Abstract
Multiple subtypes were found to be epidemic in the Shenzhen men who have sex with men (MSM) population, which always predicts the emergence of a unique recombinant. In 2012, CRF55_01B was first reported, which later was proven to have originated in MSM in Shenzhen city. In this study, we reported a unique recombinant form (URF) of HIV-1 identified in a man who has had sex with men in Shenzhen city. The strain showed a genomic schematic map similar to CRF55_01B with subtype C segments inserted in the gag and pol genes. The full-length genome was amplified in two halves with 1-kb overlap regions. The PCR products were cloned and sequenced. A recombination detection program showed that two subtype C fragments and two subtype B fragments were inserted into the CRF01_AE backbone genome in the gag and pol regions. In the phylogenetic tree, the subtype C fragments clustered with CRF07_BC variants and the other segments grouped with CRF55_01B strains except for one segment that clustered with CRF01_AE. Similar breakpoints between our strain and CRF65_cpx were also observed. The data suggested that the URF strain might be the recombinant form of CRF55_01B, CRF01_AE, and CRF07_BC. This is the first report of a third generation of recombination of HIV-1 that originated from CRF55_01B in China. The identification of the URF indicated the severity of the HIV epidemic in Shenzhen MSM and the urgent need for epidemiological surveillance of the new recombination.
Recombination contributed greatly to the genetic diversity of human immunodeficiency viruses (HIV-1). To now, 72 circulating recombinant forms (CRFs) (www.hiv.lanl.gov\content\hiv-db\CRFs\CRFs.html) have been reported, which is responsible for more than 20% of HIV infections in the global AIDS epidemic.1 Ten CRFs, including CRF01_AE, CRF07_BC, CRF08_BC, CRF55_01B, CRF57_BC, CRF59_01B, CRF61_BC, CRF62_BC, CRF64_BC, and CRF65_cpx, were first identified in China. Among them, CRF07_BC was the most dominant CRF in China, followed by CRF01_AE, CRF08_BC, and other CRFs.2 CRF55_01B was first reported in 2012 in men who have sex with men (MSM) in China and further proved to have originated in Shenzhen city.3, 4 Subsequently, the strain was identified in MSM residing in other cities, suggesting that it was spreading rapidly.4
Shenzhen, locating in the southern coast of China and bordering Hong Kong, is the earliest open city. With a postmodern culture and humanistic environment, Shenzhen attracted a large number of young people from all over the country to work and live. Among the various populations in Shenzhen, 80% consisted of a floating population that favored the spread of HIV. The first HIV-positive case was reported in 1992 in Shenzhen city.5 Since then, HIV spread quickly in the MSM population in the city. By the end of 2013, more than 7,000 HIV infections were confirmed by the Shenzhen center for disease control and prevention (unpublished data released by the Shenzhen center for disease control and prevention; http://epaper.xkb.com.cn/view/900381). In only one year, 2013, more than 1,300 cases were reported, which is 19.2% higher than 2012 (unpublished data released by the Shenzhen center for disease control and prevention; www.sznews.com/news/content/2013-11/30/content_8820942.htm).
Several subtypes, including subtype B, CRF07_BC, and CRF01_AE, were all found in Shenzhen with CRF01_AE as the dominant strain.4,5 In 2014, CRF55_01B become one of the popular strains in MSM in Shenzhen, together with CRF01_AE, CRF07_BC, CRF08_BC, and subtype B.4 The prevalence of more than one subtype in the same population at the same time always predicts the emergence of novel mosaic strains.6 In this study, we characterized a novel recombinant that originated from CRF55_01B and CRF07_BC, in which subtype B and C fragments were inserted into the CRF01_AE genome backbone.
A peripheral blood specimen was collected from a 24-year-old male patient shortly after he was confirmed to be HIV positive in 2011 from Shenzhen, China with informed consent. The patient was infected through homosexual contact. No antiretroviral drugs were taken. DNA was extracted using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The HIV full-length genome was amplified in two halves with 1-kb overlapping regions using cellular genomic DNA as the template. Specific primers were edited to fulfill the seminested PCR to obtain the full-length HIV DNA provirus (Table 1).7,8 The two partial molecules of the entire S15 genome were amplified with the Platinum Taq DNA polymerase High Fidelity kit (Invitrogen, Carlsberg, CA).
Table 1.
