Significance
In a cell that is not actively moving, the shape of the nucleus and its central position are robustly maintained in a mechanical homeostasis. How nuclear shape and position are maintained despite the various types of intracellular forces that act on the nuclear surface is an open question. Dysregulated positioning and irregular nuclear shape are associated with a variety of human pathologies. In this paper we estimate the magnitude of the forces that are required to displace and deform the cell nucleus and identify the main molecules that resist these forces. Our results are important because they reveal how the nucleus has such a stable shape and position in the cell, despite the different forces acting on it.
Keywords: nuclear forces, cytoskeleton, nuclear positioning, nuclear mechanics, nuclear shape
Abstract
How cells maintain nuclear shape and position against various intracellular and extracellular forces is not well understood, although defects in nuclear mechanical homeostasis are associated with a variety of human diseases. We estimated the force required to displace and deform the nucleus in adherent living cells with a technique to locally pull the nuclear surface. A minimum pulling force of a few nanonewtons—far greater than typical intracellular motor forces—was required to significantly displace and deform the nucleus. Upon force removal, the original shape and position were restored quickly within a few seconds. This stiff, elastic response required the presence of vimentin, lamin A/C, and SUN (Sad1p, UNC-84)-domain protein linkages, but not F-actin or microtubules. Although F-actin and microtubules are known to exert mechanical forces on the nuclear surface through molecular motor activity, we conclude that the intermediate filament networks maintain nuclear mechanical homeostasis against localized forces.
The nucleus in a cultured cell such as a fibroblast is close to the center of the cell and typically has a smooth, regular shape. It is known that the migrating cell moves the nucleus by transferring cytoskeletal forces through connections between the cytoskeleton and the nuclear surface (1, 2). Even in a stationary cell, the nuclear shape and central position are stably maintained in mechanical homeostasis at defined locations in the cell, despite the fact that the dynamic cytoskeleton continues to generate constantly fluctuating forces on the nucleus (3, 4).
The source of fluctuating forces on the nucleus includes nuclear-embedded microtubule motors such as dynein and kinesin (5–7) and actomyosin forces that push on and pull the nucleus (2, 8, 9). The nucleus is also exposed to extracellular forces, such as those applied to adhesion receptors, which can be transmitted through the cytoskeleton onto the nuclear surface (10–12). How nuclear shape and position are maintained in mechanical homeostasis despite the different types of forces that act on the nucleus is an open question. This is particularly important because deregulated positioning and irregular nuclear shapes are associated with a variety of human pathologies (reviewed in ref. 13).
Here we describe a method to apply forces directly to nuclei in cultured, living, adherent cells. With this method, we estimate a minimum pulling force of a few nanonewtons—far greater than typical intracellular motor forces—is required to significantly displace and deform the nucleus. Although F-actin and microtubules are known to exert mechanical forces on the nuclear surface through molecular motor activity, we show that the intermediate filament networks maintain nuclear mechanical homeostasis.
Results
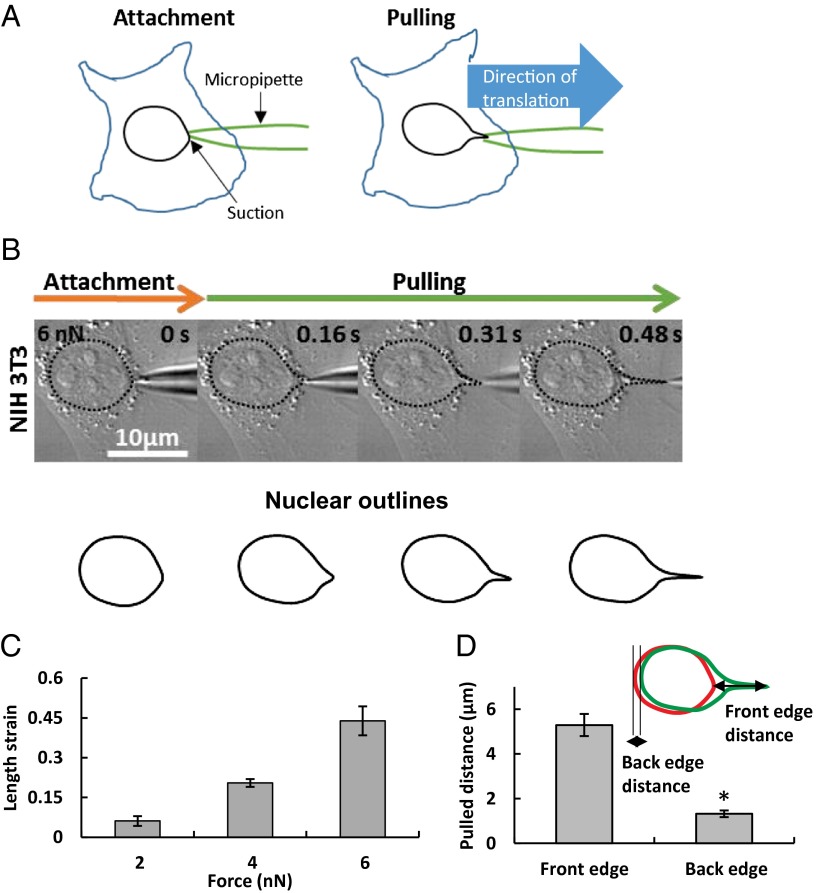
