Significance
Proper animal development requires the robust execution of cell fates under stressful conditions. In Caenorhabditis elegans, reciprocal interactions between heterochronic genes and the p38 innate immune pathway help to coordinate development with pathogen defense. Importantly, the robustness of developmental cell fate expression during infection depends on functional redundancy among genes encoding microRNAs (miRNAs) of the let-7 family. These findings underscore the importance of miRNA pathways in conferring robustness to developmental programs under stressful conditions and highlight roles for heterochronic genes not only as developmental timers but also as modulators of innate immune responses. The let-7 family miRNAs and p38 innate immune pathway are evolutionarily conserved; therefore, this study presents implications for similar integration of these two pathways in other animal systems.
Keywords: let-7 family microRNAs, p38, Pseudomonas aeruginosa, developmental timing, innate immunity
Abstract
Animals maintain their developmental robustness against natural stresses through numerous regulatory mechanisms, including the posttranscriptional regulation of gene expression by microRNAs (miRNAs). Caenorhabditis elegans miRNAs of the let-7 family (let-7-Fam) function semiredundantly to confer robust stage specificity of cell fates in the hypodermal seam cell lineages. Here, we show reciprocal regulatory interactions between let-7-Fam miRNAs and the innate immune response pathway in C. elegans. Upon infection of C. elegans larvae with the opportunistic human pathogen Pseudomonas aeruginosa, the developmental timing defects of certain let-7-Fam miRNA mutants are enhanced. This enhancement is mediated by the p38 MAPK innate immune pathway acting in opposition to let-7-Fam miRNA activity, possibly via the downstream Activating Transcription Factor-7 (ATF-7). Furthermore, let-7-Fam miRNAs appear to exert negative regulation on the worm’s resistance to P. aeruginosa infection. Our results show that the inhibition of pathogen resistance by let-7 involves downstream heterochronic genes and the p38 MAPK pathway. These findings suggest that let-7-Fam miRNAs are integrated into innate immunity gene regulatory networks, such that this family of miRNAs modulates immune responses while also ensuring robust timing of developmental events under pathogen stress.
During development, animals routinely encounter environmental, physiological, and nutritional challenges that threaten to compromise the robust execution of developmental programs. Therefore, the genetic programming of development includes mechanisms to ensure that developmental events occur flawlessly despite stressful conditions (1, 2). Several studies indicate that microRNAs (miRNAs) are used to maintain the robustness of biological processes under stress conditions (3–10). miRNAs are endogenous, noncoding, small RNAs that posttranscriptionally regulate gene expression primarily through binding to the 3′UTR of target mRNAs, which results in translation inhibition and/or mRNA degradation (11). miRNAs with the same seed sequence (nucleotides 2–7 of the mature miRNA sequence), which are predicated potentially to share the same set of targets (12), are grouped into a family.
The miRNA lin-4 and the miRNAs of the let-7 family (let-7-Fam) are central to the regulation of pluripotency and differentiation in many animal systems, including mammals (13–18). Four Caenorhabditis elegans let-7-Fam miRNAs, let-7, mir-48, mir-84, and mir-241, function in concert to repress key heterochronic gene targets, including daf-12, lin-41, and hbl-1, to stage-specifically regulate the timing of the hypodermal seam cell fates (16, 19–22). In C. elegans, the temporal patterns of cell division and cell fate during larval development are exquisitely invariant, even though larvae develop as free-living inhabitants of a changing environment in soil and decaying plant materials.
In the wild, worms encounter a variety of bacteria species as food sources. Several of these species are proven to be pathogenic to C. elegans (23), thus representing environmental stressors. Interestingly, recent studies have shown that let-7-Fam miRNAs seem not only to regulate developmental events but also to regulate the antibacterial and inflammatory response in several animal systems (24–28). At least two of the C. elegans let-7-Fam miRNAs are shown to regulate the worm’s survival to Pseudomonas aeruginosa infection (27). These previous findings suggest that let-7-Fam miRNAs could possibly coordinate developmental timing and innate immune responses so as to contribute to the robustness of development during pathogen infection.
Here, we show that let-7-Fam miRNAs are engaged in reciprocal interactions with innate immune pathways. Using genetically sensitized backgrounds, we find that the developmental timing phenotypes of let-7-Fam miRNA mutants are modified by their bacterial diet, particularly by growth on pathogenic P. aeruginosa. The let-7-Fam miRNA activity is negatively regulated on P. aeruginosa by the p38 MAPK pathway. Moreover, let-7-Fam miRNAs exhibit negative regulation on pathogen resistance, possibly through several pathways, including p38 MAPK signaling pathway, and also through downstream heterochronic genes, particularly the let-7-Fam miRNA targets lin-41 and hbl-1. Our findings suggest that genetic redundancy among let-7-Fam miRNAs enables these miRNAs to control the expression of developmental cell fates robustly, while also modulating innate immune responses according to the pathogenicity of the worm’s bacterial diet.
Results
Growth on P. aeruginosa Aggravates the Heterochronic Phenotypes of let-7-Fam miRNA Mutants.
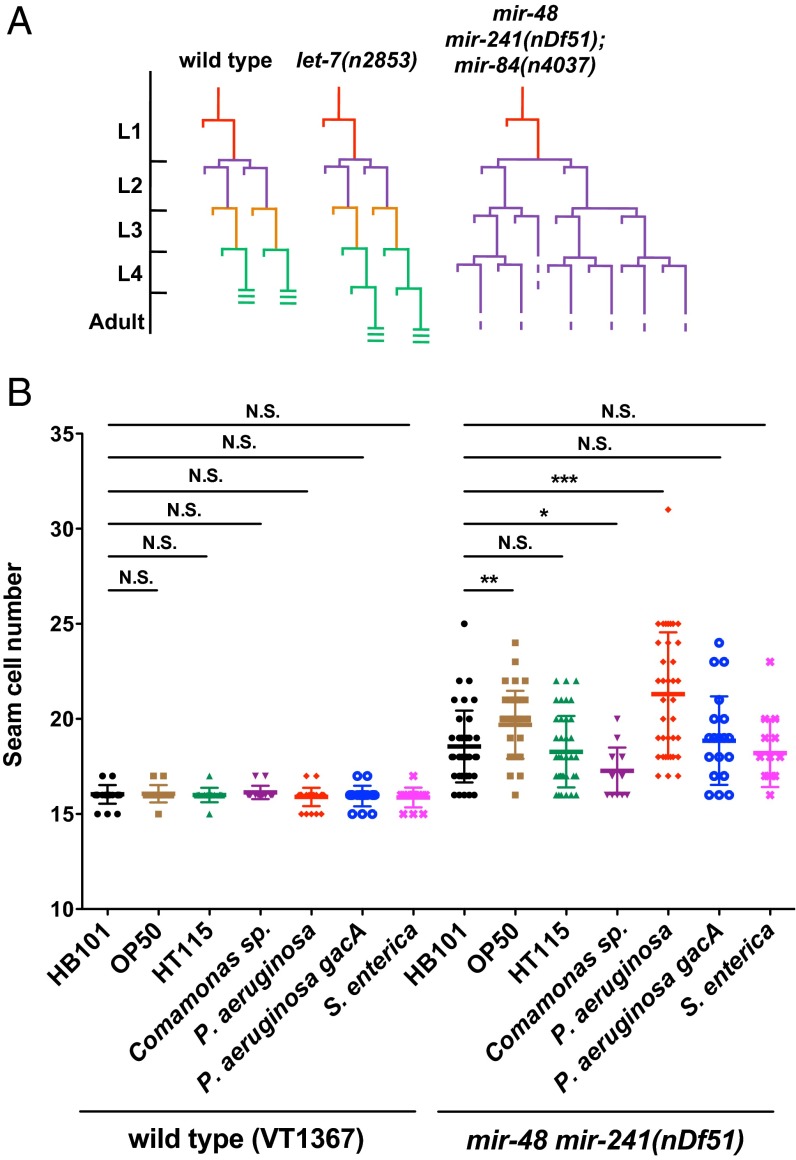
The let-7-Fam miRNAs act semiredundantly in controlling the developmental timing of certain stage-specific hypodermal seam cell fates in C. elegans. Loss of let-7-Fam miRNAs results in reiterations of early larval seam cell division patterns at later stages, and many seam cells in these mutants also fail to differentiate adult-specific cuticular structures (called adult alae) properly at the larval stage 4 (L4) molt (15, 16) (Fig. 1A). These heterochronic let-7-Fam miRNA mutant phenotypes are easily quantified by using microscopy to measure the number of seam cells and to score for the formation of adult alae in young adults. To investigate whether different bacterial food sources could have an impact on the regulation of developmental timing by let-7-Fam miRNAs, we used a genetically sensitized let-7-Fam miRNA mutant strain [mir-48 mir-241(nDf51)] that exhibits a partially penetrant heterochronic phenotype. Wild type (WT) animals have an average number of 16 seam cells, and 100% of the animals have complete adult alae at the young adult stage, whereas mir-48 mir-241(nDf51) animals display an average of 18.5 seam cells, and ∼60% of the animals exhibit incomplete adult alae.
