Significance
Proteins fold to their native structure in significantly shorter timescales than expected for a random search mechanism. Rapid folding, which is likely to be important to optimize yield and avoid misfolded, aggregated states, has been proposed to result from specific folding pathways. However, mapping such pathways and understanding the principles by which they are selected remain central problems in protein science. Repeat proteins, containing tandem repeated structural units, greatly simplify folding studies. Here, we exploit the linear architecture of a β-strand-containing leucine-rich repeat protein and discover that folding is highly polarized toward the C terminus. By measuring the energy distribution, we observe that polarization is guided by local stability.
Keywords: PP32, LRR proteins, on-pathway intermediate, folding pathway selection, polarized transition state
Abstract
The leucine-rich repeat domain of PP32 is composed of five β-strand-containing repeats anchored by terminal caps. These repeats differ in sequence but are similar in structure, providing a means to connect topology, sequence, and folding pathway selection. Through kinetic studies of PP32, we find folding to be rate-limited by the formation of an on-pathway intermediate. Destabilizing core substitutions reveal a transition state ensemble that is highly polarized toward the C-terminal repeat and cap. To determine if this nucleus for folding corresponds to the most stable region of PP32, we monitored amide hydrogen exchange by NMR spectroscopy. Indeed, we find the highest protection to be biased toward the C terminus. Sequence manipulations that destabilize the C terminus spread out the transition state toward the middle of the protein. Consistent with results for helical ankyrin repeat proteins, these results suggest that local stabilities determine folding pathways.
Almost 50 y ago, Cyrus Levinthal suggested that protein folding is a guided process, rather than a random search (1). Since then, it has been proposed that rapid folding can be accomplished via a single preferred pathway (2–4). However, contrasting models of funneled, nonspecific folding capture key aspects of the folding process, including specific intermediates and transient stable structures (5, 6). To what extent does protein folding follow a single pathway, and to the extent that a single pathway dominates, what determines that pathway?
For some globular proteins, specific kinetic intermediates have been identified with features of thermodynamically stable substructures of the native state. This observation suggests specific pathways that correspond to low energy folding routes (7–12). However, directly testing this thermodynamic control of folding pathway selections requires mapping of local stability. Hydrogen exchange methods have provided the most detailed energy maps of native proteins (13), but sequence-distant contacts and irregular tertiary structures of globular proteins tend to blur the boundaries of local stability.
In contrast to globular proteins, elongated repeat proteins comprise tandem repeated secondary structural units, giving rise to regular topology lacking sequence-distant contacts. As a result, the contributions made by different regions of a repeat protein to its folding thermodynamics and kinetics can be easily dissected and compared. If local stability dictates folding, a preferred pathway should be observed, because repeats differ in sequence. If topology drives folding, multiple parallel pathways would be observed, because the repeats are similar in structure. For designed consensus α-helical ankyrin repeat constructs, with repeats of nearly identical sequence, folding proceeds via parallel pathways (14). In contrast, naturally occurring ankyrin repeat proteins with repeats of high sequence variation fold through preferred pathways (15–18). Regions that initiate folding correspond to low-energy structures, based on energy landscapes determined at single repeat resolution, consistent with low-energy folding routes for α-helical repeat proteins (16, 19–21).
Much less is known about the folding mechanisms of β-strand-containing repeat proteins, and particularly, leucine-rich repeat (LRR) proteins. To date, the folding pathways for only two LRR proteins have been elucidated. Like the ankyrin repeat domains, the LRR domain of Internalin B (InlB) also folds through a somewhat polarized pathway, with the transition state ensemble involving the N-terminal capping motif and first three repeats (22). The folding of the mRNA exporter protein TAP (TAPLRR) is more complicated, with a triangular mechanism and a diffuse transition state involving most of the protein (23). However, how these pathways are selected remains unknown.
To test if LRR proteins fold via low-energy pathways, we investigated the folding mechanism and stability distribution of the LRR domain of PP32. This domain is composed of a linear array of five repeats, each containing a β-strand followed by an extended structure, a 310-helix, or a short α-helix (Fig. 1) (24). The N and C termini of PP32 are capped by a helix–loop–helix motif and a β-hairpin, respectively. Deletion and mutational analysis showed that whereas the N-terminal cap contributes stability, the C-terminal cap is critical for stability: without the C-terminal cap, PP32 is entirely unfolded (25). Here we investigate the effects of destabilizing substitutions on folding kinetics and find that PP32 folds via an on-pathway intermediate with a highly polarized C-terminal pathway. We also map residue-specific stabilities throughout the LRR domain of PP32 using amide hydrogen exchange by NMR spectroscopy and find that the C-terminal repeat and cap have the highest protection factors. Thus, the region to first become structured during refolding corresponds to the most stable part of the protein. Destabilizing the C terminus of PP32 causes the highly polarized folding pathway to become much more diffuse. These results highlight the importance of local stabilities in determining the folding mechanisms of LRR proteins, as observed with α-helical repeat proteins.
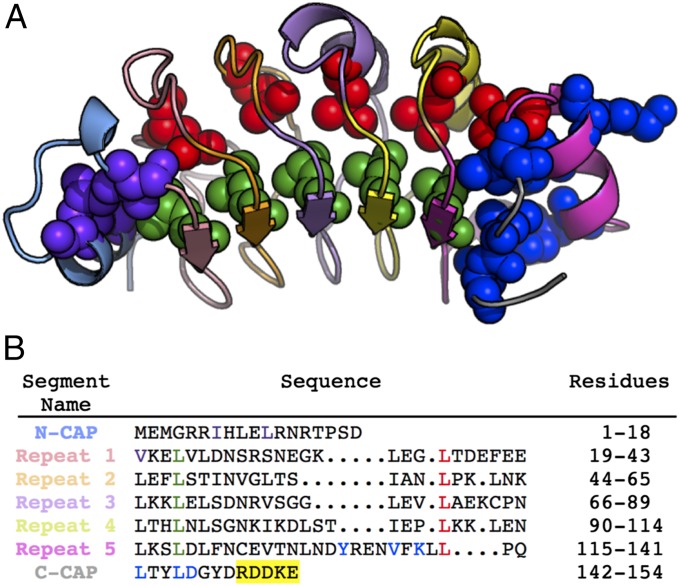
Fig. 1.
Structure and sequence of PP32. (A) Ribbon representation of the crystal structure of the LRR domain of PP32 (24). The N-cap, repeats 1–5, and the C-cap are colored as in B. The residues substituted for Φ-value analysis are shown in sphere representation. (B) Sequence alignment of the five LRRs of PP32. The five C-terminal residues (150–154) included in this study but not present in the crystal structure are highlighted in yellow.
Results
Spectroscopic Changes upon PP32 Unfolding.
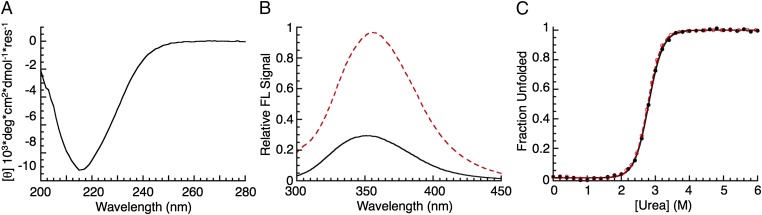
To study the folding of PP32, we monitored structural changes by circular dichroism (CD) at 220 nm and by fluorescence, which monitor β-sheet structure and the tryptophan residue at the C terminus, respectively (Fig. 2). Urea-induced equilibrium unfolding monitored by both techniques resulted in the same Cm and values, suggesting that CD and fluorescence capture the folding reaction. For the remainder of this work, we present data for equilibrium unfolding of PP32 and variants by CD and folding kinetics by fluorescence.
Fig. 2.
Solution spectroscopy and equilibrium unfolding of PP32. (A) Far-UV CD spectrum showing characteristic β-strand signal with a minimum at 217 nm. (B) Tryptophan fluorescence emission spectra in buffer (continuous black line) and 6 M urea (broken red line). (C) Urea-induced denaturation monitored by CD at 220 nm (filled black circle, continuous black line) and fluorescence (open red circle, broken red line). Lines result from fitting a two-state unfolding model to the data. The CD data are adapted from ref. 25.
Urea Dependence of Folding and Unfolding Kinetics.
