Significance
The constant generation of olfactory sensory neurons throughout life makes the system appealing for studies of the mechanisms of axon extension and connectivity. Understanding the mechanisms leading to the genesis of these new neurons is fundamental for the development of therapeutic treatments. We provide here, to our knowledge, the first detailed analysis of the sequential steps leading toward fully differentiated sensory neurons. We show that odorant receptors are not involved in the initial steps of differentiation, but only later in the final process of maturation. Moreover, the proteins that we studied here also have been implicated in normal and pathophysiological events ranging from kidney function to cancer development, making our data valuable across different disciplines.
Keywords: olfactory epithelium, axon guidance, tamoxifen, olfactory marker protein, Ascl1
Abstract
Odorant receptors (OR) are strongly implicated in coalescence of olfactory sensory neuron (OSN) axons and the formation of olfactory bulb (OB) glomeruli. However, when ORs are first expressed relative to basal cell division and OSN axon extension is unknown. We developed an in vivo fate-mapping strategy that enabled us to follow OSN maturation and axon extension beginning at basal cell division. In parallel, we mapped the molecular development of OSNs beginning at basal cell division, including the onset of OR expression. Our data show that ORs are first expressed around 4 d following basal cell division, 24 h after OSN axons have reached the OB. Over the next 6+ days the OSN axons navigate the OB nerve layer and ultimately coalesce in glomeruli. These data provide a previously unidentified perspective on the role of ORs in homophilic OSN axon adhesion and lead us to propose a new model dividing axon extension into two phases. Phase I is OR-independent and accounts for up to 50% of the time during which axons approach the OB and begin navigating the olfactory nerve layer. Phase II is OR-dependent and concludes as OSN axons coalesce in glomeruli.
In the mouse olfactory system, olfactory sensory neurons (OSNs) extend their axons from the olfactory epithelium (OE) to the olfactory bulb (OB), where they converge to form glomeruli. Each OSN expresses only 1 of ∼2,400 candidate odorant receptor (OR) alleles. OSNs expressing the same OR can be widely dispersed in the OE, yet their axons converge in only two to three molecularly specific glomeruli of a possible 3,700 (1). It was first recognized almost 20 y ago that substituting an OR-coding region with that of a different OR resulted in the glomerular convergence of axons at an ectopic location relative to that of the native ORs (2). This led to the suggestion that ORs have an instructive role in the extension and glomerular coalescence of OSN axons, most likely mediated by homophilic fasciculation (3–5).
Postnatal OSNs are derived from self-renewing precursors located proximal to the deep basal lamina of the OE (6). Following the division of globose basal stem cells, OSN neuroblasts transiently express Achaete-scute homolog 1 (Ascl1) followed by two phases of differentiation (6–8). Initially, they express growth-associated protein–43 (GAP-43), a marker of immature cells. Subsequently, the OSNs down-regulate GAP-43 and express olfactory marker protein (OMP), a universal marker of mature OSNs.
Although there is a consensus on the involvement of ORs in OSN axon glomerular convergence, when ORs exert that influence following basal cell division or axon extension is not known. Moreover, the developmental progression of GAP-43 to OMP, as a measure of OSN differentiation and maturation, or of adenylate cyclase 3 (AC3), a downstream signaling molecule also implicated in axon extension, has not been considered in the context of OR expression or OSN dynamics. Here, we determined when ORs exert their influence on OSN axons. We assessed two fundamental questions: (i) When do OSNs express ORs relative to progenitor cell division and the expression of GAP-43, OMP and AC3? (ii) How does the extension of OSN axons correlate with OR onset and the molecular differentiation of OSNs?
Results
OSN Expression of GAP-43, OMP, and AC3.
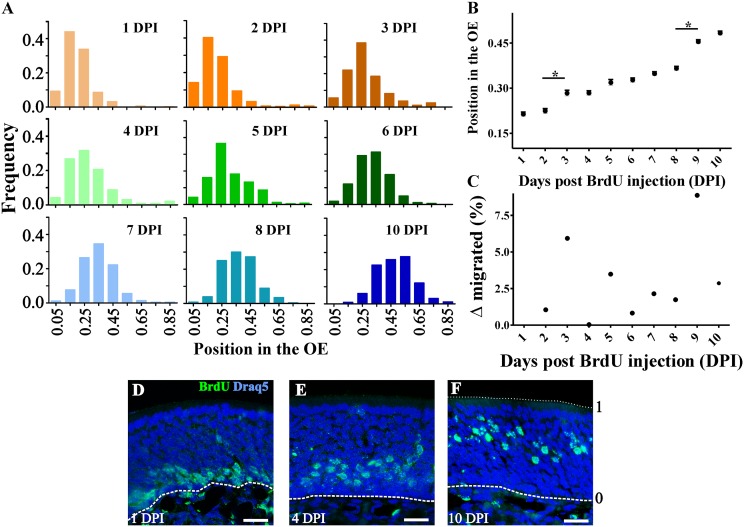
Following globose basal cell mitosis, OSNs migrate from the stem cell niche at the basal lamina toward the apical surface of the OE. To determine how molecular differentiation correlates with radial migration of OSNs through the OE, we injected BrdU at postnatal day (PND) 7 and examined the position of BrdU+ nuclei at 24-h intervals from 1 to 10 d postinjection (DPI). At 1 DPI, BrdU+ nuclei were restricted to the OE immediately adjacent to the basal lamina. By 10 DPI, BrdU+ nuclei had migrated to the upper lamina of the OE (Fig. 1A). The migratory rate of OSNs in the OE was constant, <3.5% of the OE width/day, except between 2–3 DPI (∼6%) and 8–9 DPI (∼9%) (Fig. 1 B and C).
Fig. 1.
OSNs migrate radially in the OE. (A) Histogram distributions showing BrdU+ cells relative frequency in the OE. The basal lamina is considered as 0 and the surface of the OE as 1 (shown in F). Relative position of BrdU+ cells expressed as mean ± SEM (B; *P < 0.001) and migrated distance (C). (D–F) Representative images of BrdU+ (green) labeling at 1 (D), 4 (E), and 10 (F) DPI. Nuclear marker Draq-5 is shown in blue. Dotted and dashed lines delineate the surface and the basal lamina of the OE, respectively. (Scale bars, 20 μm.)