To determine the forces that are required for moving and deforming the nucleus, and to identify the cellular components that oppose motion and deformation, we devised a method to apply forces directly to nuclei in cultured, living, adherent cells. We first sealed a micropipette tip (0.5-µm diameter) to the nuclear surface in well-spread cells with a specified suction pressure. We then translated the pipette tip away from the nucleus at a known speed (Fig. 1A). The nucleus deformed and moved during the translation of the pipette (Fig. 1B), and finally released from the pipette tip. At the point that the nucleus releases from the micropipette tip, the resistance force to nuclear motion and deformation should balance the force on the outer nuclear surface created by the suction. Although the suction pressure in the micropipette is known, this pressure is larger than the actual pressure on the nuclear surface owing to flow across the pores in the nuclear membrane (14, 15). However, as shown in SI Text, the resistance to flow through the nuclear pores is much larger than the resistance to flow through the micropipette. Consequently, the actual force on the nucleus at the point of release should be essentially equal to the suction force (defined as the suction pressure × the cross-sectional area), which we report in our results.
Fig. 1.
Deformation of the nucleus in a living, adherent cell. (A) Schematic of how nuclear forces were applied in a living, adherent cell. A narrow micropipette tip (∼0.5-µm diameter) was attached to the nuclear surface and forces were applied using capillary suction pressure at the contact site. The pipette was then pulled at a known rate and the nuclear response was observed. (B) DIC images show result of translation of 0.5-µm-tip micropipette sealed to the nuclear surface with a 6 nN suction force in an NIH 3T3 fibroblast cell. Translation of the micropipette pulled and deformed the nucleus as evident from the outlined shapes. (C) Nuclear deformation as quantified by length strain increased with the applied suction force (all values are different from each other at P < 0.05; n = 6 for 2 nN, n = 7 for 4 nN, n = 14 for 6 nN). (D) The deformation was much larger than the motion of the nucleus, as can be seen from the overlay of the outlines of the initial shape (red) and deformed shape (green). The extent of nuclear motion was quantified from the translation of the back edge and the extent of nuclear deformation measured as the distance moved by the front edge. The plot shows that the motion of the front edge was significantly larger than the back edge (*P < 0.05 according to Student’s t test; n = 10). Values are the mean ± SEM.
The extent of nuclear motion and deformation at the point of release depended on the suction pressure. At zero suction pressure, the nucleus separated from the pipette without any measurable change in shape or position, suggesting that there is negligible adhesion between the tip and the nuclear surface. At higher suction pressures, the nucleus deformed noticeably (Fig. 1B). The front edge typically formed a nuclear protrusion in the direction of the micropipette translation. As shown in Fig. 1C, the degree of deformation, quantified by the length strain (ε = L/L0 − 1, where L and L0 are the lengths of the nucleus at maximum deformation and initially, respectively) increased with the suction force.
By performing these experiments in the presence of cell membrane-impermeable dyes, we confirmed that damage to the cell membrane was confined only to a thin lipid tether that formed due to the motion of the micropipette; the cells were confirmed to be viable throughout the experiment (Fig. S1). We note that the extent of nuclear deformation was independent of the loading rate, which implies a predominantly elastic (nonviscous) resistance to the pulling force (Fig. S2).