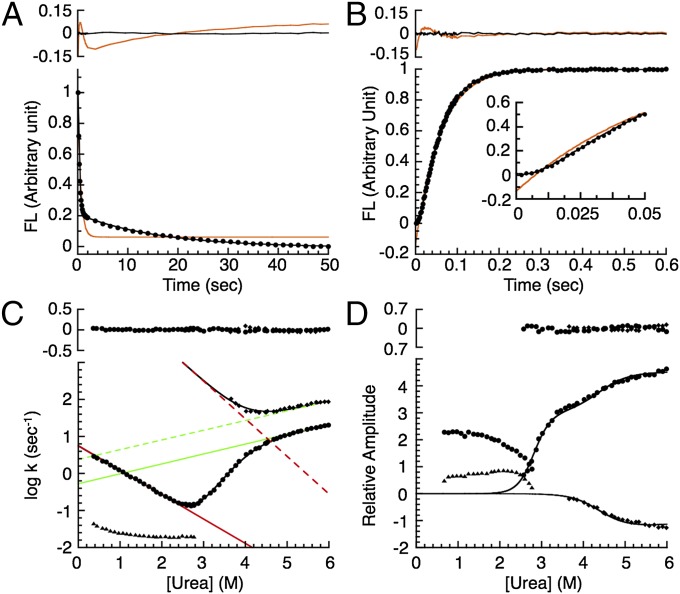
To gain insights into the folding pathway of PP32, we monitored fluorescence changes upon rapid dilution of native and denatured protein to various urea concentrations by stopped flow. Both refolding and unfolding reactions appear to be multiphasic. The data are poorly fitted by a single-exponential model (red lines in Fig. 3 A and B), resulting in large, nonrandom residuals (Fig. 3 A and B, Upper). In contrast, a double-exponential model fits both folding and unfolding transients quite well (black lines in Fig. 3 A and B), resulting in small, random residuals (Fig. 3 A and B, Upper). For refolding, a fast major phase is followed by a slow minor phase. Amplitudes for both refolding phases have the same sign, in the refolding direction. For unfolding, an early lag is followed by a slow major phase (Fig. 3B, Inset). Amplitudes for the unfolding phases have opposite signs: the amplitude of the major phase is in the unfolding direction, whereas that for the minor phase is in the refolding direction.
Fig. 3.
Refolding and unfolding kinetics of PP32. (A) Fluorescence-detected refolding (circles) from 4.6 to 0.42 M urea and (B) unfolding to 6 M urea. Red and black lines show single- and double-exponential fits, respectively. (Upper) Residuals. (C) Urea dependence of rate constants and (D) associated amplitudes for folding. Circles, major refolding and unfolding phases; triangles, minor refolding phase; diamonds, minor unfolding phase. The black lines result from fitting of (C) rate constants (excluding the minor proline-limited refolding phase) and (D) unfolding amplitudes using a sequential three-state model (Experimental Procedures). Straight lines in C show fitted folding (red) and unfolding (green) rate constants for conversion between D and I (continuous) and between I and N (dashed). (C and D, Upper) Residuals from the three-state model.
PP32 has five prolines, each in trans configuration (Fig. S1A). Cis–trans prolyl isomerization reactions are known to produce a slow, denaturant-insensitive phase during refolding, with a rate constant between 0.01 and 0.1 s−1 (26–38). The magnitude and denaturant dependence of the rate constant for the slow refolding phase in PP32 (triangles in Fig. 3C) are consistent with prolyl isomerization. To further explore whether the slow refolding phase results from prolyl isomerization, we carried out interrupted unfolding double-jump experiments, in which the protein is unfolded, held in unfolding conditions for various delay times, and then refolded (27, 34, 35, 39). If this slow phase is due to prolyl isomerization, the associated amplitudes would be small at short delay times, because most of the prolines have yet to convert to the cis conformation upon unfolding. Indeed, for PP32, the ratio of the slow phase amplitude to the total amplitude increases as a function of delay time (Fig. S1B), further supporting the slow refolding phase as prolyl isomerization phase.
In contrast, the major (fast) phase for refolding has a significant linear denaturant dependence, likely corresponding to a major conformational step in folding. This refolding phase joins up to the major unfolding phase at the equilibrium unfolding midpoint to form a V-shaped chevron plot (circles in Fig. 3C). Unlike the refolding phases, both unfolding phases show significant denaturant dependencies. Moreover, the major phase of the unfolding arm shows a rollover at 4.4 M urea. This type of nonlinearity is commonly observed for proteins folding through an intermediate (40). For PP32, an intermediate in unfolding is supported by the observation of a second kinetic phase (squares in Fig. 3C). The rollover and minor phase in unfolding are consistent with a populated on-pathway kinetic intermediate (39). This type of unfolding intermediate has been observed in other linear repeat proteins, including the ankyrin domains of the Notch receptor (39) and IκBα (41), and in an ankyrin consensus series (14).
To obtain stepwise rate constants and their urea dependences, we globally fit a sequential three-state model (D⇌I⇌N; Experimental Procedures) to the major refolding rate constants, the major and minor unfolding rate constants, and the associated unfolding amplitudes (black lines in Fig. 3 C and D). These data are well-fitted by the model, as illustrated by the small, random residuals (Fig. 3 C and D, Upper). Moreover, both the kinetically deduced (calculated as −RT*ln{kDIkIN/kNIkID}) and m-value (calculated as mDI – mID + mIN − mNI) agree well with corresponding equilibrium values (Table 1), strongly supporting the on-pathway intermediate mechanism for PP32 folding. This good agreement between kinetic and equilibrium values also suggests that the kinetic parameters are well-determined from the fit. Although there is considerable uncertainty in the extrapolated value of the rate constant for the second step in folding (kIN; Table 1), it is much greater than kDI. Thus, formation of the intermediate from the denatured state is the rate-limiting step in folding.
Table 1.
