Abstract
Mapping gene expression as a quantitative trait (eQTL mapping) can reveal local and distant associations with functionally important genetic variation informative for disease. Recent studies are reviewed which have demonstrated that this approach is particularly informative when applied to diverse immune cell populations and situations relevant to infection and immunity. Context-specific eQTL have now been characterised following endotoxin activation, induction with interferons, mycobacteria, and influenza, together with genetic determinants of response to vaccination. The application of genetical genomic approaches offers new opportunities to advance our understanding of gene–environment interactions and fundamental processes in innate and adaptive immunity.
Introduction
Individuals vary in susceptibility to infectious disease and only a minority develop the most severe complications [1]. Responses to pathogen invasion and immunity to infection require coordinated regulation of gene expression. At any given locus gene expression may vary markedly between individuals. This variation has a strong genetic component, with polymorphisms associating with gene expression known as expression quantitative trait loci (eQTL) [2,3]. Genome-wide association studies (GWAS) of disease susceptibility demonstrate the majority of identified risk loci involve non-coding single nucleotide variants (SNVs) and these are thought to exert primarily regulatory effects on gene expression [4•]. The identification of regulatory genetic variants and eQTL is therefore of significant biomedical interest. Whilst variance in expression is highly gene specific, eQTL contribute to phenotypic diversity with regulatory variation proposed as the predominant driver of local population adaptive changes [5]. Quantitative changes in gene expression influence the outcomes of immune responses and genes implicated in immune-related disease demonstrate higher heritability for expression, as do those under balancing selection, reflecting the selective pressure of infectious disease [6•]. In this review we highlight recent insights from eQTL analysis in settings relevant to immunity and infection. This work has illustrated the extent of context-specific associations, informing our understanding of gene–environment interactions and population-wide variance in expression of key genes implicated in immunity to infection that may help refine our understanding of immunogenetics.
Informativeness of eQTL mapping
The eQTL approach builds upon the principles of genetical genomics [7••], whereby analysis of intermediate phenotypes (such as transcript abundance) between those at the organismal level (i.e. disease trait) and underlying DNA variation provides insights into the functional correlates of associated variation. This is particularly useful for non-Mendelian immune and infectious disease phenotypes where interaction between polygenic variants and environmental factors may be required for disease phenotype manifestation. In brief, eQTL can be subdivided into those that show association with gene expression locally (typically within 1Mb region, likely cis-acting) and those that demonstrate association to distant genes — known as trans eQTL. Such associations identify putative functional regulatory genetic variants and the specific genes or gene networks that may be modulated. trans eQTL have the potential to provide unbiased discovery of novel pathways and processes, especially those that influence multiple genes (discussed further below) although effect sizes are typically smaller and, given genome-wide testing, trans associations are harder to resolve when accounting for Type-1 error rates. Recent meta-analyses involving large cohorts for whom peripheral blood eQTLs have been mapped [8••] has made notable progress into the identification of reproducible trans eQTL however. There is also evidence that induction of gene expression with immune stimulants may enrich for trans-effects by harmonizing expression patterns and reducing variance [9••]. The intersection of eQTL and GWAS has been conducive to hypothesis generation regarding the identity of causative genes underlying disease associations [10]. Although local eQTL effect sizes increase as distance to the gene transcription start site falls, it is increasingly appreciated many local eQTL act over longer ranges through distal enhancer effects, making resolution of GWAS difficult [8••,11]. This is illustrated by recent work demonstrating that obesity risk polymorphisms located in the FTO gene have functional effects across over 400 kb on the homeobox gene IRX3, proposed to be the causative mechanism for obesity susceptibility [12•].
Trans eQTL provide unbiased insights into immunological pathways
trans eQTL, whilst typically more difficult to detect, often leave a signature upon expression across multiple different loci — so called ‘master regulatory regions’. By resolving local effects of these polymorphisms on gene expression, the likely upstream causative gene can be identified permitting unbiased delineation of physiologically relevant downstream pathways and avoiding the use of genomic interference technologies such as gene knockouts. An illustrative example being recently identified in blood where a lupus associated variant associated in trans with several interferon response genes [8••]. By combining sequencing and ChIP-seq data the same SNV was found to have local effects upon expression of the transcription factor IKZF1 with the genes in trans being enriched for IKZF1 binding. In addition to mechanistically resolving a role for IKZF1 in lupus predisposition, it also suggested that many genes implicated in lupus pathogenesis are subtly dysregulated in healthy carriers of a predisposing susceptibility variant. Similarly, many trans associations involve SNVs within the Major Histocompatibility Complex (MHC) [8••,9••,13•,14•], which is 10-fold enriched for such associations [14•], with evidence these are context specific and correlate to MHC class II gene expression [9••]. Another intriguing master-regulatory trans eQTL resolved only in B-cells has local effects upon expression of the transcription factor KLF4, a gene implicated in pluripotency and breast cancer susceptibility [15] and robustly expressed in monocytes. Carriage of the minor allele of rs61414050 was associated with upregulation of several innate pro-inflammatory genes and, whilst it should be noted that this allele was relatively rare (minor allele frequency in population of European ancestry 4.2%) and therefore replication of this effect is required, such widespread changes in expression are likely to have functional outcomes.
eQTL vary in effect size across tissue and cell type
The majority of early human eQTL studies focused on Epstein-Barr Virus transformed lymphoblastoid cell lines (LCLs) which, although informative [16•], have significant limitations due to immortalization and physiological relevance. Notably, immune cells are characterised by a high degree of inter-cellular cross-talk and antigen presentation. Use of LCL monocultures fails to capture the effects of this cross-talk on expression and also diminishes the detection of other physiologically relevant context-specific effects. The latter may depend on the specific cell and tissue type analysed [13•,17••,18•], population [19•], sex [20], age [20,21], geography [22], disease [23] and environmental context [9••,24,25••], all of which may impact immunity.