Primers Used for Gene Amplification
| Genes | Primers | Position in HXB2 | Primer sequence (5′-3′) | Direction |
|---|---|---|---|---|
| 5′-half | U-LTR5-AE-new | 1–34 | 5′-GCGCCGAATTCTGGATGGGCTAATTTACTCCARGARAAGACAAGA-3′ | Forward (outer and inner) |
| 07Rev8-new | 5219–5185 | 5′-CCTARTGGGATRTGTACTTCTGARCTTACYTTTGG-3′ | Reverse (outer) | |
| Rev11 | 5066–5041 | 5′-ATCATCACCTGCCATCTGTTTTCCAT-3′ | Reverse (inner) | |
| 3′-half | D-LTR3-AE | 9656–9691 | 5′-GGTCTGAGGGATCTCTAGTTACCAGAGTCC–3′ | Reverse (outer and inner) |
| 07For7 | 4875–4912 | 5′-CAAATTAYAAAAATTCAAAATTTTCGGGTTTATTACAG-3′ | Forward (inner) | |
| VIF1 | 4900–4923 | 5′-GGGTTTATTACAGGGACAGCAGAG-3′ | Reverse (inner) |
The same conditions were used in two rounds of amplifications of two halves as follows: 94°C for 2 min, and then three cycles of 94°C for 30 s, 60°C for 1 min, and 68°C for 5 min 30 s; then 32 cycles of 94°C for 15 s, 60°C for 30 s, and 68°C for 5 min; followed by 68°C for 10 min. The PCR product was purified and cloned into the pEASY-T1 vector followed by sequencing with a variety of internal specific primers (available on request).
Both overlapping subgenomic DNA fragments were successfully amplified and cloned into pEASY-T1 vector. All sequenced fragments were edited and then assembled into contiguous sequences on a minimum overlap of 30 bp with a 99–100% minimal mismatch with ContigExpress software, which is a component of the Vector NTI Suite 6.0. A full-length genome of 9,709 bp length was obtained. A Basic Local Alignment Search Tool (BLAST) search against the HIV-1 sequence database (http://hiv-web.lanl.gov/content/index) and all of the sequence collections in the laboratory was used to check for potential contamination; no evidence of sample contamination was observed. HIV-1 Sequence Quality Analysis (www.hiv.lanl.gov/content/sequence/QC/index.html) showed that the gene structure of the full-length genome sequence was normal with the nine open reading frames (ORFs) intact and opened.
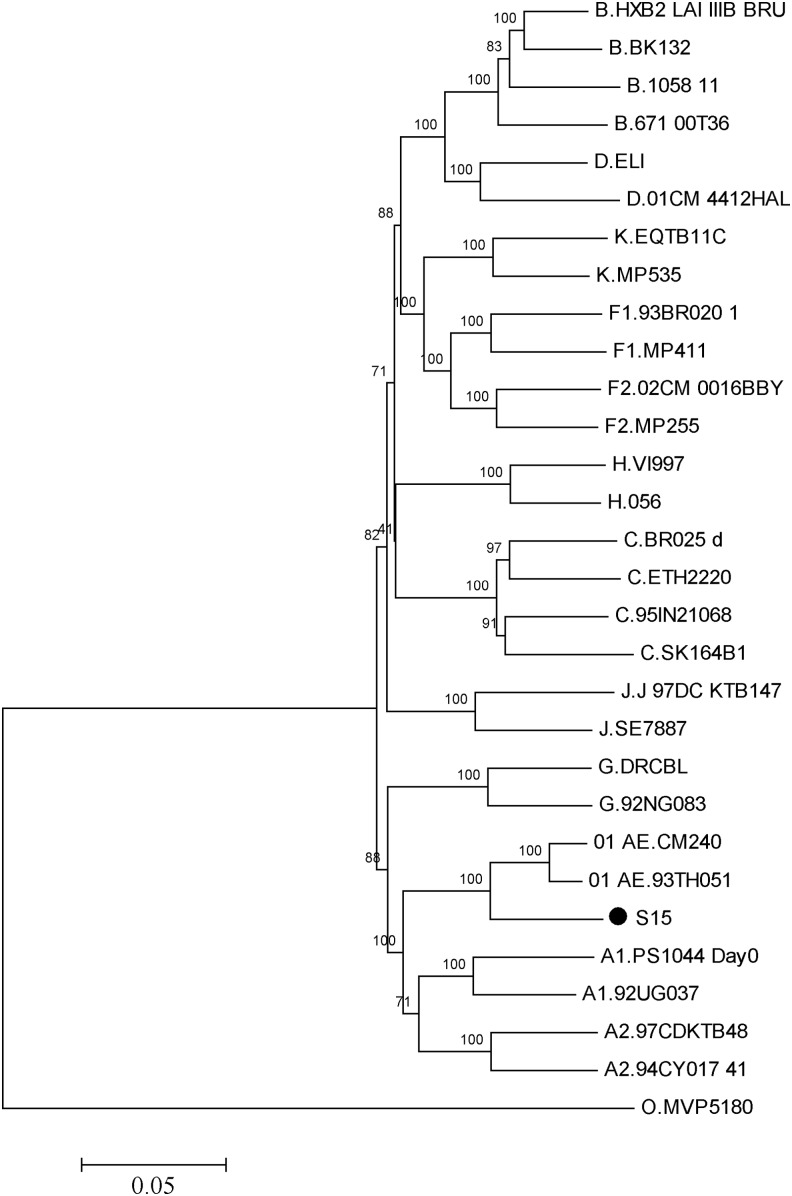
The HIV genotype was determined using the national center for biotechnology information viral genotyping tool (www.ncbi.nih.gov/projects/genotyping/formpage.cgi) and further confirmed by phylogenetic analysis with reference sequences using the neighbor-joining method in MEGA5.0 software and the maximum likelihood (ML) method in PhyML software.9 For phylogenetic analysis, the S15 full-length genome sequence was aligned with HIV-1 strains of various reference