Fig. 1.
Effects of bacterial food on the heterochronic phenotype of a let-7-Fam partial loss-of-function mutant. (A) Diagrams of seam cell V lineage in WT (N2), let-7(n2853), and mir-48 mir-241(nDf51); mir-84(n4037) animals. L1 to L4 are the four larval stages in C. elegans postembryonic development. (B) Scatter plot of seam cell number for WT (VT1367) and mir-48 mir-241(nDf51) animals raised on seven different bacteria: E. coli (HB101, OP50, and HT115), Comamonas sp. (DA1877), P. aeruginosa (PA14), P. aeruginosa (PA14 gacA), and S. enterica (SL1344). WT animals in this experiment are the strain of VT1367 that carries an integrated col-19 (an adult-specific collagen) transcriptional reporter (maIs105) to assist in the quantification of seam cell numbers. The maIs105 is also present in the genetic background of all of the other stains used in heterochronic phenotype analysis. E. coli HB101 was used as a control. In this and all subsequent scatter plots, the line in the middle is the mean value and error bars represent SDs. N.S., not significant. *P < 0.05, **P < 0.01, ***P < 0.001; two-tailed t test (n ≥ 15).
We scored seam cell numbers and adult alae formation for WT and mir-48 mir-241(nDf51) animals after development on six different bacterial diets, including three strains of Escherichia coli that are typically used as laboratory food sources (HB101, OP50, and HT115) and three other bacterial species (Comamonas sp. DA1877, P. aeruginosa PA14, and Salmonella enterica SL1344) that have been shown to have a significant effect on the physiology of C. elegans (23, 29). As expected, WT animals exhibited no evidence of developmental timing abnormalities regardless of the bacterial diet (Fig. 1B and Table 1). However, mir-48 mir-241(nDf51) animals showed a quantitatively different seam cell phenotype, dependent upon the bacterial food source (Fig. 1B). Notably, when grown on P. aeruginosa, mir-48 mir-241(nDf51) animals exhibited an enhanced seam cell phenotype compared with growth on HB101 (Fig. 1B). Consistent with this enhanced seam cell phenotype, mir-48 mir-241(nDf51) animals also exhibit an enhanced adult alae phenotype on P. aeruginosa (Table 1). These results suggest that bacterial food source can modulate the activity of the heterochronic gene pathway. Because animals with all let-7-Fam miRNAs intact showed no heterochronic phenotypes on any of the bacterial diets, these results indicate that let-7-Fam miRNAs act redundantly to maintain the robustness of developmental timing under the influence of dietary stress.
Table 1.
Adult alae phenotypes of WT and let-7-Fam miRNA mutants cultured on different bacterial food sources
| Percentage of animals with different adult alae phenotypes | ||||||
| Genotype | Treatment | Complete | Gapped | No alae | n | Test1 |
| WT (VT1367) | E. coli HB101 (control) | 100 | 0 | 0 | 30 | |
| E. coli OP50 | 100 | 0 | 0 | 15 | N.S. | |
| E. coli HT115 | 100 | 0 | 0 | 15 | N.S. | |
| Comamonas sp. DA1877 | 100 | 0 | 0 | 15 | N.S. | |
| P. aeruginosa PA14 | 100 | 0 | 0 | 30 | N.S. | |
| P. aeruginosa PA14 gacA | 100 | 0 | 0 | 15 | N.S. | |
| S. enterica SL1344 | 100 | 0 | 0 | 15 | N.S. | |
| mir-48 mir-241(nDf51) | E. coli HB101 | 40 | 60 | 0 | 42 | |
| E. coli OP50 | 28 | 72 | 0 | 39 | N.S. | |
| E. coli HT115 | 60 | 40 | 0 | 40 | N.S. | |
| Comamonas sp. DA1877 | 40 | 60 | 0 | 15 | N.S. | |
| P. aeruginosa PA14 | 8 | 92 | 0 | 37 | *** | |
| P. aeruginosa PA14 gacA | 30 | 70 | 0 | 21 | N.S. | |
| S. enterica SL1344 | 27 | 73 | 0 | 15 | N.S. | |
| mir-48(n4097); mir-84(n4037) | E. coli HB101 | 100 | 0 | 0 | 31 | |
| P. aeruginosa PA14 | 93 | 7 | 0 | 30 | N.S. | |
| let-7(n2853) | E. coli HB101 | 0 | 77 | 23 | 30 | |
| P. aeruginosa PA14 | 0 | 42 | 58 | 26 | ** | |
The χ2 test was used for comparison between E. coli HB101 (control) and other bacterial treatment for the same genotype. N.S., not significant. **P < 0.01; ***P < 0.001.
The most dramatic effect of diet on the developmental timing phenotypes of mir-48 mir-241(nDf51) animals was from growth on the pathogenic bacterium P. aeruginosa. Therefore, we focused our further studies on the effects of P. aeruginosa. The heterochronic phenotype enhancement in mir-48 mir-241(nDf51) animals on P. aeruginosa suggests that the activity of the remaining family members (chiefly let-7 and mir-84) may be decreased upon exposure to P. aeruginosa. To examine whether this decrease of let-7-Fam miRNA activity is restricted to certain members of the family, we tested a series of other let-7-Fam miRNA mutants for their developmental timing phenotypes upon P. aeruginosa treatment. Two other let-7-Fam miRNA mutants showed enhancement in their heterochronic phenotypes on P. aeruginosa. Specifically, mir-48(n4097); mir-84(n4037) animals showed an enhancement in seam cell phenotype (Fig. S1A), whereas let-7(n2853) animals exhibited an adult alae phenotype enhancement (Table 1). These results suggest that the activity of mir-241 and mir-48 could also be decreased by P. aeruginosa treatment. In conclusion, the activities of all four let-7-Fam miRNAs appear to be decreased upon growth on P. aeruginosa.
Modulation of let-7-Fam miRNA Activity by the p38 MAPK Pathway in Response to P. aeruginosa Infection.
The observed effects of bacterial diet on the developmental timing phenotypes of let-7-Fam miRNA mutants could be caused by various properties of the bacterial food, such as nutritional quality and pathogenic toxicity. This issue is of particular interest in the case of P. aeruginosa, which can support C. elegans larval development as a sole food source yet is also a pathogen capable of infecting C. elegans. To determine if the pathogenicity of P. aeruginosa is required for the modulation of heterochronic phenotypes elicited in let-7-Fam miRNA mutants, we cultured mir-48 mir-241(nDf51) larvae on the gacA mutant of P. aeruginosa PA14. gacA is an important regulator of the cell density-dependent gene expression in P. aeruginosa, and it is required for the production of exoenzymes and secondary metabolites (30). The pathogenicity of the PA14 gacA strain is dramatically decreased compared with the WT PA14 (31). We observed that the enhancement of let-7-Fam miRNA mutant phenotypes was substantially reduced for larvae grown on PA14 gacA compared with larvae grown on WT PA14 (Fig. 1B and Table 1). This observation suggests that the pathogenicity of P. aeruginosa is crucial for the modulation of let-7-Fam miRNA activity.