We quantified nuclear movement by measuring the displacement of the trailing edge of the nucleus (Fig. 1D). The extent of nuclear protrusion was greater than the net displacement of the trailing edge (Fig. 1D). This observation that the nucleus moves less than it deforms suggests a tight integration between the nucleus and its surroundings. This clear protrusion and translation was consistently observed in several types of adherent living mammalian cells [see Fig. S3 for examples in mouse embryonic fibroblasts (MEFs), human SW13 epithelial cells, and human MCF10A mammary cells; NIH 3T3 fibroblasts are shown in Fig. 1B].
Following release of the nucleus from the micropipette, the nuclear shape and position recovered partially. The relaxation time scale of the recovery of the nuclear deformation and its displacement reflects the relative magnitude of elastic versus viscous resistance in nuclear mechanics. We used fluorescence-based imaging to precisely track the deformed nuclear shape. Although GFP-histone H1.1 is a commonly used live nuclear fluorescence marker, we surprisingly found that nuclei labeled with GFP-histone H1.1 showed markedly shorter protrusions than unlabeled nuclei (Fig. S4). Overexpression of histone H1.1 might alter the bulk mechanical properties of the nucleus owing to increased chromatin compaction. Because Hoechst stains may alter nuclear properties as well (16), we tested the effect of the dye SYTO 59, a DNA minor-groove binding molecule (17) specifically developed for live-cell imaging. Here we found no differences in the mechanical response of stained and unstained nuclei (Fig. S4); we therefore used SYTO 59 for nuclear tracking purposes.
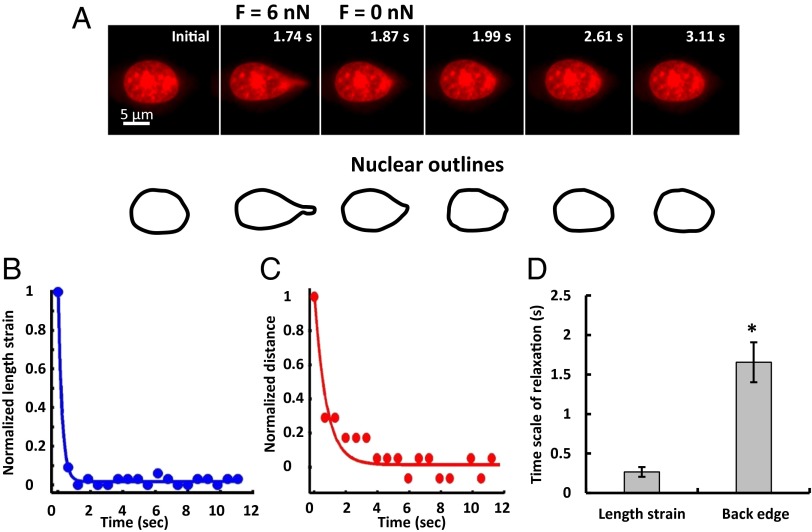
We monitored the shapes of the pulled, fluorescent nuclei as a function of time (Fig. 2A and Movie S1) from which we calculated (i) a length strain εt = Lt/L0 − 1, where Lt and L0 are the lengths of the nucleus at time t and initially, respectively, and (ii) displacement of the trailing edge of the nucleus in the pulled direction. From the length strain and displacement dynamics, we estimated the time scales of shape and position relaxation. Upon detachment, the nucleus relaxed back close to its original shape remarkably quickly (<1 s, Fig. 2A). Nuclear length strain relaxed much more rapidly (<500 ms) than the recovery of nuclear position (1.7 ± 0.3 s, Fig. 2 B–D). The large stiffness and rapid relaxation of the shape and position may reflect the strongly elastic nature of the nuclear lamina and the perinuclear space that resists nuclear deformation and motion. Therefore, we focused next on identifying the main contributors to nuclear mechanical behavior.
Fig. 2.