Fitted parameters for two-state equilibrium unfolding and three-state kinetic refolding and unfolding PP32 variants
| Variant | ||||||||||||
| Wild-type | 5.60 | 0.53 | 3.47 × 105 | 2.40 | −0.88 | 0.36* | −1.36 | 0.35* | 7.93 ± 0.18 | 8.29 ± 0.01 | 2.86 ± 0.02 | 2.95 ± 0.02 |
| I7A | 6.38 | 0.80 | 3.08 × 105 | 36 | −0.88 | 0.45* | −1.48 | 0.16* | 5.86 ± 0.06 | 6.48 ± 1.87 | 2.85 ± 0.04 | 2.97 ± 0.04 |
| L11A | 6.93 | 2.01 | 3.83 × 104 | 4.78 | −0.91 | 0.36† | −1.22 | 0.35† | 5.92 ± 0.10 | 5.95 ± 0.04 | 2.95 ± 0.04 | 2.85 ± 0.02 |
| V19A | 6.55 | 1.55 | 2.53 × 105 | 4.57 | −0.89 | 0.37* | −1.52 | 0.33* | 6.69 ± 0.17 | 7.20 ± 0.06 | 3.05 ± 0.13 | 3.11 ± 0.02 |
| L22A | 6.99 | 2.52 | 6.66 × 102 | 2.51 | −0.92 | 0.36† | −0.72 | 0.35† | 3.88 ± 0.05 | 3.84 ± 0.01 | 2.36 ± 0.03 | 2.34 ± 0.02 |
| L37A | 6.52 | 2.76 | 5.35 × 102 | 2.23 | −0.93 | 0.36† | −0.72 | 0.35† | 3.68 ± 0.15 | 3.69 ± 0.08 | 2.28 ± 0.07 | 2.36 ± 0.11 |
| L47A | 7.51 | 3.27 | 5.52 × 102 | 1.68 | −0.87 | 0.36† | −1.00 | 0.35† | 2.98 ± 0.17 | 3.86 ± 0.06 | 2.22 ± 0.06 | 2.58 ± 0.13 |
| L60A | 5.79 | 0.49 | 6.61 × 102 | 0.88 | −0.80 | 0.36† | −1.79 | 0.35† | 4.06 ± 0.10 | 5.29 ± 0.01 | 2.61 ± 0.01 | 3.31 ± 0.01 |
| L69A | 6.28 | 0.79 | 3.20 × 103 | 0.29 | −0.88 | 0.60* | −1.74 | 0.57* | 4.99 ± 0.13 | 6.63 ± 0.01 | 3.02 ± 0.03 | 3.79 ± 0.02 |
| L83A | 9.40 | 0.97 | 8.40 × 101 | 0.89 | −1.39 | 0.36† | −0.83 | 0.35† | 4.46 ± 0.02 | 3.97 ± 0.01 | 3.25 ± 0.02 | 2.93 ± 0.04 |
| L93A | 5.26 | 0.88 | 6.07 × 102 | 0.98 | −1.11 | 0.36† | −1.41 | 0.35† | 4.95 ± 0.19 | 4.79 ± 0.04 | 3.47 ± 0.11 | 3.23 ± 0.18 |
| L109A | 0.89 | 0.65 | 5.23 × 102 | 0.57 | −1.13 | 0.36† | −1.35 | 0.35† | 4.56 ± 0.08 | 4.15 ± 0.01 | 3.37 ± 0.17 | 3.18 ± 0.02 |
| L118A | 0.71 | 1.07 | 3.13 × 102 | 1.14 | −1.48 | 0.36† | −0.85 | 0.35† | 3.60 ± 0.12 | 3.03 ± 0.03 | 3.26 ± 0.12 | 3.03 ± 0.03 |
| Y131F | 12.68 | 0.64 | 2.93 × 104 | 2.82 | −1.02 | 0.36† | −1.30 | 0.35† | 6.97 ± 0.09 | 7.13 ± 0.03 | 3.05 ± 0.04 | 3.03 ± 0.06 |
| V135G | 0.03 | 0.65 | 3.29 × 104 | 1.12 | −1.21 | 0.36† | −1.46 | 0.35† | 4.07 ± 0.19 | 4.13 ± 0.60 | 3.61 ± 0.16 | 3.38 ± 0.04 |
| K137G | 1.27 | 0.57 | 6.72 × 105 | 5.41 | −0.92 | 0.36† | −1.39 | 0.35† | 7.26 ± 0.12 | 7.30 ± 1.24 | 3.04 ± 0.17 | 3.02 ± 0.03 |
| L139A | 0.03 | 0.66 | 1.01 × 104 | 0.64 | −0.77 | 0.36† | −1.45 | 0.35† | 4.40 ± 0.16 | 3.85 ± 0.24 | 3.36 ± 0.11 | 2.93 ± 0.03 |
| L142A | 0.03 | 0.67 | 9.16 × 103 | 0.70 | −1.03 | 0.36† | −1.41 | 0.35† | 3.86 ± 0.19 | 3.77 ± 0.17 | 3.29 ± 0.15 | 3.15 ± 0.02 |
| L145A | 0.33 | 0.69 | 1.04 × 103 | 0.73 | −1.33 | 0.36† | −1.22 | 0.35† | 4.01 ± 0.18 | 3.79 ± 0.02 | 3.37 ± 0.09 | 3.26 ± 0.04 |
| D146L | 1.03 | 0.74 | 1.45 × 102 | 0.80 | −1.30 | 0.36† | −0.84 | 0.35† | 3.49 ± 0.26 | 3.22 ± 0.04 | 2.77 ± 0.22 | 2.85 ± 0.08 |
| Y131F/D146L | 4.53 | 0.60 | 1.41 × 103 | 109 | −0.98 | 0.36† | −0.59 | 0.35† | 4.72 ± 0.14 | 4.01 ± 0.09 | 2.69 ± 0.14 | 2.28 ± 0.14 |
| ΔNCap PP32 | 6.93 | 2.57 | 4.36 × 103 | 2.31 | −0.9 | 0.36† | −1.10 | 0.35† | 4.13 ± 0.09 | 4.97 ± 0.10 | 2.37 ± 0.04 | 2.65 ± 0.12 |
Rate constants k in s−1; in kcal⋅mol−1; m-values in kcal⋅mol−1⋅M−1.
For these constructs, parameters were fitted simultaneously to major and minor observed rate constants and unfolding amplitudes.
For these variants, which show only one unfolding phase, mID and mNI were fixed at the WT values. Errors for and are the SD of three independent experiments. Errors for and (mDI – mID + mIN – mNI) are propagated from standard propagation formulas.
Structure Formation Along the PP32 Folding Pathway.
To determine the structure of PP32 along the folding pathway, we investigated the equilibrium and kinetic effects of point substitutions along the molecule, with the idea that substitutions slowing down folding kinetics are in regions that are structured in the transition states. We made substitutions in the N- and C-terminal caps, as well as the conserved leucines on the β-strand and convex side of the repeats (Fig. 1). Except for K137G, D146L, and Y131F, all substitutions are structurally conservative, removing only part of a hydrophobic side chain, and should not introduce new interactions. All variants are destabilized by at least 1 kcal⋅mol−1 (except for K137G), but remain largely folded (Fig. S2 and Table 1), allowing for accurate determination of Φ-values (42).
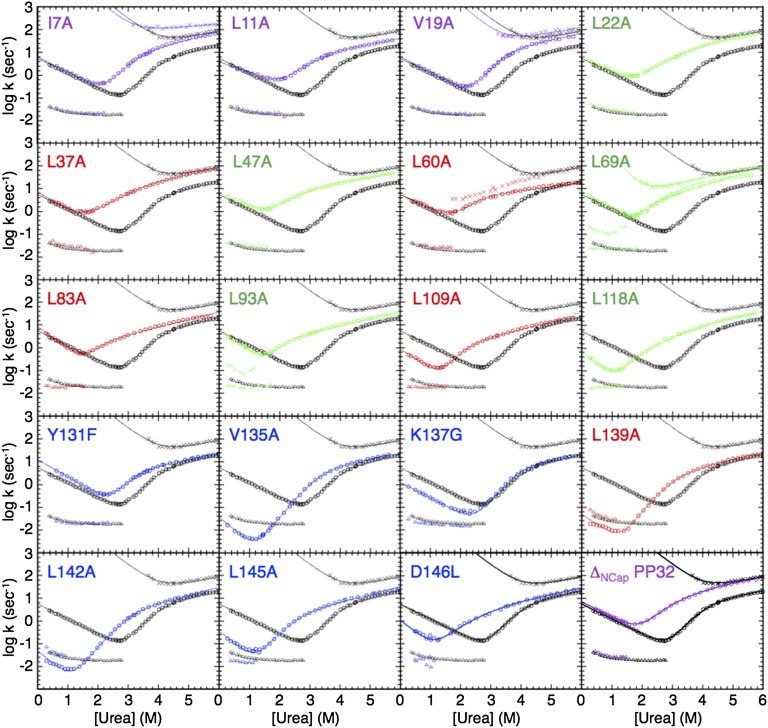
As with WT PP32, the refolding kinetics of all variants are biphasic (Fig. 4). The slow minor phase of each variant is similar to that of WT, consistent with it being a proline isomerization phase, rather than a folding reaction. The fast refolding phase is unaffected by substitutions from the N-terminal helical cap to repeat three (L93A), but is significantly slowed down by substitutions in the C terminus (repeats four and five, and capping motif).
Fig. 4.
Chevron plots for variants of PP32. Urea dependence of fluorescence-monitored rate constants for the major refolding and unfolding phases (circles), the minor refolding phase (triangles), and the minor unfolding phase (diamonds) of WT PP32 (black chevron in each panel), and variants (colors are as in Fig. 4). Lines result from fitting a sequential three-state kinetic model to the data.
In contrast to WT PP32, for most variants (except I7A, V19A, L60A, and L69A), only a single unfolding phase is observed (Fig. 4), suggesting that the on-pathway intermediate is no longer populated to high levels. However, rollovers at high urea concentrations persist in the chevron plots of all variants, suggesting that the intermediate continues to influence the kinetics of unfolding. Whereas the unfolding reactions of the N-terminal variants (I7A, L11A, V19A, L22A, L37A, L47A, L60A, and L69A) significantly speed up, those of variants in the central repeats (L83A, L93A, L109A, and L118A) are slightly faster, and those of the C-terminal variants (Y131A, V135G, K137G, L139A, L142A, L145A, and D146L) remain largely unaffected.
