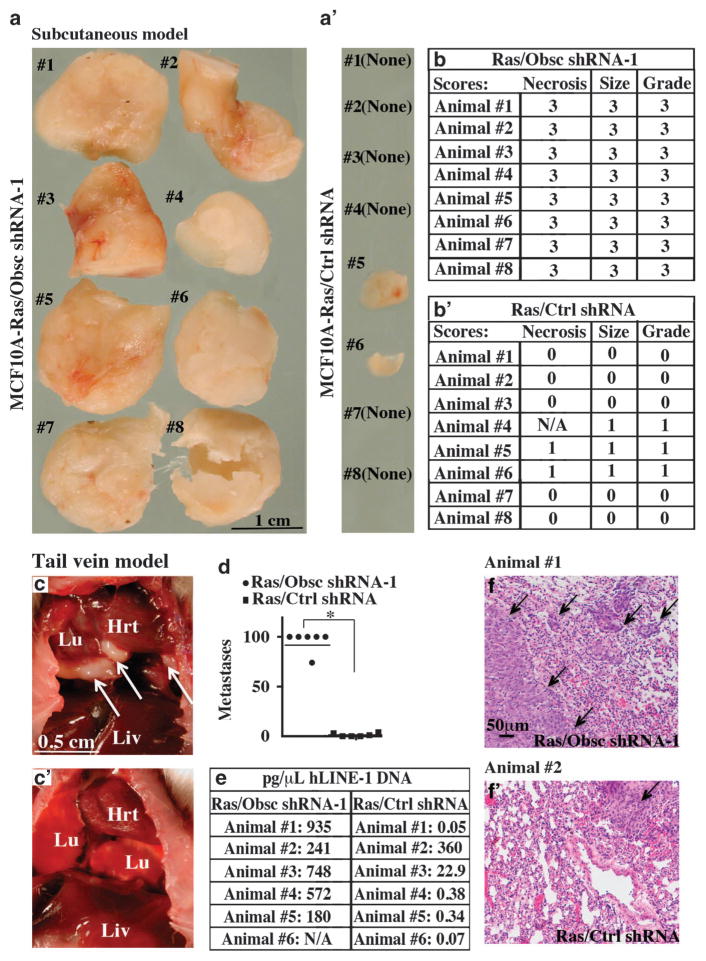
Because loss of giant obscurins in MCF10A cells is associated with increased growth and stemness as well as induction of EMT and increased motility and invasiveness in vitro, we examined its effects on tumorigenicity and distant colonization in vivo. As the MCF10A mammary epithelial cell line is highly resistant to tumor development, multiple genetic alterations are typically needed to promote robust tumor formation.19–21 For this reason, we used an MCF10A cell line that stably expresses an activated form of K-Ras to provide a weakly tumorigenic background to test the specific contribution of obscurins (or lack thereof) to tumor progression.19–21 We first assessed the tumorigenic potential of MCF10A cells stably expressing active K-Ras oncogene and obscurin shRNA-1 (Ras/Obsc shRNA-1) or K-Ras and scramble control shRNA (Ras/ctrl shRNA) in a subcutaneous model; notably, the presence of K-Ras did not affect the expression levels of giant obscurins in either control shRNA or obscurin shRNA-1-transduced cells as shown by immunoblotting analysis (Supplementary Figure S5). We detected large (>1 cm3) primary tumors of high grade (grade 3) in all (8/8) mice injected with Ras/Obsc shRNA-1 cells 9 weeks postinjection, as evidenced by gross morphological (Figure 6a) and histological (Figure 6b) analysis. In contrast, six out of the eight mice (6/8) subcutaneously transplanted with Ras/ctrl shRNA cells were devoid of tumors, while two out of the eight mice (2/8) developed small (<0.1 cm3) low-grade (grade 1) tumors (Figures 6a′ and b′).
Figure 6.
MCF10A cells stably transduced with obscurin shRNA-1 and the K-Ras oncogene form primary and distant tumors in a subcutaneous and a tail vein injection model, respectively. (a, a′) Images of primary tumors formed 9 weeks posttransplantation of MCF10A Ras/Obsc shRNA-1 cells; all 8 mice (8/8) developed >1 cm3 tumors. In contrast, 6 out of 8 (6/8) mice transplanted with MCF10A Ras/Ctrl shRNA developed no tumors, and the remaining 2 out of 8 mice (2/8) developed small (<0.1 cm3) tumors. (b, b′) Scoring of tumors following transplantation of Ras/Obsc shRNA-1 or Ras/Ctrl shRNA MCF10A cells in adult NSG mice. Scores for necrosis were determined by histological evaluation and correspond to 0 =no tumor, 1 = <5% necrosis, 2 =5–25% necrosis and 3 =>25% necrosis. Scores for tumor size were based on digital caliper measurements and correspond to 0 =no tumor, 1 = < 0.1 cm3, 2 =0.1–1 cm3 and 3 =>1 cm3. Finally, scores of tumor grade were calculated from the sum of the necrosis and tumor size scores and correspond to grade 0 =total score 0, grade 1 =total score 1–2, grade 2 =total score 3–4 and grade 3 =total score 5–6. NA, not available, due to the small size of the tumor. (c, c′) Gross morphology of the internal organs of mice injected via the tail vein with Ras/Obsc shRNA-1 (c) or Ras/Ctrl shRNA (c′) cells showing the presence of large tumors (arrows) in the lungs of the former but not the latter. (d) Quantification of the number of micrometastases present in the lungs of mice after tail vein injection of MCF10A cells expressing K-Ras and obscurin shRNA-1 or K-Ras and scramble control shRNA; n = 6 mice per group, error bars =s.d., *P<0.03; t-test. (e) Quantification of the presence of human DNA in the lungs of mice injected with Ras/Obsc shRNA-1 or Ras/Ctrl shRNA cells using quantitative PCR for hLINE-1; NA, not available due to death of the animal. (f, f′) Representative images of histological evaluation of the lungs of animals injected with Ras/Obsc shRNA-1 (f; Animal no. 1) and Ras/Ctrl shRNA (f′; Animal no. 2) cells. A single lobe from each animal was fixed, stained with hematoxylin and eosin (H&E) and examined for signs of lung micrometastases, indicated by arrows.