Although C. elegans can develop throughout postembryonic development and into the adult stage with P. aeruginosa as a sole food source, adults succumb to P. aeruginosa infection within a few days. A key pathway that enables larvae to survive on P. aeruginosa is the conserved p38 MAPK cascade (TIR-1/NSY-1/SEK-1/PMK-1) that initiates the innate immune response for antibacterial defenses (32, 33). We found that genetic removal of any p38 MAPK pathway component blocks the enhancement of the let-7-Fam miRNA mutant phenotypes on P. aeruginosa (Fig. 2 A and C). We interpret this result to indicate that p38 MAPK pathway activation in larvae grown on P. aeruginosa is required for the down-regulation of let-7-Fam miRNA activity. Furthermore, one of the major downstream transcription factors of the p38 MAPK pathway is ATF-7 (34). We therefore tested whether ATF-7 is involved in the modulation of let-7-Fam miRNA mutant phenotypes. We observed that mir-48 mir-241(nDf51) animals homozygous for a null mutation of ATF-7 [atf-7(qd22qd130)] or for a phosphorylation-defective allele of ATF-7 [atf-7(qd22)] (34) did not exhibit enhanced heterochronic phenotypes on P. aeruginosa (Fig. 2 B and C). Interestingly, even for animals grown on E. coli, loss of ATF-7 led to a significant enhancement of heterochronic phenotypes in mir-48 mir-241(nDf51) animals (Fig. 2B). Therefore, ATF-7 seems to exhibit a positive regulation on let-7-Fam miRNA activity when animals are grown on E. coli, and upon P. aeruginosa infection, this regulation could be altered depending on the phosphorylation potential of ATF-7. Models for the effects of different atf-7 alleles on let-7-Fam miRNA activity are shown in Fig. S2 B–D.
Fig. 2.
p38 MAPK pathway and its downstream transcription factor atf-7 are required for the suppression of let-7-Fam miRNA activity on P. aeruginosa. (A) Deletion of any p38 MAPK pathway component blocks the enhancement of mir-48 mir-241(nDf51) animals’ seam cell phenotype on P. aeruginosa. (B) Seam cell number of atf-7(qd22qd130); mir-48 mir-241(nDf51) and atf-7(qd22); mir-48 mir-241(nDf51) animals treated with either HB101 (control) or PA14. qd22qd130 is a putative null allele of atf-7. ***P < 0.001, two-tailed t test (n ≥ 15). (C) Adult alae phenotype for double mutants of mir-48 mir-241(nDf51) and p38 MAPK pathway components. ***P < 0.001; χ2 test (n > 15).
Effects of P. aeruginosa Infection and p38 Signaling on let-7-Fam miRNA Gene Expression.
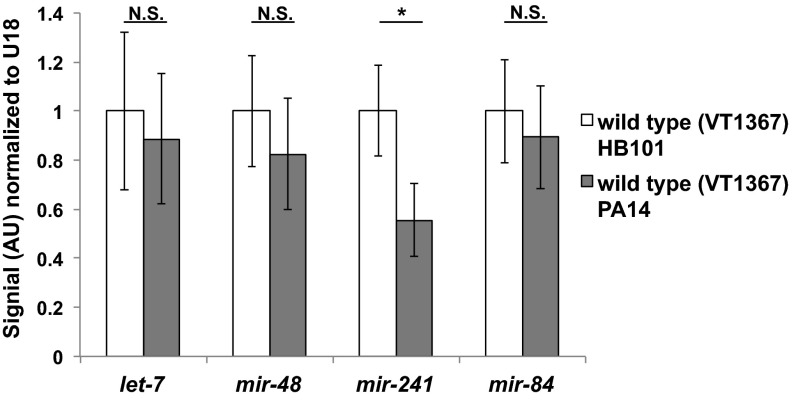
To investigate possible mechanisms for the apparent reduction of let-7-Fam miRNA activity upon P. aeruginosa infection, we first measured the mature let-7-Fam miRNA levels in animals exposed to P. aeruginosa throughout larval development to young adults. We observed an approximately twofold decrease in mir-241 but no significant change in the levels of other let-7-Fam miRNAs on P. aeruginosa (Fig. 3). Because we did not observe any reduction of mir-84 or let-7 levels on P. aeruginosa, the enhanced heterochronic phenotypes of mir-48 mir-241(nDf51) animals cannot simply be accounted for by a reduction of let-7-Fam miRNA levels in whole animals. Upon P. aeruginosa infection, it is possible that let-7-Fam miRNAs are regulated through modulation of the potency of their action, without affecting their overall levels. However, because our miRNA quantitation was performed on total RNA from whole worms, we would not necessarily detect tissue-specific changes in let-7-Fam miRNA levels.
Fig. 3.
Mature let-7-Fam miRNA levels upon P. aeruginosa infection. Total RNA from whole animals was used to perform Firefly miRNA assays. Animals (VT1367) were raised on either HB101 (control) or PA14 from the synchronized L1 stage to young adult stage at 20 °C. Error bars represent SDs. *P < 0.05; two-tailed t test. AU, arbitrary unit.
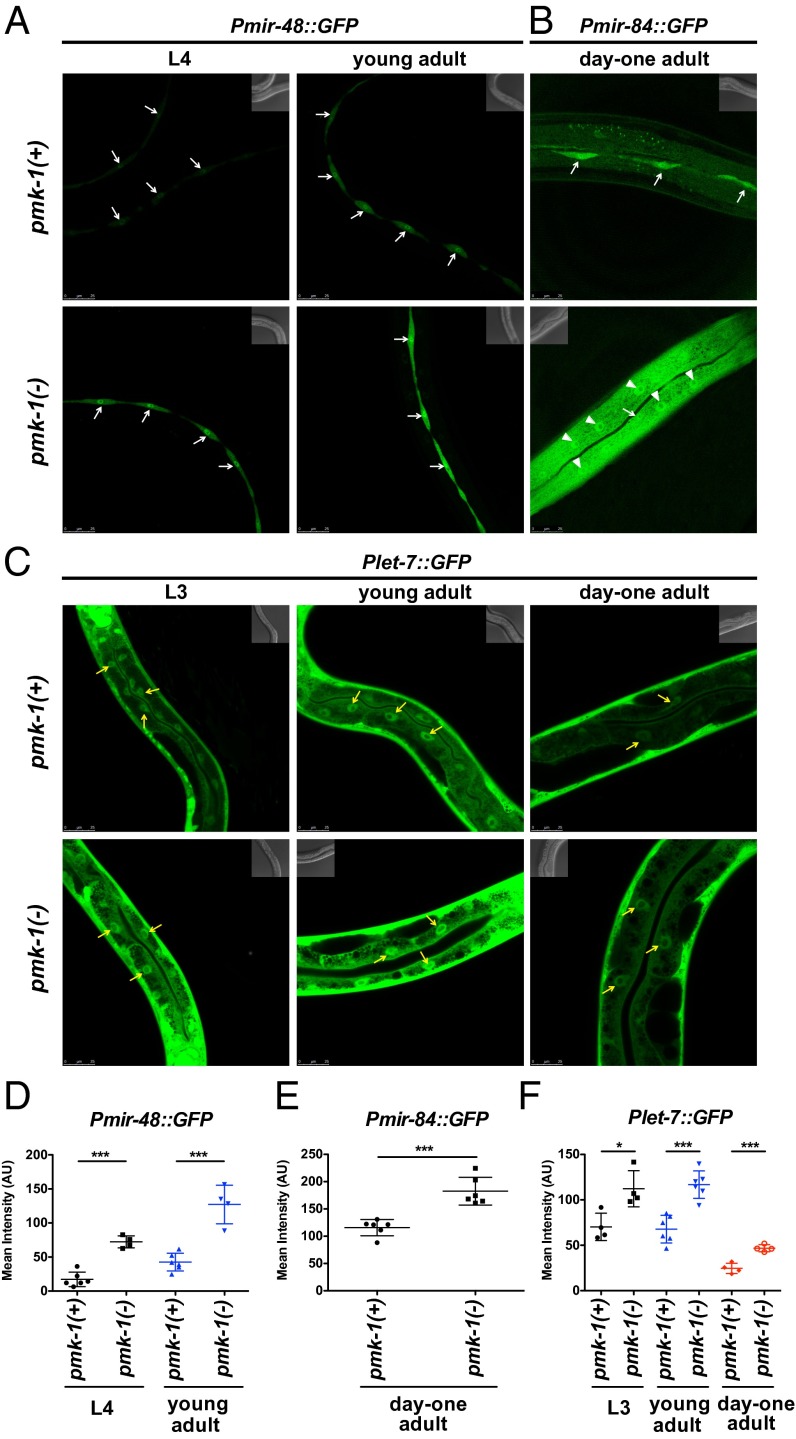
The results of our phenotypic analysis indicate that the activities and/or levels of let-7-Fam miRNAs are modulated by the p38 MAPK pathway. Because the p38 MAPK pathway is known to regulate gene expression transcriptionally, we examined whether p38 loss of function could affect the transcriptional activities of let-7-Fam miRNA genes. For these studies, we used transgenic worms carrying transcriptional reporters expressing GFP from let-7-Fam miRNA gene promoters. We observed stage- and tissue-specific increases of GFP expression for Pmir-48::GFP, Pmir-84::GFP, and Plet-7::GFP in the pmk-1 mutant compared with WT animals (Fig. 4). The developmental stages at which we detected differences in reporter activities between WT and pmk-1 mutant animals were L4 and young adult stages for Pmir-48::GFP; day 1 adult for Pmir-84::GFP; and L3, young adult, and day 1 adult for Plet-7::GFP. For mir-48 and mir-84, up-regulation of these reporters by loss of pmk-1 was observed in the hypodermis, suggesting a cell-autonomous effect of pmk-1 signaling on let-7-Fam miRNA transcription (Fig. 4 A, B, D, and E). Also, loss of pmk-1 resulted in detectable Pmir-84::GFP expression in the hyp7 syncytium compared with essentially undetected expression of Pmir-84::GFP in hyp7 for WT animals. For the let-7 transcriptional reporter, an increase of reporter activity in the absence of pmk-1 was apparent in the intestine but not in the hypodermis (Fig. 4 C and F). These data suggest that the p38 MAPK pathway negatively regulates the transcription of let-7-Fam miRNAs and that this regulation is tissue- and stage-specific.