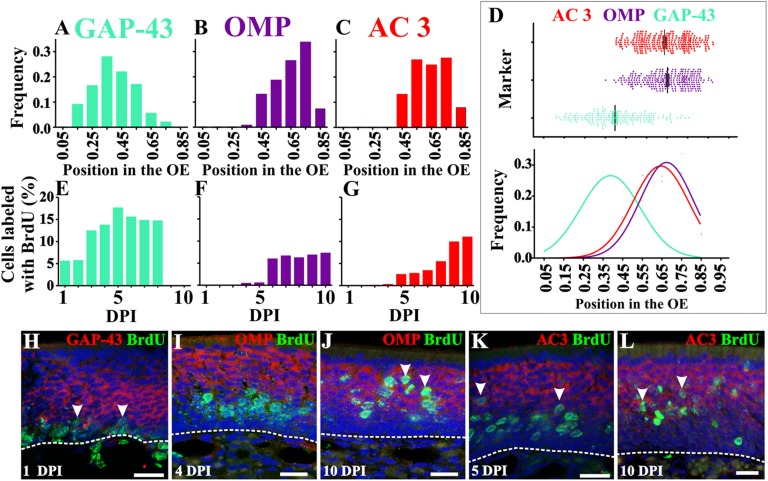
We then analyzed the spatial distribution of the OSN markers: GAP-43 for immature OSNs, OMP for mature OSNs, and AC3, an OR downstream signaling molecule. Using in situ hybridization, we determined that GAP-43+ cells were preferentially located in the lower half of the OE (Fig. 2 A, D, and H), whereas OMP+ cells were restricted to the upper half of the OE (Fig. 2 B, D, I, and J). The laminar distribution of AC3+ cells did not overlap with the GAP-43+ cells, but overlapped almost perfectly with OMP+ cells (Fig. 2 C, D, K, and L), although the AC3+ cells did have a more basal distribution (slight shift to the left in Fig. 2D), indicating that expression may precede that of OMP.
Fig. 2.
Timeline of GAP-43, OMP, and AC3 expression. Distribution of GAP-43+ (A), OMP+ (B), and AC3+ (C) OSNs in the OE. (D) Scatter plot for GAP-43, OMP, and AC3 showing their relative position in the OE (Top) and normal distributions associated with histograms (Bottom). Although largely restricted to the lower lamina of the OE, GAP-43 cell bodies exhibit a wider distribution than OMP or AC3. Quantification of cells double-labeled with BrdU and GAP-43 (E), OMP (F), or AC3 (G). Although some OMP+ and AC3+ cells were also BrdU+ at earlier days, these were very few (<0.05%) and inconsistent across subjects. (H–L) BrdU+ staining in green and in situ hybridization in red for GAP-43 (H), OMP (I and J), and AC3 (K and L). Arrowheads indicate double-labeled cells. Dashed lines delineate OE basal lamina. Draq-5 is in blue. (Scale bars, 20 μm.)
OR Expression Relative to GAP-43, OMP, and AC3.
Our next goal was to assess OSN differentiation and maturation by determining the spatial-temporal expression of GAP-43, OMP, and AC3 as a function of basal cell division. Based upon the data thus far, we hypothesized that the temporal expression would be GAP-43 > AC3 > OMP. We first tested this hypothesis with probability theory to predict colocalization based on our data from location of BrdU+ nuclei in the OE and the spatial distribution of OSN markers. This analysis suggested that GAP-43/BrdU cells appear at 1 DPI, AC3/BrdU at 5–6 DPI, and OMP/BrdU at 6+ DPI (SI Materials and Methods and Fig. S1 A–G). We then tested these predictions using in situ hybridization for GAP-43, OMP, or AC3 combined with BrdU immunohistochemistry to establish time of cell division. As our model predicted, we detected double-labeled GAP-43+/BrdU+ OSNs at 1 DPI (Fig. 2 E and H and Fig. S2) although not all BrdU+ cells were GAP-43+ at this time. Therefore, an increasing number of colabeled cells were detected in the following days (from 5% at 1 DPI to >17% at 5 DPI). Also, as our model predicted, BrdU+ cells that colocalized with OMP+ were first evident at 6 DPI, and their frequency remained stable in subsequent days (Fig. 2 F and J and Fig. S2). Finally, double-labeled AC3+/BrdU+ cells consistently appeared at 5 DPI (∼2.5%), also in agreement with our predictive model (Fig. 2 G and K and Fig. S2). These data suggest that the earliest time point at which an OSN can be labeled as “mature,” at least based on OMP expression, is 6 d postdivision.
Despite compelling evidence in the literature that ORs instruct OSN axons, the timing of OR expression relative to OSN differentiation and axon extension is not known.