subtypes (A–D, F–H, J, and K) and CRF01_AE obtained from the Los Alamos HIV Database (http://hiv-web.lanl.gov/) using Clustal W software. The alignment was then edited manually in BioEdit software (version 7.0.0; T. Hall, North Carolina State University, Raleigh, NC) and converted into Fasta format. A neighbor-joining (NJ) tree was constructed using the Kimura two-parameter method including both transitions and transversions with MEGA3.1. An ML tree was constructed using the GTR+I+G substitution model. The reliability of topologies was estimated by performing bootstrap analysis with 500 replicates for the NJ tree and 100 replicates for the ML tree. The S15 sequence clustered together with CRF01_AE subtype reference sequences but set up a distinct branch (Fig. 1).
FIG. 1.
Phylogenetic tree analysis. A neighbor-joining tree was created with full-length genomes of our S15 strain (labeled with a black dot) sequence and the reference sequences of subtype A–D, F–H, J, K, CRF01_AE, and group O (http://hiv-web.lanl. gov/). Each reference sequence is labeled with the HIV-1 subtype, followed by the sequence name. The bootstrap probability (more than 70%) based on 500 replicates is shown on the nodes. The scale bar represents 5% genetic distance (0.05 substitution per site).
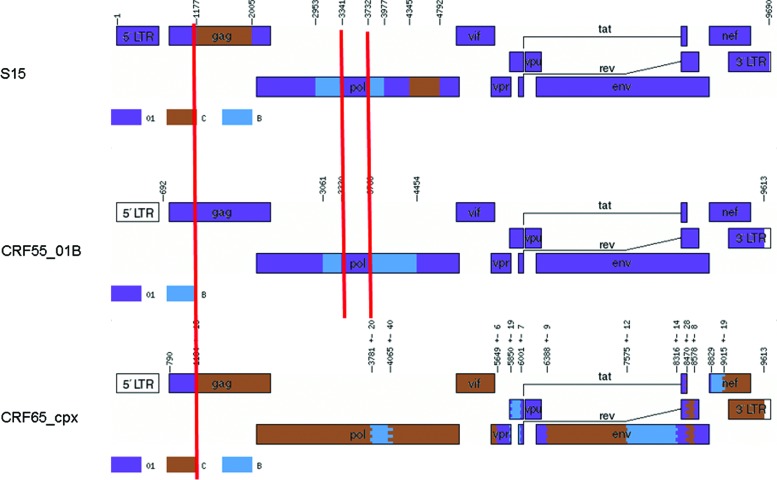
The possible intertype mosaicism was screened with the online Recombination Identification Program (version 3.0; http://hiv-web.lanl.gov). Online software jpHMM-HIV10 was further performed to determine the breakpoints. The Map-Draw Tool available at the Los Alamos HIV sequence database was used to generate the genomic map of S15 (Fig. 2). The results showed that two subtype B fragments and two subtype C fragments were inserted into the CRF01_AE backbone. The subtypes of each of the segments were further verified using phylogenetic analysis with the reference sequences of subtype A–D, F–H, J, and K (http://hiv-web.lanl.gov/) and gave the same results as the RIP and jpHMM analysis (data not shown). CRF65_cpx was the only CRF involving CRF01_AE, B, and C reported in China.11 Therefore the similarities of breakpoints between our strain and CRF65_cpx were further compared, which showed that both of them share one recombination breakpoint at the 1209 site according to the HXB2 calibrator (Fig. 2). The similar genomic structure from the 5′ end to the 2005 site (HXB2 calibrator) between the S15 strain and CRF65_cpx suggested that they might share a common ancestor.