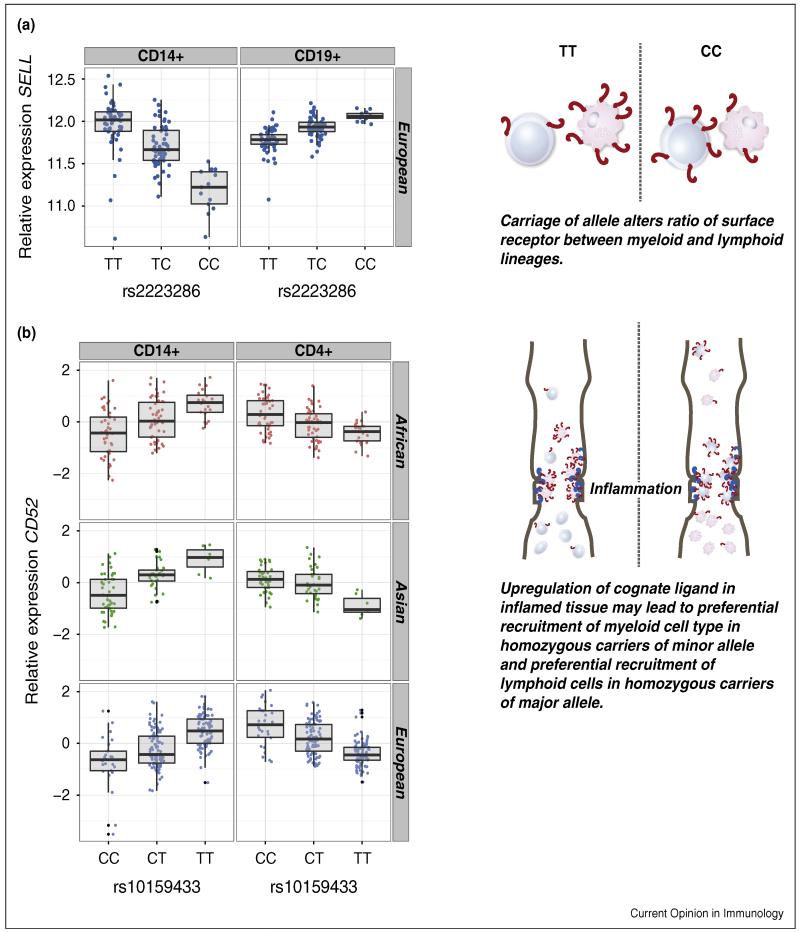
Peripheral blood mononuclear cells (PBMCs) form an easily accessibly primary substrate for eQTL analysis. However, they represent a heterologous assortment of multiple cell subsets, each with markedly different expression profiles and separate roles in immunity; the proportional composition varying inter-individually and intra-individually over time. eQTL for one cell subtype, which may only form a small proportion of total cell count, are at risk of not being detected upon analysis of whole blood or PBMCs. This is a primary motivator behind methods to identify cell specific eQTL. These involve attempts to bioinformatically deconvolute the signal of specific subtypes from whole blood [26], or use physical methods of separation. To date only a modest number of studies have been published interrogating different primary immune cell types from peripheral blood and tissues (Table 1), although this will be advanced by ongoing efforts such as the ImmVar project (www.immvar.org). From these initial studies it is apparent that specific analysis of lymphoid and myeloid cell types demonstrates widespread variation in eQTL activity according to cell type. Intriguingly, eQTL frequently demonstrate marked difference in effect sizes between different peripheral blood cell types, even when the gene expression is similar. In a few circumstances this effect is opposing in nature between cells, with polymorphisms simultaneously associated with increased expression in one cell type and reduced expression in an alternative cell type. Just as eQTL can elicit changes in the balance of gene expression between immune effector and regulatory genes within one cell type, this differential activity of immune eQTL may result in the altered balance of gene expression between different arms of the immune system. Where the gene encodes a surface receptor, differential expression might be anticipated to result in variation in proportion of cell subset types recruited by the respective marker ligand. Whilst this is currently untested, recent studies have demonstrated such effects for polymorphisms associated with contrasting effects between monocytes and T-cells on expression of CD52, encoding the target of the immunosuppressant Alemtuzumab, and the gene SELL encoding L-selectin, a protein which plays a crucial role in leukocyte trafficking to areas of inflammation [27] between B cells and monocytes (Figure 1). The CD52 observation is of significance in the field of immunosuppression in organ transplantation and in the management of multiple sclerosis where Alemtuzumab is used to deplete T-cells. A polymorphism regulating expression of surface CD52 between T-cells and monocytes might be expected to alter the proportional depletion of T-cells and monocytes. Carriage of the minor allele of rs2223286, a SNP intronic to SELL that disrupts a consensus motif for FOS and is located in a c-Fos ChIP-seq peak is associated with increased expression of SELL in B-cells and markedly reduced expression in monocytes [13•]. This opposing effect on expression from an eQTL has also been observed across tissues [51] and across contexts (discussed below). Study of such polymorphisms suggests that some alleles associate with increased variability in gene expression over different cell types and conditions, essentially regulating the relative genomic permissiveness of a polymorphism to affect expression (Figure 2).