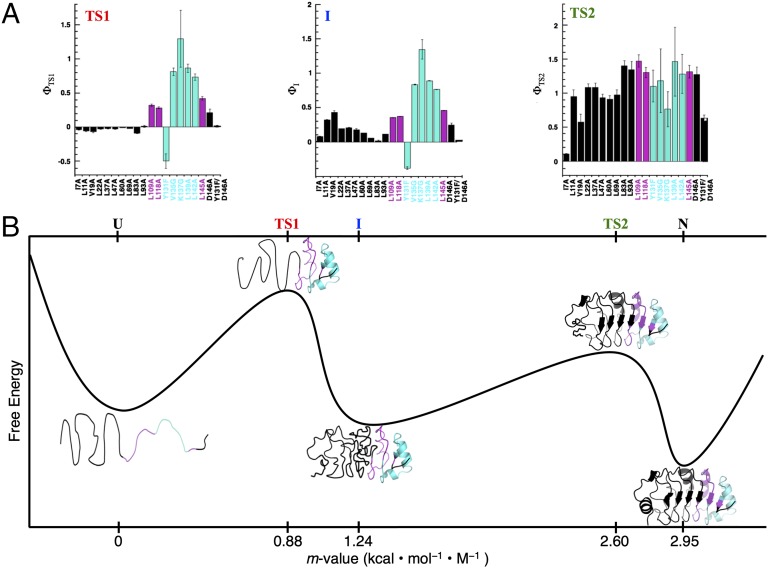
To quantitatively map out the folding pathway, we fit a sequential three-state model to the chevron plot of each variant (Table 1 and solid lines in Fig. 4). For variants that show two unfolding phases (I7A, V19A, and L69A), the rate constants and amplitudes are well-determined by the model. For the variants without an observed minor unfolding phase, there is not enough information to determine the full parameter set. To constrain these fits, we set mNI and mID to WT values. The fitted curves describe the data accurately and the resulting parameters are well-determined. Using equilibrium values and the fitted kDI, kIN, kNI, kID values for WT PP32 and variants in the absence of urea, we calculated Φ-values for TS1 (D to I), the on-pathway intermediate, and TS2 (I to N). The Φ-value reflects the extent to which a substitution affects a rate constant relative to a related equilibrium constant (43, 44). A low Φ-value indicates that the site of substitution is not structured at a specific stage in folding (e.g., TS1, I, TS2), whereas a high Φ-value indicates high structure. For TS1, which is formed in the rate-limiting step, Φ-values are low in the N-terminal cap and first three repeats, slightly increase in the fourth repeat, and drastically increase in the fifth repeat and C-terminal cap (Fig. 5A). At residue 146, the most C-terminal residue we substituted, the Φ-value decreases to an intermediate level. These Φ-values suggest that the C-terminal repeat and cap are structured in TS1, whereas the rest of the molecule remains largely unfolded (Fig. 5B). For the intermediate state, Φ-values in the N-terminal repeats increase to moderate values, indicating partial structure formation. For TS2, Φ-values are high at most positions, indicating structure formation along the entire LRR domain. Only the residues at the very beginning of the N-terminal cap appear to become structured after TS2.
Fig. 5.
Φ-values and the folding pathway of PP32. (A) Folding Φ-values for the first (rate-limiting) transition state (TS1), intermediate (I), and second transition state (TS2). Residues that are structured (Φ > 0.5), partially structured (0.25 < Φ < 0.5), and unstructured (Φ < 0.25) in TS1 are in cyan, magenta, and black, respectively. (B) A reaction coordinate for folding of PP32. The structural models of TS1, I, and TS2 are derived from the Φ-values in A. The location of each successive state (from D to N) is based on kinetic m-values (the sum of the m-values of preceding steps, as determined from the sequential three-state fit of the WT PP32 kinetic data; Table 1).
This Φ-value-based map of the folding pathway is in good agreement with kinetic m-values from the sequential three-state analysis of the WT PP32 domain (Table 1 and Fig. 5). The kinetic m-value for forming the rate-limiting TS1 is ∼30% (0.88/2.95) of the total m-value for folding, consistent with structure formation in repeats four and five. The kinetic m-value for forming I from TS1 is small, which coincides with partial structure formation in some of the N-terminal repeats, suggesting a fairly high level of solvent-accessible surface area in these N-terminal repeats. The kinetic m-value for forming TS2 from I is large (Table 1), corresponding to substantial organization of the N-terminal repeats (Fig. 5A).
If PP32 folds via a polarized C-terminal pathway, removing the N-terminal region should not affect the first step of folding. To test this prediction, we measured folding kinetics of a PP32 construct that lacks the N-terminal α-helical capping motif (ΔNCap PP32) (25). The urea-dependent refolding phases of ΔNCap PP32 are similar to those of WT PP32 (Fig. 4), whereas the slow unfolding phase significantly speeds up. These observations are consistent with the results from Φ-value analysis, confirming that the N-terminal cap is not involved in the first step of folding.
Residue-Specific Hydrogen Exchange of PP32.
To map out the distribution of stability along PP32, we monitored exchange rates of backbone amide hydrogens with solvent using NMR. Hydrogen exchange (HX) provides residue-specific information on the distribution of stability within the native state (13). The amide hydrogens in PP32 exchange over very different timescales: many peaks disappear before acquisition of the first spectrum, whereas others persist for over 3 mo (Fig. 6 A and B). For peaks with measurable exchange profiles, rate constants for exchange (kex) were fitted from peak heights, and residue-specific protection factors (PFs) were calculated as the ratio of the observed kex values to those for exchange from fully solvent-exposed unstructured state (kint; Experimental Procedures).
Fig. 6.
Residue-specific hydrogen exchange of PP32. (A) Heteronuclear single quantum coherence (HSQC) spectra after PP32 is exchanged from H2O to D2O buffer for various times: 21 min (black), 19 h (blue), 10 d (yellow), and 69 d (red). (B) Exponential decays of peak heights of representative amide protons with very different exchange rates. The lines show single-exponential fits to the data. (C) Protection factors (kex/kint) for main-chain amides with quantifiable protection. The β-sheet residues are red-filled circles; non–β-sheet residues are black circles. The solid curve results from fitting a cosine-modulated Gaussian distribution to the data (Materials and Methods). The N terminus shows uniformly low protection. The region with highest protection factors is toward the C terminus. (D) Mapping of protection factors onto the structure of PP32. For residues with main-chain amides that have quantifiable protection and are not exposed to solvent, Cα’s are shown in spheres, and colored from white to blue, with blue having the highest protection factors.
Plotting the PFs as a function of sequence and mapping the PFs onto PP32 structure reveal increasing protection from the N terminus to the C terminus (Fig. 6 C and D). This trend (at pD 6.7) is also observed for pD 6.2 and pD 7.2, indicating that exchange is limited by stability, and not by opening kinetics. Therefore, the C-terminal region appears to have much a higher local stability than the N-terminal region, consistent with our previous findings that PP32 completely unfolds upon removal of part of the C-terminal cap, but retains C-terminal structure upon removal the N-terminal α-helical cap (25). In addition, there appears to be substantial variation of protection factors within each repeat. The β-strands and neighboring residues show the greatest protection (red points in Fig. 6C), and the sequences opposite the β-strand show the greatest exchange. To attempt to capture both the high C-terminal protection and the local variation within repeats, we fitted protection factors using a normal distribution (shifted toward the C terminus) modulated by a cosine function (solid curve in Fig. 6C; Experimental Procedures). Although this model does not capture all of the variation in protection factors (especially the local variation at the C terminus), the asymmetric distribution of stability is reproduced, as is the periodicity of the local variation. In particular, the fitted frequency of the cosine term–22 residues per cycle–matches the periodicity of the sequence repeats (Fig. 1).
