Abstract
Over several decades, various forms of genomic analysis of the human major histocompatibility complex (MHC) have been extremely successful in picking up many disease associations. This is to be expected, as the MHC region is one of the most gene-dense and polymorphic stretches of human DNA. It also encodes proteins critical to immunity, including several controlling antigen processing and presentation. Single-nucleotide polymorphism genotyping and human leukocyte antigen (HLA) imputation now permit the screening of large sample sets, a technique further facilitated by high-throughput sequencing. These methods promise to yield more precise contributions of MHC variants to disease. However, interpretation of MHC-disease associations in terms of the functions of variants has been problematic. Most studies confirm the paramount importance of class I and class II molecules, which are key to resistance to infection. Infection is likely driving the extreme variation of these genes across the human population, but this has been difficult to demonstrate. In contrast, many associations with autoimmune conditions have been shown to be specific to certain class I and class II alleles. Interestingly, conditions other than infections and autoimmunity are also associated with the MHC, including some cancers and neuropathies. These associations could be indirect, owing, for example, to the infectious history of a particular individual and selective pressures operating at the population level.
Keywords: MHC, HLA, polymorphism, antigen presentation, antigen processing, imputation
INTRODUCTION
The major histocompatibility complex (MHC) has been studied for more than 60 years, and its early history is well documented (67). Serological typing uncovered associations between the MHC and many interesting immune phenotypes long before the cloning of class I and class II genes and determination of the structures of their encoded proteins. The molecular nature of the various immune response phenotypes mapping to the MHC was obscure until the 1980s, when DNA cloning and MHC class I structures emerged. The concept of T cell recognition of peptides in grooves was extremely appealing. It prefaced a number of findings that simplified a hitherto complex and confusing field: Diverse MHC phenotypes, from mixed lymphocyte reactions to suppressor T cells, all relate to the activities of a small number of class I and class II molecules.
The MHC region is associated with more diseases (mainly autoimmune and infectious) than any other region of the genome (93). Many associations were first detected by human leukocyte antigen (HLA) typing for either class I or class II (81). The extensive linkage disequilibrium meant that in some cases either class I or class II could be used to detect disease association, although pinpointing causative genes was more difficult. For example, hemochromatosis, a recessive iron storage disorder, is affected by mutations in the class I–related gene HFE (35). This condition was initially associated with HLA-A*03, which led some laboratories to start sequencing patients around HLA-A. Extensive analysis of cosmids later showed that the HFE gene is related to class I but maps to a location several megabases telomeric of the MHC.
The high gene density, extreme polymorphism, and clustering of genes with related functions, in addition to the strong linkage disequilibrium, continue to make it difficult to tease apart effects of individual loci. With some exceptions, diseases associate most strongly with various alleles of classical class I and class II loci, with weaker contributions from other MHC loci (50). Given this information, with modern molecular tools arising from genomics and immunology, we should be in a good position to understand the mechanisms underpinning MHC-associated disease. One difficulty is that, at the genomic level, it is still a major undertaking to fine map associations in this region and establish causal coding or regulatory variants. In addition, at the functional level, MHC class I and class II molecules are involved in so many diverse immune cell interactions that it has proved difficult to determine the stage at which the disease-associated allelic products act.
Most disease-associated variation in the MHC concerns subtle effects of common alleles—in other words, normal variation. Rare individuals without functional class I or class II molecules do exist and are severely immunocompromised, as is the case in bare lymphocyte syndrome types I and II (27, 110). There is still a poor understanding of (a) what drives MHC polymorphism; (b) which precise variants amid the sea of variation are functionally important, primarily in terms of resolving specific amino acid changes associated with disease; and (c) what the disease mechanism is at the molecular level. It is sobering to reflect that, after several decades, progress has really taken place only on the second of those items, and even then the progress has tended to be refinement more than novel insight. The classic papers on amino acid position 57 in HLA-DQB in type 1 diabetes appeared 25 years ago (114). The main genetic links to autoimmune diseases were traced to discrete changes in groove residues. At the same time, the machinery for loading peptides onto MHC molecules was revealed (59), components of which, such as transporter associated with antigen processing (TAP), were encoded within the MHC. These findings, remarkable though they were, have not led to an understanding of how peptides in MHC grooves lead to autoimmune disease and drive the unprecedented polymorphism (82).
FUNCTIONS OF MHC CLASS I AND CLASS II MOLECULES
The functions of MHC class I and class II are generally considered separately, although these molecules most likely have a common evolutionary history (38). Both sets of proteins bind peptides and present them to receptors on T cells. Class I molecules are additionally sensed by receptors on natural killer (NK) cells [such as killer cell immunoglobulin-like receptors (KIRs)] and by cells of the monocyte lineages [leukocyte immunoglobulin-like receptors (LILRs)] (14, 119). Some of these interactions are sensitive to peptides (33). It was initially believed that specific peptides from pathogens were presented by HLA molecules and recognized by T cells. Because there is a relatively small pool of T cell receptors, it is difficult to envisage how specificity is achieved in immune recognition. This is particularly intriguing because a single MHC molecule may in principle bind more than a million different peptides (133). In fact, the range of peptides that MHC molecules can bind may be more important than the specific peptides (82). Class I and class II functions were initially thought to be distinct, although CD74 (the invariant chain, Ii), which is classically associated with class II molecules, in some circumstances also binds with class I molecules. For a long time this was considered a curiosity, but it may now help to explain the rerouting of class I molecules for cross-presentation, the mechanism by which some cells appear to take up and process extracellular antigens on class I molecules for presentation to cytotoxic CD8 T cells (7).
GENES AND ALLELES
The IMGT/HLA Database (http://www.ebi.ac.uk/imgt/hla) contains a compilation of sequences and tools for logging, comparing, and naming HLA alleles. It is regularly updated, and an international committee meets periodically to standardize the nomenclature (102). The nomenclature has undergone radical updating in recent years to accommodate the large number of new alleles. In brief, each locus has its own designation followed by an asterisk (such as HLA-A* and HLA-DRB1*). These are followed by allele designations, which are unique numbers comprising up to four sets of digits separated by colons. The first four digits are most commonly used and are referred to as four-digit typing. The first two digits refer to the type, which in many cases corresponds to the serological antigen carried by an allotype (such as HLA-A*02). This is then separated by a colon from the next set of digits, which denotes the subtypes that differ in amino acid sequence (such as HLA-A*02:01 and HLA-A*02:02). Following another colon, a third set of digits may be used for synonymous (silent) nucleotide substitutions, and a fourth set may be used for changes in intronic regions. A suffix may also be added to indicate proteins with null (N) or low (L) expression and those that are secreted (S), cytoplasmic (C), aberrant (A), or questionable (Q).
ORIGIN AND EVOLUTION
Adaptive immunity may have appeared approximately 450 Mya in an ancestor of jawed vertebrates, after the introduction of a RAG transposon into an IgSF-encoding gene of a vertebrate ancestor. Duplication events then led to segmented Ig- and TCR-encoding genes (5). IgSF domains can be grouped into V, I, C1, and C2, and MHC molecules have a V-C1 arrangement that gave rise to class I and II. Which appeared first is debated (38). A prototypic MHC region that lacks class I and class II genes has been identified in amphioxus (16). A primitive MHC region may have been the birthplace of sets of genes in the immune system, as a center of an immunological “big bang” that seeded other immune system genes (3, 22). This idea is supported by the finding that β2-microglobulin, which associates with the heavy chain of class I molecules, is linked to class I and class II genes in the nurse shark (90). Approximately four regions of the human genome contain genes paralogous to those in the MHC. This led to the proposal that the MHCs of jawed vertebrates developed along with ancient chromosomal duplications to produce large-scale duplications of genome fragments, in accordance with the 2R hypothesis, which invokes two cycles of genome duplication during vertebrate evolution that were then followed by gene loss and rearrangement (60). The chicken MHC is held as an example of a “minimal, essential MHC” that contains a set of genes common to the MHCs of most other species without any additional loci (1, 62).
The MHC region appears to have undergone intermittent gene duplication and deletion in different species, generating related loci that produce similar proteins. The human class I region contains three genes, HLA-A, HLA-B, and HLA-C, which are not orthologous with genes in other species, including mice. Gene duplication produces genes that eventually acquire new functions. In some species, gene duplication has been prolific; pygmy mice, for example, have more than 100 class I genes, mostly pseudogenes (26).
Classical class I molecules are present in all classes of jawed vertebrates. Several class I–related genes have appeared at different stages of evolution. Class I–related NKG2D ligands such as MIC and ULBP have been found only in placental and marsupial mammals (132). CD1- and EPCR-encoding genes have a more ancient origin and are found in chickens and reptiles (80). Other class I–related sequences, including genes encoding HFE, neonatal Fc receptor, zinc-α2-glycoprotein, and MR1, have been identified only in mammals.
MHC ORGANIZATION
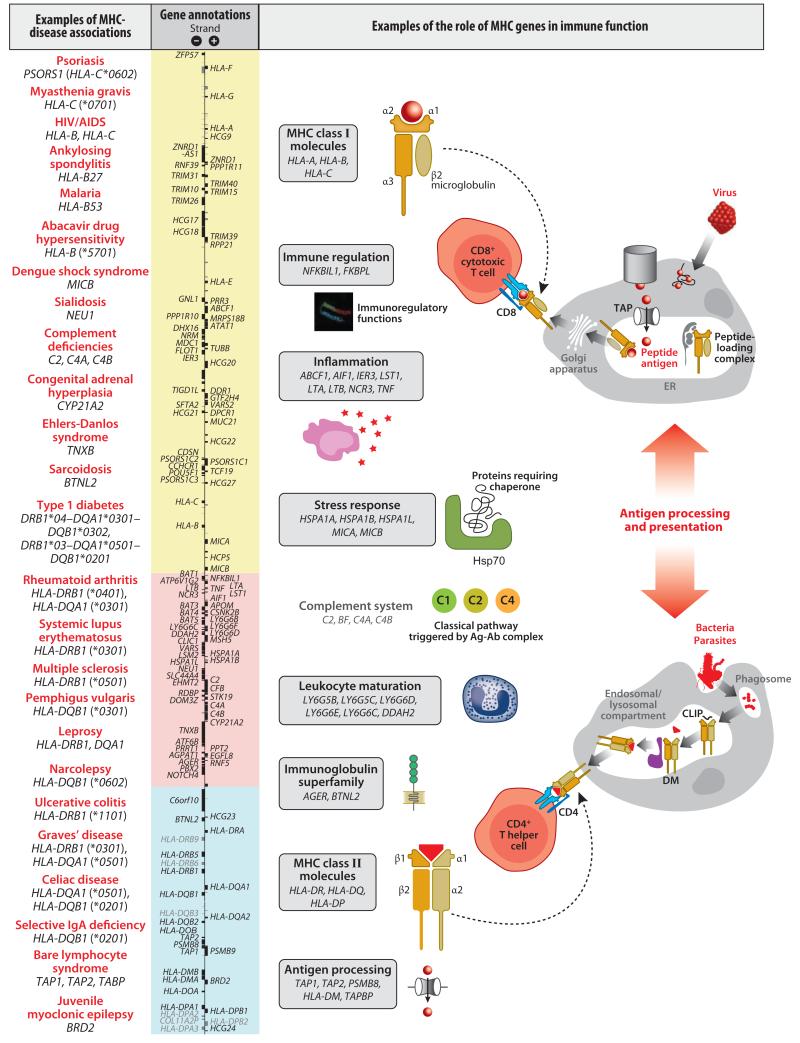
The clustering of antigen-processing and antigen-presenting genes in the MHC is consistent with the idea that the region evolved from a block of duplicated immune system genes. The human MHC was one of the first large genomic regions to be fully sequenced; it contains ~260 genes in a ~4-Mb span on chromosomal region 6p21.3 (Figure 1). Now that all of the genes have been clearly identified, other genomic features are of interest, including microRNAs (miRNAs) that may modulate gene expression (http://www.mirbase.org) as well as other features of the functional genomic landscape (Figure 1).