We then examined the ability of Ras/Obsc shRNA-1 or Ras/ctrl shRNA MCF10A cells to form lung tumors in a lung colonization (tail vein injection) model. Indeed, all (6/6) mice injected with Ras/Obsc shRNA-1 cells via their tail vein exhibited high levels of lung micrometastases (68 to >100 foci per section) by 9 weeks postinjection (Figures 6c, d and f). In contrast, of the six mice injected with Ras/ctrl shRNA cells, three (3/6) had no micro-metastases, while three (3/6) developed few lung metastatic foci (1–4 foci per section) (Figures 6c′). Human long interspersed nuclear element-1 (hLINE-1) analysis22–24 quantitatively confirmed the presence of human DNA in the lungs of all mice injected with Ras/Obsc shRNA-1 cells (Figure 6e). On the contrary, only two out of the six mice (2/6) injected with Ras/ctrl shRNA cells displayed appreciable levels of human DNA (Figure 6e). Collectively, our findings indicate that loss of giant obscurins in the presence of an active oncogene markedly potentiates the tumorigenic and metastatic potentials of breast epithelial cells.
DISCUSSION
Cancer is a disease caused by alterations in multiple genes, which promote cell growth, decrease apoptosis and increase cell motility and invasion.25 Recently, we identified that downregulation of giant obscurins in breast epithelial cells results in increased cell survival following exposure to DNA stress due to decreased apoptosis, implicating giant obscurins as potential tumor suppressors.11 We herein demonstrate that loss of giant obscurins also provides mammary epithelial cells with a growth advantage as seen by the ability of obscurin-KD cells to robustly form both primary and secondary mammospheres. Notably, ~ 40% of the primary mammosphere population bears the CD44+/CD24−signature, which is indicative of cell stemness, thereby suggesting that loss of giant obscurins leads to enrichment of a cell population with tumor-initiating potential. Consistent with these observations, giant obscurins exhibit markedly reduced expression in higher grade (grade ≥2) human breast cancer biopsies of ductal and lobular differentiation, suggesting that their loss may correlate with undifferentiated, more aggressive and metastasis-prone tumors. Interestingly, when residual obscurins, representing truncated and/or mutant forms, were detected, they exhibited a cytoplasmic punctate distribution, potentially corresponding to lysosomal accumulations.
A critical step in cancer progression is the disruption of AJs, resulting in decreased cell–cell adhesion, which promotes motility and contributes to the invasion and metastasis of cancer cells.26–28 In our prior work, we observed that obscurin-KD cells displayed increased cell scattering commensurate with a more mesenchymal appearance compared with control cells, which formed epithelial sheets.11 We herein demonstrate that this phenotypic alteration is accompanied by reduced expression and disrupted localization of major epithelial junctional proteins like N-cadherin, β-catenin, connexin-43, plakoglobin and claudin-1. Disruption of the E-cadherin/β-catenin complex increases the pool of cytoplasmic and nuclear β-catenin, which can induce the initiation of aberrant Wnt signaling.29,30 Nuclear β-catenin, in conjunction with T-cell factor and lymphocyte enhancer transcription factor, promotes the transcription of multiple genes, which in turn can lead to abnormal cell cycle progression and uncontrollable growth.31 KD of giant obscurins in MCF10A cells resulted in near loss of β-catenin from cell–cell junctions that was accompanied by accumulation of residual protein in the nucleus, suggesting that loss of giant obscurins may result in increased cell survival and growth due to deregulated β-catenin-mediated transcription. It is noteworthy that most of the changes we observed in the expression levels of junctional proteins do not correspond to similar changes in their transcript levels (Supplementary Figure S2d), indicating that giant obscurins may have key roles in maintaining and stabilizing proteins in the cell–cell junctions. This is reminiscent of the scaffolding role of giant obscurins in striated muscle cells, where they provide binding sites to diverse cytoskeletal and membrane-associated proteins via their tandem adhesion and signaling motifs.1,32,33
The disruption of AJs occurring at the onset of cancer metastasis is followed by EMT, a process through which epithelial cells dedifferentiate, and acquire a mesenchymal phenotype that increases their motility and invasiveness.14,34 Loss of giant obscurins from MCF10A cells results in a significant increase of the amounts of major mesenchymal proteins, such as N-cadherin and vimentin, and of the transcriptional regulators Slug and Twist. Interestingly, Slug and Twist are known transcriptional repressors of E-cadherin expression,35,36 suggesting that their upregulation may be contributing to changes seen in the E-cadherin levels. Our findings thus reveal that giant obscurins are critical to maintaining the junctional organization, polarity and stability of breast epithelial cells, therefore precluding their dedifferentiation followed by the accumulation of mesenchymal proteins.