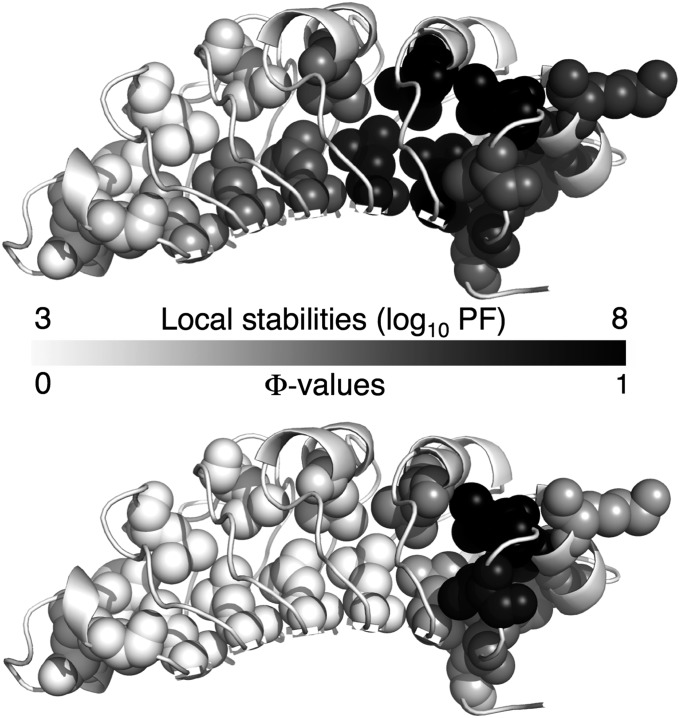
Transition state structure can be directly mapped to local stability by comparing Φ-values with PFs at the same sites (Fig. 7). The amide hydrogens of these residues are buried or involved in hydrogen bonds. Moreover, the two substituted leucines within each repeat are structurally conserved, providing direct comparison of different regions of PP32. The stability of PP32 increases gradually toward the C terminus and is highest for repeats four and five and part of the C-terminal cap. Although the stability map is more spread out than the Φ-value map, both are polarized toward the C terminus, suggesting that the folding pathway of PP32 is determined by local stability.
Fig. 7.
Folding pathway of PP32. Ribbon representation, with Φ-value substitutions shown as spheres. Residues are shaded from white to black, with black having the highest local stability (Upper) and Φ-value (Lower). For direct comparison, only sites with both Φ-value and local stability data are shaded. Based on the coincidence of high protection factors and Φ-values, folding is initiated at the most stable region of PP32.
Consequences of Destabilizing the C Terminus on the Folding Pathway of PP32.
To further test if stability dictates the location of the transition state ensemble, we determined the folding pathway of a C-terminally destabilized PP32 variant, Y131F/D146L (YD). Because the structure of YD is highly similar to that of WT PP32 (25), the destabilizing effect should be localized to the C terminus. Although YD is significantly destabilized, it retains sufficient stability for Φ-value analysis (Fig. S2D and Table 1). For direct comparison, we made the same substitutions along the YD construct as we made in WT PP32 (Fig. 8A). The YD variants are highly destabilized, to about the same extent as in the WT background. Complete unfolding transitions are observed for all but two variants (L93A and L139A; Fig. S2E).
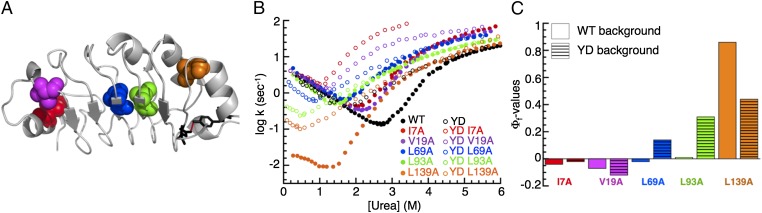
Fig. 8.
Effects of destabilizing the C terminus to the folding pathway of PP32. (A) Ribbon representation of PP32. Residues Y131 and D146, which we have substituted to destabilize the C terminus, are shown as black sticks. Residues substituted in Φ-value analysis in the Y131F/D146L (YD) background are shown in sphere representation. (B) Urea dependence of fluorescence-monitored rate constants for the major refolding and unfolding phases of variants in WT and YD backgrounds. (C) Φ-values for TS1 in the WT and YD backgrounds. The transition state for the YD construct is less polarized than for WT PP32.
To determine the effects of the substitutions on folding kinetics as the C terminus is destabilized, we compare the major phases of the chevron plots for the variants in the WT and YD backgrounds (Fig. 8B). In the YD background, we observe increased sensitivity to substitutions at the N terminus and decreased sensitivity to substitutions at the C terminus, compared with WT PP32. To quantitatively assess the effects of destabilizing the C terminus on the structure of transition state ensemble for the rate-limiting step in refolding of PP32, we compare Φ-values for the five variants in the YD and WT backgrounds (Fig. 8C). The Φ-values for L139A are the highest in both backgrounds, indicating that both fold via their C-termini. However, whereas Φ-values in WT PP32 show a highly polarized transition state restricted to repeat five, the significant positive Φ-values in repeat three and four of YD indicate a more dispersed transition state. Thus, destabilizing the C terminus (the preferred folding route in WT PP32) spreads out the transition state, switching from a highly polarized structure to a more extended region.
Discussion
The linear LRR domain of PP32 is well-suited for mechanistic studies of folding. It has sufficient stability to accommodate destabilizing point substitutions, permitting Φ-values to be determined reliably (Fig. 2 and Table 1). Its linear, repetitive architecture allows the results of Φ-values analysis to be evaluated in the context of local stability variation in structurally conserved fragments.
Overall Folding Mechanism of PP32.
Kinetic analysis of PP32 refolding and unfolding reveals an on-pathway intermediate. On-pathway intermediates are also observed for the α-helical ankyrin repeat domains, including Notch (16), p16 (37), and IκBα (41), but not for the two LRR proteins InlB (28) and TAPLRR (23) studied to date. For PP32, formation of this intermediate from the denatured state is rate-limiting in refolding at low to moderate urea concentrations. Although the rate constant for the I to N step (kIN) cannot be determined with great precision, given the large extrapolation in the chevron plot, it is clear that kIN exceeds the rate constant for the D to I step (kDI) by several orders of magnitude (Table 1), and may be as large as estimated “speed limits” for folding (45, 46).
In contrast, the rate constants for the two unfolding steps (kNI and kID) are similar to each other at all urea concentrations, with the first unfolding step (from N to I) about fivefold faster than the second (from I to D) (Table 1 and Fig. 3C). The similarity of these two rate constants produces a lag in unfolding at high denaturant concentrations, where I is more stable than N, and thus becomes transiently populated during unfolding. Below 4.4 M urea, I is less stable than N; thus, unfolding shifts to a single-exponential conversion from N to D, but is rate-limited by the transition state between I and D. As a result, at the urea midpoint for the reaction between N and I (see minimum in minor phase chevron, Fig. 3C), there is a rollover in the unfolding arm of the chevron for the major phase. This rollover reflects a shift from a modest denaturant dependence above this midpoint (resulting from surface area changes between I and TS1; Fig. 5B) to a more severe denaturant dependence below this midpoint (resulting from surface area changes between N and TS1).
The Discrete C-Terminal Folding Pathway Is Selected Based on Local Stability.
Φ-value analysis for 20 substitutions along PP32 reveals a highly polarized rate-limiting transition state (D to I) involving only the C-terminal repeat and cap (Figs. 4 and 5 and Table 1). Kinetic studies of a construct without the N-terminal cap (ΔNCap PP32) further support the C-terminal polarized folding pathway of PP32. The degree to which the folding nucleus of PP32 is localized is striking in comparison with other naturally occurring repeat proteins, such as InlB and the ankyrin domains of Notch, p16, and IκBα. Structure in the rate-limiting transitions states for these proteins includes roughly half of each protein (16, 18, 22, 41).
Through amide hydrogen exchange measurements monitored by NMR, we have found that PP32 is more stable toward the C terminus (Fig. 6). The correspondence between local stability and high Φ-values is consistent with stability dictating folding pathway selection. However, whereas the Φ-values for the TS1 are near zero for most of the protein, and approach the limiting value of one for a small cluster of residues (135–142), indicative of a highly polarized pathway, the stability distribution determined by HX is more spread out (Fig. 7). This difference results from the fact that native-state hydrogen exchange and Φ-value analysis probe different parts of the energy landscape. Φ-value analysis probes the minimum structural unit required to lead to downhill folding from the denatured state. In contrast, HX studies observe unfolding reactions in the context of the folded state, such that structural modules (foldons) (47) can stabilize one another. Work on consensus α-helical repeat proteins (48, 49) and on deletion studies of LRR proteins (25, 50) have shown that the interfacial interactions between repeats are strongly stabilizing. Thus, we expect the highly stable C terminus of PP32 to protect neighboring repeats from exchange, broadening the observed stability map toward the N terminus.