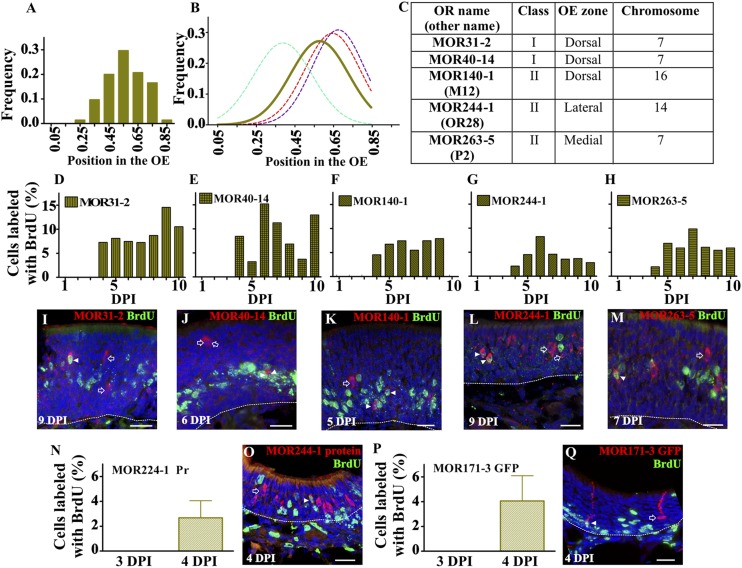
Therefore, we asked: When are ORs expressed during OSN maturation? We chose to use in situ hybridization to determine the earliest time that ORs can be translated following cell division because it is a more conservative approach compared with immunohistochemical detection of ORs, given that mRNA expression precedes protein expression. ORs can be grouped according to class (I and II), broad OE domains of expression (dorsal, medial, and lateral), and chromosome location. We assessed a total of five ORs that included both class I and class II, each of the OE domains (including a dorsal class II OR), and three different chromosomes. We first analyzed the laminar distribution in the OE of four OR-expressing cell populations (MOR31-2, MOR40-14, MOR140-1, and MOR263-5). The expression of MOR244-1 in the lateral domain where the thickness of the OE is narrow precluded us from including this OR in the laminar analysis. The laminar distribution showed OR-expressing cells in the upper lamina of the OE; hence, the spatial distribution of ORs falls apical to GAP-43 and basal to OMP and AC3 (Fig. 3 A and B). This distribution profile with our probability theory predicts that staining for ORs and BrdU will colocalize around 3–4 DPI (Fig. S1 H and I). To test this prediction, we determined the onset of OR expression following cell division using in situ hybridization for each of the five ORs coupled with BrdU immunohistochemistry (Fig. 3C). The earliest time that we observed cells double-labeled with an OR and BrdU was 4 DPI (Fig. 3 C–M and Fig. S3). We then examined the translation of two OR mRNAs using an antibody specific to MOR244-1 and a transgenic mouse in which we detected MOR171-3 OSNs via GFP expression. In both cases translation was first detected at 4 DPI, in agreement with the in situ data (Fig. 3 N–Q and Fig. S4). Hence, the onset of both mRNA and protein was consistent across all of the ORs tested. This strongly suggests that the onset of OR expression relative to time of cell division is independent of OR class, epithelial domain, or chromosome location.
Fig. 3.
ORs are expressed beginning at 4 DPI. (A) Distribution of MOR31-2, MOR40-14, MOR140-1, and MOR263-5 in the OE. (B) Normal distribution of ORs (solid line) in relation to the three classical markers (dotted lines). (C) OR classification based on class, OE zones of expression, and chromosome location. (D–H) Quantification of cells double-labeled with BrdU and MOR31-2 (D), MOR40-14 (E), MOR140-1 (F), MOR244-1 (G), and MOR263-5 (H) shows OR onset at 4 DPI. (I–M) BrdU staining in green with in situ hybridization in red for MOR31-2 (I), MOR40-14 (J), MOR140-1 (K), MOR244-1 (L), and MOR263-5 (M). Open arrows show cells that are OR+/BrdU−, and arrowheads identify OSNs double-labeled, OR+/BrdU+. Quantification of cells double-labeled with BrdU, MOR244-1 protein (N), and MOR171-3 GFP (P) shows translation of OR onset at 4 DPI. BrdU staining in green with immunohistochemistry in red for MOR244-1 (O) and MOR171-3 GFP (Q). Dashed lines delineate OE basal lamina. Draq-5 in blue. (Scale bars, 20 μm.)
OSN Axon Extension and Molecular Differentiation.
Our data thus far show that OSNs express ORs after GAP-43 and a minimum of 24 and 48 h before AC3 and OMP detection, respectively. We next wanted to understand the spatial-temporal expression of these developmentally regulated genes relative to extension of OSN axons to the OB. To begin addressing this gap in our knowledge, we devised two methods to assess OSN axon extension. First, we injected a fluorescent dextran into the most anterior part of the OB. At the time of injection, OSN axons that had reached the anterior OB retrogradely transported the dextran, allowing us to determine the laminar location of OSN somata in the OE. Using this approach, dextran-labeled somata were primarily located in the upper lamina of the OE. The spatial distribution of the dextran+ cells overlapped with the expression of ORs, AC3, and OMP (Fig. S5 A and B). Triple staining for GAP-43, OMP, and dextran showed that >70% of the dextran+ cells were also OMP+ whereas <10% were GAP-43+ (Fig. S5 C–E). These data suggest that under the conditions of this experimental protocol the majority of axons that are dextran+ and that project to the dorsal portion of the OB are those of mature OSNs.