The acquisition of EMT is accompanied by increased cell motility and invasion14 and is thought to enable tumor cells to migrate from the primary tumor to distant secondary sites.37 Obscurin-KD MCF10A cells exhibit increased and persistent migration, as evidenced by wound-healing and single-cell migration assays. Metastasizing tumor cells are also capable of degrading and invading through the surrounding basement membrane of distant organs and tissues.38 Indeed, downregulation of giant obscurins promotes cell invasion through a Matrigel chamber. It therefore appears that loss of giant obscurins from breast epithelium may contribute to multiple steps during tumor formation and metastasis.
Enhanced cell migration is accompanied by increased formation of actin-rich protrusions, such as filopodia, at the leading edge of the cell, which are required for cell propulsion.39 The formation of these protrusions is used by cells to extend forward into their surroundings and is accompanied by increased actin dynamics.40 Loss of giant obscurins in mammary epithelial cells resulted in the formation of numerous filopodia-like protrusions near the regions of cell–cell contact. FRAP analysis revealed increased actin dynamics in obscurin-KD cells relative to controls. Thus the increased migratory potential of obscurin-KD cells appears to be due to altered cell–cell junctions and increased actin dynamics. The Rho family of GTPases and their regulators are involved in cell motility.41 Giant obscurins contain a RhoGEF motif, which specifically activates RhoA, a major regulator of actin microfilaments.42 It is therefore plausible that loss of giant obscurins leads to deregulated RhoA signaling, which has been intimately associated with the development and progression of metastatic breast cancer.
Loss of giant obscurins in the presence of active K-Ras imparts tumorigenic, migratory and invasive capabilities to breast epithelial cells, as demonstrated by the rapid formation of large primary tumors in a subcutaneous model and lung trapping in a lung colonization model. Importantly, Ras is insufficient to transform MCF10A cells to a fully tumorigenic and metastatic phenotype in mice.20 However, co-expression of Ras and Bmi-1, a stemness-inducing factor, promotes MCF10A tumorgenicity and metastasis in vivo.19 Similarly, our findings demonstrate that concomitant loss of giant obscurins and expression of active Ras result in the rapid formation of primary and distant tumors, which exhibit morphological (that is, size >1 cm3) and histological (that is, grade 3) characteristics indicative of less differentiated and thus more aggressive tumors.
In summary, our studies demonstrate that loss of giant obscurins from breast epithelial cells results in major cytoskeletal and signaling alterations, which are essential for both local tumor formation and distant colonization, suggesting that giant obscurins may have tumor- and metastasis-suppressive roles.
MATERIALS AND METHODS
Materials
Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich (St Louis, MO, USA).
Cell culture
MCF10A stable clones expressing obscurin shRNA-1, obscurin shRNA-2 or control shRNA plasmids were generated and maintained as described.11 Stable clones of obscurin shRNA-1 MCF10A cells were transiently transfected with HiPerfect using siRNA oligonucleotides for human TWIST1 (5′-TGGGATCAAACTGGCCTGCAA-3′), human SNAI2/SLUG (5′-CACACTGAGTGACGCAATCAA-3′) or Control-AllStars-1 (Qiagen, Hilden, Germany). Protein lysates were collected 24 h (for SNAI2) and 4 days (for TWIST1) posttransfection, at which time points we observed the highest down-regulation for Snai2/Slug and Twist proteins, respectively. MCF10A stable clones expressing K-Ras were transfected with obscurin shRNA-1 or scramble control shRNA using Lipofectamine 2000 (Invitrogen, Grand Island, NY, USA); 1.5 μg/ml puromycin was added to the medium to select for stably transfected cells 3 days posttransfection. Chemically immortalized 184 A1 HMEC cells43 were maintained in Mammary Epithelial Cell growth media supplemented with 0.005 mg/ml transferrin and 1 ng/ml cholera toxin at 37 °C, 5% CO2. Cells were transiently transfected with HiPerfect using either an siRNA oligonucleotide (5′-AGAUUAUACUGG UUCAAAGAUGGCC-3′) targeting human obscurin Ig60 or a scramble control siRNA (Origene Technologies, Rockville, MD, USA). Protein lysates were collected at 72 h posttransfection, at which time point we consistently observed >70% downregulation of the expression levels of giant obscurins.
Antibodies
The antibodies used were as follows; rabbit polyclonal: obscurin Ig65/66 (600 ng/ml),33 obscurin-Ig58/59 (2 μg/ml) that was generated using a mouse GST-Ig58/59 fusion protein (amino acids 5218–5390, accession number: NP_954603), plakoglobin (11146-1-AP, Proteintech Group Inc., Chicago, IL, USA), myosin IIA (3403, Cell Signaling Technology, Danvers, MA, USA), twist (sc-15393, Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA), claudin-1 (4933, Cell Signaling Technology) and ZO-1 (5406, Cell Signaling Technology); mouse monoclonal: β-catenin (sc-7963, Santa Cruz Biotechnology), connexin-43 (13-8300, Invitrogen), p120 catenin (ab11508, Abcam, Cambridge, MA, USA), glyceraldehyde 3-phosphate dehydrogenase (AM4300, Applied Biosystems, Carlsbad, CA, USA); rabbit monoclonal: E-cadherin (3195, Cell Signaling Technology), α-catenin (ab51032, Abcam), vimentin (5741, Cell Signaling Technology) and slug (9585, Cell Signaling Technology).