Figure 1.
Genomic landscape of the MHC. The classical MHC is shown on the short arm of chromosome 6 (base pair positions 29,640,000–33,120,000 from the Genome Reference Consortium Human Build 37, hg19), comprising the class I, II, and III regions. Transcription and chromatin states are illustrated for CD20+ normal human B cells using data from the ENCODE project (29). Constitutive expression of many MHC genes occurs in this cell type. Transcribed regions are shown by strand orientation for polyA+ RNA >200 nucleotides long from whole cells quantified by RNA-seq (red) based on short reads generated by the Illumina GAIIx platform. Separate tracks are shown for short total RNA (20–200 nucleotides long) (blue), with directional reads from the 5′ ends sequenced on an Illumina GAIIx. Chromatin accessibility is shown for the same cells based on DNase I hypersensitivity analyzed by DNase-seq (black), and is a useful guide to the location of putative regulatory regions. Data are also shown for a specific chromatin modification (H3K27ac) (green) for these cells analyzed by ChIP-seq. H3K27ac is an activating acetylation mark useful, for example, in identifying active enhancers. In terms of the recombination landscape of the MHC, data are shown for the deCODE recombination map (69) (dark brown), representing calculated rates of recombination (sex-averaged) using 10-kb windows. Vertebrate conserved elements are shown based on analysis of 46 species with prediction using PhastCons (107) (light brown). Sequence-level variation is shown for simple nucleotide polymorphisms, that is, single-nucleotide substitutions and small insertions and deletions (indels) found with at least 1% frequency in dbSNP. Variants are denoted in black except those in coding regions with synonymous variants (green), nonsynonymous variants (red), splice-site variants (red), and untranslated-region variants (blue). Remarkably high levels of polymorphism are seen, notably in classical HLA genes where variation is enriched in coding exons involved in defining the antigen-binding cleft. Structural genomic variants are also shown from the Database of Genomic Variants (54) involving segments of DNA larger than 1 kb. Copy number variants (CNVs) and indels are illustrated relative to the reference where gain in size (blue), loss in size (red), or both gain and loss in size (brown) have been reported. Structural variation is common in the MHC, including the RCCX module in the MHC class III region (comprising a number of genes, including RP-C4A/B-CYP21-TNXB), which may be duplicated or triplicated and present in different configurations, including two versions of the C4 gene (50). Other structurally complex sites include the HLA-DRB1 hypervariable region, which has five major haplogroups comprising variable numbers of functional genes and pseudogenes. All data tracks were downloaded from the UCSC Genome Browser (http://genome.ucsc.edu) (64).
The human MHC is generally used as the canonical arrangement, divided into three regions. The highly polymorphic classical class I genes HLA-A, HLA-B, and HLA-C and two clusters of nonclassical class I genes define the class I region. The class III region is the most gene-dense region in the human genome. It comprises many nonimmune genes as well as genes encoding critical mediators of innate immunity, including the tumor necrosis factor (TNF) and complement gene loci. With the possible exception of BRD2 (RING3), all genes in the class II region have functions related to antigen processing or presentation. In many species, this region houses the antigen-processing TAP-, LMP-, and tapasin-encoding genes.
As expected, nonhuman primate MHCs are similar to the human MHC, although they have different numbers of MIC-related genes associated with class I. Chimpanzees have a single fused MICA/MICB gene, and MIC genes may not be orthologous and are duplicated in less related primates. Interestingly, mice lack MIC-related genes but have other NKG2D ligands encoded on different chromosomes. In humans, only two MIC genes are functional, MICA and MICB, both centromeric to HLA-B. MIC genes are highly polymorphic (102). The MICA*008 allele is quite common in some populations, although it encodes a truncated protein with altered basolateral sorting (113). Human cytomegalovirus (HCMV) UL142 prevents surface expression of MICA molecules on infected cells, and MICA*008 is resistant to downregulation, which may explain the prevalence of the allele (131).
The mouse MHC has a similar organization to the human MHC, except for an extra classical class I locus centromeric to the class II region. Different mouse strains have altered numbers of additional class I loci.
Other species can differ widely in the number and arrangement of genes (63). Cats lack DQ genes but have expanded DR genes. Similarly, cows and sheep have replaced DP genes with DI/DY and have variable numbers of DQ loci. All mammals have MHC class I, but the lack of orthology suggests multiple rounds of duplication and gene loss in evolution. In contrast, the orthology of the main class II sequences is obvious. The chicken MHC, the B locus, is extremely small (92 kb) and contains only ~19 genes—the “minimal, essential MHC” (62). The class III region is outside the class I region and includes a histone-encoding gene and C4 loci. The tapasin-encoding gene (TAPBP) is located within the chicken class II region. Chicken TAP genes are close to class I, providing evidence of functional coevolution. In rats there also appears to be integration of functions of allelic products encoded in cis. In other words, alleles of peptide transporters are linked in cis on haplotypes with appropriate class I allotypes for receiving peptides pumped by that version of the transporter (92). There seems to be coevolution of TAP and class I alleles, as is also postulated for polymorphic chains of heterodimeric molecules (13). In both cases, it may be advantageous to encode the interacting partners in the same genetic cluster so that they can evolve in a concerted fashion. MHC-linked proteasome genes are missing in chickens, but the MHC contains putative NK cell receptor–related genes that may form a genetically linked ligand/receptor pair. Other class II α genes are separate from the MHC in chickens. The MHCs in fish, which are probably the most abundant vertebrates, also differ from the human and mouse gene arrangements, particularly in regard to the separation of class I and class II. Interestingly, cod appear to have lost class II functions altogether (109).
Class I genes appear to have evolved from different lineages in different species. This is clearly evident in monotremes, marsupials, and eutherians (placental mammals). Class II genes, in contrast, appear to have arisen from a common ancestor in the main mammal groups (9).
In summary, the MHC exhibits great diversity among different species by loss and gain of loci, and there are many different models: In mole rats, the class II DR loci are deleted and the DP genes are duplicated; cats lack the DQ subregion and do not express DP but have expanded DR genes; ruminants lost the DP subregion but have novel class II DI/DY genes; and cod have lost class II altogether. It has been proposed that MHC regions have undergone large-scale duplication events, leaving paralogous sets of genes on chromosomes 1, 9, and 19 in humans.
IMMUNE SYSTEM GENES IN THE MHC
The MHC contains many genes with putative immune functions (Figure 2). It is possible that linkage of these genes to MHC class I and class II is of functional importance in tuning immune responses. For example, the TNF gene is encoded within the class III region, flanked by lymphotoxin α– and lymphotoxin β–encoding genes (LTA, LTB). TNF is an important proinflammatory cytokine that affects cell proliferation and differentiation; it regulates a broad range of biological activities, particularly inflammation, and is implicated in both innate and adaptive immune responses. The variation in TNF genes occurs in regulatory regions, such as transcription factor and enhancer binding sites, which influence expression levels of TNF and thus the serum level (96). Many studies have indicated a link between variation in the TNF locus and disease (56), but the field remains controversial, and establishing causal relationships between alleles and functions independent of linked variants has been difficult.
Figure 2.
The MHC, disease, and immune function. The MHC shows associations with almost all known autoimmune diseases as well as many inflammatory and infectious diseases. Major disease associations are listed by trait. Recent high-resolution SNP typing has defined specific SNP markers in some instances, but associations defined by HLA type remain robust; examples of top associations are shown. Extensive linkage disequilibrium has made fine mapping such associations challenging. In some instances, associated haplotypes span several megabases of the classical MHC. Examples of the role of MHC genes in immune function are illustrated, including the key role in antigen presentation and processing as well as inflammation, the complement cascade, and stress response (51). The tapasin-encoding gene (TAPBP), which is involved in peptide loading onto MHC class I and in the association of MHC class I with TAP, is not shown here but is just outside the class II region shown.
Other examples include components of the complement cascade encoded by the C2, BF, C4A, and C4B genes together with the TAP and LMP genes encoding the immunoproteasome. Knockout of both the latter genes and the other immunoproteasome subunit encoded outside the MHC results in changes in antigen presentation (66). Genes involved in the stress response include HSPA1A and HSPA1B (encoding the molecular chaperone heat shock protein 70) and the MICA and MICB genes, which, as discussed above, encode stress-inducible ligands for NKG2D.
POLYMORPHISM
Haldane first drew attention to the role of infection in driving polymorphism (73). Most genes in the genome have a modest number of variants—perhaps two or three major alleles, with the remainder being uncommon or rare variants. Contrast this with HLA-B, the most polymorphic human MHC gene, which as of 2012 was known to comprise more than 2,000 alleles (http://www.ebi.ac.uk/imgt/hla), several orders of magnitude greater than the number of alleles for the vast majority of genes. Most of the variation under positive selection encodes the peptide-binding grooves of HLA molecules, and most classical MHC class I and class II genes are polymorphic in this region (Figure 1), with the exception of HLA-DRA.