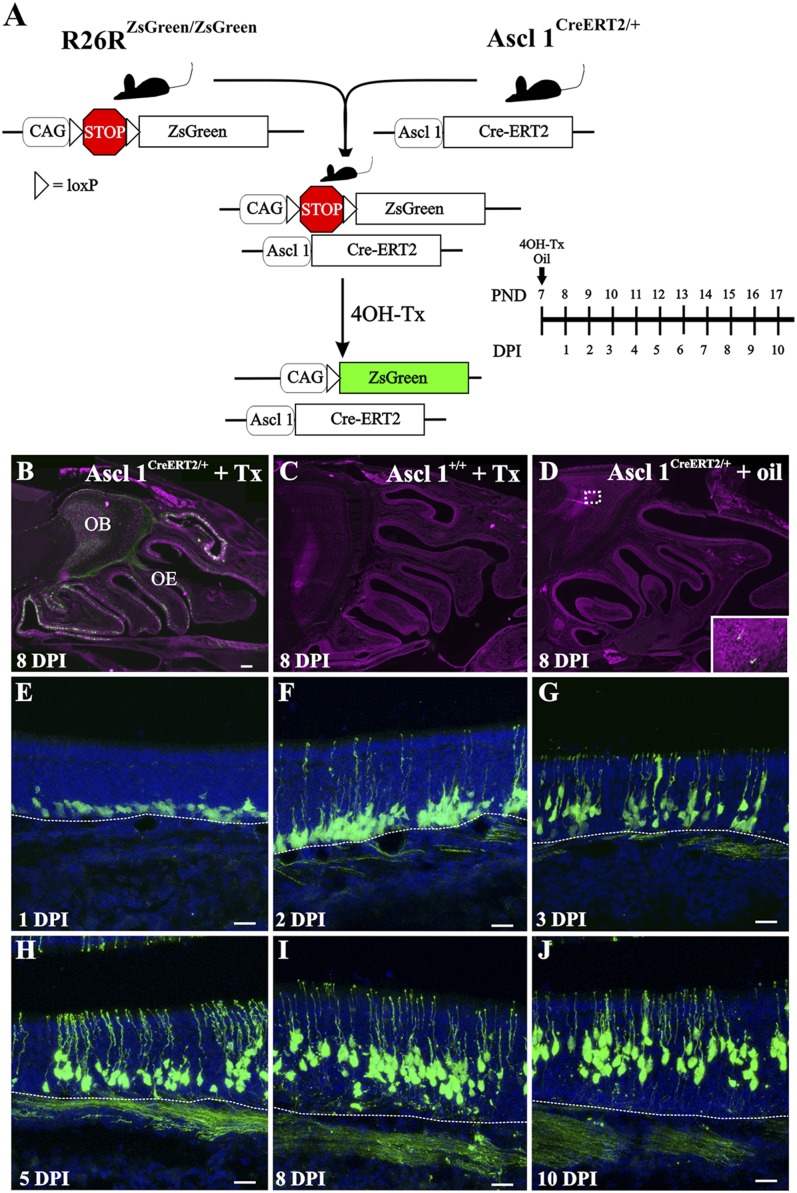
These experiments revealed which OSN axons had reached or extended beyond the point of dextran injection in the OB. The dextran labeling did not provide any insight into the progression of OSN axon development or the location of the GAP-43+ axons. Therefore, we developed an innovative strategy to assess OSN axon extension. By devising an in vivo genetic fate-mapping strategy, we could examine the spatial-temporal characteristics of OSN axon extension from the OE into the OB. We used the Asc11 promoter, which is expressed in globose basal cells in the OE, linked to an inducible Cre enzyme. Ascl1CreERT2/+ mice (9) were crossed with R26RZsGreen/ZsGreen reporter mice. The Cre enzyme fused to the estrogen receptor (CreERT2) remains inactive in the cytoplasm. Following 4OH-Tamoxifen (4OH-Tx) injection, CreERT2 translocates into the nucleus, allowing for recombination and ZsGreen expression. We first tested the specificity of this model by injecting both experimental (Ascl1CreERT2/+; R26RZsGreen/-) and control (Ascl1+/+; R26RZsGreen/-) mice at PND 7 with 4OH-Tx or sunflower oil (vehicle) (Fig. 4A). Ascl1CreERT2/+; R26RZsGreen/- mice showed extensive expression of recombined ZsGreen+ cells in the OE and axons in the OB by 8 DPI (Fig. 4B) Importantly, this recombination was dependent on CreERT2 expression and 4OH-Tx as the control Ascl1+/+; R26RZsGreen/- mice injected with 4OH-Tx showed no recombined ZsGreen+ cells (Fig. 4C). Similarly, the experimental Ascl1CreERT2/+; R26RZsGreen/- mice did not have any significant ZsGreen+ cells scattered in the OE or the OB when injected with oil (Fig. 4D). Hence, we restricted further analyses to the experimental Ascl1CreERT2/+; R26RZsGreen/- mice and examined them at 24-h intervals following a single injection of 4OH-Tx.
Fig. 4.
Ascl1CreERT2 mice track OSNs in the OE following basal cell division. (A) Strategy for labeling a newly generated subpopulation of OSNs. Timeline shows the relation between DPI and PND. (B–D) Experimental Ascl1CreERT2/+;R26RZsGreen/- mice show extensive ZsGreen expression in OE and OB at 8 DPI after 4HO-Tx (B) and sparse labeling (fewer than four cells per section) after oil injection (D). Control Ascl1+/+;R26RZsGreen/- mice show no labeling after 4HO-Tx (C). (E–J) ZsGreen+ cells are immediately adjacent to the basal lamina (E). Labeled cell bodies migrate radially toward the lumen as they develop (F–J), suggesting that Ascl1 cells do not or very rarely divide. Apical dendrites and axons could be seen from 2 DPI onward. Dashed lines delineate OE basal lamina. Draq-5 is in magenta (B–D) and blue (E–J). [Scale bars, 200 μm (B–D); 20 μm (E–J).]
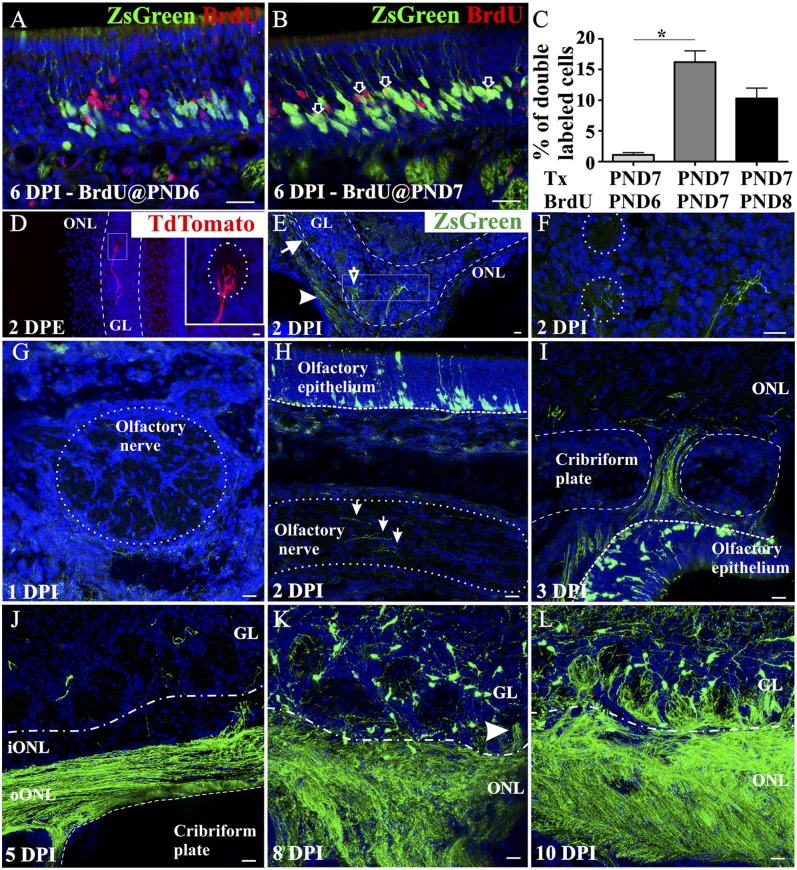
To determine if ZsGreen expression following tamoxifen and BrdU labeling are equivalent measures of cell fate, we carried out the following experiments. We administered 4OH-Tx at PND 7 and injected BrdU 24 h earlier (PND 6), simultaneously (PND 7) or 24 h later (PND 8). When administered together, the percentage of ZsGreen+ OSNs colabeled with BrdU was significantly higher than when BrdU was administered 24 h earlier (Fig. 5 A–C). The injection of BrdU 24 h after 4OH-Tx showed a decrease in the number of double-labeled cells, suggesting that some of the recombined cells can undergo mitosis in the 24-h period following 4OH-TX. Nevertheless, of relevance for our analysis, the earliest and highest number of double-labeled cells were obtained when both injections were done simultaneously, indicating that both BrdU and 4OH-Tx–induced recombination are labeling the same population of cells within a narrow time window of basal cell division.