Human samples and immunohistochemistry
Human breast cancer tumor microarray slides containing paraffin-embedded invasive ductal breast carcinoma and age-, sex-, and grade-matched normal tissue were examined (US Biomax Inc., Rockville, MD, USA). In particular, we analyzed 10 biopsies of IDC grade-1, 35 biopsies of IDC grade-2, 6 biopsies of IDC grade-3 and 3 biopsies of invasive lobular carcinoma grade-2. Tissue sections were deparaffinized in xylene, rehydrated with graded alcohol washes and subjected to antigen retrieval using 10 mM sodium citrate. Following washes in TBS (Tris borate saline) containing 0.025% Triton X-100 (TBST), sections were blocked with 10% normal goat serum and 1% bovine serum albumin (BSA) in TBS for 2 h. Slides were incubated with the rabbit polyclonal obscurin Ig58/59 antibody diluted in TBS containing 1% BSA and incubated overnight at 4 °C. Sections were subsequently washed with TBST and incubated for 2 h with goat anti-rabbit Alexa Fluor-568 secondary antibody (LifeTechnologies, Carlsbad, CA, USA). Following extensive washes in TBST, slides were mounted with ProLong Gold Antifade Reagent with 4,6-diamidino-2-phenylindole (LifeTechnologies) and analyzed in a LSM510 confocal microscope (Carl Zeiss MicroImaging, Thornwood, NY, USA) under × 40 magnification.
Generation of protein lysates and western blotting
Age-, stage- and sex-matched human normal and breast cancer biopsies were commercially obtained (ILSbio, Chestertown, MD, USA). Tissue homogenates were prepared on ice with a hand-homogenizer in radioimmuno-precipitation assay buffer supplemented with a cocktail of protease inhibitors (Roche, Mannheim, Germany) and phosphatase inhibitors (200 nM Imidazole, 100mM Sodium Flouride, 115 mM Sodium Molybdate, 100 mM Sodium Orthovanadate, 400 mM Sodium Tartrate Dihydrate, 100 mM β-Glycerophosphate, 100 mM Sodium Pyrophosphate and 10 mM EGTA), as previously described.44 Protein lysates were electrophoresed on sodium dodecyl sulfate-NuPAGE gel (Invitrogen), transferred to nitrocellulose membranes and probed with primary antibodies as specified in the text. Alkaline phosphatase-conjugated anti-mouse or anti-rabbit IgG (1:3,000; Jackson ImmunoResearch, West Grove, PA, USA) were used, and immunoreactive bands were visualized with a chemiluminescence detection kit (Applied Biosystems). The relative abundance of immunoreactive bands was determined with densitometry using the Image J software (NIH, Bethesda, MD, USA).
Mammosphere culture
Single MCF10A cells stably transduced with obscurin shRNA-1, obscurin shRNA-2 or scramble control shRNA were plated in ultralow attachment plates (Corning, Lowell, MA, USA) at a density of 10 000 viable cells/ml in serum-free growth media (Dulbecco’s modified Eagle’s medium/F12 with GlutaMAX), supplemented with insulin (10 μg/ml), hydrocortisone (0.5 μg/ml), cholera toxin (100 ng/ml), epidermal growth factor (20 ng/ml), 1% penicillin–streptomycin and puromycin (1.5 μg/ml). Cultures were supplemented with serum-free growth media every other day for 14 days at which time point primary spheres were measured and those >100 μm were counted as tumor spheres. Mammospheres were then collected by gentle centrifugation (800 r.p.m.) and dissociated enzymatically (2 min in 0.25% trypsin-EDTA (1 ×); Invitrogen) and mechanically using a sterile pipette. To evaluate secondary mammosphere-forming efficiency, dissociated cells obtained from primary mammospheres were seeded at one cell per well into 96-well ultralow attachment plates (Corning). Cells were grown as described above for 14 days, and the number of secondary mammospheres per well with sizes of 50–100 μm was counted. Sphere-forming efficiency was calculated as the number of spheres formed divided by the original number of single cells seeded and were expressed as percentage.
Fluorescence activated cell sorting
CD24 and CD44 expression was analyzed in MCF10A cells stably expressing obscurin shRNA-1 grown in a monolayer or in mammospheres. Cells were dissociated following incubation in trypsin/EDTA combined with manual trituration using a Pasteur pipette and pelleted by centrifugation at 1000 g for 10 min. Cells were resuspended in phosphate-buffered saline (PBS) with 0.1% BSA (wt/vol). Cell suspensions were incubated with mouse anti-human CD44-APC antibody (BD Pharmigen, San Jose, CA, USA) and mouse anti-human CD24-PE antibody (BD Pharmigen) for 30 min at 4 °C. All flow cytometry assays were performed using a BD FACSCalibur, and data analysis was accomplished using the BD CellQuest Pro software (BD Biosciences, Bedford, MA, USA).
AlamarBlue assay
Cell growth of MCF10A obscurin-KD and control cells was measured using the AlamarBlue assay (LifeTechnologies). AlamarBlue reagent was added to the cell culture at 10% v/v and incubated for 16 h at 37 °C, 5% CO2. The percentage (%) of AlamarBlue reduction was determined by measuring absorbance at 550 and 620 nm. Data are presented as the percentage of AlamarBlue reduced per number of cells.