Rapid nuclear relaxation upon force release. (A) A typical nucleus is shown at the indicated times. After the nucleus deformed when the micropipette tip sealed to the nuclear surface with a suction force of 6 nN was translated, it rapidly relaxed back to its original shape and position on detachment from the micropipette tip; steady state after detachment was achieved in a few seconds. (B and C) The time-dependent recovery of the length strain (which is a measure of nuclear deformation, blue circles) and back edge distance (red circles) could be described by single exponential fits (solid lines), with the length strain relaxation being much faster than the back edge relaxation; quantification is shown (D). *P < 0.05, n = 10.
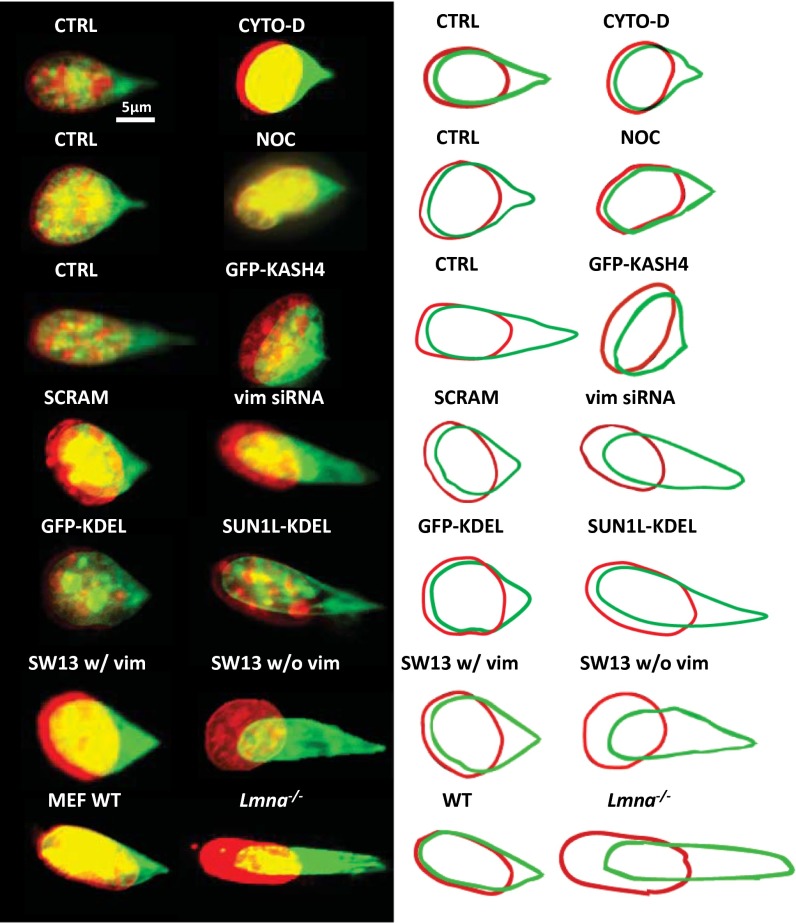
Because our method allowed us to locally pull on the nucleus in adherent, spread cells, we asked whether the cytoskeleton contributed to the observed resistance to nuclear deformation and motion. Because F-actin is thought to pull on the nuclear surface through physical connections maintained by transmembrane actin-associated nuclear lines (18), we depolymerized F-actin with cytochalasin-D treatment (Fig. S5A), but this caused no significant difference in the extent of nuclear deformation or translation under the same force (Fig. 3 and Table 1, and see also Fig. S5C). Similarly, no significant difference was observed upon depolymerizing microtubules with nocodazole (Fig. 3, Fig. S5 B and C, and Table 1). However, when F-actin or microtubules were disrupted, nuclear position did not recover completely following these treatments (Table 1). These results suggest that neither the F-actin network nor the microtubule cytoskeleton is required for the fibroblast nucleus to resist local forces, but they are required for rapid repositioning of the decentered nucleus.
Fig. 3.