FIG. 2.
Comparison among the mosaic structures of CRF55_01B, CRF65_cpx, and our S15 strain. The Map-Draw Tool available at the Los Alamos HIV sequence database was used to generate the mosaic structure. The same breakpoints in different genome schematics were labeled with a red line. The breakpoints of the S15 strain were calculated with jpHMM-HIV software (http://jphmm.gobics.de/), while the breakpoints of CRF55_01B and CRF65_cpx were cited from the Los Alamos HIV sequence database. Color images available online at www.liebertpub.com/aid
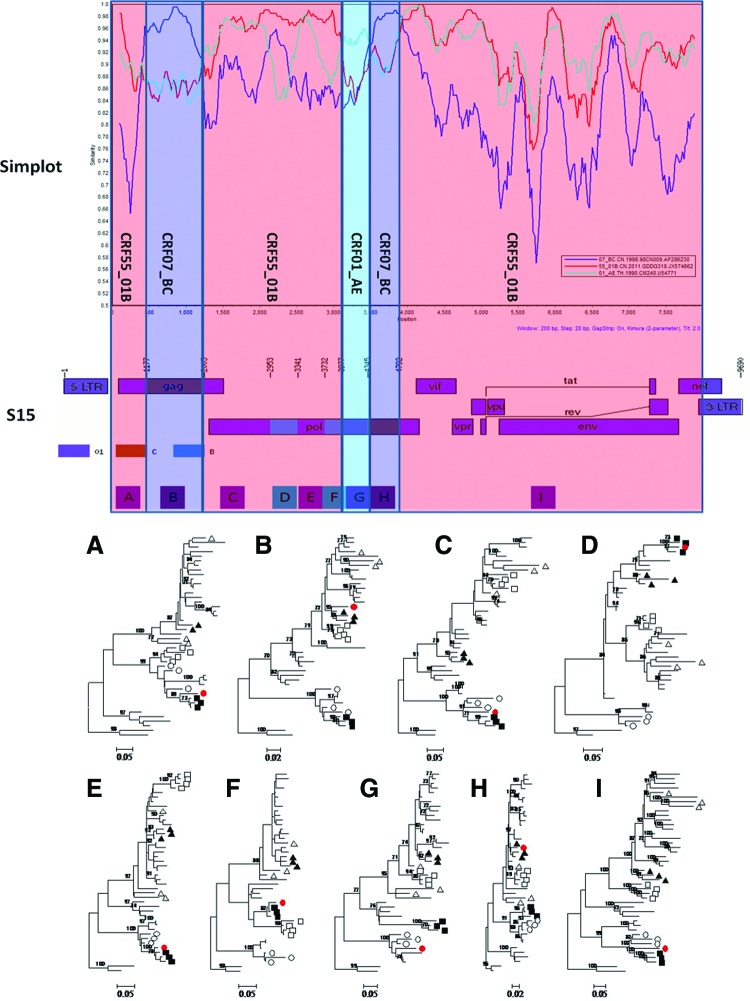
To explore the phylogenetic relationship of each subtype segment delimited for the S15 strain, all of the segments were analyzed and compared to all of the CRFs identified in China and subtype B, C, and J reference strains through the ML phylogenetic tree (Fig. 3). Among nine mosaic segments of S15 strains, six segments (A, C–F, and I) clustered with CRF55_01B strains, two segments (B and H) clustered with CRF07_BC strains, while one segment (G) clustered with CRF01_AE and CRF59_01B strains (Fig. 3). The results were further proven by similarity plot analysis (version 3.5.1; S. Ray, Johns Hopkins University, Baltimore, MD; http://sray.med.som.jhmi.edu/RaySoft/SimPlot/) using reference strains of subtype CRF55_01B, CRF07_BC, and CRF01_AE after removing all of gaps in the alignment using online Gapstreeze software (www.hiv.lanl.gov/content/hiv-db/GAPSTREEZE/ strip_ready.html) (Fig. 3). The data suggested that the S15 strain might be the recombinant form of CRF55_01B, CRF07_BC, and CRF01_AE. Although segments A and B of the S15 strain shared a structure similar to CRF65_cpx, neither of them clustered with the CRF65_cpx strain in the phylogenetic tree, which suggested that the 1209 sites might be the hot sites suitable for CRF01_AE and B subtype recombination. Analysis of more recombinant forms composed of CRF01_AE and B segments will be helpful to illustrate this.
FIG. 3.