Fig. 5.
Tracking OSN axons in Ascl1CreERT2 mice. Experimental mice were injected with 4OH-Tx at PND 7 and with BrdU at PND 6 (A and C), at PND 7 (B and C), or at PND 8 (C) (n = 4 for each group). Higher levels of double-labeled cells are observed when both injections are done simultaneously (C), suggesting that BrdU and ZsGreen label the same population of OSNs within the narrow time window of basal cell division. The single asterisk indicates statistically significant (P < 0.05). Within 48 h following a postnatal electroporation of a Tdtomato-expressing plasmid, fluorescent axons can be detected branching in the glomerular layer (D). (Inset) A higher magnification of the square. Electroporation of a CreERT2 plasmid in R26RZsGreen/- mice, followed by 4OH-Tx 24 h later, shows ZsGreen expression in the outer ONL (oONL, arrowhead in E), the inner ONL (iONL, arrow in E), and the glomeruli (open arrow in E). Higher magnification of the square in E is shown in F. (G–L) ZsGreen+ axons are not observed in axon fascicles at 1 DPI (G), and only a few labeled axons are present by 2 DPI (arrows in H). At 3 DPI labeled axons cross the cribriform plate (I) and at 5 DPI (J) remain in the oONL and rarely transition into the iONL. By 8 DPI, ZsGreen+ axons enter the ventral glomerular layer (GL, arrowhead in K) and by 10 DPI innervate most ventral glomeruli (L). Draq-5 in blue. (Scale bars, 20 μm.)
Analysis of the recombined OSNs revealed that by 1 DPI ZsGreen+ cells were located in the most basal part of the OE immediately adjacent to the basal lamina (Fig. 4E). From 1 DPI onward the location of the ZsGreen+ OSN somata showed the same radial migration toward the more superficial lamina of the OE that we described for BrdU+ cells. In contrast to the BrdU labeling, ZsGreen labeled the OSNs in their entirety so that the development of both the apical dendrites and axons could be followed. At 1 DPI, apical processes extending toward the surface of the OE were occasionally seen (Fig. S6A). By 2 DPI, many apical dendrites reached the surface of the OE and terminated in a dendritic knob within the nasal cavity (Fig. 4F). Those processes that had not yet reached the lumen of the nasal cavity terminated in enlarged elliptical tips, consistent with dendritic growth cones (Fig. S6C′′′, arrowheads). By 3 DPI, the frequency of apical dendrites reaching the surface of the OE increased (Fig. 4G). As noted above, the recombined ZsGreen+ OSN somata continued their radial migration in the OE (Fig. 4 G–J), suggesting that Ascl1-mediated recombination occurred in a cell population that has limited, if any, capacity for further division.
Because the speed of axonal transport could influence the interpretation of our data, before analyzing OSN axon extension we performed two control experiments to assess the axonal transport of fluorescent proteins in OSNs. Postnatal electroporation of the OE labels OSNs, independent of their birthdate. Therefore, mature OSNs whose axons had already reached the OB can be labeled, and we can assess the time required for a fluorescent marker to appear in the OB. We first electroporated the OE with a Tdtomato-expressing plasmid in control mice and looked for fluorescently labeled axons 48 h later (Fig. 5D). In a second set of controls, we electroporated R26RZsGreen/- mice with a CreERT2-expressing plasmid, induced the expression with 4OH-Tx 24 h later, and analyzed the expression of ZsGreen in the OB 48 h later (Fig. 5 E and F). In both experiments, fluorescently labeled OSN axons were observed within 48 h in the outer and inner olfactory nerve layer and the glomerular layer where mature axonal arbors were evident within the glomerular neuropil.
Finally, we turned our attention to the behavior of 4OH-Tx–induced ZsGreen-labeled OSN axons while they navigate toward the OB. At 1 DPI, no ZsGreen+ axons were observed in nerve fascicles in the lamina propria (Fig. 5G and Fig. S7 A–C). The first ZsGreen+ axons were found in the olfactory nerve fascicles in the lamina propria deep in the OE at 2 DPI and were capped by distinctive enlargements most plausibly identified as axonal growth cones (Fig. 5H, arrows and Fig. S6 B′′, C′′, and C′′′′, arrows). However, at 2 DPI no axons were observed in the foramina of the cribriform plate or in the OB olfactory nerve layer (ONL) (Fig. S7D). The first evidence of OSN axons crossing the cribriform plate and joining the ONL came at 3 DPI (Fig. 5I). Given that in the control electroporation experiments we observed fluorescent OSN axons in the glomeruli within 48 h, we conclude that 4OH-Tx induction clearly reveals the extension of axons and not simply the speed of axoplasmic transport of fluorescent proteins. At 4 and 5 DPI, OSN axons were restricted to the outer ONL (10) (Fig. 5J and Fig. S7E). It was not until 6 DPI that ZsGreen+ axons entered the inner ONL (Fig. S7F). The first ZsGreen+ axons reached the glomerular layer at 8 DPI and were double-labeled for OMP (Fig. 5K, arrowhead and Fig. S7I), although only a few ventral glomeruli contained ZsGreen+ axons (Fig. S7G). By 10 DPI, glomeruli in the ventral part of the OB were robustly innervated by ZsGreen+ axons (Fig. 5L). By contrast, most of the anterior and dorsal glomeruli showed no evidence of innervation by recombined OSN axons at 10 DPI (Fig. S7H). The differential innervation between the dorsal and ventral glomeruli is consistent with our dextran experiments. Following dextran injections into the dorsal OB, predominately OMP+ OSNs were retrogradely labeled, suggesting that it may take longer for dorsally directed OSN axons to coalesce and converge into glomeruli.