Previous studies have shown that changes to local stability can affect transition state structures. For the coiled coil GCN4, destabilizing point substitutions at one end of the molecule shift the folding pathway away from the site of destabilization (51), whereas site-specific metal-induced stabilization shifts the folding pathway to the site of stabilization (52). For the four-ankyrin repeat protein myotrophin, destabilizing C-terminal substitutions shift the folding pathway from the C terminus to the N terminus (17). In contrast, a set of consensus substitutions stabilizing the last two repeats of the Notch ankyrin domain shifts the folding pathway from the middle repeats to the C-terminal repeats (20, 21). Here, we show that the C-terminally destabilized variant Y131F/D146L folds via a much more diffuse transition state than WT PP32 (Fig. 8), providing additional evidence that folding pathways are determined by local energetics. However, unlike the ankyrin domains, C-terminal repeats of PP32 still participate in the transition state despite local destabilization, consistent with the C terminus being significantly more stable than the rest of PP32. To reroute the folding pathway of PP32 so that the C-terminal repeats fold after the transition state ensemble, a combination of substitutions destabilizing the C terminus and stabilizing the N-terminal repeats may be required.
Possible Determinants for Folding Pathway Selections in Other LRR Proteins.
The folding mechanisms of three LRR proteins whose pathways have been elucidated to date are very different from one another. Like PP32, the structure of the transition-state ensemble for InlB is also localized, although to the N terminus instead of the C terminus (28). In contrast, TAPLRR folds via a diffuse transition state in which the first three (of four) repeats are structured and the N-terminal cap makes nonnative contacts with the first repeat (23). It is possible that pathway selections for both InlB and TAPLRR are also based on local stability, as observed for PP32, in which case, the N-terminal capping motif and first three repeats would be the most stable region of InlB, whereas the repeats of TAPLRR would have more uniform stabilities.
Secondary structure might also contribute to the variations in folding pathways among these three LRR proteins. α-helices have been suggested to fold faster than β-stranded structure (53). Although PP32, InlB, and TAPLRR all have β-sheets spanning their concave surfaces, they differ significantly in helix content. The C-terminal LRRs of PP32 have α-helices on their convex surfaces (Fig. 1A), but the N-terminal LRRs do not. Thus, these helices may help initiate folding of the C-terminal LRRs. The transition state for folding of InlB includes the N-terminal cap, which is the only region with α-helices. The uniform distribution of α-helices along the TAPLRR domain may contribute to its diffuse transition state and may lead to parallel folding pathways, as recently reported for an α-helical ankyrin repeat protein series (14). However, helical structure alone is not enough to determine folding routes because preferred pathways are observed for α-helical repeats proteins, where repeats all have a high helix content (15–18, 21).
Biological Implications.
PP32 is primarily a nuclear protein. Nuclear import is mediated by a nuclear localization signal in its acidic region, C-terminal to the LRR domain (54). However, PP32 can also shuttle back to the cytoplasm through binding of the LRR domain to nuclear export protein Crm1 (55). Crm1 recognizes its cargo through leucine-rich nuclear export signals with a consensus sequence of x-Φ0-x2-Φ1-x2,3-Φ2-x2,3-Φ3-x-Φ4, where hydrophobic residues Φ (most commonly leucine) are separated by 1–3 residues x (often polar or charged; 56, 57). This consensus sequence is found in PP32 LRRs 1–4, at least one of which is likely to interact with Crm1. Structures of Crm1 complexes have shown that the conserved Φ’s of the consensus sequence dock into the pockets of the hydrophobic cleft of Crm1 (58, 59). Because the conserved Φ’s of PP32 make up its core and are not solvent-exposed, binding to Crm1 is likely to require partial unfolding of PP32. Our HX result showing that the N terminus is much less stable than the C terminus suggests the N-terminal repeats (LRRs 1–3) as likely binding motifs for Crm1.
Experimental Procedures
Subcloning, Protein Expression, and Purification.
The gene encoding PP32 was a kind gift from the laboratory of Cynthia Wolberger (Johns Hopkins University School of Medicine, Baltimore, MD). Constructs encoding PP32, ΔNCap PP32, and PP32 variants I7A, V19A, L69A, L93A, L139A, Y131F, D146L, and Y131F/D146L have been describe previously (25). Additional point substitutions were made using QuikChange (Stratagene). Protein were expressed and purified as described (25).
Fluorescence Spectroscopy.
Fluorescence emission spectra were collected on an Aviv ATF 105 spectropolarimeter in a 1.0-cm path-length cuvette. The protein concentration was 3 μM, in storage buffer [20 mM NaPO4, 150 mM NaCl, 0.1 mM tris(2-carboxyethyl)phosphine (TCEP) (pH 7.8)] with 0 or 5.4 M urea, at 20 °C.
Equilibrium Unfolding.
Urea-induced unfolding was monitored by CD (Aviv Model 400 CD spectrometer) at 220 nm or fluorescence by exciting at 295 nm and recording emission at 324 nm. Urea was deionized by chromatography over mixed-bed resin (Bio-Rad). Urea concentration was determined by refractometry. Urea titrations were carried out using a computer-controlled Microlab syringe titrator (Hamilton). Samples contained 2–4 μM protein, 20 mM sodium phosphate, 150 mM NaCl, 0.1 mM TCEP (pH 7.8). At each urea concentration, samples were equilibrated for 5 min at 20 °C and signal-averaged for 30 s. Two-state analysis of equilibrium unfolding transitions was carried out as described by Street et al. (60).
Kinetic Folding Studies.
Fluorescence-detected unfolding and refolding kinetic measurements were made on an Applied Photophysics SX18MV-R stopped-flow rapid mixing device. Emission was monitored using a 320 cutoff filter, following excitation at 280 nm to monitor changes in the environment surrounding the single tryptophan at the C terminus. Final protein concentrations were 1–3 μM. Experiments were done in 20 mM sodium phosphate, 150 mM NaCl, 0.1 mM TCEP (pH 7.8) and at 20 °C. Unfolding and refolding amplitudes and rate constants were determined using nonlinear least-squares to fit the following equation to the individual progress curves:
Where represents the fluorescence at equilibrium, and and represent the change in fluorescence signal contributed by the ith phase and the rate constant for the ith phase, respectively. Two phases were necessary and sufficient to describe PP32 refolding and unfolding kinetics of WT PP32 and variants I7A, V19A, L60A, and L69A. Only one phase was sufficient to describe the unfolding for the rest of the variants.
To capture this complexity in refolding and unfolding, data were fitted with a sequential three-state model:
where the denatured (D) state is converted to the native (N) state through a single on-pathway intermediate (I) (41, 61). Microscopic rate constants from the sequential three-state model and their urea dependences were obtained from the global fitting of the observed rate constants for refolding and unfolding and, when applicable, the associated amplitudes for major and minor unfolding phases, all as a function of urea. The program Profit 6.1.16 (Quantum Soft) was used for fitting, with scripts kindly provided by Thomas Kiefhaber (Technische Universität München, Garching, Germany) and Andreas Moeglich (Humboldt-Universität zu Berlin, Berlin, Germany).
Φ-values were calculated from the following relationships:
where the rate constants and free energy changes were extrapolated to zero molar denaturant and mut and wt indicate values for variants and WT PP32, respectively. Free energies of unfolding from N to D were determined from equilibrium urea unfolding experiments.
Hydrogen Exchange of WT PP32.
15N-labeled PP32 [800 μM in 20 mM NaPO4, 50 mM NaCl, 0.2 mM TCEP (pH 6.8)] was exchanged into D2O buffer (20 mM NaPO4, 50 mM NaCl, 0.1 mM EDTA, 0.2 mM TCEP, pDs 6.2, 6.7, or 7.2) by centrifugal gel filtration as described by (62). A total of 2 mL of preswollen Sephadex G-25 Fine (GE Healthcare) was placed into a 3-mL spin column (Fisher Scientific), washed 4–5 times with 2 mL of D2O buffer by low-speed spinning (tabletop swing bucket centrifuge for 3 min at 3000 × g), followed by 560 mL of protein sample. Final protein concentration was ∼650 μM in 95% D2O sample buffer.