Immunofluorescence combined with confocal microscopy
Cells were fixed with freshly prepared 3.7% paraformaldehyde, permeabilized with 0.25% Triton X-100 in PBS containing 1 mg/ml BSA (PBS/BSA) and blocked with 5% normal goat serum. Cells were subsequently incubated overnight at 4 °C with primary antibodies as specified in the text, counterstained with goat anti-rabbit Alexa Fluor-568 or goat anti-mouse Alexa Fluor-568 IgG (LifeTechnologies) at a 1:200 dilution and analyzed in a LSM510 confocal microscope (Carl Zeiss) under × 63 magnification.
RNA isolation, cDNA synthesis and PCR Amplification
Total RNA was isolated from cultured cells with the TRIzol reagent (Invitrogen) and reverse-transcribed using the SuperScript III First-Strand Synthesis Kit (Invitrogen). PCR amplification was performed with GoTaq Green Master Mix (Promega), using the primer sets listed in Supplementary Table S1. All amplicons were purified and sequenced.
Fluorescence recovery after photobleaching
FRAP was used to analyze actin molecule dynamics in living cells. Cells were transfected with LifeAct-GFP using Lipofectamine 2000 (Invitrogen) and re-plated onto collagen-coated glass coverslips with polydimethylsiloxane (PDMS) barriers to retain cell media. After allowing the cells to adhere and form a monolayer for 24 h, coverslips with cells were positioned on the stage of a Zeiss LSM 510 META laser scanning confocal microscope (Carl Zeiss). A circular region of interest at the cell–cell borders with 4-μm diameter was photobleached using a 488-nm laser at 100% power. Images were captured at maximum rate (2.15 second intervals) before and after photobleaching using a × 63/1.4 NA oil objective and the LSM software (version 4.2, Carl Zeiss). Image J was used to analyze fluorescence intensity in the photobleached region as a function of time using the ‘Plot Z-axis profile’ function. In non-fluorescent regions of the image, background fluorescence as a function of time was measured to be zero using the same method. At least 20 images of the cell–cell borders per cell group were analyzed. Intensity within the photobleached region was normalized such that an intensity value of one corresponded to the mean fluorescence intensity of five prebleach images, while an intensity value of zero corresponded to the fluorescence intensity just after photobleaching. Data for fluorescence intensity as a function of time (I(t)) were fit to the following standard equation for binding kinetics: I(t) = Ifinal(1 − e−τt), where Ifinal is the mobile fraction of actin molecules, and τ is related to the half-life (τ1/2) by the following formula: τ1/2 = −ln(0.5)/τ.
Migration in 2-dimensional substratum using LifeAct-GFP
To image actin dynamics live during cell migration, MCF10A cells were transfected with LifeAct-GFP using Lipofectamine 2000. Two days after transfection, cells were plated onto type 1 collagen-coated glass coverslips (20 μg/ml in PBS), allowed to adhere overnight and imaged at 2-min intervals for ~ 4 h using an automated Nikon fluorescence microscope (×20 objective) (Nikon, Tokyo, Japan).
Wound-healing and invasion assays
Wound healing was measured by growing confluent cell monolayers as described above in six-well tissue culture dishes (Corning) that were uncoated or precoated with human fibronectin (BD Biosciences) or collagen type I (BD Biosciences). A scrape was made through the monolayer with a sterile plastic pipette tip, and fresh media was added. Images were taken with an inverted microscope (×10 objective) at time zero and after a 12-h incubation period at 37 °C, 5% CO2. Migration was expressed as the average of the difference between the measurement at time zero and 12 h obtained from three independent experiments.
Invasion was measured by adding 250 000 cells suspended in 0.5 ml growth media to the upper chamber of a Matrigel-coated Invasion Chamber (BD Biosciences). The lower chamber contained growth media with 10% fetal bovine serum. The inserts were incubated at 37 °C, 5% CO2 for 16 h. At the end of the 16-h incubation period, the cells that had invaded the lower chamber were fixed and stained with 0.5% crystal violet in 20% methanol. The number of invaded cells was quantified by counting at least six random fields from three independent experiments under an inverted light microscope (Olympus IX51, Center Valley, PA, USA) with a × 10 objective.
Fabrication of the microchannel device
The microchannel device was fabricated by standard lithography, as previously described.15–18 Briefly, SU-8 photoresist was spin coated onto a silicon wafer to a thickness of 10 μm and cross-linked by exposure to ultraviolet light through a photomask with variable transparent separation distances, ranging from 3 to 50 μm. Developer was used to remove non-crosslinked photoresist. This process was repeated with a 50-μm-thick layer of SU-8 and a second mask containing two 400-μm-wide channels spaced 200-μm apart. PDMS was prepared at a 10:1 ratio with curing agent poured over the silicon master and degassed in a vacuum chamber for 2 h. The PDMS was baked at 85 °C for 1 h, peeled off the master, cut to appropriate size and pierced to form inlet and outlet ports. The PDMS device and 75-mm glass coverslips were treated with oxygen plasma for 3 min and irreversibly sealed together upon contact, forming four-walled micro-channels. The surface of the PDMS microchannels was functionalized by adding type-1 collagen (20 μg/ml in PBS) to the ports of the device and incubating for 1 h at 37 °C.