Discussion
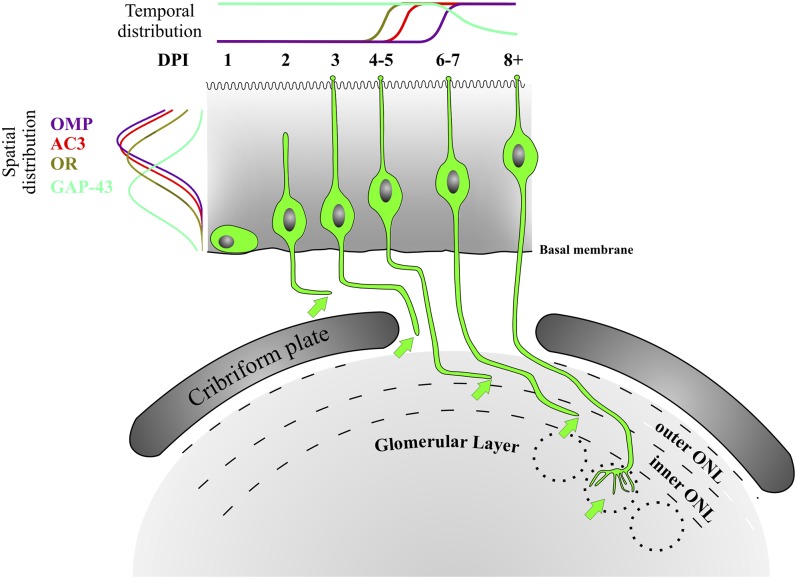
The most significant finding to emerge from these studies is the late onset of OR expression in OSN neuroblasts following basal cell division. Our data establish that OR onset occurs around 4 d following cell division, after the OSN axons have reached the olfactory bulb and begun to integrate into the olfactory nerve layer. An additional 4–10 d are required for OSN axons to navigate the olfactory nerve layer and coalesce in a glomerulus. Our fate-mapping strategy for OSNs provides, to our knowledge, the first detailed description of OSN axon navigation from the OE to the OB. Moreover, we correlate OSN axon extension with the time of cell division, expression of OSN genes, migration of OSNs within the OE, and extension of apical dendrites (Fig. 6). Our fate mapping, coupled with BrdU birthdating, establishes that, at the earliest, GAP-43 is detected within 24 h following division, OR mRNA and protein appear at 4 d, AC3 at 5 d, and OMP at 6 d postdivision. In designing these studies, we specifically selected PND 7 for the labeling to assay OSN axon growth when the primary targets in the OB were in place and the number of OSNs was rapidly increasing. To complement our findings, additional studies in the embryo, when the first axons approach the OB, and in the adult, after the pathway is fully established, will be important.
Fig. 6.
OSN differentiation with spatial-temporal gene profiles. The spatial distribution of OSN cell bodies expressing GAP-43 (cyan), ORs (light green), AC3 (red), and OMP (purple) within the olfactory epithelium is on the left. The temporal expression profile of these genes is at the top. The morphology of olfactory sensory neurons, in green, is correlated with the corresponding DPI. Arrows indicate the ending of the axon at each developmental stage. Dashed lines demarcate edges of the inner and outer ONL. Dotted circles represent glomeruli.
These results strongly suggest that OSN axons approach the OB and integrate into the outer ONL independently of OR expression. We previously reported that sorting of homophilic OSN axons (those expressing the same OR) was most evident in the inner ONL, proximal to their final convergence in a glomerulus (11). Similarly, adult OR reporter mice revealed that OSN axons expressing the same OR showed little or no homophilic fasciculation until they were proximal to the site of glomerular convergence (e.g., fig. 3 in ref. 1 and fig. 2D in ref. 3). Recent reports suggest that OSNs can switch to a different OR (12, 13) or switch between alleles (13, 14) before stably expressing one OR. Our data show that the earliest an OR can be detected by in situ hybridization or immunohistochemistry is 4 d following basal cell division. In some OSNs the initial expression of an OR may occur even later. In this case, the final OR choice and stabilization would not occur until the axons enter the inner ONL and initiate the OR-homophilic apposition that leads to glomerular convergence. Late onset of OR expression may represent a checkpoint to ensure that the axons have reached a critical point such as the cribriform plate. Similarly, a delay could prevent premature fasciculation of OSN axons expressing the same OR.
In addition to ORs, cAMP and downstream members of the transduction cascade can also influence axonal convergence, glomerular formation, and positioning in the OB (15–18). Specifically, blocking the interaction of the OR with Golf prevents OSN axons from entering the inner ONL (see fig. 1 in ref. 16). Interestingly, our data show that before moving into the inner ONL, AC3 expression is up-regulated in OSNs, suggesting that AC3, and therefore cAMP production, may be a prerequisite for OSN axons transitioning from the outer to the inner ONL. Similarly, we showed earlier that deletion of the HCN1 subunit, a direct downstream target of cAMP, resulted in a disruption in the organization of the inner/outer ONL (17). Collectively, these data suggest that the timing of AC3 expression is important for the dynamic behavior of OSN axons within the ONL.
The timing of OR expression that we show here advocates revisiting methods commonly used to study OR selection and OSN axon outgrowth. We and others have used the OMP promoter to force the expression of OR genes (19–24) as well as to knock out genes via Cre-recombinase (25–27). However, our current results show that OMP is expressed, at the earliest, 48 h after OR expression is initiated and after axons have reached the OB and drawn proximal to the point of glomerular convergence. The data presented here may explain why expression of one OR using the OMP promoter had little effect on OB architecture, but early induction of OR expression (using the Gγ8 promoter that is expressed in immature OSNs) altered glomeruli morphology (22). Although the function of OMP remains elusive, it was suggested that OMP expression is initiated by retrograde signals when OSN axons establish synaptic contacts (28). However, we show here that OMP is expressed at least 2 days before the earliest axons have converged into a glomerulus. Thus, OMP expression does not appear to be dependent on the OSN axons establishing synapses.