Several areas of the MHC, such as the class I region, appear to have undergone repeated duplication and deletion, possibly owing to retroviral activity. The number of loci in the class II region, particularly the number of HLA-DRB loci, varies for different haplotypes. In the class III region, a unit called RCCX contains complement component C4 as well as cytochrome P450 21-hydroxylase–encoding genes (CYP21). The C4 copy number ranges from two to six. Systemic lupus erythematosus risk increases when haplotypes carry two copies of C4 and decreases when they carry more than five copies (134).
Regarding the generation of the observed genetic diversity, it seems unlikely that point mutation would be common, because it depends on chance. A more likely mechanism is allele conversion. Here, variation in peptide-binding pockets may be exchanged between allotypes by recombination. A single crossover between two alleles will result in a hybrid allele. A double crossover could exchange a small part of the groove, even a single pocket. These mechanisms could efficiently combine sections of existing variation in novel ways. Another mechanism that has been described by studying the so-called bm mutants of mice is gene conversion, where class I genes appear to donate stretches of sequence to other, related class I loci (88). The mechanism here could be misalignment at meiosis of related sequences, allowing nonallelic homologous recombination. Some of the resulting novel sequences could become established in the population by genetic drift. However, the high level of nonsynonymous mutations in MHC sequences encoding the peptide-binding grooves suggests strong selection.
There are two main models of the selection mechanism: balancing selection/heterozygote advantage and frequency-dependent selection (91). The first model is understandable in the context of a virus, such as HIV, that produces escape variants at a significant frequency. A heterozygous HLA-B*27/*57 individual would present two obstacles to escape from as compared with either homozygote. In the second model, as a virus escapes presentation by a common allotype, individuals possessing a rare type would be at a selective advantage and would expand in number. Once this in turn becomes the most common allotype, individuals possessing it would also eventually be the more likely target of escape. According to classic genetic theory, selection takes place at the level of the individual. However, MHC variation in a population may provide a form of herd immunity, as the larger the number of variants there are, the less likely it is that an infection will spread.
Resistance to disease is thought to drive MHC variation, but evidence for this in humans is limited. There are relatively few good examples of infectious diseases associated with MHC markers, whereas there are numerous examples of associated autoimmune conditions. This may be explained partially by the fact that microorganisms comprise many proteins, which therefore exhibit many different peptide epitopes for binding to MHC grooves. Any specific MHC molecule may not necessarily be an obvious disease-resistant allele given that many different peptides are available for presentation. Some of the best models for the association of the MHC with resistance to disease have come from studies of chickens. The peptide-binding specificity of the dominantly expressed class I molecule in different chicken haplotypes correlates with resistance to tumors caused by Rous sarcoma virus, and cell surface expression level correlates with susceptibility to tumors caused by Marek’s disease virus (61).
In spite of the proposed importance of the MHC for disease resistance, some species appear to be relatively monomorphic. Cheetahs are a celebrated example, although whether they are actually monomorphic has been questioned (17). It is not clear whether they have more effective ways of resisting disease, whether their lifestyle and habitat preclude infectious disease spread, whether they are indeed vulnerable to infection and extinction, or whether the data are wrong.
There are some suggestions that the polymorphism is driven by mechanisms other than resistance to infection. Data suggest that female sticklebacks prefer males with a large number of MHC alleles (101). In more recent work, the introduction of different parasites into stickleback populations resulted in reduced numbers of fish with certain MHC haplotypes in the next generation. Curiously, selection was not obviously associated with the parental generation, as there was no reduction in the numbers of fish with vulnerable haplotypes. This work appears to support the notion that the MHC serves as a cue to promote outbreeding. The proposal that the MHC leads to olfactory cues that aid mate choice in mice, and even in humans, is an old one. A number of alternative functions have been described for MHC molecules. A case can be made that both reproductive fitness and resistance to infection exert selection on MHC variation. MHC class I molecules have been proposed as playing a role in neuronal plasticity (105) and olfaction (45), best demonstrated so far in experimental animals that lack class I molecules or β2-microglobulin. It is difficult to rule out effects of cryptic infection in mice with class I knocked out. Interestingly, class I molecules also appear to govern maternal-fetal interactions in humans through interaction of KIRs with fetal polymorphic HLA-C molecules (20).
There is considerable additional variation in regions flanking the class I and class II polymorphic regions (117). Indeed, the divergence of some haplotypes in areas of class I and class II may be >20-fold higher than the divergence in other areas of the genome. This is consistent with independent haplotype evolution long before human speciation (41). Areas of extreme divergence also tend to flank polymorphic class I and class II loci, which is proposed to be due to hitchhiking of neighboring neutral variation with strong selection for variant peptide-binding groove sequences. These ideas led to the proposal that the flanking regions accumulate deleterious mutations, with consequences for MHC-linked disease (106). An attractive hypothesis to explain this variation was expressed as “Associative Balancing Complex evolution” (123). Essentially, the idea is that the MHC is relatively rarely expressed in a homozygous state, leading to weak purifying selection. Groove variation is certainly not the only consideration, and regulatory mutations affecting expression levels or differential splicing could play a role. The best example of this is HLA-C levels in HIV infection (72).
HAPLOTYPES
The marked linkage disequilibrium in the MHC led to the concept of “polymorphic frozen blocks” (24). These could be explained by several different mechanisms. First, they could have arisen simply by recent expansion of large families in isolated populations combined with insufficient time for recombination. Alternatively, recombination may be unfavorable because of sequence restraints. A mechanism has been proposed that invokes reduced pairing at meiosis of genomic regions exhibiting marked variation. Overall recombination is lower in the MHC region compared with the rest of the genome. Where recombination has occurred between divergent haplotypes, it may juxtapose highly diverged blocks next to highly conserved blocks. This is clearly useful for disease mapping. Another attractive explanation for allelic blocks is clustering of alleles of proteins that work together—in other words, maintenance of functionally coordinated sets of alleles. The classic example is TAP transporters linked to class I alleles they serve best, as exemplified by chickens and rats (92, 126). As a number of MHC genes have immune-related functions, epistasis is likely.
In some cases, these haplotypic blocks may be very long, spanning several megabases across the classical MHC (sometimes referred to as ancestral or extended MHC haplotypes) (86, 127, 135). In European populations, a number of such haplotypes are present at a relatively high allele frequency, suggesting a potential selective advantage, although in current human populations possession of such haplotypes appears to be predominantly deleterious and associated with a number of autoimmune diseases. The full sequence for eight such disease risk haplotypes is available through the MHC Haplotype Project (50) and discussed in more detail below.
DISEASES ASSOCIATED WITH THE MHC
Several Mendelian disorders have been detected through HLA typing. These have been explained largely as genes within or linked to the MHC region. For example, congenital adrenal hyperplasia is due to mutations in the class III gene CYP21 (130) (Figure 2).
The key question is which infections are, or have been, responsible for driving MHC variation. This group would include ancient diseases, although some of the best evidence for a direct link between infection and polymorphism comes from a more modern condition, AIDS. Several papers have pointed to escape variants of HIV-1 through the generation of peptide sequences that evade T cell recognition or influence KIRs or LILRs (15, 53, 75, 87). To understand how diseases drive MHC polymorphism at the genetic level, the issue may be divided into two aspects: generation of the variation and selection for the variants in the population. These simple models, which are not mutually exclusive, raise some interesting considerations.
Some viruses, as well as many tumors, employ strategies to downmodulate HLA expression so that they escape from T cell recognition. This may also underlie the curious phenomenon of infectious transfer of canine and Tasmanian devil tumors from animal to animal without immune rejection (8). The HLA-C locus is not downmodulated by HIV. This locus is only modestly polymorphic, is expressed at a lower level than HLA-A and HLA-B, and exhibits interaction with a set of receptors on NK cells, the two domain KIRs. Genome-wide association study (GWAS) screens for low HIV-1 viral load identified the region near the HLA-C gene as being critically important. Variation 35 kb upstream of HLA-C [single-nucleotide polymorphism (SNP) rs9264942, −35 C/T] was associated with control of HIV-1. Possession of this SNP correlated with levels of HLA-C mRNA transcripts and cell surface expression. It turned out, however, that the −35 SNP is not a causal variant; rather, it is in linkage disequilibrium with another variation that more directly influences HLA-C expression. The variation in question is in the 3′ end of the HLA-C gene and influences binding of a miRNA, has-miR-148a (71). Binding of the miRNA to its target site results in lower surface expression, adding another level of diversity in addition to the polymorphism of HLA-C over its peptide-binding groove. Contrary to expectations, HLA-C expression level, rather than specific alleles, may have the greatest influence on HIV control (5a). Recent work indicates that many HLA-B alleles are also resistant to downregulation by HIV Nef (97).
When considering associations of MHC markers with infections, it is important that in many cases the disease phenotype being measured is not necessarily susceptibility; rather, it may be a complication following infection. An example is hypovolemic shock (dengue shock syndrome), a life-threatening complication of dengue, which is associated with MICB variation (65). The HLA haplotype also has a significant effect on the outcome of human T-lymphotrophic virus type I (HTLV-1) infection (57). These effects are not surprising, given the complex and far-reaching effects the MHC has on immune responses (44).
GWAS for autoimmune conditions have implicated many genes and markers, although few associations are as significant as those with the MHC, and the effect sizes observed here are considerably greater. Many of these conditions are associated with a particular set of class I or class II alleles, consistent with the involvement of specific peptides, although in most cases the peptides have not been identified. Genomic analysis is generally consistent with direct implication of class I and class II, although in many cases this does not relate to a single allele. One of the best examples is narcolepsy, which causes disabling daytime sleepiness. The condition is associated with HLA-DR15 in all populations studied, and HLA-DQB1*0602 has been identified as the primary association (18, 85). The disease is characterized by destruction of a small set of hypothalamic neurons associated with the peptide hypocretin/orexin, which is important for sleep.
In most other cases it has been difficult to identify the key genetic association, for at least three reasons: the density of MHC genes, the strong linkage disequilibrium, and the effects of multiple HLA loci. These problems have confounded accurate disease mapping in the HLA region for several decades. Larger studies now generate sufficient statistical power to uncover independent HLA class I and class II associations, as has been done for type 1 diabetes, multiple sclerosis, and systemic lupus erythematosus. The precise MHC genes responsible for the inflammatory skin disease psoriasis have proved more difficult to pursue. Psoriasis was associated with HLA-C more than 30 years ago. A possible explanation of why this disease was difficult to analyze is that other MHC genes are involved in addition to HLA-C, including the nearby HLA-B and C6orf10 genes. Evidence has also been found for interaction involving HLA-C and a non-MHC gene, ERAP1, such that variants in ERAP1 exert an effect only in people with psoriasis who carry the HLA-C risk allele (112). This is biologically plausible because ERAP1 encodes an endoplasmic reticulum aminopeptidase involved in peptide trimming before HLA class I presentation. Interaction with ERAP1 was also found for ankylosing spondylitis, in which the disease association is restricted to individuals with HLA-B27 (31).