Role of the cytoskeleton in mediating nuclear homeostasis. Overlay images of a typical nucleus before (red) and after (green) forcing normal cells and in cells with perturbed cytoskeletal elements (the nucleus was stained with SYTO 59 dye; the colors are to aid visualization and correspond to the same dye). The corresponding outlines are shown on the right to aid visualization. CTRL, NIH 3T3 cells; CYTO-D, NIH 3T3 cells treated with cytochalasin-D; GFP-KASH4, NIH 3T3 cells overexpressing GFP-KASH4; GFP-KDEL, NIH 3T3 cells inducibly expressing SS-GFP-KDEL; Lmna−/−, MEFs lacking lamin A/C; NOC, NIH 3T3 cells treated with nocodazole; SCRAM, NIH 3T3 cells transfected with scrambled siRNA; SUN1L-KDEL, NIH 3T3 cells inducibly expressing SS-HA-SUN1L-KDEL; SW13 w/vim, SW13 cells containing vimentin; SW13 w/o vim, SW13 cells lacking vimentin; vim siRNA, NIH 3T3 cells transfected with vimentin siRNA; WT, wild-type MEFs.
Table 1.
Nuclear response to pulling in NIH 3T3 fibroblasts, SW13 adrenal carcinoma epithelial cells, and MEFs
| Just before detachment | At steady state after detachment | ||||
| Condition | n | Maximum nuclear deformation, % | Maximum nuclear translation, µm | Relaxation of nuclear deformation, % | Recovery of nuclear position, % |
| NIH 3T3 fibroblasts | |||||
| Control | 12 | 26 ± 3 | 1.32 ± 0.15 | 69 ± 4 | 121 ± 14 |
| CYTO-D | 10 | 28 ± 3 | 1.77 ± 0.19 | 65 ± 4 | 65 ± 4* |
| NOC | 10 | 26 ± 4 | 1.73 ± 0.31 | 60 ± 7 | 68 ± 2* |
| GFP-KASH4 | 10 | 26 ± 4 | 1.68 ± 0.29 | 56 ± 5 | 49 ± 6* |
| SCRAM | 13 | 21 ± 3 | 1.74 ± 0.35 | 67 ± 6 | 85 ± 6 |
| Vim siRNA | 25 | 38 ± 4† | 2.89 ± 0.57† | 76 ± 2 | 72 ± 5 |
| SS-GFP-KDEL | 12 | 22 ± 3 | 1.02 ± 0.24 | 61 ± 1 | 86 ± 11 |
| SS-HA-SUN1L-KDEL | 10 | 39 ± 6# | 1.75 ± 0.46 | 64 ± 3 | 66 ± 6 |
| SW13 adrenal carcinoma cells | |||||
| With vim | 11 | 20 ± 2 | 1.16 ± 0.27 | 59 ± 7 | 91 ± 22 |
| Without vim | 13 | 34 ± 4†† | 2.07 ± 0.33†† | 56 ± 3 | 62 ± 7†† |
| Without vim rescued | 8 | 20 ± 2 | 1.02 ± 0.17 | 50 ± 5 | 94 ± 5 |
| MEFs | |||||
| WT | 10 | 20 ± 2 | 0.97 ± 0.15 | 56 ± 6 | 102 ± 1 |
| Lmna−/− | 13 | 50 ± 5** | 2.37 ± 0.53** | 53 ± 3 | 49 ± 8** |
| GFP-KASH4 in WT | 9 | 15 ± 2 | 0.51 ± 0.04** | 57 ± 6 | 58 ± 7* |
Maximum nuclear deformation and the recovery of nuclear deformation was calculated with the approach in Fig. S10A. Maximum nuclear translation was calculated as the distance traveled by the back edge until nuclear detachment from the micropipette tip. Values are mean ± SEM. Force = 6 nN for NIH 3T3 fibroblasts and 4 nN for SW13 adrenal carcinoma cells and MEFs. Statistically significant differences between control and condition are indicated by symbols: *P < 0.05 relative to control; †P < 0.05 relative to SCRAM; **P < 0.05 relative to WT; #P < 0.05 relative to SS-GFP-KDEL; ††P < 0.05 relative to With vim. Control, NIH 3T3 cells; CYTO-D, NIH 3T3 cells treated with cytochalasin-D; GFP-KASH4, NIH 3T3 cells overexpressing GFP-KASH4; GFP-KASH4 in WT, wild-type MEFs overexpressing GFP-KASH4; Lmna−/−, MEFs lacking lamin A/C; NOC, NIH 3T3 cells treated with nocodazole; SCRAM, NIH 3T3 cells transfected with scrambled siRNA; SS-GFP-KDEL, NIH 3T3 cells inducibly expressing SS-GFP-KDEL; SS-HA-SUN1L-KDEL, NIH 3T3 cells inducibly expressing SS-HA-SUN1L-KDEL; Vim siRNA, NIH 3T3 cells transfected with vimentin siRNA; With vim, SW13 cells containing vimentin; Without vim, SW13 cells lacking vimentin; Without vim rescued, SW13 cells lacking vimentin on transfection with CFP-vimentin; WT, wild-type MEFs.