Our analyses of OMP, AC3, GAP-43, and ORs have used either in situ hybridization or immunohistochemistry, neither of which is entirely suitable for quantifying levels of expression. Future studies may benefit from the use of FACS to isolate ZsGreen-labeled cells for qPCR to determine if there are fluctuations in absolute levels during axon extension.
In summary, our results demonstrate that OSN axons approach the olfactory bulb and integrate into the outer olfactory nerve layer independently of odorant receptor expression. We therefore propose a new model in which the navigation of OSN axons occurs in two phases. The first phase, in which the axons exit the epithelium and enter the outer ONL, is OR-independent. The second phase, in which the axons begin to coalesce and then converge into glomeruli, is OR-dependent.
Materials and Methods
Animals.
CD-1 mice (Charles River) at PND 7 were injected intraperitoneally with the thymidine analog BrdU (50 mg/kg) twice, 2 h apart. Ascl1tm1.1(Cre/ERT2)Jejo/J mice (The Jackson Laboratory stock # 012882) were crossed with B6.Cg-Gt(ROSA)26Sortm6(CAG-ZsGreen1)Hze/J reporter mice (The Jackson Laboratory stock # 007906). Ascl-1 (Achaete-scute homolog 1) was formerly known as Mash-1. Pups were injected at PND 7 with BrdU as indicated above and 4HO-tamoxifen (40 mg/kg body weight) or sunflower oil. At different days postinjection, mice were anesthetized with pentobarbital and perfused transcardially with PBS [PBS: 0.1 M phosphate buffer (PB) and 0.9% NaCl, pH 7.4] with 1 unit/mL of heparin, followed by 4% (wt/vol) paraformaldehyde in PBS at 4 °C. Tissue was postfixed for 2 h in the same fixative, transferred to PBS, embedded, and kept at −20 until use. Postinjection times ranged from 1 to 10 DPI. M72-GFP mice were a kind gift of Peter Mombaerts, Max Planck Research Unit for Neurogenetics, Frankfurt. To determine if tamoxifen-induced ZsGreen expression and BrdU labeling are equivalent measures of cell fate, we administered 4OH-Tx at PND 7; injected BrdU 24 h earlier (PND 6), simultaneously (PND 7), or 24 h later (PND 8); and killed the animals at PND 13. All animal care and use was approved by the Yale University Animal Care and Use Committee. The thickness of the OE is variable, and to quantify the location of BrdU+ nuclei and GAP-43–, OMP-, OR-, or AC3-expressing cells, we normalized the data by designating the basal lamina = 0 and the apical surface of the OE = 1.
In Situ Hybridization.
Tissue was processed as previously described (1). Probes were obtained by in vitro transcription using the digoxigenin-labeling kit (Roche). MOR140-1 (M12), MOR244-1 (OR28), and MOR263-5 (P2) OR plasmids were gifts from P. Mombaerts, Max Planck Research Unit for Neurogenetics, Frankfurt. Adenylyl cyclase III (ACIII) plasmid was a gift from J. Pluznick, Johns Hopkins University, Baltimore. MOR31-2 and MOR40-14 plasmids were gifts from T. Bozza, Northwestern University, Evanston, IL. Other probes were obtained by RT-PCR and cloned into TOPO-TA II vector using the following primers: OMP—5′ TTCTGGCGCAAGGAAGAC 3′–5′ CTGGCCGTTTGCATCTCT 3′; GAP-43—5′ AACTCCCCGTCCTCCAAG 3′–5′ AACCGGGGTACAGTGCAA 3′; sense probe showed no signal. Double immunohistochemistry for BrdU and in situ hybridization was done following previously published protocols (1) and developed with HNPP/Fast Red (Roche). Images were acquired with a Leica confocal microscope. Digital images were color-balanced using Adobe Photoshop CS2 (Adobe Systems). The original 8-bit images were used for quantitative analyses. Figures were constructed using Corel Draw ×3 (Corel).
Immunohistochemistry.
Sections were stained with antibodies as previously described (29). The antibodies and detailed protocols are found in SI Materials and Methods.
Postnatal Electroporation.
Postnatal day 5 mouse pups were anesthetized by chilling on ice. DNA solutions (3 μg/μL) in TE containing 0.1% fast green were injected into one side of the nasal cavity using a Picospritzer with a glass pipette under a dissecting microscope. After DNA injection, tweezer-type electrodes (model 520, 5-mm diameter, BTX), briefly soaked, were placed to softly hold the heads of the pups, and three 100-V square pulses of 50-ms duration with 950-ms intervals were applied using a pulse generator [ECM830 (BTX)]. Animals were then warmed and placed back in the cage with the mother. Mice were processed 48 h later as described above. Plasmids used were pCAG-tdtomato and pCAG-ERT2CreERT2 (30).
Supplementary Material
Acknowledgments
We thank Courtney Rubin, Christine Kaliszewski, and Dolores Montoya for assistance; Dr. Lawrence Hsieh for providing the plasmids; and Dr. Mariano Viapiano (Department of Neurosurgery, Harvard Medical School) for discussion. This work was supported by NIH Grants DC000210 and DC012441 (to C.A.G.), DC010894 (to D.J.R.-G.), DC012130 (to A.S.M.), and DC011134 (to F.I.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1417955112/-/DCSupplemental.