Table 1. Examples of primary human cell types and tissues for which eQTL analyses have been reported.
| Study | Cell or tissue type | Condition | n | Ethnicity | Cohort | Ref. |
|---|---|---|---|---|---|---|
| Peripheral blood | ||||||
| Fehrmann et al. (2011) | Whole blood | Venesection | 1469 | European | UC (49); CeD (111); COPD (453); ALS cases and controls (856) | [14•] |
| Mehta et al. (2013) | Whole blood | Venesection | 322a, 740 + 653b | European | Population cohorts | [46] |
| Westra et al. (2013) | Whole blood | Venesection | 5311c, 2775b | European | 9 cohorts | [8••] |
| Goring et al. (2007) | PBMCs | Freshly isolated | 1240 | Mexican-American | San Antonio Family Heart Study | [47] |
| Zeller et al. (2010) | CD14+ monocytes | Freshly isolated | 1490 | European | Gutenberg Heart Study | [48•] |
| Fairfax et al. (2012) | CD14+ monocytes | Freshly isolated | 283 | European | Healthy volunteers | [13•] |
| Fairfax et al. (2014) | CD14+ monocytes | Unstimulated, LPS, IFNγ | 432 | European | Healthy volunteers | [9••] |
| Raj et al. (2014) | CD14+ monocytes | Freshly isolated | 401 | Multi-ethnic | Healthy volunteers | [41] |
| Fairfax et al. (2012) | CD19+ B cells | Freshly isolated | 283 | European | Healthy volunteers | [13•] |
| Raj et al. (2014) | CD4+ T cells | Freshly isolated | 407 | Multi-ethnic | Healthy volunteers | [41] |
| Ferraro et al. (2014) | CD4+ T cells | Freshly isolated | 168 | European | T1D (60), T2D (30), healthy controls (78) | [49] |
| Ferraro et al. (2014) | CD4 + FOXP3+ regulatory T cells | Freshly isolated | 168 | European | T1D (60), T2D (30), healthy controls (78) | [49] |
| Dimas (2013) | Mixed T cell population | PHA stimulated (from umbilical cord blood) | 75 | European | Newborns | [17••] |
| Lee (2014) | Monocyte-derived dendritic cells | Unstimulated, LPS, influenza A, IFNβ | 534 | European | Healthy volunteers | [25••] |
| Other cell and tissue types | ||||||
| Dimas et al. (2013) | Fibroblasts | From umbilical cord blood | 75 | European | Newborns | [17••] |
| Wagner et al. (2014) | Fibroblasts | From forearm skin | 62 | European | Healthy donors | [50] |
| Zhong et al. (2010) | Adipose tissue (omental) | Surgical resection | 916 | European | Gastric bypass surgery patients | [51] |
| Zhong et al. (2010) | Adipose tissue (subcutaneous) | Surgical resection | 870 | European | Gastric bypass surgery patients | [51] |
| Fu et al. (2012) | Adipose tissue (visceral) | Surgical resection | 77 | European | Elective bariatric surgery patients | [52] |
| Fu et al. (2012) | Adipose tissue (subcutaneous) | Surgical resection | 83 | European | Elective bariatric surgery patients | [52] |
| Ding et al. (2010) | Skin | Biopsy | 110 | European | Psoriasis patients (53), healthy controls (57) | [53] |
| Schadt et al. (2008) | Liver | Postmortem and surgical resection | 427 | European | Liver collections at tissue resource centres | [54] |
| Zhong et al. (2010) | Liver | Surgical resection | 707 | European | Gastric bypass surgery patients | [51] |
| Innocenti et al. (2011) | Liver | Surgical | 266 | European and African | Donor | [55] |
| Fu et al. (2012) | Liver | Surgical resection | 74 | European | Elective bariatric surgery patients | [52] |
| Myers et al. (2007) | Brain | Postmortem | 193 | European | Neuropathologically normal human brain samples | [56] |
| Hao et al. (2012) | Lung | Lung resection (non neoplastic sample pulmonary parencyhma) | 1111 | European | Patients undergoing lung surgery for cancer | [57] |
| Fu et al. (2012) | Skeletal muscle | Surgical resection | 83 | European | Elective bariatric surgery patients | [52] |
n, number of individuals in eQTL analysis. PBMCs, peripheral blood mononuclear cells; UC, ulcerative colitis; CeD, celiac disease; COPD, chronic obstructive pulmonary disease; ALS, amyotrophic lateral sclerosis; T1D, type 1 diabetes; T2D, type 2 diabetes.
Discovery.
Replication.
Meta-analysis.
Figure 1. Cellular specificity in observed eQTL.
(a) cis eQTL for SELL, encoding the cell surface adhesion molecule selectin L (CD62L) was found to show association with a clear directional effect between CD14+ monocytes and CD19+ B cells [13•]. (b) cis eQTL for CD52 show directionality between CD14+ monocytes and CD4+ T cells [41].