Fig. 4.
pmk-1 regulates the transcriptional activity of let-7-Fam miRNA genes. Confocal images show the transcriptional reporters of mir-48 (A; maIs150), mir-84 (B; maIs138), and let-7 (C; maIs137) in WT [pmk-1(+)] and pmk-1(km25) [pmk-1(−)] animals. Animals were raised on E. coli HB101 at 20 °C. Stages of the animals are as indicated. White arrows indicate seam cells, white arrowheads indicate hyp7 nuclei, and yellow arrows point to representative intestine nuclei. (D–F) Mean intensities of the intended tissues in A–C. Error bars represent SDs. *P < 0.05, ***P < 0.001; two-tailed t test.
let-7-Fam miRNAs Exhibit Negative Roles in Resistance to P. aeruginosa Infection.
Our results indicate that during larval development, worms actively down-regulate their let-7-Fam miRNA activity upon exposure to P. aeruginosa. Additionally, we found that pmk-1 regulates the transcription of let-7 in the intestine, a major tissue for pathogen resistance in C. elegans (35). Therefore, we hypothesized that this inhibition of let-7-Fam miRNA activity may reflect a survival response to the presence of pathogen during larval development. Central to this hypothesis is the premise that let-7-Fam miRNAs may negatively regulate pathogen resistance; hence, the down-regulation of their activity would promote survival. Accordingly, we examined the survival capacity of several let-7-Fam miRNA mutants placed on P. aeruginosa beginning as L4-stage animals.
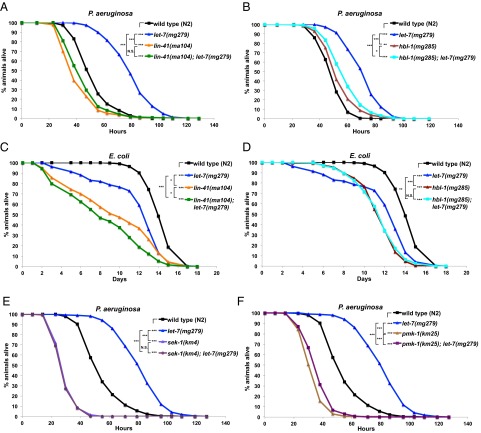
First, we measured the survival of animals homozygous for let-7(mg279), a weak loss-of-function allele (with very weak developmental timing phenotypes), and animals homozygous for let-7(n2853), a stronger loss-of-function allele (with developmental timing phenotypes resembling the let-7 null mutant) on P. aeruginosa. We observed that let-7(mg279) animals displayed a dramatic improvement in survival against P. aeruginosa infection compared with WT animals (Fig. 5A), whereas let-7(n2853) animals displayed the opposite, a decreased survival on P. aeruginosa (Fig. 5B). Interestingly, populations of the double-mutant mir-48 mir-241(nDf51) and mir-48(n4097); mir-84(n4037) animals exhibited biphasic survival curves (Fig. 5A). Apparently, for each of these double mutants, a portion of the population dies faster than WT, whereas the remainder of the population survives longer than WT on P. aeruginosa.
Fig. 5.
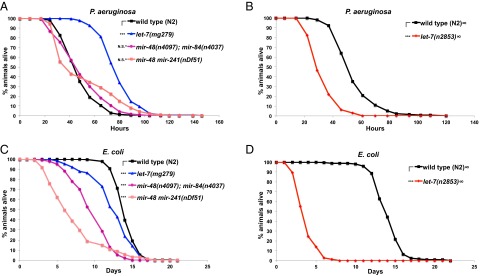
let-7-Fam miRNAs regulate pathogen resistance against P. aeruginosa and lifespan on E. coli. (A) Survival of WT (N2), let-7(mg279), mir-48(n4097); mir-48(n4037), and mir-48 mir-241(nDf51) animals on P. aeruginosa. (B) Survival of WT (N2) and let-7(n2853) animals on P. aeruginosa. (C) Lifespan of WT (N2), let-7(mg279), mir-48(n4097); mir-48(n4037), and mir-48 mir-241(nDf51) animals on E. coli HB101. (D) Lifespan of WT (N2) and let-7(n2853) animals on E. coli HB101. Animals were usually raised at 20 °C before being transferred to a P. aeruginosa killing assay and lifespan assay at 25 °C, except for experiments with let-7(n2853) (noted with ∞ next to the strain names) in which strains were raised at 15 °C (to reduce the number of bursting adults) before being transferred to 25 °C for the P. aeruginosa killing assay and lifespan assay. ***P < 0.001; log-rank test. For the let-7-Fam miRNA mutants that exhibit different survival kinetics on P. aeruginosa compared with WT, Fisher’s exact tests were applied (data in Table S2). ^Statistically significant outcome in Fisher’s exact test.
To confirm that the opposite pathogen survival phenotypes of distinct let-7 mutants are attributable to the reduction of let-7 gene activity, we tested the ability of a single-copy let-7 transgene (maIs380), which fully rescues the developmental phenotypes of both let-7 alleles (Fig. S3 A and B), to also rescue the characteristic survival phenotypes of let-7(mg279) and let-7(n2853) mutants. maIs380 rescued the extended survival phenotype of let-7(mg279) animals (Fig. S3C) and the shortened survival phenotype of let-7(n2853) animals on P. aeruginosa (Fig. S3D). Interestingly, the let-7 rescuing transgene also rescued the biphasic survival phenotype of mir-48 mir-241(nDf51) and mir-48(n4097); mir-84(n4037) animals (Fig. S3 E and F). These results support the conclusions that let-7-Fam miRNAs function redundantly to regulate survival on P. aeruginosa and that all of the survival phenotypes of let-7-Fam miRNA mutants on P. aeruginosa result from different degrees of let-7-Fam miRNA loss of function.
To determine whether the complex survival phenotypes of let-7-Fam miRNA mutants on P. aeruginosa could reflect underlying effects of let-7-Fam miRNAs on overall fitness of the animals (regardless of diet), we examined the longevity of these mutants on E. coli HB101. All of the let-7-Fam miRNA mutants we tested displayed a degree of shortened lifespan compared with WT animals (Fig. 5 C and D), which indicates that let-7-Fam miRNAs function positively to regulate longevity in C. elegans. Therefore, the decreased survival phenotype of the stronger loss-of-function mutant let-7(n2853) animals on P. aeruginosa could simply be due to reduced longevity of these animals. However, importantly, the weak loss-of-function mutant let-7(mg279) animals exhibit improved survival against P. aeruginosa infection despite the fact that these animals have a shortened lifespan. These results suggest that let-7-Fam miRNAs negatively modulate pathogen resistance while also positively regulate longevity, such that a mild reduction in let-7-Fam miRNA activity [by let-7(mg279)] promotes survival on P. aeruginosa because the longevity of these animals is not severely compromised. With regard to the biphasic survival phenotype of mir-48 mir-241(nDf51) and mir-48(n4097); mir-84(n4037) animals on P. aeruginosa, we propose that there are variations in the extent of reduced let-7-Fam miRNA activity within the population in these mutants, such that animals with mildly reduced let-7-Fam miRNA activity exhibit enhanced pathogen resistance, whereas others with greater reduced let-7-Fam miRNA activity show decreased survival on P. aeruginosa possibly due to reduced longevity in general.