Sarcoidosis is a further example of an MHC-associated autoimmune disease in which linkage disequilibrium has made resolving specific variants and causal genes challenging. Several reports of an association between this disease and BTNL2 are confounded by the proximity of HLA-DR3 (HLA-DRB1*03), which is very strongly associated (120).
These issues make genetic analysis of the MHC more problematic than screening for markers on other chromosomes. Most autoimmune conditions are multifactorial and involve environmental triggers as well as many gene variants. Multiple sclerosis provides a good example of the complexity of analyzing these factors. Multiple sclerosis is associated with HLA-DRB1*1501, and susceptibility may be increased by low exposure to sunlight, resulting in low levels of vitamin D. Interestingly, vitamin D response elements crop up around the HLA-DRB1 gene. HLA-DR expression may be influenced by vitamin D because of this element, linking genetic and environmental influences (43, 98).
Imputation of classical HLA type to four-digit resolution based on high-density SNP genotyping is an accurate, fast, and cost-effective alternative to conventional serological testing that is suitable for analyzing large cohorts (74), and is discussed in more detail below. This should greatly facilitate ongoing efforts to fine map disease associations involving the MHC and allow for integration with genome-wide association analysis. This is important for polygenic diseases notably involving more than one MHC gene (for example, type 1 diabetes and multiple sclerosis) and potential interactions with non-MHC genes.
Interestingly, some acute drug reactions are associated with specific HLA allotypes (11). Perhaps the most critical relationship is between abacavir and HLA-B*57:01. HLA-B*15:02 is associated with carbamazepine sensitivity, which is linked to Stevens-Johnson syndrome in Han Chinese. These reactions are particular to single MHC specificities. Recent work has provided insight into the nature of these drug sensitivities. HLA-B*57:01 was purified from cells treated with abacavir (55). Unmodified abacavir purified along with this HLA molecule and peptides eluted from it, with noncanonical residues at position 2. It was proposed that abacavir bound specifically to the antigen-binding cleft of HLA-B*57:01, resulting in stimulation of a novel set of diverse T cell clones. In contrast, T cells from carbamazepine-induced sensitivity were reported to invoke a narrow range of specific T cells (68). A crystal structure of abacavir in combination with HLA-B*57:01 with peptide showed that the drug noncovalently binds to the class I molecule (55). These data indicate that small-molecule drugs noncovalently bind to specific HLA molecules, altering the peptide repertoire. In this study, the authors pointed out that the findings could have implications for understanding the initiation of autoimmunity by breaking tolerance. A similar mechanism could apply in relation to HLA-DR-restricted, gold-specific T cells in rheumatoid arthritis patients treated with the element (103). This may also apply to the link between the genetic association and T cell activation in beryllium disease and sensitization (12, 21).
Some MHC specificities are associated with cancers of a suspected viral etiology, including nasopharyngeal carcinoma and Hodgkin’s lymphoma. There is considerable evidence that MHC class I plays a role in controlling cancer. Viral escape from MHC class I presentation is important, and class I expression plays a role in cancer surveillance (8, 70).
Other conditions have been linked to the MHC region but are not obviously due to peptide presentation. The associated genes include BRD2, which is linked to type 2 diabetes (129). There have been reports of schizophrenia being associated with MHC class I, which may be related to the findings that some class I molecules are associated with neural plasticity (105). Some forms of Parkinson’s disease are associated with class II alleles, and studies have demonstrated associations with independent markers in the class II region that are not in strong linkage disequilibrium with one another (42, 47). The region is also linked to other psychoneurological conditions that are not obviously related to immunity, such as smoking behavior (39). In the latter case, the linkage may be associated with odorant receptor genes outside the MHC region (104). These disorders are clearly affected by many factors, including infection, which may be associated with conditions such as schizophrenia and autism (116). Coronary artery disease has also been associated with the HLA-C region (23). One way to view these interesting links is that the infectious history of an individual is critical for the development of his or her immune repertoire, which may have a knock-on effect on other conditions, from aging to neurological conditions, later in life. Some cases of neurological disorder may relate to infection, autoimmunity, or an inflammatory immune response.
CIS- AND TRANS-REGULATORY EFFECTS OF HLA ALLELES
There are major haplotype-specific differences reported for MHC gene expression (122). This is most significant in relation to the zinc-finger protein–encoding gene ZFP57. This work also demonstrated marked haplotype-specific differences in splicing as well as in transcription from intergenic regions in the MHC.
Regulatory variants may be specifically mapped by looking for association between genotype and gene expression. The idea is that genetic variants that affect expression of genes mapping in their vicinity [cis–expression quantitative trait loci (cis-eQTLs)] can be used to identify the true disease gene and separate it from associated variation. The MHC is enriched for such associations (121), although sequence variation in this region may confound cis-eQTLs from conventional microarrays when it occurs in regions to which probes hybridize.
In addition to these cis effects, SNPs may also affect expression of unlinked genes, including genes on different chromosomes—so-called trans-eQTLs. Fehrmann et al. (36), for example, noted that 48% of trans-acting SNPs associated with quantitative traits mapped within the HLA region, and Fairfax et al. (34) found that specific HLA alleles exhibited trans-association with the expression of specific genes in monocytes. Both groups found a striking example in AOAH, encoding acyloxyacyl hydrolase lipase, which degrades bacterially derived lipopolysaccharide associated with the class II region. Fairfax et al. determined that expression of this gene in monocytes is related to the presence of HLA-DRB1*04, *07, and *09, which are on haplotypes that contain the DR53 gene. Another gene, ARHGAP24, involved in actin remodeling, is also influenced by the same haplotypes. These effects may be very indirect, but they nevertheless suggest a central role for individual differences in the MHC in controlling responses and inflammatory processes over and above direct effects of antigen presentation to T cells.
RECEPTORS ON CELLS OTHER THAN T CELLS
Class I and class II molecules are sentinels of infection that report to receptors on T cells. In addition to these receptors, proteins on the surfaces of NK cells and cells of the myeloid lineage have specific receptors for class I. NK cell surface receptors include both inhibitory and activating molecules. Most are expressed in a variegated, stochastic fashion, allowing for different patterns of expression on different NK clones. NK cells are able to discriminate targets expressing different class I ligands, creating a repertoire of functionally different NK cells without a need for the somatic rearrangement typical of T cell receptors. Engagement of these receptors by intact class I molecules inhibits further action, but if class I is missing, as in some viral infections and tumors, then activation takes place, leading to cytokine production and in some cases the death of the target (125).
Human class I receptors on NK cells are from two classes of protein. The C-type lectin receptors are NKG2/CD94 heterodimers that recognize the nonclassical HLA-E molecule. KIRs recognize classical class I molecules. These receptors may be activating or inhibitory in character. According to the missing-self hypothesis, cells that express a class I ligand for these receptors are resistant to NK killing, whereas loss of class I after viral infection or tumorigenesis to escape T cell recognition results in NK activation (58). The response of the NK cells seems to be controlled by the balance of engagement of different activating and inhibitory receptors. The LILRs on myeloid/monocyte lineages also engage class I ligands to control macrophage or dendritic cell modulation (14). Both KIRs and LILRs may be found on some cells.
Interestingly, KIRs and LILRs are encoded in a complex array of genes, the leukocyte receptor complex (LRC) (6). This complex is on chromosomal region 19q13.4, so alleles of these highly polymorphic genes are inherited independently of HLA ligands, which may have consequences for disease association. Combinations of HLA and KIR alleles have been associated with viral infection, malaria, autoimmune disease, cancer, transplantation, and complications of pregnancy (46, 48, 77, 79, 124). Similarly, HLA/LILR combinations are associated with the outcome of HIV infection (52). LRC haplotypes exhibit significant variation in sequence and particularly in copy number, which may be generated by nonallelic homologous recombination (89, 118). There is evidence for coevolution of combinations of KIR and HLA variants (108).
SNPs AND STATISTICAL IMPUTATION MAY REPLACE CLASSICAL HLA TYPING FOR SOME APPLICATIONS
Early studies of MHC disease associations relied on laborious serological typing that used sera from multiparous women and later replaced some of the reagents with monoclonal antibodies. Sequence analysis revealed that the painstaking serology, which involved international networks, had accurately distinguished tiny allelic differences. DNA typing took over from serology, and today most clinical HLA typing, to four digits, is based on polymerase chain reaction (PCR). These methods remain costly and laborious. SNP typing was introduced to replace classical typing, although if applied to the MHC, as it was in GWAS screens, it had some limitations (25, 78, 86).
More recently, genotype imputation may overcome these problems. The goal is to access large reference panels where SNP genotypes and classical alleles have been determined. An algorithm is then used that effectively relates SNP patterns to HLA alleles. This is particularly challenging in the MHC because of the large amount of genetic variation and the extensive linkage disequilibrium in relation to different populations (32). Leslie et al. (74) pointed out four ways in which conventional SNP tagging falls short for HLA typing: (a) Many HLA alleles are rare, so combinations of common SNPs do not provide sufficient resolution; (b) some HLA alleles are embedded in different haplotypes; (c) the sheer number of alleles calls for typing a large number of SNP tags; and (d) SNP tags fitted in small, focused studies may not transfer to future projects, particularly with different populations. They proposed that the haplotype structure and linkage disequilibrium may be used to advantage because if two haplotypes share extensive SNP identity over the locus, then they are identical by descent and would most likely share the same HLA allele. Simulations indicated that each allele may be predicted from the combination of haplotypes on which it occurs. The simulations used training data from samples with European and African ancestry—an important consideration, as some extended HLA haplotypes and specific HLA alleles are notoriously population specific. The method was then validated on another large sample set. Approximately 100 SNPs were needed to predict HLA alleles to two digits with ~95% accuracy.