We next tested whether intermediate filaments mediate nuclear homeostasis. In cells transfected with siRNA that decreased vimentin expression and reduced the number of discernible vimentin intermediate filaments (VIFs) (Fig. S6 A and B and ref. 19), the degree of translation and deformation of the nucleus was much greater than the control (Fig. 3, Table 1, and Fig. S7). We reproduced these results in SW13 adrenal carcinoma clones that do not express vimentin, and the large nuclear deformation and motion in these cells was rescued by expression of CFP-vimentin. Together, these results suggest that vimentin intermediate filaments are the primary cytoskeletal system that resists nuclear deformation and motion.
By comparing deformation between wild-type and Lmna−/− MEFs, we found that lamin A/C, which is the key component of the nuclear lamina encoded by the Lmna gene, is required for the nucleus to resist deformation, similar to the requirement for VIFs (Fig. 3, Table 1, and Fig. S7). The mechanical role of lamin A/C found here is consistent with the results of Discher and coworkers (20). Given that lamin A/C is a nuclear protein, it was anticipated to mediate only shape changes and not translation of the nucleus. Surprisingly, the nuclear translation was greater and recovered less in Lmna−/− MEFs than in wild-type cells (Table 1).
The LINC complex (for linker of nucleoskeleton to the cytoskeleton) physically connects F-actin, microtubules, and intermediate filaments to the nuclear surface (21–23). We therefore tested whether the LINC complex was required for F-actin and microtubules to reposition a decentered nucleus by disrupting the LINC complex with overexpression of GFP-KASH4 (Klarsicht, Anc-1, Syne homology). KASH4 is a domain of nesprin-4 that binds to sun1/2 (Sad1p, UNC-84) proteins in the inner nuclear membrane; overexpression of GFP-KASH4 competitively inhibits the endogenous KASH4 domain linkages with sun1/2 and hence disrupts the linkages with the cytoskeleton (24). As a result, it competitively inhibits the endogenous nesprin proteins that connect the cytoskeleton to the Sun1/2 proteins. The nucleus did not recover its original position after release of force in these cells, suggesting that microtubules and F-actin require nuclear linkages to fully restore nuclear position (Table 1, Fig. 3, and Fig. S8). We confirmed these results with GFP-KASH4 expression in wild-type MEFs (Table 1).
We next performed pulling experiments in two NIH 3T3 cell lines that we created: one inducibly expressing SS-GFP-KDEL and the other inducibly expressing SS-HA-SUN1L-KDEL, a dominant-negative protein that breaks the connections between endogenous KASH and SUN-domain proteins (22). Perturbation of the LINC complex by SS-HA-SUN1L-KDEL (signal sequencing-hemaglutinin-SUN1 protein lumenal domain-ER retrieval amino acid sequence) in our cells was verified by loss of nesprin-2 from the nuclear envelope (NE) as observed by immunofluorescence (Fig. S9). We found that SUN1L-KDEL nuclei deformed more compared with the GFP-KDEL control nuclei (Fig. 3 and Table 1). Because we did not find large differences in the organization of VIFs between control and SUN1L-KDEL cells (Fig. S6D), we speculate that SUN1L-KDEL–based LINC-complex perturbation may change the properties of the lamina through as yet unknown mechanisms.
Discussion
By directly applying a local pulling force on the nuclear surface with a micropipette, we estimated the magnitude of the forces that are required to deform and translate the nucleus in living, adherent cells. If the nucleus were unconstrained in the cell, it would translate with the micropipette without much change in shape, but we found that the nucleus deformed more than it translated in response to the pulling force. The fact that nucleus deforms more than it translates directly demonstrates a tight mechanical integration between the nucleus and its surroundings.