In summary, let-7-Fam miRNAs exhibit negative regulation on animals’ resistance to P. aeruginosa infection, as evidenced by the increased survival on P. aeruginosa for let-7(mg279) animals. On the other hand, some let-7-Fam miRNA mutants exhibited decreased survival on P. aeruginosa, which probably results from an epistatic function of let-7-Fam miRNAs in promoting longevity.
Downstream Heterochronic Genes May Mediate the Enhanced Survival of let-7(mg279) Animals on P. aeruginosa.
To investigate further the mechanism by which let-7-Fam miRNAs negatively regulate pathogen resistance, we focused on understanding the enhanced survival phenotype of let-7(mg279) animals. This emphasis was for several reasons. First, let-7-Fam miRNAs contribute redundantly to survival on P. aeruginosa. Hence, let-7 can serve as a proxy for the other let-7-Fam miRNA genes in our genetic analyses. Additionally, let-7(mg279) animals exhibit a striking survival phenotype on P. aeruginosa that we expected would be more tractable for genetic analysis than the complex survival phenotypes of the other let-7-Fam miRNA mutants. Finally, among the let-7-Fam mutants tested here, let-7(mg279) animals exhibited the least reduced lifespan on E. coli. Therefore, using let-7(mg279) animals to explore the mechanism of let-7-Fam miRNAs in pathogen resistance could minimize potentially confounding effects of lifespan in our analysis.
The let-7-Fam miRNAs regulate a set of specific target gene mRNAs during larval development to control developmental timing. For let-7, the most prominent targets are lin-41 and hbl-1 (20–22). To determine if lin-41 and hbl-1 are downstream of let-7 in the regulation of pathogen resistance, we tested the survival of let-7(mg279) animals on P. aeruginosa in the presence of lin-41 or hbl-1 loss-of-function mutations. Interestingly, animals with partial loss of function for lin-41 exhibited decreased survival upon P. aeruginosa infection, and lin-41 also suppressed the enhanced survival phenotype of let-7(mg279) animals (Fig. 6A). These findings suggest that aside from its developmental timing role downstream of let-7, lin-41 is also possibly one of the targets of let-7 for the regulation of pathogen resistance.
Fig. 6.
lin-41, hbl-1, and p38 MAPK pathway modulate the enhanced pathogen resistance of let-7(mg279) animals. lin-41(ma104) (A) and hbl-1(mg285) (B) suppress the enhanced survival phenotype of let-7(mg279) animals on PA14. (C) Lifespan of strains used in A on E. coli HB101. (D) Lifespan of strains used in B on E. coli HB101. sek-1 (E) and pmk-1 (F) are also required for the enhanced survival phenotype of let-7(mg279) animals on PA14. *P < 0.05, **P < 0.01, ***P < 0.001; log-rank test.
hbl-1(mg285) also suppressed the enhanced survival phenotype of let-7(mg279) on P. aeruginosa, but unlike lin-41(ma104), the hbl-1(mg285) mutation alone did not cause any obvious survival phenotype (Fig. 6B). Because hbl-1(mg285) is only a partial loss-of-function mutation, epistasis cannot be interpreted unequivocally, but these results are consistent with both hbl-1 and lin-41 functioning downstream of let-7 for pathogen sensitivity. We note that hbl-1 has been shown to regulate the transcription of let-7 negatively (36). Therefore, the suppression of let-7(mg279) enhanced survival phenotype by hbl-1(mg285) could reflect a derepression of let-7 gene transcription.
In addition, we examined the longevity on E. coli HB101 of lin-41(ma104) and hbl-1(mg285) animals along with animals mutant for lin-41(ma104) or hbl-1(mg285) in combination with let-7(mg279) (Fig. 6 C and D). The results suggest that lin-41 and hbl-1 positively regulate lifespan on E. coli. However, because hbl-1(mg285) animals exhibit similar survival on P. aeruginosa compared with WT animals (Fig. 6B), the suppression of hbl-1(mg285) on let-7(mg279) animals’ enhanced survival phenotype on P. aeruginosa is likely caused by a role of hbl-1 in pathogen resistance, rather than longevity. On the other hand, the suppression of let-7(mg279) animals’ survival phenotype on P. aeruginosa by lin-41 could be contributed, in part, by the role of lin-41 in longevity and, in part, by a role in pathogen resistance.
The p38 MAPK Pathway Is Required for the Prolonged Survival Phenotype of let-7(mg279) Animals on P. aeruginosa.
In addition to targeting lin-41 and hbl-1, let-7 could (directly or indirectly) regulate genes in the innate immune response pathways to regulate pathogen resistance negatively. Interestingly, in the p38 MAPK pathway, tir-1, nsy-1, and sek-1 are all predicted targets of let-7-Fam miRNAs (37). Therefore, we hypothesized that let-7 could function upstream of the p38 MAPK pathway to regulate survival on P. aeruginosa. Accordingly, we examined the survival of animals doubly mutant for let-7(mg279) and loss-of-function mutations in p38 MAPK pathway components. We observed that loss of any p38 MAPK pathway component could suppress the enhanced survival phenotype of let-7(mg279) animals (Fig. 6 E and F and Fig. S4). Also, previous studies indicate that the p38 MAPK pathway does not affect longevity in C. elegans (32, 38). Therefore, these results indicate that let-7(mg279) animals’ enhanced pathogen resistance phenotype requires the p38 MAPK pathway.
To examine whether let-7 may regulate factors upstream of p38 in the MAPK pathway, we tested for an elevated level of phosphorylated p38 in protein extracts of let-7(mg279) animals compared with extracts of WT animals. These experiments did not conclusively reveal an impact of let-7 on p38 phosphorylation (Fig. S5). However, because these assays for phosphorylated p38 were performed on extracts of whole animals, it is possible that let-7 may function upstream of the p38 MAPK pathway in a tissue-specific manner. Although, genetically, the p38 MAPK pathway is downstream of let-7 in regulating survival on P. aeruginosa, we cannot rule out the possibility that it might function in parallel with lin-41 and hbl-1 for the prolonged survival of let-7(mg279) animals on P. aeruginosa.
let-7(mg279) Animals Exhibit Reduced Accumulation of Outer Membrane Vesicles in the Intestine During P. aeruginosa Infection.
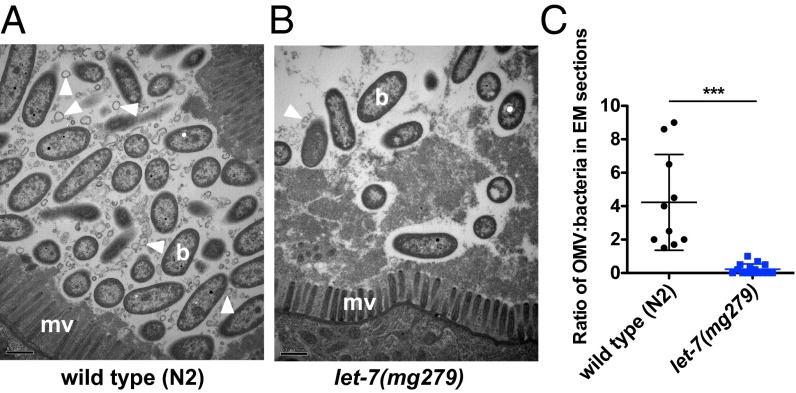
To explore further the basis for why let-7(mg279) animals survive better than WT animals on P. aeruginosa, we examined the intestines of infected worms using transmission EM. At 8 or 24 h after infection, we did not observe any difference in the intestinal cytopathology between WT and let-7(mg279) animals (Fig. S6). However, after 48 h of infection, we observed a significant reduction of outer membrane vesicles (OMVs) in the intestinal lumen of let-7(mg279) animals compared with WT animals (Fig. 7). OMVs are secreted by P. aeruginosa and known to function as a virulence factor and toxin delivery platform (39). These results suggest that the prolonged survival of let-7(mg279) animals on P. aeruginosa reflects an enhanced countervirulence activity in let-7(mg279) animals compared with WT animals.
Fig. 7.
let-7(mg279) animals exhibit reduced abundance of bacterial OMVs in their intestinal lumen during P. aeruginosa infection. Transmission electron micrographs of transversal midbody sections of a WT (N2) animal (A) and let-7(mg279) animal (B) infected with P. aeruginosa PA14 for 48 h. b, bacterial cell; mv, microvilli. Arrowheads point to representative OMVs. (Scale bars: 0.5 μm.) (C) Ratio of OMVs to bacteria in electron micrographs. The electron micrographs were randomly sampled with a 1-μm square five times, and the numbers of OMVs and bacteria within the square were counted. Two animals were tested for both the WT and let-7(mg279) groups. Error bars represent SDs. ***P < 0.001; two-tailed t test.