This pioneering study was relatively small. The authors suggested that 10 copies of an allele in a database would be necessary. Because there are well over 2,000 unique class I and class II alleles, a database of ~22,000 individuals would be needed to cover 10 copies of each allele. In practice, however, given the low frequency of some alleles, 2,000 samples chosen to globally represent major alleles might suffice. Clearly, there are some limitations to using SNPs. One problem concerns the propensity of HLA alleles to undergo gene conversion: Rare haplotypes may exhibit an identical SNP pattern while harboring alleles that differ over a critical short sequence in a peptide-binding pocket. Another problem is that some rare alleles may be missing from the database. A modification of the original algorithm, HLA*IMP, with improved SNP selection and parallelized training on 2,500 individuals provides accuracy of 92–98% at the four-digit level and 97% at the two-digit level (28). Although imputation sacrifices some accuracy, this is outweighed by the advantage of being able to type very large sets of DNA at low cost. It may not be reliable for donor matching in, for example, hematopoietic transplantation.
These new technologies allow screening of large panels of samples, which provide much improved statistical rigor in genetic analysis. Such techniques are necessary to overcome the problems associated with linkage disequilibrium in the MHC region. So far, there have been only a small number of large-scale studies, which have supported and extended previous findings. For example, Raychaudhuri et al. (99) examined 5,018 rheumatoid arthritis patients and 14,974 controls using a large reference panel to impute classical HLA alleles. The data confirmed the association with amino acids 70–74 of DRB1, the so-called shared epitope; an association was also confirmed with residue 11, further along the groove. Amino acid 11 of DRB1 is also implicated in ulcerative colitis (4). These approaches undoubtedly signal the vanguard of many other studies employing large disease cohorts of mixed ancestry. High-density SNP mapping was used to identify multiple independent susceptibility loci for severe IgA deficiency (37). The primary association mapped to HLA-DQB1*02, with additional effects from other class II DRB1 alleles and haplotypes. Despite the strong population-specific frequencies of HLA alleles, there was a good correlation across different ethnic backgrounds.
HIGH-THROUGHPUT SEQUENCING
Early attempts at MHC sequencing were laborious by today’s standards, using Sanger sequencing of bacterial artificial chromosome clones. The first complete sequences appeared in 1999, listing 224 genes, 128 of which were predicted to be expressed (84). There followed a more systematic approach of eight common haplotypes forming consanguineous cell lines (50, 111, 117). These studies identified more than 44,000 SNPs, which have been invaluable for disease studies. Two related haplotypes were compared in considerable detail: HLA-A26-B18-Cw5-DR3-DQ2 and HLA-A1-B8-Cw7-DR3-DQ2, which share the same HLA-DRB1, HLA-DQA1, and HLA-DQB1 alleles. The two sequences exhibited high levels of variation, similar to those for other HLA-disparate haplotypes. The exception was a 158-kb segment encompassing the HLA-DRB1, HLA-DQA1, and HLA-DQB1 genes. There was very limited polymorphism in this region, compatible with identity by descent and relatively recent common ancestry, estimated at <3,400 generations. These findings were consistent with early ideas that shuffling of ancestral blocks distributes certain DR-DQ allelic combinations on different haplotypes (100). This may also occur for blocks of sequence in the class I genes. In this way, favored combinations of alleles spread across haplotypes and populations without necessarily being separated by recombination. The data imply that recombination takes place more frequently in regions flanking the blocks than within them, or that certain allele combinations are maintained by natural selection. These studies identified regions that differed by well over 60 base pairs per kilobase, spanning the whole of the gene and its flanking regions, not only those sequences encoding the peptide-binding groove regions of class I and class II genes. As mentioned above, this extensive variation flanking HLA genes may accumulate by hitchhiking, which could lead to accumulation of deleterious mutations and contribute to disease susceptibility (106). Another proposed mechanism for accumulation of mutations is that selection is less frequent in HLA genes than it is in other regions of the genome, and because there is so much variation, homozygosity is relatively rare (111, 123).
High-throughput sequencing is an obvious approach to determining HLA genotype. A pioneering early attempt at deep sequencing provided some insight into the way the genomic structure of the MHC has been shaped by selection, recombination, and gene-gene interaction (100). So far, it has been exploited mostly to target polymorphic regions of classical HLA loci. Several groups have typed alleles using massively parallel sequencing platforms like the Roche/454 systems (10, 30, 40, 49). These studies amplified separate exons for multiplex sequencing.
For MHC sequencing, a major problem with the short reads generated by high-throughput sequencing systems such as the Illumina GAIIx relates to the difficulties of read mapping, given the extent of sequence level and structural polymorphism together with potential confounding by choice of MHC reference sequence. There are significant advantages to amplifying longer read lengths encompassing more than one exon. Lind et al. (76) used this approach; similarly, Wang et al. (128) used even longer-range PCR amplification of genomic DNA to sequence over multiple coding exons of HLA-A, HLA-C, and HLA-DRB1. The advantage of this technique is that long stretches of alleles can be identified, allowing more accurate data and apparently a 99% concordance rate.
Another approach is to use microarray sequence capture and targeted enrichment followed by next-generation sequencing of the products. Pröll et al. (94) used this method to sequence 3.5 Mb of the MHC and identified 3,025 variant SNPs in a single experiment. Bait design for such sequence capture is challenging in the MHC if it is to be comprehensive and avoid bias. The frequency of errors generated in the amplification of polymorphic HLA sequences remains to be accurately determined.
EPIGENETICS
Epigenetic mechanisms are known to be important in immune function (notably in defining cell identity and function during development) and provide an important interface with environmental factors that may be important in disease. A variety of epigenetic mechanisms that do not result from changes in DNA sequence have been proposed, including methylation, acetylation, and histone modification. These mechanisms are often invoked when phenomena arise that are difficult to explain by conventional genetics. The MHC was one of the first regions to undergo methylation analysis and to have a number of tissue-specific differentially methylated regions defined (115).
A potential example of an epigenetic effect involving the MHC is the parent-of-origin effect reported in multiple sclerosis—the finding that offspring of affected mothers are more likely to develop the disease than those of affected fathers. The effect has been localized to HLA-DRB1*15, the major risk allele for multiple sclerosis (19).
Epigenetic modifications such as DNA methylation are critical to the regulation of gene expression, and mapping specific chromatin modifications can be valuable in resolving regulatory regions. For the MHC, such maps are being generated by the Encyclopedia of DNA Elements (ENCODE) project (29) and other initiatives (Figure 1), and it will be interesting to see how these can be resolved in an allelic or haplotypic fashion to inform efforts to resolve functionally important genetic variation. For specific genes important in the control of MHC gene expression, such as CIITA, which encodes the HLA class II master activator, complex epigenetic events are key to controlling cellular and stimulus specificity of expression (83).
POPULATION MIGRATION AND SELECTION
Because it is so polymorphic, the MHC is highly useful in tracing population migration as well its potential relationship to pathogen-mediated selection. Prugnolle et al. (95) showed that HLA diversity contours relate to distance from African origins. Single et al. (108) proposed that KIRs for HLA class I could be traced similarly and, moreover, that polymorphic receptors and ligands exhibit a coevolutionary relationship.
ADMIXTURE WITH ARCHAIC HUMANS
MHC polymorphism is so ancient that it transcends species barriers. As new species are generated from a group of territorially isolated individuals, a set of diverse MHC alleles is passed on. Novel alleles may then be generated in the emerging species, but their provenance can be traced by sequence analysis. In some cases, distinct species may have identical alleles, according to the transspecies hypothesis (67). Admixture of closely related species can occasionally result in sequence capture. This mechanism has been proposed in human evolution to explain the HLA-B*73 allele, which in sequence terms is an outlier in relation to other human HLA-B alleles. Comparison of human MHC sequences with those of Denisovans, who were related to Neandertals, suggested that HLA-B*73 was acquired in western Asia through admixture (2). The argument for this was somewhat indirect, but it supported the idea that certain haplotypes introgressed into modern Eurasian and Oceanian populations and only much later were introduced into African populations.
CONCLUSIONS
Decades of research on the MHC has established the region as genetically unique in terms of its polymorphism and association with disease. The DNA cloning era clearly established that the most important loci in terms of disease were class I and class II, and these have received unprecedented attention. Nevertheless, resolving specific functional variants has remained a daunting challenge, and it is clear that other genes embedded in the MHC make a contribution to human diseases, although this remains to be analyzed fully. Most of the noncoding DNA and the significance of genetic variation involving such regions also remain to be explored. Gene-gene interactions, gene-environment interactions, and complex epigenetic mechanisms are likely to be important in determining disease risk, requiring new approaches and context-specific analysis. It is clear, however, that this remarkable region of the genome, which has taught us so much about the nature of the variable genome and its function, still has many important secrets to be revealed.