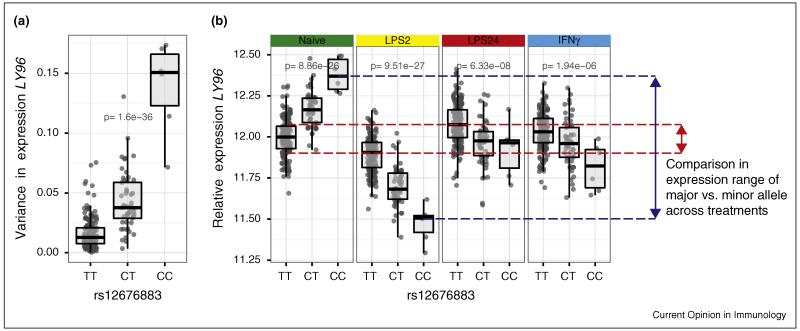
Figure 2. Genomic permissiveness in gene expression for an innate immune co-receptor gene.
LY96 (also known as MD2) encodes a co-receptor for LPS crucial for TLR4 activity. Variance in monocyte LY96 expression across treatments was analysed as a quantitative trait and correlated with underlying genotype. (a) rs1267688 forms a peak eQTL for LY96 expression. (b) This SNV is also most strongly associated with variance in expression over different states of induction of monocytes with LPS (2 or 24 h) or with IFNγ (24 h). A broad range is seen in expression levels for the minor allele dependent on environmental stimulus [9••] providing genomic permissivity in the population.
eQTL are dependent upon cellular context
Just as the activity of a regulatory variant may change across cell type, it can similarly vary within the same cell type according to cellular context. This property of eQTL was first shown in yeast with detection of trans effects interacting with environmental conditions [28•]. Immune cells, irrespective of function, are characterised by a continual high-degree of inter-cellular cross-talk and the capacity to mount prominent phenotypic changes in response to environmental cues such as encountering pathogens or inflammatory stimuli. These properties are predicated upon the dynamic regulation of genome-wide gene expression over short time-frames, often in orthogonal directions dependent upon the nature of stimuli. Immune related eQTL are therefore particularly likely to demonstrate context specificity and they may impact overall immune response.
This paradigm was illustrated in a study of 92 genetically diverse inbred mouse strains, which mapped genetic determinants of the transcriptomic response to inflammatory stimuli including lipopolysaccharide (LPS) applied to macrophages [29•]. This defined gene–environment interactions including context-specific expression hotspots in which variants modulated large number of genes in trans. Such analysis enabled identification of novel regulators of inflammation. In addition, mapping local eQTL facilitated identification of candidate genes within murine immune-related trait intervals including for atherosclerosis and susceptibility to Salmonella typhimurium.
Recent work in primary human immune cells has similarly highlighted the extent of local and distant genetic associations with gene expression that are specific to innate immune activation. We mapped induced eQTL for LPS and for interferon-gamma (IFNγ) in CD14+ monocytes from 432 healthy Europeans, demonstrating the temporal and stimulus specificity of regulatory variants [9••]. The analysis found that the majority of local eQTL were observed only in activated monocytes and included key nodal genes and effector molecules in canonical innate immune response signaling pathways. Stimulus-induced trans networks were identified, putatively driven by diverse mechanisms, including the transcription factor IRF2 subsequently validated by mapping IRF2 binding genome-wide, and a local eQTL for IFNB1 modulating an IFNβ cytokine network of genes developing in trans over time post LPS activation. Trans-associations were also found to map to coding variants in genes known to affect enzymatic activity and ligand binding. This approach additionally implied novel roles for transcription factors with NFE2L3 expression associating with an IFNγ inducible trans network composed almost entirely of proteasomal genes [10]. Stimulus induced eQTL were furthermore notably enriched for GWAS reported disease variants related to immunity and inflammation including bacterial infection.
Similar stimulus specificity was observed in human dendritic cells (DCs) in work by Lee and colleagues who studied genomic modulators of 415 differentially expressed genes in response to LPS, influenza A and IFNβ for a cohort of 534 individuals from different ethnic groups [25••]. The work resolved stimulus induced eQTL with evidence that this commonly involved allelic effects on activated transcription factors. Exemplar DC gene networks were revealed, as illustrated by a stimulus specific local eQTL for IRF7, which was found to be associated in trans with an interferon enriched gene set, validated with overexpression studies. Of particular relevance to immunity to infection, this study also resolved a local eQTL for NOD2 found to involve a SNP previously associated with leprosy after IFNβ exposure.
Insights from human disease: eQTL and Mycobacterium tuberculosis
For infectious disease, the ability to study response to exposure to particular pathogens ex vivo provides a tractable approach to define eQTL. Barreiro and colleagues illustrated this by mapping genetic modulators of the transcriptomic response to a virulent strain of Mycobacterium tuberculosis (MTB) in human DCs, cells that play a key role in anti-mycobacterial immunity [30]. Exposure to MTB for 18 h significantly induced immune response genes and, despite limited power (n = 65), identified 198 context-specific response eQTL that were enriched for key immune signaling genes such as RIPK2 and TNFRSF11A while also showing overlap with variants associated with pulmonary tuberculosis on GWAS including the MAPK phosphatase gene DUSP14.