References
- 1.Richard MB, Taylor SR, Greer CA. Age-induced disruption of selective olfactory bulb synaptic circuits. Proc Natl Acad Sci USA. 2010;107(35):15613–15618. doi: 10.1073/pnas.1007931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mombaerts P, et al. Visualizing an olfactory sensory map. Cell. 1996;87(4):675–686. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- 3.Feinstein P, Bozza T, Rodriguez I, Vassalli A, Mombaerts P. Axon guidance of mouse olfactory sensory neurons by odorant receptors and the beta2 adrenergic receptor. Cell. 2004;117(6):833–846. doi: 10.1016/j.cell.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 4.Feinstein P, Mombaerts P. A contextual model for axonal sorting into glomeruli in the mouse olfactory system. Cell. 2004;117(6):817–831. doi: 10.1016/j.cell.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 5.Maritan M, et al. Odorant receptors at the growth cone are coupled to localized cAMP and Ca2+ increases. Proc Natl Acad Sci USA. 2009;106(9):3537–3542. doi: 10.1073/pnas.0813224106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwob JE. Neural regeneration and the peripheral olfactory system. Anat Rec. 2002;269(1):33–49. doi: 10.1002/ar.10047. [DOI] [PubMed] [Google Scholar]
- 7.Beites CL, Kawauchi S, Crocker CE, Calof AL. Identification and molecular regulation of neural stem cells in the olfactory epithelium. Exp Cell Res. 2005;306(2):309–316. doi: 10.1016/j.yexcr.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 8.Murdoch B, Roskams AJ. Olfactory epithelium progenitors: Insights from transgenic mice and in vitro biology. J Mol Histol. 2007;38(6):581–599. doi: 10.1007/s10735-007-9141-2. [DOI] [PubMed] [Google Scholar]
- 9.Kim EJ, Ables JL, Dickel LK, Eisch AJ, Johnson JE. Ascl1 (Mash1) defines cells with long-term neurogenic potential in subgranular and subventricular zones in adult mouse brain. PLoS ONE. 2011;6(3):e18472. doi: 10.1371/journal.pone.0018472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Au WW, Treloar HB, Greer CA. Sublaminar organization of the mouse olfactory bulb nerve layer. J Comp Neurol. 2002;446(1):68–80. doi: 10.1002/cne.10182. [DOI] [PubMed] [Google Scholar]
- 11.Treloar HB, Feinstein P, Mombaerts P, Greer CA. Specificity of glomerular targeting by olfactory sensory axons. J Neurosci. 2002;22(7):2469–2477. doi: 10.1523/JNEUROSCI.22-07-02469.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bader A, Bautze V, Haid D, Breer H, Strotmann J. Gene switching and odor induced activity shape expression of the OR37 family of olfactory receptor genes. Eur J Neurosci. 2010;32(11):1813–1824. doi: 10.1111/j.1460-9568.2010.07458.x. [DOI] [PubMed] [Google Scholar]
- 13.Fuss SH, Zhu Y, Mombaerts P. Odorant receptor gene choice and axonal wiring in mice with deletion mutations in the odorant receptor gene SR1. Mol Cell Neurosci. 2013;56:212–224. doi: 10.1016/j.mcn.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Lyons DB, et al. An epigenetic trap stabilizes singular olfactory receptor expression. Cell. 2013;154(2):325–336. doi: 10.1016/j.cell.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chesler AT, et al. A G protein/cAMP signal cascade is required for axonal convergence into olfactory glomeruli. Proc Natl Acad Sci USA. 2007;104(3):1039–1044. doi: 10.1073/pnas.0609215104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imai T, Suzuki M, Sakano H. Odorant receptor-derived cAMP signals direct axonal targeting. Science. 2006;314(5799):657–661. doi: 10.1126/science.1131794. [DOI] [PubMed] [Google Scholar]
- 17.Mobley AS, et al. Hyperpolarization-activated cyclic nucleotide-gated channels in olfactory sensory neurons regulate axon extension and glomerular formation. J Neurosci. 2010;30(49):16498–16508. doi: 10.1523/JNEUROSCI.4225-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakashima A, et al. Agonist-independent GPCR activity regulates anterior-posterior targeting of olfactory sensory neurons. Cell. 2013;154(6):1314–1325. doi: 10.1016/j.cell.2013.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fleischmann A, Abdus-Saboor I, Sayed A, Shykind B. Functional interrogation of an odorant receptor locus reveals multiple axes of transcriptional regulation. PLoS Biol. 2013;11(5):e1001568. doi: 10.1371/journal.pbio.1001568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fleischmann A, et al. Mice with a “monoclonal nose”: Perturbations in an olfactory map impair odor discrimination. Neuron. 2008;60(6):1068–1081. doi: 10.1016/j.neuron.2008.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen MQ, Zhou Z, Marks CA, Ryba NJ, Belluscio L. Prominent roles for odorant receptor coding sequences in allelic exclusion. Cell. 2007;131(5):1009–1017. doi: 10.1016/j.cell.2007.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen MQ, Marks CA, Belluscio L, Ryba NJ. Early expression of odorant receptors distorts the olfactory circuitry. J Neurosci. 2010;30(27):9271–9279. doi: 10.1523/JNEUROSCI.1502-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma L, et al. A developmental switch of axon targeting in the continuously regenerating mouse olfactory system. Science. 2014;344(6180):194–197. doi: 10.1126/science.1248805. [DOI] [PubMed] [Google Scholar]
- 24.Tsai L, Barnea G. A critical period defined by axon-targeting mechanisms in the murine olfactory bulb. Science. 2014;344(6180):197–200. doi: 10.1126/science.1248806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pluznick JL, et al. Renal cystic disease proteins play critical roles in the organization of the olfactory epithelium. PLoS ONE. 2011;6(5):e19694. doi: 10.1371/journal.pone.0019694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murdoch B, Roskams AJ. A novel embryonic nestin-expressing radial glia-like progenitor gives rise to zonally restricted olfactory and vomeronasal neurons. J Neurosci. 2008;28(16):4271–4282. doi: 10.1523/JNEUROSCI.5566-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephan AB, et al. The Na(+)/Ca(2+) exchanger NCKX4 governs termination and adaptation of the mammalian olfactory response. Nat Neurosci. 2012;15(1):131–137. doi: 10.1038/nn.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farbman AI, Margolis FL. Olfactory marker protein during ontogeny: Immunohistochemical localization. Dev Biol. 1980;74(1):205–215. doi: 10.1016/0012-1606(80)90062-7. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez-Gil DJ, Greer CA. Wnt/Frizzled family members mediate olfactory sensory neuron axon extension. J Comp Neurol. 2008;511(3):301–317. doi: 10.1002/cne.21834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuda T, Cepko CL. Controlled expression of transgenes introduced by in vivo electroporation. Proc Natl Acad Sci USA. 2007;104(3):1027–1032. doi: 10.1073/pnas.0610155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.