ACKNOWLEDGMENTS
J.T. is supported by the Medical Research Council (MRC) and the Wellcome Trust, with partial support from the National Institute of Health Research (NIHR) Cambridge Biomedical Research Centre. J.C.K. is supported by the European Research Council (ERC) under the European Commission 7th Framework Programme (FP7/2007–2013) (281824), the MRC (98082), the NIHR Oxford Biomedical Research Centre, and the Wellcome Trust (075491/Z/04 to core facilities, Wellcome Trust Centre for Human Genetics).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Abi-Rached L, Gilles A, Shiina T, Pontarotti P, Inoko H. Evidence of en bloc duplication in vertebrate genomes. Nat. Genet. 2002;31:100–5. doi: 10.1038/ng855. [DOI] [PubMed] [Google Scholar]
- 2.Abi-Rached L, Jobin MJ, Kulkarni S, McWhinnie A, Dalva K, et al. The shaping of modern human immune systems by multiregional admixture with archaic humans. Science. 2011;334:89–94. doi: 10.1126/science.1209202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abi-Rached L, McDermott MF, Pontarotti P. The MHC big bang. Immunol. Rev. 1999;167:33–44. doi: 10.1111/j.1600-065x.1999.tb01380.x. [DOI] [PubMed] [Google Scholar]
- 4.Achkar JP, Klei L, Bakker PI, Bellone G, Rebert N, et al. Amino acid position 11 of HLA-DRβ1 is a major determinant of chromosome 6p association with ulcerative colitis. Genes Immun. 2012;13:245–52. doi: 10.1038/gene.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–51. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 5a.Apps R, Qi Y, Carlson JM, Chen H, Gao X, et al. Influence of HLA-C expression level on HIV control. Science. 2013;340:87–91. doi: 10.1126/science.1232685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrow AD, Trowsdale J. The extended human leukocyte receptor complex: diverse ways of modulating immune responses. Immunol. Rev. 2008;224:98–123. doi: 10.1111/j.1600-065X.2008.00653.x. [DOI] [PubMed] [Google Scholar]
- 7.Basha G, Omilusik K, Chavez-Steenbock A, Reinicke AT, Lack N, et al. A CD74-dependent MHC class I endolysosomal cross-presentation pathway. Nat. Immunol. 2012;13:237–45. doi: 10.1038/ni.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belov K. Contagious cancer: lessons from the devil and the dog. BioEssays. 2012;34:285–92. doi: 10.1002/bies.201100161. [DOI] [PubMed] [Google Scholar]
- 9.Belov K, Deakin JE, Papenfuss AT, Baker ML, Melman SD, et al. Reconstructing an ancestral mammalian immune supercomplex from a marsupial major histocompatibility complex. PLoS Biol. 2006;4:e46. doi: 10.1371/journal.pbio.0040046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bentley G, Higuchi R, Hoglund B, Goodridge D, Sayer D, et al. High-resolution, high-throughput HLA genotyping by next-generation sequencing. Tissue Antigens. 2009;74:393–403. doi: 10.1111/j.1399-0039.2009.01345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bharadwaj M, Illing P, Theodossis A, Purcell AW, Rossjohn J, McCluskey J. Drug hypersensitivity and human leukocyte antigens of the major histocompatibility complex. Annu. Rev. Pharmacol. Toxicol. 2012;52:401–31. doi: 10.1146/annurev-pharmtox-010611-134701. [DOI] [PubMed] [Google Scholar]
- 12.Bowerman NA, Falta MT, Mack DG, Kappler JW, Fontenot AP. Mutagenesis of beryllium-specific TCRs suggests an unusual binding topology for antigen recognition. J. Immunol. 2011;187:3694–703. doi: 10.4049/jimmunol.1101872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braunstein NS, Germain RN, Loney K, Berkowitz N. Structurally interdependent and independent regions of allelic polymorphism in class II MHC molecules. Implications for Ia function and evolution. J. Immunol. 1990;145:1635–45. [PubMed] [Google Scholar]
- 14.Brown D, Trowsdale J, Allen R. The LILR family: modulators of innate and adaptive immune pathways in health and disease. Tissue Antigens. 2004;64:215–25. doi: 10.1111/j.0001-2815.2004.00290.x. [DOI] [PubMed] [Google Scholar]
- 15.Carlson JM, Listgarten J, Pfeifer N, Tan V, Kadie C, et al. Widespread impact of HLA restriction on immune control and escape pathways of HIV-1. J. Virol. 2012;86:5230–43. doi: 10.1128/JVI.06728-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castro LF, Furlong RF, Holland PW. An antecedent of the MHC-linked genomic region in amphioxus. Immunogenetics. 2004;55:782–84. doi: 10.1007/s00251-004-0642-9. [DOI] [PubMed] [Google Scholar]
- 17.Castro-Prieto A, Wachter B, Sommer S. Cheetah paradigm revisited: MHC diversity in the world’s largest free-ranging population. Mol. Biol. Evol. 2011;28:1455–68. doi: 10.1093/molbev/msq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chabas D, Taheri S, Renier C, Mignot E. The genetics of narcolepsy. Annu. Rev. Genomics Hum. Genet. 2003;4:459–83. doi: 10.1146/annurev.genom.4.070802.110432. [DOI] [PubMed] [Google Scholar]
- 19.Chao MJ, Barnardo MC, Lincoln MR, Ramagopalan SV, Herrera BM, et al. HLA class I alleles tag HLA-DRB1*1501 haplotypes for differential risk in multiple sclerosis susceptibility. Proc. Natl. Acad. Sci. USA. 2008;105:13069–74. doi: 10.1073/pnas.0801042105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chazara O, Xiong S, Moffett A. Maternal KIR and fetal HLA-C: a fine balance. J. Leukoc. Biol. 2011;90:703–16. doi: 10.1189/jlb.0511227. [DOI] [PubMed] [Google Scholar]
- 21.Dai S, Murphy GA, Crawford F, Mack DG, Falta MT, et al. Crystal structure of HLA-DP2 and implications for chronic beryllium disease. Proc. Natl. Acad. Sci. USA. 2010;107:7425–30. doi: 10.1073/pnas.1001772107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Danchin EG, Pontarotti P. Towards the reconstruction of the bilaterian ancestral pre-MHC region. Trends Genet. 2004;20:587–91. doi: 10.1016/j.tig.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 23.Davies RW, Wells GA, Stewart AF, Erdmann J, Shah SH, et al. A genome-wide association study for coronary artery disease identifies a novel susceptibility locus in the major histocompatibility complex. Circ. Cardiovasc. Genet. 2012;5:217–25. doi: 10.1161/CIRCGENETICS.111.961243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dawkins R, Leelayuwat C, Gaudieri S, Tay G, Hui J, et al. Genomics of the major histocompatibility complex: haplotypes, duplication, retroviruses and disease. Immunol. Rev. 1999;167:275–304. doi: 10.1111/j.1600-065x.1999.tb01399.x. [DOI] [PubMed] [Google Scholar]
- 25.de Bakker PI, McVean G, Sabeti PC, Miretti MM, Green T, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat. Genet. 2006;38:1166–72. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delarbre C, Jaulin C, Kourilsky P, Gachelin G. Evolution of the major histocompatibility complex: a hundred-fold amplification of MHC class I genes in the African pigmy mouse Nannomys setulosus. Immunogenetics. 1992;37:29–38. doi: 10.1007/BF00223542. [DOI] [PubMed] [Google Scholar]
- 27.de la Salle H, Hanau D, Fricker D, Urlacher A, Kelly A, et al. Homozygous human TAP peptide transporter mutation in HLA class I deficiency. Science. 1994;265:237–41. doi: 10.1126/science.7517574. [DOI] [PubMed] [Google Scholar]
- 28.Dilthey AT, Moutsianas L, Leslie S, McVean G. HLA*IMP—an integrated framework for imputing classical HLA alleles from SNP genotypes. Bioinformatics. 2011;27:968–72. doi: 10.1093/bioinformatics/btr061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.ENCODE Proj. Consort. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Erlich RL, Jia X, Anderson S, Banks E, Gao X, et al. Next-generation sequencing for HLA typing of class I loci. BMC Genomics. 2011;12:42. doi: 10.1186/1471-2164-12-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Evans DM, Spencer CC, Pointon JJ, Su Z, Harvey D, et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat. Genet. 2011;43:761–67. doi: 10.1038/ng.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evseeva I, Nicodemus KK, Bonilla C, Tonks S, Bodmer WF. Linkage disequilibrium and age of HLA region SNPs in relation to classic HLA gene alleles within Europe. Eur. J. Hum. Genet. 2010;18:924–32. doi: 10.1038/ejhg.2010.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fadda L, Borhis G, Ahmed P, Cheent K, Pageon SV, et al. Peptide antagonism as a mechanism for NK cell activation. Proc. Natl. Acad. Sci. USA. 2010;107:10160–65. doi: 10.1073/pnas.0913745107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fairfax BP, Makino S, Radhakrishnan J, Plant K, Leslie S, et al. Genetics of gene expression in primary immune cells identifies cell type–specific master regulators and roles of HLA alleles. Nat. Genet. 2012;44:502–10. doi: 10.1038/ng.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, et al. A novel MHC class I–like gene is mutated in patients with hereditary haemochromatosis. Nat. Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 36.Fehrmann RS, Jansen RC, Veldink JH, Westra HJ, Arends D, et al. Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS Genet. 2011;7:e1002197. doi: 10.1371/journal.pgen.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferreira RC, Pan-Hammarstrom Q, Graham RR, Fontan G, Lee AT, et al. High-density SNP mapping of the HLA region identifies multiple independent susceptibility loci associated with selective IgA deficiency. PLoS Genet. 2012;8:e1002476. doi: 10.1371/journal.pgen.1002476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flajnik MF, Canel C, Kramer J, Kasahara M. Which came first, MHC class I or class II? Immunogenetics. 1991;33:295–300. doi: 10.1007/BF00216688. [DOI] [PubMed] [Google Scholar]
- 39.Fust G, Arason GJ, Kramer J, Szalai C, Duba J, et al. Genetic basis of tobacco smoking: strong association of a specific major histocompatibility complex haplotype on chromosome 6 with smoking behavior. Int. Immunol. 2004;16:1507–14. doi: 10.1093/intimm/dxh152. [DOI] [PubMed] [Google Scholar]
- 40.Gabriel C, Danzer M, Hackl C, Kopal G, Hufnagl P, et al. Rapid high-throughput human leukocyte antigen typing by massively parallel pyrosequencing for high-resolution allele identification. Hum. Immunol. 2009;70:960–64. doi: 10.1016/j.humimm.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 41.Gaudieri S, Leelayuwat C, Tay GK, Townend DC, Dawkins RL. The major histocompatability complex (MHC) contains conserved polymorphic genomic sequences that are shuffled by recombination to form ethnic-specific haplotypes. J. Mol. Evol. 1997;45:17–23. doi: 10.1007/pl00006194. [DOI] [PubMed] [Google Scholar]