Discussion
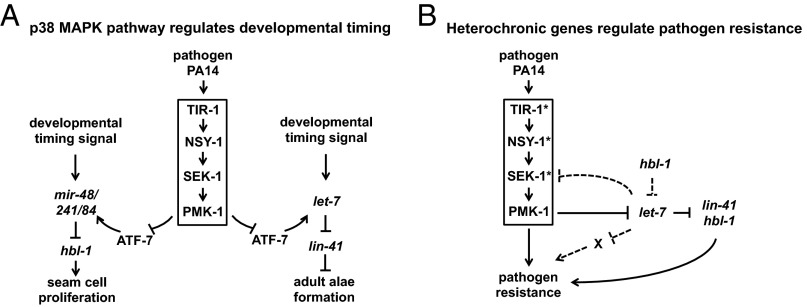
Animals are challenged by diverse physiological and environmental stresses during development but they nevertheless execute temporal and spatial patterns of developmental events with remarkable robustness. The let-7-Fam miRNAs function in the heterochronic gene pathway to regulate the specification and execution of stage-specific cell fates during C. elegans larval development (15, 16). Here, we report evidence that the activity of let-7-Fam miRNAs is regulated by the p38 innate immune response pathway during growth on the bacterial pathogen P. aeruginosa (Fig. 8A). Furthermore, we show that let-7-Fam miRNAs and several other heterochronic genes also function in the modulation of the worm's resistance against P. aeruginosa infection (Fig. 8B). Our findings uncover fundamental connections between the heterochronic gene pathway and innate immune response pathway, and suggest that these connections could serve to optimize the coordination of temporal cell fate specification and antibacterial responses in the developing larvae.
Fig. 8.
Model. (A) Model for the regulation of developmental timing by P. aeruginosa and the p38 MAPK pathway in C. elegans. (B) Model for the roles of heterochronic genes in pathogen resistance on P. aeruginosa. *Predicated targets of let-7-Fam miRNAs in the p38 MAPK pathway. X represents other possible pathways that are regulated by let-7 to promote pathogen resistance on P. aeruginosa. The dotted line indicates proposed regulatory interactions not yet tested experimentally.
An important implication of our findings is that the collective function of let-7-Fam miRNAs provides robustness to the specification of temporal fates in WT C. elegans larvae, especially when they are challenged by pathogenic bacteria. This conclusion is based on a few interesting findings in our study. First, growth on pathogen affects temporal seam cell fate phenotypes only in sensitized genetic backgrounds where the full complement of let-7-Fam miRNAs is compromised by mutation. For WT, most of the single mutants, and let-7(mg279) mir-84(n4037) animals, developmental timing phenotypes were not affected by P. aeruginosa infection (Fig. S1A), which indicates that the down-regulation of let-7-Fam miRNA activity caused by P. aeruginosa is tolerated in these animals (without compromising temporal cell fate specification) due to the genetic redundancy of let-7-Fam miRNAs. Moreover, we also observed an increase in the variation of seam cell number in let-7-Fam miRNA mutants as the activity of this miRNA family declines, and the variation in seam cell number is enhanced by P. aeruginosa infection in those same let-7-Fam miRNA mutants where we observed P. aeruginosa-induced heterochronic phenotype enhancement (Fig. S1B). This variation of seam cell number could reflect noisy target gene expression caused by reduction of let-7-Fam miRNA activity, and could also represent a breakdown in the robustness of seam cell fate determination. These data (Fig. S1B) further support the idea that let-7-Fam miRNAs function to protect the robustness in the temporal patterns of the seam cell program, especially in the face of P. aeruginosa infection.
We observed that certain other bacterial food sources, besides P. aeruginosa, can also modulate the seam cell phenotype in the let-7-Fam miRNA sensitized genetic background, albeit with less potency than P. aeruginosa (Fig. 1B). One surprising result is that E. coli OP50, a routine food source for most laboratories, also enhances the seam cell numbers of mir-48 mir-241(nDf51) animals (although this enhancement is very mild compared with the effects of P. aeruginosa and there is no change in the adult alae phenotype). Interestingly, pmk-1(km25) partially suppresses the enhanced heterochronic phenotypes of mir-48 mir-241(nDf51) animals on OP50 (Fig. S1 C–E), consistent with previous findings that OP50 could be slightly pathogenic to worms (40). Therefore, the enhanced seam cell phenotype of the let-7-Fam miRNA mutant on OP50 could be partially due to the slight pathogenicity of the bacteria. However, other properties of OP50 could contribute to the phenotype as well because pmk-1(km25) did not fully suppress all of the enhanced heterochronic phenotype of mir-48 mir-241(nDf51) animals on OP50 (Fig. S1E). Nevertheless, it is noteworthy that WT larvae exhibit robustly normal developmental timing regardless of the bacterial diets tested here, supporting our conclusion that the full let-7-Fam miRNA genotype underlies the robustness of developmental timing to dietary and/or pathogenic stress.
Our findings that P. aeruginosa elicits phenotypic modulations of let-7-Fam miRNA mutants reveal a regulatory circuit connecting the p38 MAPK innate immunity pathway and the heterochronic gene developmental timing pathway via let-7-Fam miRNAs. The role of the p38 pathway in this process is supported by our observation that loss-of-function mutations that disable the p38 MAPK pathway block the phenotypic enhancement of let-7-Fam miRNA mutants on P. aeruginosa. Also in support of this conclusion is our result that constitutive activation of p38 by knocking down of a negative regulator of p38 signaling, vhp-1 (the p38 phosphatase) (41, 42), phenocopied the P. aeruginosa-induced heterochronic phenotype enhancement in let-7-Fam miRNA mutants (Fig. S2 F and G).
Although our findings show that the p38 MAPK pathway is involved in regulation of let-7-Fam miRNA activity, certain aspects of this regulatory circuit differ in interesting ways from canonical p38 innate immune signaling. First, the tir-1(ok1052) allele that lacks the N-terminal Heat/Armadillo motif of the protein blocks the enhancement of let-7-Fam miRNA mutant phenotypes (Fig. S2A), even though this motif has been shown to be dispensable for pathogen resistance (43, 44). This result suggests that activation of the p38 MAPK pathway is necessary for the regulation of let-7-Fam miRNA activity upon P. aeruginosa infection but that p38 signaling alone without the activity of the Heat/Armadillo motif of TIR-1 is not sufficient. Moreover, we observed that ATF-7 exerts complex and allele-specific modes of regulation on let-7-Fam miRNA activity with and without P. aeruginosa infection, presumably depending on its phosphorylation potential (Fig. S2 B–E). This finding is consistent with previously published observations that upon phosphorylation by PMK-1 in the face of P. aeruginosa infection, ATF-7 switches its mode of regulation on pmk-1/p38–mediated gene expression (34). More intriguingly, the atf-7(gk715) allele enhances the phenotypes of the let-7-Fam miRNA mutant even more than the atf-7 null allele does, and the phenotype is suppressed when animals were exposed to P. aeruginosa (Fig. S2A). This atf-7(gk715) allele removes the first exon from two atf-7 gene isoforms, leaving the remaining isoforms unaffected, suggesting that there is an activation domain in the N-terminal region of ATF-7 protein. Removal of this region in the gk715 allele results in a reversal of function for the protein. Previous findings have suggested that the N-terminal region of mammalian ATF-7 homologs is essential for transcriptional activity (45, 46), a situation that, according to our results, appears to be evolutionally conserved in C. elegans. Although further studies are required to understand these noncanonical functions of TIR-1 and ATF-7 on let-7-fam miRNA activity, nevertheless our results provide a basis for uncovering the mechanisms for how the p38 MAPK pathway regulates let-7-fam miRNAs.
Our experiments using let-7-Fam miRNA gene transcriptional reporters in WT and pmk-1 mutant growing on E. coli suggest that p38 signaling could inhibit let-7-Fam miRNA activity, at least in part, at the transcriptional level. Thus, upon P. aeruginosa infection, activated p38 could further reduce let-7-Fam miRNA gene expression relative to growth on E. coli, which correlates with the enhanced heterochronic phenotype of let-7-Fam miRNA mutants on P. aeruginosa. However, we were not able to explore the GFP reporter activity of let-7-Fam miRNA genes upon P. aeruginosa infection directly due to apparent nonspecific degradation of GFP (possibly caused by cellular autophagy and/or necrosis) when animals were treated with P. aeruginosa. Additionally, it should be noted that the transcriptional regulation by p38 of let-7-Fam miRNAs could be more complex than observed here, because the reporter transgenes used in our study may not have necessarily contained all relevant regulatory elements (47).