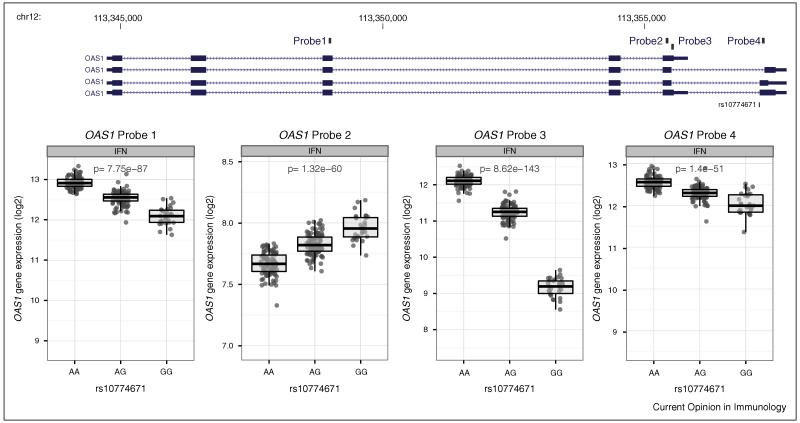
To address the extent to which associations with transcript abundance manifested at the protein level (pQTLs) the authors assayed 19 cytokines in cell culture supernatant. Whilst they did not find significant overlap, the application of new high-throughput approaches using mass spectrometry-based proteomic methods should empower investigators to more comprehensively investigate pQTLs [31]. This is of particular relevance in further investigating the effects of eQTL on cell surface receptor expression. Alternative splicing is critical to generation of proteomic diversity and may be modulated by underlying genetic variation. Such mechanisms are important to consider when defining eQTL (Figure 3) and will be facilitated by the increased resolution provided by RNA-seq.
Figure 3. Association with isoform specific gene expression for OAS1, a key IFN inducible gene involved in anti-viral defence.
In human monocytes exposed to IFNγ, a highly significant cis eQTL shows directional association by allele dependent on the interrogating Illumina expression probe [9••]. The associated SNP, rs10774671 is reported to modulate alternative splicing [42] and to be a risk allele for West Nile virus [43], antibody levels to measles mumps and rubella vaccination [44] and Dengue outcomes [45].
MicroRNAs (miRNAs) bind complimentary mRNA sequences, acting as post-transcriptional regulators with consequences for antimicrobial defence. miRNA expression is induced and evidence suggests genetic modulators of miRNA abundance [32-34,35••] have context dependent downstream outcomes. In a follow up study Barreiro and colleagues observed that 40% of 346 miRNAs assayed were regulated by MTB although only 3% of had evidence of local eQTL [36]. This relatively lower proportion of miRNAs modulated by regulatory variants compared to mRNA is similar to that observed in LCLs [35••], consistent with the proposal that miRNAs have a critical function in maintaining cellular and biological function in the face of internal or external perturbations [37].
Genetics of response to vaccination
Collection of longitudinal samples encompassing both the pre-morbid and the disease states can enable definition of context-specific effects. This approach is challenging however, although clinically relevant controlled interventions do exist, such as responses to drugs or vaccination, that can be studied in patients and healthy individuals. A key concern in vaccinology is identification of individuals who fail to respond to a vaccine and do not develop immunity. Vaccine response has a heritable component and context specific eQTL in key immune genes may underpin inter-individual variation to vaccine response. Franco and colleagues recently reported genomic determinants of response to inactivated trivalent influenza vaccine for a discovery and replication cohort (n = 119 and 128 respectively), recruited in sequential years on whom transcriptomic profiling of whole blood was performed on day 0, 1, 3 and 14 together with analysis of antibody titre response [38]. This enabled an integrated analysis in which the transcriptional response to vaccination, modulation by genotype and consistency of effect at the level of antibody response could be assessed. Multiple genes (n = 20) were identified with strong evidence of genotypic effects on immune response to vaccination. These included genes involved in antigen presentation and membrane trafficking. Such studies offer the promise of improving the understanding of determinants of vaccine immunogenicity and potential identification of predictive biomarkers of response together with enhanced vaccine development.
Conclusions
Immune related genes show high levels of inter-individual variation in expression, much of which is attributable to underlying heritable factors. eQTL analysis of primary immune cell populations from peripheral blood has been highly informative, although has underlined the context-specificity of such associations. Further work to consider at high resolution the immune cell phenotype being studied is required. Moreover, defining and modulating the environmental context appears critical given that stimulus specific or response eQTLs are common and informative for disease. Whilst a variety of algorithms exist to attempt to deconvolute underlying cell specific signatures, we believe that these will inevitably underestimate the true nature of eQTL and that investment is needed in efforts to continue to explore purified cell populations and extend this approach using single cell transcriptomics [39]. The widespread adoption of next generation sequencing technologies, notably RNA-seq, permits a greater dynamic range and resolution to be achieved in quantification of gene expression, including at splice isoform resolution, and definition of allele-specific differences in expression [35••]. In parallel, it is also possible to resolve non-coding RNAs and the epigenetic landscape within which regulatory variants act [40], as well as the extent of genetic diversity in both host and pathogen. The application to patient samples to resolve effects in the disease context will be important but challenging given disease heterogeneity. Nonetheless, integrating eQTL analysis with defined clinical interventions and longitudinal analysis may prove highly informative as highlighted by the successful application of eQTL analysis to understand inter-individual variation in response to vaccination.