1H-15N HSQC spectra were immediately collected after solvent exchange on a Bruker 600 MHz spectrometer equipped with a cryoprobe at 20 °C. About 20 spectra were collected during the first 12 h. Subsequently, one spectrum was collected every few hours, everyday, then every few days for the next 3 mo. Spectra were processed using NMRPipe (63), and displayed and analyzed with Sparky (64). Assignments for amide resonances of PP32 are from ref. 25. Peak heights were determined in Sparky. To compare the exchange rates of different residues of PP32, we calculated protection factors (PF) with the following equation:
where kint is the rate constant for exchange from amino acids in random coil (65), and kex is the observed exchange rate constant obtained from fitting a single-exponential decay function to the change in the heights of amide cross-peaks in the series of HSQC spectra.
Protection factors were fitted as a function of n, the residue number (Fig. 6C) with a cosine-modulated Gaussian function:
| [1] |
The fitted parameters are the amplitude a, mean μ, and SD σ of the Gaussian function, and the amplitude b, repeat period nrep, and phase c of the cosine modulation. Fitted values and confidence intervals are given in Table S1.
Supplementary Material
Acknowledgments
We thank Dr. Cynthia Wolberger for providing us with the gene encoding the LRR domain PP32, and Dr. Carlos A. Castañeda for assistance in analysis of HX data. We thank The Johns Hopkins University Biomolecular NMR Center for providing facilities and resources. This research is supported by National Institutes of Health Grant GM068462 (to D.B.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1412165112/-/DCSupplemental.
References
- 1.Levinthal C. Are there pathways for protein folding? J Chim Phys. 1968;65:44–45. [Google Scholar]
- 2.Kim PS, Baldwin RL. Intermediates in the folding reactions of small proteins. Annu Rev Biochem. 1990;59(1):631–660. doi: 10.1146/annurev.bi.59.070190.003215. [DOI] [PubMed] [Google Scholar]
- 3.Matthews CR. Pathways of protein folding. Annu Rev Biochem. 1993;62(1):653–683. doi: 10.1146/annurev.bi.62.070193.003253. [DOI] [PubMed] [Google Scholar]
- 4.Myers JK, Oas TG. Mechanism of fast protein folding. Annu Rev Biochem. 2002;71(1):783–815. doi: 10.1146/annurev.biochem.71.110601.135346. [DOI] [PubMed] [Google Scholar]
- 5.Oliveberg M, Wolynes PG. The experimental survey of protein-folding energy landscapes. Q Rev Biophys. 2005;38(3):245–288. doi: 10.1017/S0033583506004185. [DOI] [PubMed] [Google Scholar]
- 6.Wolynes PG, Onuchic JN, Thirumalai D. Navigating the folding routes. Science. 1995;267(5204):1619–1620. doi: 10.1126/science.7886447. [DOI] [PubMed] [Google Scholar]
- 7.Hoang L, Bédard S, Krishna MMG, Lin Y, Englander SW. Cytochrome c folding pathway: Kinetic native-state hydrogen exchange. Proc Natl Acad Sci USA. 2002;99(19):12173–12178. doi: 10.1073/pnas.152439199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu W, et al. Stepwise protein folding at near amino acid resolution by hydrogen exchange and mass spectrometry. Proc Natl Acad Sci USA. 2013;110(19):7684–7689. doi: 10.1073/pnas.1305887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jennings PA, Wright PE. Formation of a molten globule intermediate early in the kinetic folding pathway of apomyoglobin. Science. 1993;262(5135):892–896. doi: 10.1126/science.8235610. [DOI] [PubMed] [Google Scholar]
- 10.McCallister EL, Alm E, Baker D. Critical role of β-hairpin formation in protein G folding. Nat Struct Biol. 2000;7(8):669–673. doi: 10.1038/77971. [DOI] [PubMed] [Google Scholar]
- 11.Nauli S, Kuhlman B, Baker D. Computer-based redesign of a protein folding pathway. Nat Struct Biol. 2001;8(7):602–605. doi: 10.1038/89638. [DOI] [PubMed] [Google Scholar]
- 12.Raschke TM, Marqusee S. The kinetic folding intermediate of ribonuclease H resembles the acid molten globule and partially unfolded molecules detected under native conditions. Nat Struct Biol. 1997;4(4):298–304. doi: 10.1038/nsb0497-298. [DOI] [PubMed] [Google Scholar]
- 13.Krishna MMG, Hoang L, Lin Y, Englander SW. Hydrogen exchange methods to study protein folding. Methods. 2004;34(1):51–64. doi: 10.1016/j.ymeth.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Aksel T, Barrick D. Direct observation of parallel folding pathways revealed using a symmetric repeat protein system. Biophys J. 2014;107(1):220–232. doi: 10.1016/j.bpj.2014.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Löw C, et al. Folding mechanism of an ankyrin repeat protein: Scaffold and active site formation of human CDK inhibitor p19(INK4d) J Mol Biol. 2007;373(1):219–231. doi: 10.1016/j.jmb.2007.07.063. [DOI] [PubMed] [Google Scholar]
- 16.Bradley CM, Barrick D. The notch ankyrin domain folds via a discrete, centralized pathway. Structure. 2006;14(8):1303–1312. doi: 10.1016/j.str.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 17.Lowe AR, Itzhaki LS. Rational redesign of the folding pathway of a modular protein. Proc Natl Acad Sci USA. 2007;104(8):2679–2684. doi: 10.1073/pnas.0604653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang KS, Fersht AR, Itzhaki LS. Sequential unfolding of ankyrin repeats in tumor suppressor p16. Structure. 2003;11(1):67–73. doi: 10.1016/s0969-2126(02)00929-2. [DOI] [PubMed] [Google Scholar]
- 19.Mello CC, Barrick D. An experimentally determined protein folding energy landscape. Proc Natl Acad Sci USA. 2004;101(39):14102–14107. doi: 10.1073/pnas.0403386101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tripp KW, Barrick D. Enhancing the stability and folding rate of a repeat protein through the addition of consensus repeats. J Mol Biol. 2007;365(4):1187–1200. doi: 10.1016/j.jmb.2006.09.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tripp KW, Barrick D. Rerouting the folding pathway of the Notch ankyrin domain by reshaping the energy landscape. J Am Chem Soc. 2008;130(17):5681–5688. doi: 10.1021/ja0763201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Courtemanche N, Barrick D. The leucine-rich repeat domain of Internalin B folds along a polarized N-terminal pathway. Structure. 2008;16(5):705–714. doi: 10.1016/j.str.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly SE, et al. Diffuse transition state structure for the unfolding of a leucine-rich repeat protein. Phys Chem Chem Phys. 2014;16(14):6448–6459. doi: 10.1039/c3cp54818j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huyton T, Wolberger C. The crystal structure of the tumor suppressor protein pp32 (Anp32a): Structural insights into Anp32 family of proteins. Protein Sci. 2007;16(7):1308–1315. doi: 10.1110/ps.072803507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dao TP, Majumdar A, Barrick D. Capping motifs stabilize the leucine-rich repeat protein PP32 and rigidify adjacent repeats. Protein Sci. 2014;23(6):801–811. doi: 10.1002/pro.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bradley CM, Barrick D. Effect of multiple prolyl isomerization reactions on the stability and folding kinetics of the notch ankyrin domain: Experiment and theory. J Mol Biol. 2005;352(2):253–265. doi: 10.1016/j.jmb.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 27.Brandts JF, Halvorson HR, Brennan M. Consideration of the Possibility that the slow step in protein denaturation reactions is due to cis-trans isomerism of proline residues. Biochemistry. 1975;14(22):4953–4963. doi: 10.1021/bi00693a026. [DOI] [PubMed] [Google Scholar]
- 28.Courtemanche N, Barrick D. Folding thermodynamics and kinetics of the leucine-rich repeat domain of the virulence factor Internalin B. Protein Sci. 2008;17(1):43–53. doi: 10.1110/ps.073166608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamen DE, Woody RW. Folding kinetics of the protein pectate lyase C reveal fast-forming intermediates and slow proline isomerization. Biochemistry. 2002;41(14):4713–4723. doi: 10.1021/bi0115129. [DOI] [PubMed] [Google Scholar]