The negative regulation of let-7-Fam miRNAs upon P. aeruginosa infection is presumably beneficial to worms and can enhance their response to the pathogen. This conclusion is supported by our observation that let-7(mg279) animals with mildly reduced activity of let-7 exhibit enhanced survival in the face of P. aeruginosa infection. Even though we have shown that the p38 MAPK pathway and the let-7 targets lin-41 and hbl-1 are required for this phenotype, it is also possible that additional downstream stress response effectors, such as skn-1 (27), could mediate the regulation for pathogen resistance by let-7-Fam miRNAs. Predicted targets of let-7-Fam miRNAs include components of several pathways involved in pathogen resistance, including the p38 MAPK pathway, the unfolded protein response pathway, the oxidative stress response pathway, and the autophagy pathway (27, 32, 48, 49). We propose that let-7-Fam miRNAs could function during normal, unstressed development to dampen several stress response pathways, and that under stress conditions, such as P. aeruginosa infection, the down-regulation of let-7-Fam miRNA activity reported here would broadly augment the worm's stress response.
The intestine, hypodermis, and neurons have been shown previously to be involved in the host response to pathogen infection in C. elegans (35, 43). Although tissue-specific rescue and knock out experiments will be required to determine the anatomical site(s) of action and to test for cell autonomy of let-7-Fam miRNAs for their regulation of pathogen resistance, our findings suggest that let-7-Fam miRNAs may have an impact on pathogen resistance by acting in the intestine and possibly in the hypodermis. We did not observe any noticeable difference in the pathogen avoidance behavior between let-7(mg279) animals and WT animals (Fig. S7), arguing against an exclusively neuronal effect. However, our EM results indicate that let-7(mg279) animals actively fight off P. aeruginosa infection in the intestine. Finally, based on the transcriptional reporter results, let-7-Fam miRNAs are specifically regulated in the intestine and the hypodermis.
Implications of our findings include intriguing possibilities for evolutionarily conserved roles of let-7-Fam miRNAs in cell fate and innate immune gene regulatory networks. In mammalian cells, let-7-Fam miRNA levels have been shown to be reduced in response to infection by the bacterium Salmonella (24), the parasitic protozoan Cryptosporidium parvum (25, 26), or an inflammatory response of Src activation (28). In the latter context, let-7-Fam miRNA levels were regulated by lin-28 (28), which also functions in the heterochronic pathway with let-7-Fam miRNAs in C. elegans (16, 50). In the context of Salmonella infection, Toll-like receptor 4 and the NFκB pathway, and possibly other innate immune signals, appear to mediate the down-regulation of let-7-Fam miRNAs (51). This repression of let-7 by NFκB is analogous to the p38 MAPK pathway’s regulation of let-7-Fam miRNAs in the context of P. aeruginosa infection of C. elegans, whose genome does not contain an NFκB homolog (52, 53). It is not currently clear whether p38 could contribute to the regulation of let-7-Fam miRNAs in mammalian cells upon innate immune activation.
In conclusion, our study demonstrates that let-7-Fam miRNAs function in a feed-forward loop with the p38 MAPK pathway to promote pathogen resistance upon infection, and that the genetic redundancy among let-7-Fam miRNAs assists animals in maintaining their robust developmental programs under various stress conditions.
Materials and Methods
Nematode and Bacteria Methods.
C. elegans was cultured on nematode growth media (NGM) (54) and fed with E. coli HB101, unless otherwise noted. All of the C. elegans strains used in this study are listed in Table S1. Synchronized populations of developmentally staged worms were obtained by standard methods (55). To test different bacterial foods for effects on heterochronic phenotypes, all of the E. coli strains and Comamonas sp. were seeded onto NGM plates from saturated cultures. Procedures for culturing P. aeruginosa and S. enterica for feeding nematodes were carried out as described by Powell and Ausubel (56).
Heterochronic Phenotype Analysis.
Gravid adult animals raised at 20 °C were placed on control or treatment plates at 20 °C, and their progeny were scored at the young adult stage for adult lateral alae formation and seam cell number. Nomarski Differential Interference Contrast (DIC) microscopy and fluorescence microscopy with the maIs105 [col-19::gfp] transgene to mark lateral hypodermal cell nuclei were used to score alae formation and seam cell number, respectively.
RNA Extraction.
Animals were collected and flash-frozen in liquid nitrogen. Total RNA was extracted using TRIzol reagent (Invitrogen).
Firefly miRNA Assay.
Two micrograms of total RNA was used for Firefly miRNA assay (www.fireflybio.com/). A standard protocol provided by the manufacturer (Firefly BioWorks) was followed. Guava easyCyte 8HT (Millipore) was used for analysis. Signals (arbitrary units) were normalized to small nucleolar RNA U18 and then to the control group (HB101).
Confocal Microscopy.
Animals were mounted on glass slides with 2% (wt/vol) agarose pads and anesthetized with 10 mM levamisole. A Leica TCS SPE microsystem was used to acquire images. The mean intensities were calculated with the Leica Application Suite Advanced Fluorescence software platform.
P. aeruginosa Killing Assays and Lifespan Assays.
P. aeruginosa PA14 and E. coli HB101 were cultured in LB and seeded onto slow-killing plates (56) containing 100 μM 5-fluorodeoxyuridine (Sigma) for P. aeruginosa killing assays and lifespan assays, respectively. The seeded plates were incubated at 37 °C for 24 h and then transferred to 25 °C for 24 h before use. Assays were conducted by transferring L4-stage animals raised on E. coli HB101 to P. aeruginosa killing assay plates and lifespan assay plates at 25 °C. Animals that died prematurely due to developmental abnormalities (leading to bagging and bursting vulva) or that died after crawling off the plate were censored. Data were normalized by adjusting the numbers of live animals at each time point so as to derive a total population size of 100 for each technical replicate. Then, the normalized data for three technical replicates were averaged, and survival curves were compared using the log-rank test and Fisher’s exact test in Online Application for Survival Analysis of Lifespan Assays (OASIS) (57). All experiments were independently performed at least twice.
EM.
WT (N2) and let-7(mg279) animals were synchronized and grown on NGM plates seeded with E. coli HB101. L4-stage animals were transferred onto P. aeruginosa slow-killing plates and incubated at 25 °C for 8, 24, and 48 h. Worms were harvested and prepared for transmission EM as described (39).
Supplementary Material
Acknowledgments
We thank members of the V.R.A. laboratory for helpful discussions; S. Burke, K. McJunkin, and C. Sterling for comments on the manuscript; and the laboratories of C. Mello, A. Walker, R. Davis, and K. Matsumoto for reagents and technical assistance. Several nematode and bacteria strains used in this study were kindly provided by the Caenorhabditis Genetics Center [which is funded by the NIH Office of Research Infrastructure Programs (Grant P40 OD010440)] and the laboratories of F. Ausubel and D. Kim. The EM was performed by the core EM facility at the University of Massachusetts Medical School, supported by Award S10RR027897 from the National Center for Research Resources. Firefly BioWorks developed the firefly miRNA assay for C. elegans-specific miRNAs. This work was funded by NIH Grant R01 GM34028 (to V.R.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1422858112/-/DCSupplemental.