Acknowledgements
We thank Towfique Raj for sharing data. This work was supported by the Wellcome Trust (Grants 074318 [J.C.K.], 088891 [B.P.F.], and 090532/Z/09/Z [core facilities Wellcome Trust Centre for Human Genetics including High-Throughput Genomics Group]), the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013)/ERC Grant agreement no. 281824 (J.C.K.), the Medical Research Council (98082 [J.C.K]) and the NIHR Oxford Biomedical Research Centre.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
• of special interest
- 1.Chapman SJ, Hill AV. Human genetic susceptibility to infectious disease. Nat Rev Genet. 2012;13:175–188. doi: 10.1038/nrg3114. [DOI] [PubMed] [Google Scholar]
- 2.Battle A, Montgomery SB. Determining causality and consequence of expression quantitative trait loci. Hum Genet. 2014 doi: 10.1007/s00439-014-1446-0. 10.1007/s00439-00014-01446-00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rockman MV, Kruglyak L. Genetics of global gene expression. Nat Rev Genet. 2006;7:862–872. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- 4•.Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22:1748–1759. doi: 10.1101/gr.136127.111. [This paper demonstrates the value of integrating genetic variation with functional genomic profiling (from the ENCODE Consortium) to identify putative regulatory variants and generate hypotheses regarding mechanism of action.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fraser HB. Gene expression drives local adaptation in humans. Genome Res. 2013;23:1089–1096. doi: 10.1101/gr.152710.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6•.Wright FA, Sullivan PF, Brooks AI, Zou F, Sun W, Xia K, Madar V, Jansen R, Chung W, Zhou YH, et al. Heritability and genomics of gene expression in peripheral blood. Nat Genet. 2014;46:430–437. doi: 10.1038/ng.2951. [This paper uses a twin design to quantify expression heritability and resolve eQTL in peripheral blood.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7••.Jansen RC, Nap JP. Genetical genomics: the added value from segregation. Trends Genet. 2001;17:388–391. doi: 10.1016/s0168-9525(01)02310-1. [A landmark paper establishing the concept of genetical genomics.] [DOI] [PubMed] [Google Scholar]
- 8••.Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, Christiansen MW, Fairfax BP, Schramm K, Powell JE, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [A significant paper demonstrating hundreds of trans associations, many with disease association, through large-scale meta analysis of eQTL from peripheral blood from over 5000 individuals.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9••.Fairfax BP, Humburg P, Makino S, Naranbhai V, Wong D, Lau E, Jostins L, Plant K, Andrews R, McGee C, et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science. 2014;343:1246949. doi: 10.1126/science.1246949. [This paper highlights the extent and nature of context-specific eQTL on innate immune activation of primary human monocytes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Heyningen V, Bickmore W. Regulation from a distance: long-range control of gene expression in development and disease. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120372. doi: 10.1098/rstb.2012.0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12•.Smemo S, Tena JJ, Kim KH, Gamazon ER, Sakabe NJ, Gomez-Marin C, Aneas I, Credidio FL, Sobreira DR, Wasserman NF, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–375. doi: 10.1038/nature13138. [An elegant study demonstrating how regulatory variants may act at a distance.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13•.Fairfax BP, Makino S, Radhakrishnan J, Plant K, Leslie S, Dilthey A, Ellis P, Langford C, Vannberg FO, Knight JC. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat Genet. 2012;44:502–510. doi: 10.1038/ng.2205. [This paper highlights degree of cell type specificity in primary immune cell populations.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14•.Fehrmann RS, Jansen RC, Veldink JH, Westra HJ, Arends D, Bonder MJ, Fu J, Deelen P, Groen HJ, Smolonska A, et al. Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS Genet. 2011;7:e1002197. doi: 10.1371/journal.pgen.1002197. [An important demonstration of the informativeness of trans-eQTL with particular charactertisation of those to the MHC.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, Schmidt MK, Chang-Claude J, Bojesen SE, Bolla MK, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45:353–361. 361e351–352. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Liang L, Morar N, Dixon AL, Lathrop GM, Abecasis GR, Moffatt MF, Cookson WO. A cross-platform analysis of 14,177 expression quantitative trait loci derived from lymphoblastoid cell lines. Genome Res. 2013;23:716–726. doi: 10.1101/gr.142521.112. [One of the largest analysis of eQTL in lymphoblastoid cell lines.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17••.Dimas AS, Deutsch S, Stranger BE, Montgomery SB, Borel C, Attar-Cohen H, Ingle C, Beazley C, Gutierrez Arcelus M, Sekowska M, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [A seminal early demonstration of the extent of cell type specificity in eQTL.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18•.Nica AC, Parts L, Glass D, Nisbet J, Barrett A, Sekowska M, Travers M, Potter S, Grundberg E, Small K, et al. The architecture of gene regulatory variation across multiple human tissues: the MuTHER study. PLoS Genet. 2011;7:e1002003. doi: 10.1371/journal.pgen.1002003. [Large-scale exploration of tissue-specific eQTL in a twin cohort.