Microchannel seeding and cell migration
Cells were trypsinized, resuspended in serum-free media to neutralize the trypsin and subsequently washed in serum-free media. A suspension of 1 × 105 cells was added to the inlet port, and cells were transported along the seeding channel by pressure-driven flow. Within 5 min, the cell suspension was removed and replaced with 50 μl of serum-free media, which was also added to the upper inlets. Serum-containing media was added to the top-most inlet port, thus forming a chemoattractant gradient. A thin layer of (CO2-permeable) PDMS was placed over the chamber inlet and outlet ports to prevent media evaporation during imaging. Chambers were placed in an enclosed, humidified microscope stage at 5% CO2 and 37 °C (TIZ, Tokai Hit Co., Shizuoka, Japan). Phase-contrast time-lapse images were captured at 10-min intervals for 14 h on an inverted Nikon microscope (×10 objective) at multiple stage positions via stage automation (Nikon Elements, Nikon). Cell’s x, y position within the microchannel was identified as the midpoint between the poles of the cell body and tracked as a function of time using ImageJ. Cell velocity and chemotactic index were computed using a custom-written Matlab program (The MathWorks, Natick, MA, USA). Instantaneous cell velocity was calculated by dividing each interval displacement by the time interval (10 min), and the mean velocity for a given cell was computed by averaging instantaneous velocities for all time intervals. Chemotactic index was calculated by dividing the end-to-end displacement by the total path length of the cell. The reported velocity and chemotactic index for each condition is the mean of the pooled cells from three independent experiments.
Mice
All experimental procedures complied with guidelines provided by the Office of Laboratory Animal Welfare at the National Institutes of Health, and all protocols were approved by the Johns Hopkins University Animal Care and Use Committee. Adult NSG mice (male and female) were used as tumor recipients for mouse experiments (Stock 5557, Jackson Labs).
Subcutaneous injections
Suspensions of 2 × 106 cells in 100 μl Matrigel were injected subcutaneously into the flank of mice (eight per experimental group). Tumors were measured in three dimensions with an electronic digital caliper. Tumor volume was calculated as described (L) × (W) × (H) × π/6.45 Animals were euthanized 9 weeks postimplantation.
Tail vein injections
Mice (six per experimental group) were injected with 1 × 106 cells in a volume of 50 μl of culture media via the tail vein. Animals were euthanized 9 weeks postinjection.23
Histopathology of lung tissue
Lungs and other tissues were examined visually for lesions and metastatic foci. Lung samples for pathology were fixed in 10% buffered formalin. The left lung lobe was used for hLINE analysis, and the remaining lobes were embedded in a single cassette and sectioned for histopathological analysis of metastastatic foci.24,46 Samples were embedded in paraffin, sectioned at 5 μm and stained with hematoxylin and eosin using the standard techniques. Primary tumors were scored 0–3 based on size and 0–3 based on extent of necrosis in each tumor by histopathology. Scores were totaled, and individual tumors were graded as follows: grade 1 = total score of 1–2; grade 2 = total score of 3–4; and grade 3 = total score of 5–6. Tissue collection, histopathology analysis and grading were performed by a pathologist (DLH).
Quantification of hLINE-1 gene
DNA was extracted from mouse tissue as previously described22,24 using the DNeasy blood and tissue kit (Qiagen, Valencia, CA, USA) in a sterile biological safety cabinet to minimize the risk of human DNA contamination. Elutions were analyzed via quantitative PCR as reported previously.46 Briefly, quantitative PCR was performed in 15-μl volume with the following components: 7.5 μl iQ SYBR Green Supermix (Bio-Rad, Hercules, CA, USA), 1.5 μl of each 10 μM forward (5′-TCACTCAAAGCCGCTCAACTAC-3′) and reverse (5′-TCTGCCTTCATTTCGTTATGTACC-3′) primers and 4.5 μl of purified DNA. The reaction was monitored on a MyiQ Real Time PCR Detection System (Bio-Rad) with the following cycles: (94 °C, 2 min) × 1, (94 °C, 10 s; 67 °C, 15 s; 70 °C, 15 s) × 3, (94 °C, 10 s; 64 °C, 15 s; 70 °C, 15 s) × 3, (94 °C, 10 s; 61 °C, 15 s; 70 °C, 15 s) × 3, and (94 °C, 10 s, 59 °C, 15 s; 70 °C, 15 s) × 35. Threshold cycle number was calculated using the Bio-Rad iQTM5 software (Bio-Rad). Dilutions of human DNA purified from MCF10A breast epithelial cells were included in each plate to serve as standards.
Reproducibility and statistics
All experiments were performed in triplicates a minimum of three times, unless otherwise noted, and data are presented as mean values of independent measurements. Statistical significance was assessed using the Student’s t-test.