- 30.Kamen DE, Woody RW. Identification of proline residues responsible for the slow folding kinetics in pectate lyase C by mutagenesis. Biochemistry. 2002;41(14):4724–4732. doi: 10.1021/bi0115131. [DOI] [PubMed] [Google Scholar]
- 31.Kiefhaber T, Schmid FX. Kinetic coupling between protein folding and prolyl isomerization. II. Folding of ribonuclease A and ribonuclease T1. J Mol Biol. 1992;224(1):231–240. doi: 10.1016/0022-2836(92)90586-9. [DOI] [PubMed] [Google Scholar]
- 32.Kiefhaber T, Quaas R, Hahn U, Schmid FX. Folding of ribonuclease T1. 1. Existence of multiple unfolded states created by proline isomerization. Biochemistry. 1990;29(12):3053–3061. doi: 10.1021/bi00464a023. [DOI] [PubMed] [Google Scholar]
- 33.Kloss E, Barrick D. Thermodynamics, kinetics, and salt dependence of folding of YopM, a large leucine-rich repeat protein. J Mol Biol. 2008;383(5):1195–1209. doi: 10.1016/j.jmb.2008.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmid FX, Mayr LM, Mücke M, Schönbrunner ER. Prolyl isomerases: Role in protein folding. Adv Protein Chem. 1993;44:25–66. doi: 10.1016/s0065-3233(08)60563-x. [DOI] [PubMed] [Google Scholar]
- 35.Schmid FX, Baldwin RL. Acid catalysis of the formation of the slow-folding species of RNase A: Evidence that the reaction is proline isomerization. Proc Natl Acad Sci USA. 1978;75(10):4764–4768. doi: 10.1073/pnas.75.10.4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmid FX, Baldwin RL. The rate of interconversion between the two unfolded forms of ribonuclease A does not depend on guanidinium chloride concentration. J Mol Biol. 1979;133(2):285–287. doi: 10.1016/0022-2836(79)90536-9. [DOI] [PubMed] [Google Scholar]
- 37.Tang KS, Guralnick BJ, Wang WK, Fersht AR, Itzhaki LS. Stability and folding of the tumour suppressor protein p16. J Mol Biol. 1999;285(4):1869–1886. doi: 10.1006/jmbi.1998.2420. [DOI] [PubMed] [Google Scholar]
- 38.Zeeb M, et al. Protein folding and stability of human CDK inhibitor p19(INK4d) J Mol Biol. 2002;315(3):447–457. doi: 10.1006/jmbi.2001.5242. [DOI] [PubMed] [Google Scholar]
- 39.Mello CC, Bradley CM, Tripp KW, Barrick D. Experimental characterization of the folding kinetics of the notch ankyrin domain. J Mol Biol. 2005;352(2):266–281. doi: 10.1016/j.jmb.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 40.Baldwin RL. On-pathway versus off-pathway folding intermediates. Fold Des. 1996;1(1):R1–R8. doi: 10.1016/S1359-0278(96)00003-X. [DOI] [PubMed] [Google Scholar]
- 41.DeVries I, Ferreiro DU, Sánchez IE, Komives EA. Folding kinetics of the cooperatively folded subdomain of the IκBα ankyrin repeat domain. J Mol Biol. 2011;408(1):163–176. doi: 10.1016/j.jmb.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sánchez IE, Kiefhaber T. Evidence for sequential barriers and obligatory intermediates in apparent two-state protein folding. J Mol Biol. 2003;325(2):367–376. doi: 10.1016/s0022-2836(02)01230-5. [DOI] [PubMed] [Google Scholar]
- 43.Matthews CR. Effect of point mutations of the folding of globular proteins. In: Ray Wu LG, editor. Methods in Enzymology. Academic; New York: 1987. pp. 498–511. [DOI] [PubMed] [Google Scholar]
- 44.Otzen DE, Itzhaki LS, elMasry NF, Jackson SE, Fersht AR. Structure of the transition state for the folding/unfolding of the barley chymotrypsin inhibitor 2 and its implications for mechanisms of protein folding. Proc Natl Acad Sci USA. 1994;91(22):10422–10425. doi: 10.1073/pnas.91.22.10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kubelka J, Hofrichter J, Eaton WA. The protein folding ‘speed limit’. Curr Opin Struct Biol. 2004;14(1):76–88. doi: 10.1016/j.sbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 46.Yang WY, Gruebele M. Folding at the speed limit. Nature. 2003;423(6936):193–197. doi: 10.1038/nature01609. [DOI] [PubMed] [Google Scholar]
- 47.Englander SW, Mayne L, Krishna MMG. Protein folding and misfolding: Mechanism and principles. Q Rev Biophys. 2007;40(4):287–326. doi: 10.1017/S0033583508004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aksel T, Majumdar A, Barrick D. The contribution of entropy, enthalpy, and hydrophobic desolvation to cooperativity in repeat-protein folding. Structure. 2011;19(3):349–360. doi: 10.1016/j.str.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wetzel SK, Settanni G, Kenig M, Binz HK, Plückthun A. Folding and unfolding mechanism of highly stable full-consensus ankyrin repeat proteins. J Mol Biol. 2008;376(1):241–257. doi: 10.1016/j.jmb.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 50.Vieux EF, Barrick D. Deletion of internal structured repeats increases the stability of a leucine-rich repeat protein, YopM. Biophys Chem. 2011;159(1):152–161. doi: 10.1016/j.bpc.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moran LB, Schneider JP, Kentsis A, Reddy GA, Sosnick TR. Transition state heterogeneity in GCN4 coiled coil folding studied by using multisite mutations and crosslinking. Proc Natl Acad Sci USA. 1999;96(19):10699–10704. doi: 10.1073/pnas.96.19.10699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krantz BA, Sosnick TR. Engineered metal binding sites map the heterogeneous folding landscape of a coiled coil. Nat Struct Biol. 2001;8(12):1042–1047. doi: 10.1038/nsb723. [DOI] [PubMed] [Google Scholar]
- 53.Plaxco KW, Simons KT, Ruczinski I, Baker D. Topology, stability, sequence, and length: Defining the determinants of two-state protein folding kinetics. Biochemistry. 2000;39(37):11177–11183. doi: 10.1021/bi000200n. [DOI] [PubMed] [Google Scholar]
- 54.Matsubae M, Kurihara T, Tachibana T, Imamoto N, Yoneda Y. Characterization of the nuclear transport of a novel leucine-rich acidic nuclear protein-like protein. FEBS Lett. 2000;468(2-3):171–175. doi: 10.1016/s0014-5793(00)01218-7. [DOI] [PubMed] [Google Scholar]
- 55.Brennan CM, Gallouzi I-E, Steitz JA. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J Cell Biol. 2000;151(1):1–14. doi: 10.1083/jcb.151.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Güttler T, et al. NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1. Nat Struct Mol Biol. 2010;17(11):1367–1376. doi: 10.1038/nsmb.1931. [DOI] [PubMed] [Google Scholar]
- 57.Xu D, Farmer A, Collett G, Grishin NV, Chook YM. Sequence and structural analyses of nuclear export signals in the NESdb database. Mol Biol Cell. 2012;23(18):3677–3693. doi: 10.1091/mbc.E12-01-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dong X, et al. Structural basis for leucine-rich nuclear export signal recognition by CRM1. Nature. 2009;458(7242):1136–1141. doi: 10.1038/nature07975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Monecke T, et al. Structural basis for cooperativity of CRM1 export complex formation. Proc Natl Acad Sci USA. 2013;110(3):960–965. doi: 10.1073/pnas.1215214110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Street TO, Courtemanche N, Barrick D. Protein folding and stability using denaturants. In: Correia JJ, Detrich HW, editors. Methods in Cell Biology. Academic; New York: 2008. pp. 295–325. [DOI] [PubMed] [Google Scholar]
- 61.Khorasanizadeh S, Peters ID, Roder H. Evidence for a three-state model of protein folding from kinetic analysis of ubiquitin variants with altered core residues. Nat Struct Biol. 1996;3(2):193–205. doi: 10.1038/nsb0296-193. [DOI] [PubMed] [Google Scholar]
- 62.Jeng M-F, Englander SW. Stable submolecular folding units in a non-compact form of cytochrome c. J Mol Biol. 1991;221(3):1045–1061. doi: 10.1016/0022-2836(91)80191-v. [DOI] [PubMed] [Google Scholar]
- 63.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 64.Goddard TD, Kneller DG. 2008. SPARKY 3 (University of California, San Francisco)
- 65.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17(1):75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.