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19•.Stranger BE, Montgomery SB, Dimas AS, Parts L, Stegle O, Ingle CE, Sekowska M, Smith GD, Evans D, Gutierrez-Arcelus M, et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 2012;8:e1002639. doi: 10.1371/journal.pgen.1002639. [A key characterisation of differentiation in functional regulatory variation among diverse human populations.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao C, Joehanes R, Johnson AD, Huan T, Esko T, Ying S, Freedman JE, Murabito J, Lunetta KL, Metspalu A, et al. Sex- and age-interacting eQTLs in human complex diseases. Hum Mol Genet. 2014;23:1947–1956. doi: 10.1093/hmg/ddt582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vinuela A, Snoek LB, Riksen JA, Kammenga JE. Aging Uncouples Heritability and Expression-QTL in Caenorhabditis elegans. G3 (Bethesda) 2012;2:597–605. doi: 10.1534/g3.112.002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Idaghdour Y, Czika W, Shianna KV, Lee SH, Visscher PM, Martin HC, Miclaus K, Jadallah SJ, Goldstein DB, Wolfinger RD, et al. Geographical genomics of human leukocyte gene expression variation in southern Morocco. Nat Genet. 2010;42:62–67. doi: 10.1038/ng.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Idaghdour Y, Quinlan J, Goulet JP, Berghout J, Gbeha E, Bruat V, de Malliard T, Grenier JC, Gomez S, Gros P, et al. Evidence for additive and interaction effects of host genotype and infection in malaria. Proc Natl Acad Sci U S A. 2012;109:16786–16793. doi: 10.1073/pnas.1204945109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smirnov DA, Morley M, Shin E, Spielman RS, Cheung VG. Genetic analysis of radiation-induced changes in human gene expression. Nature. 2009;459:587–591. doi: 10.1038/nature07940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25••.Lee MN, Ye C, Villani AC, Raj T, Li W, Eisenhaure TM, Imboywa SH, Chipendo PI, Ran FA, Slowikowski K, et al. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science. 2014;343:1246980. doi: 10.1126/science.1246980. [Context specific eQTL for diverse stimuli in primary human monocyte derived dendritic cells across three populations.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Westra H-J, Arends D, Esko T, Peters MJ, Schurmann C, Schramm K, Kettunen J, Yaghootkar H, Fairfax B, Andiappan AK, et al. Cell specific eQTL analysis without sorting cells. bioRxiv. 2014 doi: 10.1371/journal.pgen.1005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wedepohl S, Beceren-Braun F, Riese S, Buscher K, Enders S, Bernhard G, Kilian K, Blanchard V, Dernedde J, Tauber R. L-selectin — a dynamic regulator of leukocyte migration. Eur J Cell Biol. 2012;91:257–264. doi: 10.1016/j.ejcb.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 28•.Smith EN, Kruglyak L. Gene-environment interaction in yeast gene expression. PLoS Biol. 2008;6:e83. doi: 10.1371/journal.pbio.0060083. [Early exploration of the genetic architecture of gene–environment interactions.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29•.Orozco LD, Bennett BJ, Farber CR, Ghazalpour A, Pan C, Che N, Wen P, Qi HX, Mutukulu A, Siemers N, et al. Unraveling inflammatory responses using systems genetics and gene–environment interactions in macrophages. Cell. 2012;151:658–670. doi: 10.1016/j.cell.2012.08.043. [An early demonstration of context-specific eQTL and gene-environment interactions for response of murine macrophages to inflammatory stimuli.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barreiro LB, Tailleux L, Pai AA, Gicquel B, Marioni JC, Gilad Y. Deciphering the genetic architecture of variation in the immune response to Mycobacterium tuberculosis infection. Proc Natl Acad Sci U S A. 2012;109:1204–1209. doi: 10.1073/pnas.1115761109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horvatovich P, Franke L, Bischoff R. Proteomic studies related to genetic determinants of variability in protein concentrations. J Proteome Res. 2014;13:5–14. doi: 10.1021/pr400765y. [DOI] [PubMed] [Google Scholar]
- 32.Borel C, Deutsch S, Letourneau A, Migliavacca E, Montgomery SB, Dimas AS, Vejnar CE, Attar H, Gagnebin M, Gehrig C, et al. Identification of cis- and trans-regulatory variation modulating microRNA expression levels in human fibroblasts. Genome Res. 2011;21:68–73. doi: 10.1101/gr.109371.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parts L, Hedman AK, Keildson S, Knights AJ, Abreu-Goodger C, van de Bunt M, Guerra-Assuncao JA, Bartonicek N, van Dongen S, Magi R, et al. Extent, causes, and consequences of small RNA expression variation in human adipose tissue. PLoS Genet. 2012;8:e1002704. doi: 10.1371/journal.pgen.1002704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gamazon ER, Ziliak D, Im HK, LaCroix B, Park DS, Cox NJ, Huang RS. Genetic architecture of microRNA expression: implications for the transcriptome and complex traits. Am J Hum Genet. 2012;90:1046–1063. doi: 10.1016/j.ajhg.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35••.Lappalainen T, Sammeth M, Friedlander MR, t Hoen PA, Monlong J, Rivas MA, Gonzalez-Porta M, Kurbatova N, Griebel T, Ferreira PG, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [A landmark paper using high-throughput RNA-sequencing data to define regulatory variation involving mRNA and miRNA in lymphoblastoid cell lines across human populations.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siddle KJ, Deschamps M, Tailleux L, Nedelec Y, Pothlichet J, Lugo-Villarino G, Libri V, Gicquel B, Neyrolles O, Laval G, et al. A genomic portrait of the genetic architecture and regulatory impact of microRNA expression in response to infection. Genome Res. 2014;24:850–859. doi: 10.1101/gr.161471.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Franco LM, Bucasas KL, Wells JM, Nino D, Wang X, Zapata GE, Arden N, Renwick A, Yu P, Quarles JM, et al. Integrative genomic analysis of the human immune response to influenza vaccination. Elife. 2013;2:e00299. doi: 10.7554/eLife.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wills QF, Livak KJ, Tipping AJ, Enver T, Goldson AJ, Sexton DW, Holmes C. Single-cell gene expression analysis reveals genetic associations masked in whole-tissue experiments. Nat Biotechnol. 2013;31:748–752. doi: 10.1038/nbt.2642. [DOI] [PubMed] [Google Scholar]