Evolutionary relationship of nine individual fragments of S15 sequences with subtype B, C, and all of the circulating recombinant forms (CRFs) identified in China. Similarity plots of the S15 strain against subtype reference strains, including CRF07_BC (98CN009), CRF55_01B (GDDG318), and CRF01_AE(CM240). The x-axis indicates the nucleotide position along the alignment (gap stripped). The y-axis denotes the distance between the sequences compared. SimPlot was performed using a window size of 200 bp and a step size of 20 bp. Maximum likelihood (ML) trees were created with A-I individual fragments of the S15 sequence (red dot) and reference sequences of subtype B, C (▵), J, CRF01_AE (○), CRF07_BC (▴), CRF08_BC, CRF55_01B (▪), CRF57_BC, CRF59_01B, CRF61_BC, CRF62_BC, CRF64_BC, and CRF65_cpx (□). The lengths of sequences used in the analysis were 1167 (A), 828 (B), 948 (C), 388 (D), 391 (E), 245 (F), 368 (G), 447 (H), and 4899 (I) base pairs, respectively, by using HXB2 as the reference genomic. Branch support values greater than 70% are indicated at the corresponding nodes of the tree. The ML tree was constructed using the GTR+I+G substitution model, which was selected with jModeltest software. Color images available online at www.liebertpub.com/aid
In this article we reported a novel recombinant strain identified in Shenzhen, China. Although there is no evidence that this particular recombinant is spreading in the city, the close phylogenetic relationship between the strain and CRF55_01B and CRF07_BC suggests that both variants are prevalent in the same population in Shenzhen and also indicates an urgent need for a genetic survey of HIV-1 in the area.
Sequence Data
The nucleotide sequence of S15 has been submitted to GenBank with the accession number KP170487 (Supplementary Sequence S1; Supplementary Data are available online at www.liebertpub.com/aid).
Supplementary Material
Acknowledgments
This work was supported by the National Key S&T Special Projects on Major Infectious Diseases (grants 2012ZX10001-002 and 2012ZX10001-006) and the National Natural Science Foundation of China (grant 81273137).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Hemelaar J, Gouws E, Ghys PD, and Osmanov S: Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS 2011;25:679–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He X, Xing H, Ruan Y, et al. : A comprehensive mapping of HIV-1 genotypes in various risk groups and regions across China based on a nationwide molecular epidemiologic survey. PLoS One 2012;7:e47289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han X, An M, Zhang W, Cai W, Chen X, Takebe Y, et al. : Genome sequences of a novel HIV-1 circulating recombinant form, CRF55_01B, identified in China. Genome Announcements 2013;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao J, Cai W, Zheng C, et al. : Origin and outbreak of HIV-1 CRF55_01B among MSM in Shenzhen, China. J Acquir Immune Defic Syndr 2014;66:e65–67 [DOI] [PubMed] [Google Scholar]
- 5.Zhao GL, Yu W, Zhang JJ, et al. : [Study on the molecular-epidemiological characteristics of HIV-1 in Shenzhen, 1992–2008]. Zhonghua Liu Xing Bing Xue Za Zhi 2012;33:82–87 [PubMed] [Google Scholar]
- 6.Takebe Y, Motomura K, Tatsumi M, et al. : High prevalence of diverse forms of HIV-1 intersubtype recombinants in Central Myanmar: Geographical hot spot of extensive recombination. AIDS 2003;17:2077–2087 [DOI] [PubMed] [Google Scholar]
- 7.Li L, Liang S, Chen L, et al. : Genetic characterization of 13 subtype CRF01_AE near full-length genomes in Guangxi, China. AIDS Res Hum Retroviruses 2010;26:699–704 [DOI] [PubMed] [Google Scholar]
- 8.Wang Z, Li J, Li L, et al. : Construction and characterization of a full-length infectious molecular clone from the HIV type 1 subtype Thai-B isolated in Henan province, China. AIDS Res Hum Retroviruses 2008;24:251–257 [DOI] [PubMed] [Google Scholar]
- 9.Guindon S. and Gascuel O: A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 2003;52:696–704 [DOI] [PubMed] [Google Scholar]
- 10.Zhang M, Schultz AK, Calef C, et al. : jpHMM at GOBICS: A web server to detect genomic recombinations in HIV-1. Nucleic Acids Res 2006;34:W463–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng Y, Wei H, Hsi J, et al. : Identification of a novel HIV type 1 circulating recombinant form (CRF65_cpx) composed of CRF01_AE and subtypes B and C in Western Yunnan, China. AIDS Res Hum Retroviruses 2014;30:598–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.