Supplementary Material
Acknowledgments
This work was supported by a Pilot Grant from The Johns Hopkins Physical Sciences-Oncology Center, The Johns Hopkins Institute for NanoBioTechnology (U54CA143868 to AK-K) and by awards from the National Science Foundation (Award NSF-1159823 to KK), the National Cancer Institute (Awards U54-CA143868 to KK; R01-CA186286 to KK; T32-CA130840 to KMS; F32-CA177756 to KMS; R01-CA154624 to SM and K01-CA166576 to MIV) and the Kleberg Foundation (to KK). Portions of this work are included in a United States patent pending, 14/221,755, filed on 21 March 2014.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
References
- 1.Kontrogianni-Konstantopoulos A, Ackermann MA, Bowman AL, Yap SV, Bloch RJ. Muscle giants: molecular scaffolds in sarcomerogenesis. Physiol Rev. 2009;89:1217–1267. doi: 10.1152/physrev.00017.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kontrogianni-Konstantopoulos A, Bloch RJ. Obscurin: a multitasking muscle giant. J Muscle Res Cell Motil. 2005;26:419–426. doi: 10.1007/s10974-005-9024-7. [DOI] [PubMed] [Google Scholar]
- 3.Perry NA, Ackermann MA, Shriver M, Hu LY, Kontrogianni-Konstantopoulos A. Obscurins: unassuming giants enter the spotlight. IUBMB Life. 2013;65:479–486. doi: 10.1002/iub.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russell MW, Raeker MO, Korytkowski KA, Sonneman KJ. Identification, tissue expression and chromosomal localization of human Obscurin-MLCK, a member of the titin and Dbl families of myosin light chain kinases. Gene. 2002;282:237–246. doi: 10.1016/s0378-1119(01)00795-8. [DOI] [PubMed] [Google Scholar]
- 5.Fukuzawa A, Idowu S, Gautel M. Complete human gene structure of obscurin: implications for isoform generation by differential splicing. J Muscle Res Cell Motil. 2005;26:427–434. doi: 10.1007/s10974-005-9025-6. [DOI] [PubMed] [Google Scholar]
- 6.Hu LY, Kontrogianni-Konstantopoulos A. The kinase domains of obscurin interact with intercellular adhesion proteins. FASEB J. 2013;27:2001–2012. doi: 10.1096/fj.12-221317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borisov AB, Raeker MO, Russell MW. Developmental expression and differential cellular localization of obscurin and obscurin-associated kinase in cardiac muscle cells. J Cell Biochem. 2008;103:1621–1635. doi: 10.1002/jcb.21551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 9.Balakrishnan A, Bleeker FE, Lamba S, Rodolfo M, Daniotti M, Scarpa A, et al. Novel somatic and germline mutations in cancer candidate genes in glioblastoma, melanoma, and pancreatic carcinoma. Cancer Res. 2007;67:3545–3550. doi: 10.1158/0008-5472.CAN-07-0065. [DOI] [PubMed] [Google Scholar]
- 10.Price ND, Trent J, El-Naggar AK, Cogdell D, Taylor E, Hunt KK, et al. Highly accurate two-gene classifier for differentiating gastrointestinal stromal tumors and leiomyosarcomas. Proc Natl Acad Sci USA. 2007;104:3414–3419. doi: 10.1073/pnas.0611373104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perry NA, Shriver M, Mameza MG, Grabias B, Balzer E, Kontrogianni-Konstantopoulos A. Loss of giant obscurins promotes breast epithelial cell survival through apoptotic resistance. FASEB J. 2012;26:2764–2775. doi: 10.1096/fj.12-205419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baum B, Georgiou M. Dynamics of adherens junctions in epithelial establishment, maintenance, and remodeling. J Cell Biol. 2011;192:907–917. doi: 10.1083/jcb.201009141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phillips TM, McBride WH, Pajonk F. The response of CD24(− /low)/CD44+ breast cancer-initiating cells to radiation. J Natl Cancer Inst. 2006;98:1777–1785. doi: 10.1093/jnci/djj495. [DOI] [PubMed] [Google Scholar]
- 14.Foroni C, Broggini M, Generali D, Damia G. Epithelial-mesenchymal transition and breast cancer: role, molecular mechanisms and clinical impact. Cancer Treat Rev. 2012;38:689–697. doi: 10.1016/j.ctrv.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 15.Balzer EM, Tong Z, Paul CD, Hung WC, Stroka KM, Boggs AE, et al. Physical confinement alters tumor cell adhesion and migration phenotypes. FASEB J. 2012;26:4045–4056. doi: 10.1096/fj.12-211441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hung WC, Chen SH, Paul CD, Stroka KM, Lo YC, Yang JT, et al. Distinct signaling mechanisms regulate migration in unconfined versus confined spaces. J Cell Biol. 2013;202:807–824. doi: 10.1083/jcb.201302132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen SH, Hung WC, Wang P, Paul C, Konstantopoulos K. Mesothelin binding to CA125/MUC16 promotes pancreatic cancer cell motility and invasion via MMP-7 activation. Sci Rep. 2013;3:1870. doi: 10.1038/srep01870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tong Z, Balzer EM, Dallas MR, Hung WC, Stebe KJ, Konstantopoulos K. Chemotaxis of cell populations through confined spaces at single-cell resolution. PLoS One. 2012;7:e29211. doi: 10.1371/journal.pone.0029211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoenerhoff MJ, Chu I, Barkan D, Liu ZY, Datta S, Dimri GP, et al. BMI1 cooperates with H-RAS to induce an aggressive breast cancer phenotype with brain metastases. Oncogene. 2009;28:3022–3032. doi: 10.1038/onc.2009.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang B, Soule HD, Miller FR. Transforming and oncogenic potential of activated c-Ha-ras in three immortalized human breast epithelial cell lines. Anticancer Res. 1997;17:4387–4394. [PubMed] [Google Scholar]