- 42.Hamza TH, Zabetian CP, Tenesa A, Laederach A, Montimurro J, et al. Common genetic variation in the HLA region is associated with late-onset sporadic Parkinson’s disease. Nat. Genet. 2010;42:781–85. doi: 10.1038/ng.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Handunnetthi L, Ramagopalan SV, Ebers GC. Multiple sclerosis, vitamin D, and HLA-DRB1*15. Neurology. 2010;74:1905–10. doi: 10.1212/WNL.0b013e3181e24124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harari A, Cellerai C, Enders FB, Kostler J, Codarri L, et al. Skewed association of polyfunctional antigen-specific CD8 T cell populations with HLA-B genotype. Proc. Natl. Acad. Sci. USA. 2007;104:16233–38. doi: 10.1073/pnas.0707570104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Havlicek J, Roberts SC. MHC-correlated mate choice in humans: a review. Psychoneuroendocrinology. 2009;34:497–512. doi: 10.1016/j.psyneuen.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 46.Hiby SE, Apps R, Sharkey AM, Farrell LE, Gardner L, et al. Maternal activating KIRs protect against human reproductive failure mediated by fetal HLA-C2. J. Clin. Investig. 2010;120:4102–10. doi: 10.1172/JCI43998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hill-Burns EM, Factor SA, Zabetian CP, Thomson G, Payami H. Evidence for more than one Parkinson’s disease-associated variant within the HLA region. PLoS ONE. 2011;6:e27109. doi: 10.1371/journal.pone.0027109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirayasu K, Ohashi J, Kashiwase K, Hananantachai H, Naka I, et al. Significant association of KIR2DL3-HLA-C1 combination with cerebral malaria and implications for co-evolution of KIR and HLA. PLoS Pathog. 2012;8:e1002565. doi: 10.1371/journal.ppat.1002565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holcomb CL, Hoglund B, Anderson MW, Blake LA, Bohme I, et al. A multi-site study using high-resolution HLA genotyping by next generation sequencing. Tissue Antigens. 2011;77:206–17. doi: 10.1111/j.1399-0039.2010.01606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horton R, Gibson R, Coggill P, Miretti M, Allcock RJ, et al. Variation analysis and gene annotation of eight MHC haplotypes: the MHC Haplotype Project. Immunogenetics. 2008;60:1–18. doi: 10.1007/s00251-007-0262-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, et al. Gene map of the extended human MHC. Nat. Rev. Genet. 2004;5:889–99. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 52.Huang J, Al-Mozaini M, Rogich J, Carrington MF, Seiss K, et al. Systemic inhibition of myeloid dendritic cells by circulating HLA class I molecules in HIV-1 infection. Retrovirology. 2012;9:11. doi: 10.1186/1742-4690-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang J, Burke PS, Cung TD, Pereyra F, Toth I, et al. Leukocyte immunoglobulin-like receptors maintain unique antigen-presenting properties of circulating myeloid dendritic cells in HIV-1-infected elite controllers. J. Virol. 2010;84:9463–71. doi: 10.1128/JVI.01009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, et al. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–51. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 55.Illing PT, Vivian JP, Dudek NL, Kostenko L, Chen Z, et al. Immune self-reactivity triggered by drug-modified HLA-peptide repertoire. Nature. 2012;486:554–58. doi: 10.1038/nature11147. [DOI] [PubMed] [Google Scholar]
- 56.Jacob CO, McDevitt HO. Tumour necrosis factor-α in murine autoimmune “lupus” nephritis. Nature. 1988;331:356–58. doi: 10.1038/331356a0. [DOI] [PubMed] [Google Scholar]
- 57.Jeffery KJ, Siddiqui AA, Bunce M, Lloyd AL, Vine AM, et al. The influence of HLA class I alleles and heterozygosity on the outcome of human T cell lymphotropic virus type I infection. J. Immunol. 2000;165:7278–84. doi: 10.4049/jimmunol.165.12.7278. [DOI] [PubMed] [Google Scholar]
- 58.Karre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319:675–78. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 59.Karttunen JT, Trowsdale J, Lehner PJ. Antigen presentation: TAP dances with ATP. Curr. Biol. 1999;9:R820–24. doi: 10.1016/s0960-9822(99)80499-0. [DOI] [PubMed] [Google Scholar]
- 60.Kasahara M. The chromosomal duplication model of the major histocompatibility complex. Immunol. Rev. 1999;167:17–32. doi: 10.1111/j.1600-065x.1999.tb01379.x. [DOI] [PubMed] [Google Scholar]
- 61.Kaufman J. The simple chicken major histocompatibility complex: life and death in the face of pathogens and vaccines. Philos. Trans. R. Soc. Lond. B. 2000;355:1077–84. doi: 10.1098/rstb.2000.0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaufman J, Milne S, Gobel TW, Walker BA, Jacob JP, et al. The chicken B locus is a minimal essential major histocompatibility complex. Nature. 1999;401:923–25. doi: 10.1038/44856. [DOI] [PubMed] [Google Scholar]
- 63.Kelley J, Walter L, Trowsdale J. Comparative genomics of major histocompatibility complexes. Immunogenetics. 2005;56:683–95. doi: 10.1007/s00251-004-0717-7. [DOI] [PubMed] [Google Scholar]
- 64.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khor CC, Chau TN, Pang J, Davila S, Long HT, et al. Genome-wide association study identifies susceptibility loci for dengue shock syndrome at MICB and PLCE1. Nat. Genet. 2011;43:1139–41. doi: 10.1038/ng.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kincaid EZ, Che JW, York I, Escobar H, Reyes-Vargas E, et al. Mice completely lacking immunoproteasomes show major changes in antigen presentation. Nat. Immunol. 2012;13:129–35. doi: 10.1038/ni.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Klein J. Natural History of the Major Histocompatibility Complex. Wiley; New York: 1986. [Google Scholar]
- 68.Ko TM, Chung WH, Wei CY, Shih HY, Chen JK, et al. Shared and restricted T-cell receptor use is crucial for carbamazepine-induced Stevens-Johnson syndrome. J. Allergy Clin. Immunol. 2011;128:1266–76. e11. doi: 10.1016/j.jaci.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 69.Kong A, Thorleifsson G, Gudbjartsson DF, Masson G, Sigurdsson A, et al. Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010;467:1099–103. doi: 10.1038/nature09525. [DOI] [PubMed] [Google Scholar]
- 70.Kraman M, Bambrough PJ, Arnold JN, Roberts EW, Magiera L, et al. Suppression of antitumor immunity by stromal cells expressing fibroblast activation protein-α. Science. 2010;330:827–30. doi: 10.1126/science.1195300. [DOI] [PubMed] [Google Scholar]
- 71.Kulkarni S, Savan R, Qi Y, Gao X, Yuki Y, et al. Differential microRNA regulation of HLA-C expression and its association with HIV control. Nature. 2011;472:495–98. doi: 10.1038/nature09914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kulpa DA, Collins KL. The emerging role of HLA-C in HIV-1 infection. Immunology. 2011;134:116–22. doi: 10.1111/j.1365-2567.2011.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lederberg J. J. B. S. Haldane (1949) on infectious disease and evolution. Genetics. 1999;153:1–3. doi: 10.1093/genetics/153.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Leslie S, Donnelly P, McVean G. A statistical method for predicting classical HLA alleles from SNP data. Am. J. Hum. Genet. 2008;82:48–56. doi: 10.1016/j.ajhg.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lichterfeld M, Yu XG. The emerging role of leukocyte immunoglobulin-like receptors (LILRs) in HIV-1 infection. J. Leukoc. Biol. 2012;91:27–33. doi: 10.1189/jlb.0811442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lind C, Ferriola D, Mackiewicz K, Heron S, Rogers M, et al. Next-generation sequencing: the solution for high-resolution, unambiguous human leukocyte antigen typing. Hum. Immunol. 2010;71:1033–42. doi: 10.1016/j.humimm.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 77.Makrigiannis AP, Parham P. The evolution of NK cell diversity. Semin. Immunol. 2008;20:309–10. doi: 10.1016/j.smim.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 78.Malkki M, Single R, Carrington M, Thomson G, Petersdorf E. MHC microsatellite diversity and linkage disequilibrium among common HLA-A, HLA-B, DRB1 haplotypes: implications for unrelated donor hematopoietic transplantation and disease association studies. Tissue Antigens. 2005;66:114–24. doi: 10.1111/j.1399-0039.2005.00453.x. [DOI] [PubMed] [Google Scholar]
- 79.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat. Genet. 2007;39:733–40. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maruoka T, Tanabe H, Chiba M, Kasahara M. Chicken CD1 genes are located in the MHC: CD1 and endothelial protein C receptor genes constitute a distinct subfamily of class-I-like genes that predates the emergence of mammals. Immunogenetics. 2005;57:590–600. doi: 10.1007/s00251-005-0016-y. [DOI] [PubMed] [Google Scholar]
- 81.McDevitt H. The discovery of linkage between the MHC and genetic control of the immune response. Immunol. Rev. 2002;185:78–85. doi: 10.1034/j.1600-065x.2002.18509.x. [DOI] [PubMed] [Google Scholar]
- 82.McMichael AJ, Jones EY. First-class control of HIV-1. Science. 2010;330:1488–90. doi: 10.1126/science.1200035. [DOI] [PubMed] [Google Scholar]
- 83.Mehta NT, Truax AD, Boyd NH, Greer SF. Early epigenetic events regulate the adaptive immune response gene CIITA. Epigenetics. 2011;6:516–25. doi: 10.4161/epi.6.4.14516. [DOI] [PubMed] [Google Scholar]
- 84.MHC Seq. Consort. Complete sequence and gene map of a human major histocompatibility complex. Nature. 1999;401:921–23. doi: 10.1038/44853. [DOI] [PubMed] [Google Scholar]
- 85.Mignot E, Kimura A, Lattermann A, Lin X, Yasunaga S, et al. Extensive HLA class II studies in 58 non-DRB1*15 (DR2) narcoleptic patients with cataplexy. Tissue Antigens. 1997;49:329–41. doi: 10.1111/j.1399-0039.1997.tb02761.x. [DOI] [PubMed] [Google Scholar]
- 86.Miretti MM, Walsh EC, Ke X, Delgado M, Griffiths M, et al. A high-resolution linkage-disequilibrium map of the human major histocompatibility complex and first generation of tag single-nucleotide polymorphisms. Am. J. Hum. Genet. 2005;76:634–46. doi: 10.1086/429393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Moore CB, John M, James IR, Christiansen FT, Witt CS, Mallal SA. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 2002;296:1439–43. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 88.Nathenson SG, Geliebter J, Pfaffenbach GM, Zeff RA. Murine major histocompatibility complex class-I mutants: molecular analysis and structure-function implications. Annu. Rev. Immunol. 1986;4:471–502. doi: 10.1146/annurev.iy.04.040186.002351. [DOI] [PubMed] [Google Scholar]
- 89.Norman PJ, Abi-Rached L, Gendzekhadze K, Hammond JA, Moesta AK, et al. Meiotic recombination generates rich diversity in NK cell receptor genes, alleles, and haplotypes. Genome Res. 2009;19:757–69. doi: 10.1101/gr.085738.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ohta Y, Shiina T, Lohr RL, Hosomichi K, Pollin TI, et al. Primordial linkage of β2-microglobulin to the MHC. J. Immunol. 2011;186:3563–71. doi: 10.4049/jimmunol.1003933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Parham P, Ohta T. Population biology of antigen presentation by MHC class I molecules. Science. 1996;272:67–74. doi: 10.1126/science.272.5258.67. [DOI] [PubMed] [Google Scholar]