- 40.Rivera CM, Ren B. Mapping human epigenomes. Cell. 2013;155:39–55. doi: 10.1016/j.cell.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raj T, Rothamel K, Mostafavi S, Ye C, Lee MN, Replogle JM, Feng T, Lee M, Asinovski N, Frohlich I, et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science. 2014;344:519–523. doi: 10.1126/science.1249547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonnevie-Nielsen V, Field LL, Lu S, Zheng DJ, Li M, Martensen PM, Nielsen TB, Beck-Nielsen H, Lau YL, Pociot F. Variation in antiviral 2′,5′-oligoadenylate synthetase (2′5′AS) enzyme activity is controlled by a single-nucleotide polymorphism at a splice-acceptor site in the OAS1 gene. Am J Hum Genet. 2005;76:623–633. doi: 10.1086/429391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lim JK, Lisco A, McDermott DH, Huynh L, Ward JM, Johnson B, Johnson H, Pape J, Foster GA, Krysztof D, et al. Genetic variation in OAS1 is a risk factor for initial infection with West Nile virus in man. PLoS Pathog. 2009;5:e1000321. doi: 10.1371/journal.ppat.1000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haralambieva IH, Ovsyannikova IG, Umlauf BJ, Vierkant RA. Shane Pankratz V, Jacobson RM, Poland GA: Genetic polymorphisms in host antiviral genes: associations with humoral and cellular immunity to measles vaccine. Vaccine. 2011;29:8988–8997. doi: 10.1016/j.vaccine.2011.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alagarasu K, Honap T, Damle IM, Mulay AP, Shah PS, Cecilia D. Polymorphisms in the oligoadenylate synthetase gene cluster and its association with clinical outcomes of dengue virus infection. Infect Genet Evol. 2013;14:390–395. doi: 10.1016/j.meegid.2012.12.021. [DOI] [PubMed] [Google Scholar]
- 46.Mehta D, Heim K, Herder C, Carstensen M, Eckstein G, Schurmann C, Homuth G, Nauck M, Volker U, Roden M, et al. Impact of common regulatory single-nucleotide variants on gene expression profiles in whole blood. Eur J Hum Genet. 2013;21:48–54. doi: 10.1038/ejhg.2012.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goring HH, Curran JE, Johnson MP, Dyer TD, Charlesworth J, Cole SA, Jowett JB, Abraham LJ, Rainwater DL, Comuzzie AG, et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 48•.Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, Castagne R, Maouche S, Germain M, Lackner K, Rossmann H, et al. Genetics and beyond — the transcriptome of human monocytes and disease susceptibility. PLoS ONE. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [An important large-scale map of eQTL in primary human monocytes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferraro A, D’Alise AM, Raj T, Asinovski N, Phillips R, Ergun A, Replogle JM, Bernier A, Laffel L, Stranger BE, et al. Interindividual variation in human T regulatory cells. Proc Natl Acad Sci U S A. 2014;111:E1111–E1120. doi: 10.1073/pnas.1401343111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner JR, Busche S, Ge B, Kwan T, Pastinen T, Blanchette M. The relationship between DNA methylation, genetic and expression inter-individual variation in untransformed human fibroblasts. Genome Biol. 2014;15:R37. doi: 10.1186/gb-2014-15-2-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhong H, Beaulaurier J, Lum PY, Molony C, Yang X, Macneil DJ, Weingarth DT, Zhang B, Greenawalt D, Dobrin R, et al. Liver and adipose expression associated SNPs are enriched for association to type 2 diabetes. PLoS Genet. 2010;6:e1000932. doi: 10.1371/journal.pgen.1000932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fu J, Wolfs MG, Deelen P, Westra HJ, Fehrmann RS, Te Meerman GJ, Buurman WA, Rensen SS, Groen HJ, Weersma RK, et al. Unraveling the regulatory mechanisms underlying tissue-dependent genetic variation of gene expression. PLoS Genet. 2012;8:e1002431. doi: 10.1371/journal.pgen.1002431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ding J, Gudjonsson JE, Liang L, Stuart PE, Li Y, Chen W, Weichenthal M, Ellinghaus E, Franke A, Cookson W, et al. Gene expression in skin and lymphoblastoid cells: Refined statistical method reveals extensive overlap in cis-eQTL signals. Am J Hum Genet. 2010;87:779–789. doi: 10.1016/j.ajhg.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Innocenti F, Cooper GM, Stanaway IB, Gamazon ER, Smith JD, Mirkov S, Ramirez J, Liu W, Lin YS, Moloney C, et al. Identification, replication, and functional fine-mapping of expression quantitative trait loci in primary human liver tissue. PLoS Genet. 2011;7:e1002078. doi: 10.1371/journal.pgen.1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Myers AJ, Gibbs JR, Webster JA, Rohrer K, Zhao A, Marlowe L, Kaleem M, Leung D, Bryden L, Nath P, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 57.Hao K, Bosse Y, Nickle DC, Pare PD, Postma DS, Laviolette M, Sandford A, Hackett TL, Daley D, Hogg JC, et al. Lung eQTLs to help reveal the molecular underpinnings of asthma. PLoS Genet. 2012;8:e1003029. doi: 10.1371/journal.pgen.1003029. [DOI] [PMC free article] [PubMed] [Google Scholar]