- 21.Ciardiello F, Gottardis M, Basolo F, Pepe S, Normanno N, Dickson RB, et al. Additive effects of c-erbB-2, c-Ha-ras, and transforming growth factor-alpha genes on in vitro transformation of human mammary epithelial cells. Mol Carcinog. 1992;6:43–52. doi: 10.1002/mc.2940060108. [DOI] [PubMed] [Google Scholar]
- 22.Dallas MR, Liu G, Chen WC, Thomas SN, Wirtz D, Huso DL, et al. Divergent roles of CD44 and carcinoembryonic antigen in colon cancer metastasis. FASEB J. 2012;26:2648–2656. doi: 10.1096/fj.12-203786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ericson K, Gan C, Cheong I, Rago C, Samuels Y, Velculescu VE, et al. Genetic inactivation of AKT1, AKT2, and PDPK1 in human colorectal cancer cells clarifies their roles in tumor growth regulation. Proc Natl Acad Sci USA. 2010;107:2598–2603. doi: 10.1073/pnas.0914018107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rago C, Huso DL, Diehl F, Karim B, Liu G, Papadopoulos N, et al. Serial assessment of human tumor burdens in mice by the analysis of circulating DNA. Cancer Res. 2007;67:9364–9370. doi: 10.1158/0008-5472.CAN-07-0605. [DOI] [PubMed] [Google Scholar]
- 25.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 26.Friedl P, Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147:992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 27.Knights AJ, Funnell AP, Crossley M, Pearson RC. Holding tight: cell junctions and cancer spread. Trends Cancer Res. 2012;8:61–69. [PMC free article] [PubMed] [Google Scholar]
- 28.Brasch J, Harrison OJ, Honig B, Shapiro L. Thinking outside the cell: how cadherins drive adhesion. Trends Cell Biol. 2012;22:299–310. doi: 10.1016/j.tcb.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gottardi CJ, Wong E, Gumbiner BM. E-cadherin suppresses cellular transformation by inhibiting beta-catenin signaling in an adhesion-independent manner. J Cell Biol. 2001;153:1049–1060. doi: 10.1083/jcb.153.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valenta T, Hausmann G, Basler K. The many faces and functions of beta-catenin. EMBO J. 2012;31:2714–2736. doi: 10.1038/emboj.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holland JD, Klaus A, Garratt AN, Birchmeier W. Wnt signaling in stem and cancer stem cells. Curr Opin Cell Biol. 2013;25:254–264. doi: 10.1016/j.ceb.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 32.Kontrogianni-Konstantopoulos A, Catino DH, Strong JC, Sutter S, Borisov AB, Pumplin DW, et al. Obscurin modulates the assembly and organization of sarcomeres and the sarcoplasmic reticulum. FASEB J. 2006;20:2102–2111. doi: 10.1096/fj.06-5761com. [DOI] [PubMed] [Google Scholar]
- 33.Kontrogianni-Konstantopoulos A, Jones EM, Van Rossum DB, Bloch RJ. Obscurin is a ligand for small ankyrin 1 in skeletal muscle. Mol Biol Cell. 2003;14:1138–1148. doi: 10.1091/mbc.E02-07-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheel C, Weinberg RA. Cancer stem cells and epithelial-mesenchymal transition: concepts and molecular links. Semin Cancer Biol. 2012;22:396–403. doi: 10.1016/j.semcancer.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drasin DJ, Robin TP, Ford HL. Breast cancer epithelial-to-mesenchymal transition: examining the functional consequences of plasticity. Breast Cancer Res. 2011;13:226. doi: 10.1186/bcr3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci USA. 2010;107:15449–15454. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiang AC, Massague J. Molecular basis of metastasis. N Engl J Med. 2008;359:2814–2823. doi: 10.1056/NEJMra0805239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bravo-Cordero JJ, Hodgson L, Condeelis J. Directed cell invasion and migration during metastasis. Curr Opin Cell Biol. 2012;24:277–283. doi: 10.1016/j.ceb.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arjonen A, Kaukonen R, Ivaska J. Filopodia and adhesion in cancer cell motility. Cell Adh Migr. 2011;5:421–430. doi: 10.4161/cam.5.5.17723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chhabra ES, Higgs HN. The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol. 2007;9:1110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- 41.Hall A. Rho family GTPases. Biochem Soc Trans. 2012;40:1378–1382. doi: 10.1042/BST20120103. [DOI] [PubMed] [Google Scholar]
- 42.Ford-Speelman DL, Roche JA, Bowman AL, Bloch RJ. The rho-guanine nucleotide exchange factor domain of obscurin activates rhoA signaling in skeletal muscle. Mol Biol Cell. 2009;20:3905–3917. doi: 10.1091/mbc.E08-10-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elenbaas B, Spirio L, Koerner F, Fleming MD, Zimonjic DB, Donaher JL, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ackermann MA, Hu LY, Bowman AL, Bloch RJ, Kontrogianni-Konstantopoulos A. Obscurin interacts with a novel isoform of MyBP-C slow at the periphery of the sarcomeric M-band and regulates thick filament assembly. Mol Biol Cell. 2009;20:2963–2978. doi: 10.1091/mbc.E08-12-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nanda A, Karim B, Peng Z, Liu G, Qiu W, Gan C, et al. Tumor endothelial marker 1 (Tem1) functions in the growth and progression of abdominal tumors. Proc Natl Acad Sci USA. 2006;103:3351–3356. doi: 10.1073/pnas.0511306103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samuels Y, Diaz LA, Jr, Schmidt-Kittler O, Cummins JM, Delong L, Cheong I, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.