- 92.Powis SJ, Deverson EV, Coadwell WJ, Ciruela A, Huskisson NS, et al. Effect of polymorphism of an MHC-linked transporter on the peptides assembled in a class I molecule. Nature. 1992;357:211–15. doi: 10.1038/357211a0. [DOI] [PubMed] [Google Scholar]
- 93.Price P, Witt C, Allcock R, Sayer D, Garlepp M, et al. The genetic basis for the association of the 8.1 ancestral haplotype (A1, B8, DR3) with multiple immunopathological diseases. Immunol. Rev. 1999;167:257–74. doi: 10.1111/j.1600-065x.1999.tb01398.x. [DOI] [PubMed] [Google Scholar]
- 94.Pröll J, Danzer M, Stabentheiner S, Niklas N, Hackl C, et al. Sequence capture and next generation resequencing of the MHC region highlights potential transplantation determinants in HLA identical haematopoietic stem cell transplantation. DNA Res. 2011;18:201–10. doi: 10.1093/dnares/dsr008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Prugnolle F, Manica A, Charpentier M, Guegan JF, Guernier V, Balloux F. Pathogen-driven selection and worldwide HLA class I diversity. Curr. Biol. 2005;15:1022–27. doi: 10.1016/j.cub.2005.04.050. [DOI] [PubMed] [Google Scholar]
- 96.Qidwai T, Khan F. Tumour necrosis factor gene polymorphism and disease prevalence. Scand. J. Immunol. 2011;74:522–47. doi: 10.1111/j.1365-3083.2011.02602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rajapaksa US, Li D, Peng YC, McMichael AJ, Dong T, Xu XN. HLA-B may be more protective against HIV-1 than HLA-A because it resists negative regulatory factor (Nef) mediated down-regulation. Proc. Natl. Acad. Sci. USA. 2012;109:13353–58. doi: 10.1073/pnas.1204199109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ramagopalan SV, Maugeri NJ, Handunnetthi L, Lincoln MR, Orton SM, et al. Expression of the multiple sclerosis-associated MHC class II allele HLA-DRB1*1501 is regulated by vitamin D. PLoS Genet. 2009;5:e1000369. doi: 10.1371/journal.pgen.1000369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Raychaudhuri S, Sandor C, Stahl EA, Freudenberg J, Lee HS, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat. Genet. 2012;44:291–96. doi: 10.1038/ng.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Raymond CK, Kas A, Paddock M, Qiu R, Zhou Y, et al. Ancient haplotypes of the HLA Class II region. Genome Res. 2005;15:1250–57. doi: 10.1101/gr.3554305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Reusch TB, Haberli MA, Aeschlimann PB, Milinski M. Female sticklebacks count alleles in a strategy of sexual selection explaining MHC polymorphism. Nature. 2001;414:300–2. doi: 10.1038/35104547. [DOI] [PubMed] [Google Scholar]
- 102.Robinson J, Mistry K, McWilliam H, Lopez R, Parham P, Marsh SG. The IMGT/HLA database. Nucleic Acids Res. 2011;39:D1171–76. doi: 10.1093/nar/gkq998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Romagnoli P, Spinas GA, Sinigaglia F. Gold-specific T cells in rheumatoid arthritis patients treated with gold. J. Clin. Investig. 1992;89:254–58. doi: 10.1172/JCI115569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Santos PS, Fust G, Prohaszka Z, Volz A, Horton R, et al. Association of smoking behavior with an odorant receptor allele telomeric to the human major histocompatibility complex. Genet. Test. 2008;12:481–86. doi: 10.1089/gte.2008.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shatz CJ. MHC class I: an unexpected role in neuronal plasticity. Neuron. 2009;64:40–45. doi: 10.1016/j.neuron.2009.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shiina T, Ota M, Shimizu S, Katsuyama Y, Hashimoto N, et al. Rapid evolution of major histocompatibility complex class I genes in primates generates new disease alleles in humans via hitchhiking diversity. Genetics. 2006;173:1555–70. doi: 10.1534/genetics.106.057034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–50. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Single RM, Martin MP, Gao X, Meyer D, Yeager M, et al. Global diversity and evidence for coevolution of KIR and HLA. Nat. Genet. 2007;39:1114–19. doi: 10.1038/ng2077. [DOI] [PubMed] [Google Scholar]
- 109.Star B, Nederbragt AJ, Jentoft S, Grimholt U, Malmstrom M, et al. The genome sequence of Atlantic cod reveals a unique immune system. Nature. 2011;477:207–10. doi: 10.1038/nature10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Steimle V, Otten LA, Zufferey M, Mach B. Complementation cloning of an MHC class II trans-activator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome) Cell. 1993;75:135–46. [PubMed] [Google Scholar]
- 111.Stewart CA, Horton R, Allcock RJ, Ashurst JL, Atrazhev AM, et al. Complete MHC haplotype sequencing for common disease gene mapping. Genome Res. 2004;14:1176–87. doi: 10.1101/gr.2188104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Strange A, Capon F, Spencer CC, Knight J, Weale ME, et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat. Genet. 2010;42:985–90. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Suemizu H, Radosavljevic M, Kimura M, Sadahiro S, Yoshimura S, et al. A basolateral sorting motif in the MICA cytoplasmic tail. Proc. Natl. Acad. Sci. USA. 2002;99:2971–76. doi: 10.1073/pnas.052701099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Todd JA, Bell JI, McDevitt HO. HLA-DQβ gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature. 1987;329:599–604. doi: 10.1038/329599a0. [DOI] [PubMed] [Google Scholar]
- 115.Tomazou EM, Rakyan VK, Lefebvre G, Andrews R, Ellis P, et al. Generation of a genomic tiling array of the human major histocompatibility complex (MHC) and its application for DNA methylation analysis. BMC Med. Genomics. 2008;1:19. doi: 10.1186/1755-8794-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Torres AR, Maciulis A, Odell D. The association of MHC genes with autism. Front. Biosci. 2001;6:D936–43. doi: 10.2741/torres. [DOI] [PubMed] [Google Scholar]
- 117.Traherne JA, Horton R, Roberts AN, Miretti MM, Hurles ME, et al. Genetic analysis of completely sequenced disease-associated MHC haplotypes identifies shuffling of segments in recent human history. PLoS Genet. 2006;2:e9. doi: 10.1371/journal.pgen.0020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Traherne JA, Martin M, Ward R, Ohashi M, Pellett F, et al. Mechanisms of copy number variation and hybrid gene formation in the KIR immune gene complex. Hum. Mol. Genet. 2010;19:737–51. doi: 10.1093/hmg/ddp538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Trowsdale J. Genetic and functional relationships between MHC and NK receptor genes. Immunity. 2001;15:363–74. doi: 10.1016/s1074-7613(01)00197-2. [DOI] [PubMed] [Google Scholar]
- 120.Valentonyte R, Hampe J, Huse K, Rosenstiel P, Albrecht M, et al. Sarcoidosis is associated with a truncating splice site mutation in BTNL2. Nat. Genet. 2005;37:357–64. doi: 10.1038/ng1519. [DOI] [PubMed] [Google Scholar]
- 121.Vandiedonck C, Knight JC. The human Major Histocompatibility Complex as a paradigm in genomics research. Brief. Funct. Genomics Proteomics. 2009;8:379–94. doi: 10.1093/bfgp/elp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vandiedonck C, Taylor MS, Lockstone HE, Plant K, Taylor JM, et al. Pervasive haplotypic variation in the spliceo-transcriptome of the human major histocompatibility complex. Genome Res. 2011;21:1042–54. doi: 10.1101/gr.116681.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.van Oosterhout C. A new theory of MHC evolution: beyond selection on the immune genes. Proc. R. Soc. B. 2009;276:657–65. doi: 10.1098/rspb.2008.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vidal SM, Khakoo SI, Biron CA. Natural killer cell responses during viral infections: flexibility and conditioning of innate immunity by experience. Curr. Opin. Virol. 2011;1:497–512. doi: 10.1016/j.coviro.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat. Immunol. 2008;9:503–10. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 126.Walker BA, Hunt LG, Sowa AK, Skjodt K, Gobel TW, et al. The dominantly expressed class I molecule of the chicken MHC is explained by coevolution with the polymorphic peptide transporter (TAP) genes. Proc. Natl. Acad. Sci. USA. 2011;108:8396–401. doi: 10.1073/pnas.1019496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Walsh EC, Mather KA, Schaffner SF, Farwell L, Daly MJ, et al. An integrated haplotype map of the human major histocompatibility complex. Am. J. Hum. Genet. 2003;73:580–90. doi: 10.1086/378101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang C, Krishnakumar S, Wilhelmy J, Babrzadeh F, Stepanyan L, et al. High-throughput, high-fidelity HLA genotyping with deep sequencing. Proc. Natl. Acad. Sci. USA. 2012;109:8676–81. doi: 10.1073/pnas.1206614109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang F, Liu H, Blanton WP, Belkina A, Lebrasseur NK, Denis GV. Brd2 disruption in mice causes severe obesity without Type 2 diabetes. Biochem. J. 2010;425:71–83. doi: 10.1042/BJ20090928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.White PC, New MI, Dupont B. Molecular cloning of steroid 21-hydroxylase. Ann. N. Y. Acad. Sci. 1985;458:277–87. doi: 10.1111/j.1749-6632.1985.tb14613.x. [DOI] [PubMed] [Google Scholar]
- 131.Wills MR, Ashiru O, Reeves MB, Okecha G, Trowsdale J, et al. Human cytomegalovirus encodes an MHC class I-like molecule (UL142) that functions to inhibit NK cell lysis. J. Immunol. 2005;175:7457–65. doi: 10.4049/jimmunol.175.11.7457. [DOI] [PubMed] [Google Scholar]
- 132.Wong ES, Sanderson CE, Deakin JE, Whittington CM, Papenfuss AT, Belov K. Identification of natural killer cell receptor clusters in the platypus genome reveals an expansion of C-type lectin genes. Immunogenetics. 2009;61:565–79. doi: 10.1007/s00251-009-0386-7. [DOI] [PubMed] [Google Scholar]
- 133.Wooldridge L, Ekeruche-Makinde J, van den Berg HA, Skowera A, Miles JJ, et al. A single autoimmune T cell receptor recognizes more than a million different peptides. J. Biol. Chem. 2011;287:1168–77. doi: 10.1074/jbc.M111.289488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yang Y, Chung EK, Zhou B, Lhotta K, Hebert LA, et al. The intricate role of complement component C4 in human systemic lupus erythematosus. Curr. Dir. Autoimmun. 2004;7:98–132. doi: 10.1159/000075689. [DOI] [PubMed] [Google Scholar]
- 135.Yunis EJ, Larsen CE, Fernandez-Vina M, Awdeh ZL, Romero T, et al. Inheritable variable sizes of DNA stretches in the human MHC: conserved extended haplotypes and their fragments or blocks. Tissue Antigens. 2003;62:1–20. doi: 10.1034/j.1399-0039.2003.00098.x. [DOI] [PubMed] [Google Scholar]