References
- 1.Kitano H. Biological robustness. Nat Rev Genet. 2004;5(11):826–837. doi: 10.1038/nrg1471. [DOI] [PubMed] [Google Scholar]
- 2.Gursky VV, Surkova SY, Samsonova MG. Mechanisms of developmental robustness. Biosystems. 2012;109(3):329–335. doi: 10.1016/j.biosystems.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 3.Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137(2):273–282. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X, Zabinsky R, Teng Y, Cui M, Han M. microRNAs play critical roles in the survival and recovery of Caenorhabditis elegans from starvation-induced L1 diapause. Proc Natl Acad Sci USA. 2011;108(44):17997–18002. doi: 10.1073/pnas.1105982108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hochbaum D, et al. DAF-12 regulates a connected network of genes to ensure robust developmental decisions. PLoS Genet. 2011;7(7):e1002179. doi: 10.1371/journal.pgen.1002179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goljanek-Whysall K, et al. MicroRNA regulation of the paired-box transcription factor Pax3 confers robustness to developmental timing of myogenesis. Proc Natl Acad Sci USA. 2011;108(29):11936–11941. doi: 10.1073/pnas.1105362108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149(3):515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cassidy JJ, et al. miR-9a minimizes the phenotypic impact of genomic diversity by buffering a transcription factor. Cell. 2013;155(7):1556–1567. doi: 10.1016/j.cell.2013.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Lu Y, Xu X-L, Gao F-B. The FTD/ALS-associated RNA-binding protein TDP-43 regulates the robustness of neuronal specification through microRNA-9a in Drosophila. Hum Mol Genet. 2013;22(2):218–225. doi: 10.1093/hmg/dds420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McJunkin K, Ambros V. The embryonic mir-35 family of microRNAs promotes multiple aspects of fecundity in Caenorhabditis elegans. G3 (Bethesda) 2014;4(9):1747–1754. doi: 10.1534/g3.114.011973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132(1):9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 14.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 15.Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 16.Abbott AL, et al. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 2005;9(3):403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulman BRM, Esquela-Kerscher A, Slack FJ. Reciprocal expression of lin-41 and the microRNAs let-7 and mir-125 during mouse embryogenesis. Dev Dyn. 2005;234(4):1046–1054. doi: 10.1002/dvdy.20599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18(10):505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Grosshans H, Johnson T, Reinert KL, Gerstein M, Slack FJ. The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Dev Cell. 2005;8(3):321–330. doi: 10.1016/j.devcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 20.Slack FJ, et al. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5(4):659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- 21.Lin S-Y, et al. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev Cell. 2003;4(5):639–650. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- 22.Abrahante JE, et al. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4(5):625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- 23.Aballay A, Ausubel FM. Caenorhabditis elegans as a host for the study of host-pathogen interactions. Curr Opin Microbiol. 2002;5(1):97–101. doi: 10.1016/s1369-5274(02)00293-x. [DOI] [PubMed] [Google Scholar]
- 24.Schulte LN, Eulalio A, Mollenkopf H-J, Reinhardt R, Vogel J. Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family. EMBO J. 2011;30(10):1977–1989. doi: 10.1038/emboj.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X-M, Splinter PL, O’Hara SP, LaRusso NF. A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J Biol Chem. 2007;282(39):28929–28938. doi: 10.1074/jbc.M702633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu G, et al. MicroRNA-98 and let-7 confer cholangiocyte expression of cytokine-inducible Src homology 2-containing protein in response to microbial challenge. J Immunol. 2009;183(3):1617–1624. doi: 10.4049/jimmunol.0804362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu F, et al. Nuclear hormone receptor regulation of microRNAs controls innate immune responses in C. elegans. PLoS Pathog. 2013;9(8):e1003545. doi: 10.1371/journal.ppat.1003545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139(4):693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacNeil LT, Watson E, Arda HE, Zhu LJ, Walhout AJM. Diet-induced developmental acceleration independent of TOR and insulin in C. elegans. Cell. 2013;153(1):240–252. doi: 10.1016/j.cell.2013.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reimmann C, et al. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol Microbiol. 1997;24(2):309–319. doi: 10.1046/j.1365-2958.1997.3291701.x. [DOI] [PubMed] [Google Scholar]
- 31.Tan MW, Rahme LG, Sternberg JA, Tompkins RG, Ausubel FM. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc Natl Acad Sci USA. 1999;96(5):2408–2413. doi: 10.1073/pnas.96.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim DH, et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science. 2002;297(5581):623–626. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- 33.Richardson CE, Kooistra T, Kim DH. An essential role for XBP-1 in host protection against immune activation in C. elegans. Nature. 2010;463(7284):1092–1095. doi: 10.1038/nature08762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shivers RP, et al. Phosphorylation of the conserved transcription factor ATF-7 by PMK-1 p38 MAPK regulates innate immunity in Caenorhabditis elegans. PLoS Genet. 2010;6(4):e1000892. doi: 10.1371/journal.pgen.1000892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shivers RP, Kooistra T, Chu SW, Pagano DJ, Kim DH. Tissue-specific activities of an immune signaling module regulate physiological responses to pathogenic and nutritional bacteria in C. elegans. Cell Host Microbe. 2009;6(4):321–330. doi: 10.1016/j.chom.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roush SF, Slack FJ. Transcription of the C. elegans let-7 microRNA is temporally regulated by one of its targets, hbl-1. Dev Biol. 2009;334(2):523–534. doi: 10.1016/j.ydbio.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hammell M, et al. mirWIP: MicroRNA target prediction based on microRNA-containing ribonucleoprotein-enriched transcripts. Nat Methods. 2008;5(9):813–819. doi: 10.1038/nmeth.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Troemel ER, et al. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2006;2(11):e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Irazoqui JE, et al. Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS Pathog. 2010;6:e1000982. doi: 10.1371/journal.ppat.1000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garigan D, et al. Genetic analysis of tissue aging in Caenorhabditis elegans: A role for heat-shock factor and bacterial proliferation. Genetics. 2002;161(3):1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim DH, et al. Integration of Caenorhabditis elegans MAPK pathways mediating immunity and stress resistance by MEK-1 MAPK kinase and VHP-1 MAPK phosphatase. Proc Natl Acad Sci USA. 2004;101(30):10990–10994. doi: 10.1073/pnas.0403546101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizuno T, et al. The Caenorhabditis elegans MAPK phosphatase VHP-1 mediates a novel JNK-like signaling pathway in stress response. EMBO J. 2004;23(11):2226–2234. doi: 10.1038/sj.emboj.7600226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pujol N, et al. Distinct innate immune responses to infection and wounding in the C. elegans epidermis. Curr Biol. 2008;18(7):481–489. doi: 10.1016/j.cub.2008.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ziegler K, et al. Antifungal innate immunity in C. elegans: PKCdelta links G protein signaling and a conserved p38 MAPK cascade. Cell Host Microbe. 2009;5(4):341–352. doi: 10.1016/j.chom.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 45.Livingstone C, Patel G, Jones N. ATF-2 contains a phosphorylation-dependent transcriptional activation domain. EMBO J. 1995;14(8):1785–1797. doi: 10.1002/j.1460-2075.1995.tb07167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Graeve F, Bahr A, Chatton B, Kedinger C. A murine ATFa-associated factor with transcriptional repressing activity. Oncogene. 2000;19(14):1807–1819. doi: 10.1038/sj.onc.1203492. [DOI] [PubMed] [Google Scholar]
- 47.Martinez NJ, et al. Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. Genome Res. 2008;18(12):2005–2015. doi: 10.1101/gr.083055.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richardson CE, Kinkel S, Kim DH. Physiological IRE-1-XBP-1 and PEK-1 signaling in Caenorhabditis elegans larval development and immunity. PLoS Genet. 2011;7(11):e1002391. doi: 10.1371/journal.pgen.1002391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zou C-G, Ma Y-C, Dai L-L, Zhang K-Q. Autophagy protects C. elegans against necrosis during Pseudomonas aeruginosa infection. Proc Natl Acad Sci USA. 2014;111(34):12480–12485. doi: 10.1073/pnas.1405032111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vadla B, Kemper K, Alaimo J, Heine C, Moss EG. lin-28 controls the succession of cell fate choices via two distinct activities. PLoS Genet. 2012;8(3):e1002588. doi: 10.1371/journal.pgen.1002588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Voinnet O. Micro-balancing innate immunity to Salmonella. EMBO J. 2011;30(10):1877–1879. doi: 10.1038/emboj.2011.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan MW, Ausubel FM. Caenorhabditis elegans: A model genetic host to study Pseudomonas aeruginosa pathogenesis. Curr Opin Microbiol. 2000;3(1):29–34. doi: 10.1016/s1369-5274(99)00047-8. [DOI] [PubMed] [Google Scholar]
- 53.Pujol N, et al. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr Biol. 2001;11(11):809–821. doi: 10.1016/s0960-9822(01)00241-x. [DOI] [PubMed] [Google Scholar]
- 54.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stiernagle T. Maintenance of C. elegans. WormBook. 2006 doi: 10.1895/wormbook.1.101.1. ed. The C. elegans Research Community, WormBook, 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Powell JR, Ausubel FM. Models of Caenorhabditis elegans infection by bacterial and fungal pathogens. Methods Mol Biol. 2008;415:403–427. doi: 10.1007/978-1-59745-570-1_24. [DOI] [PubMed] [Google Scholar]
- 57.Yang J-S, et al. OASIS: Online application for the survival analysis of lifespan assays performed in aging research. PLoS ONE. 2011;6(8):e23525. doi: 10.1371/journal.pone.0023525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.