In fibroblasts, nuclei are subjected to forces that fluctuate in space and time, yet the nucleus is not observed to undergo shape undulations and position fluctuations. Rather, it maintains a regular appearance and fixed position on the short time scales of fluctuating thermal forces and molecular motor forces. Our estimates of applied forces required to deform and move the nucleus suggest that if nuclear shape and position is to be maintained, then the magnitude of these fluctuating nuclear forces must be lower than the nanonewton scale. Forces exceeding the nanonewton scale will move and deform the nucleus as demonstrated here.
Even if there is a local force that is applied on the nanonewton scale, as soon as the force relaxes the nuclear shape will rapidly revert to its initial state. This behavior is due to the large elastic resistance to translation and deformation. The resistance primarily originates in the cytoplasmic intermediate filaments and the nuclear lamina. Precisely how the vimentin intermediate filaments help the nucleus resist forces is not clear. The fact that mechanical linkages between the VIFs and the nucleus are not required (as evident from the experiments with KASH4-expressing cells in which the nucleus deformed normally; no major change in VIF organization was seen in normal versus KASH4-expressing cells; Fig. S6C) suggests that VIFs may confer “stiffness” to the nucleus owing to being closely packed and forming a stiff filamentous “cage” around it (10). Conversely the nucleus deformed much more with expression of SUN1L-KDEL, a soluble luminal fragment of Sun1 that breaks the LINC complex by binding endogenous KASH proteins and preventing their anchoring within the NE. Given that the nuclear deformation phenotype is similar to the Lmna−/− or vimentin-deficient cells, and that intermediate filament organization was not significantly altered in SUN1L-KDEL cells, this suggests that disruption of SUN-domain protein linkages may affect the mechanical properties of the nuclear lamina (or other constituents of the nucleoskeleton).
Because F-actin and microtubule motors are known to exert forces on the nuclear surface (2, 5, 6, 18, 25–28), it is surprising that they do not contribute to mechanical homeostasis of the nucleus. It seems that these cytoskeletal networks have the primary role of generating active forces through motor activity on the nuclear surface to position it. However, it would be simplistic to assign specific roles in all circumstances to the specific types of cytoskeletal networks. For example, Yamada and coworkers (8) have shown that actomyosin contractile forces require nesprin-3 linkages with intermediate filaments for transmission of force in 3D migration.
Our experiments collectively demonstrate the utility of direct application of force to the nucleus in adherent cells with an intact cytoskeleton. Unlike other methods of mechanically manipulating the nucleus such as aspirating isolated nuclei into micropipettes, where the cytoskeleton is essentially absent, or aspirating nuclei in suspended cells, where the cytoskeleton is likely to be disassembled or organized very differently from adherent cells (20), pulling on the cytoplasm to deform the nucleus (10), or applying mechanical forces to cell-substrate adhesions (29), our method allows comparisons of nuclear shapes and displacements at the point of release from the pipette tip where the applied force can be estimated in spread, intact cells. This makes quantitative comparisons of nuclear deformations and translations under different conditions possible and reliable. Such measurements can help in the future to understand the numerous human pathologies associated with abnormal nuclear positioning and shaping.
Materials and Methods
Cell Culture.
All cell types were maintained at 37 °C in a humidified 5% (vol/vol) CO2 environment. NIH 3T3 fibroblasts were cultured in DMEM with 4.5 g/L glucose (Mediatech) supplemented with 10% (vol/vol) donor bovine serum (DBS) (Gibco) and 1% penicillin–streptomycin (Mediatech). MEFs (30) were cultured in DMEM with 4.5 g/L glucose, supplemented with 10% DBS. SW13 adrenal carcinoma cells were cultured in DMEM with 4.5 g/L glucose, supplemented with 10% DBS, 1% penicillin–streptomycin, and 25 mM Hepes buffer (Mediatech). Human breast epithelial cells (MCF 10A) were cultured in MEBM (mammary epithelial basal medium) supplemented with 20 ng/mL EGF, 0.5 mg/mL hydrocortisone, 100 ng/mL cholera toxin, 100 µg/mL insulin, and bovine pituitary extract (Lonza).
Plasmid Transfection.
Transient transfection of plasmids into cells was performed with Lipofectamine 2000 reagent (Life Technologies/Invitrogen) in OPTI-MEM media (Life Technologies/Invitrogen). EGFP-KASH4 was a kind gift of K.J.R.; CFP-vimentin was acquired from the American Type Culture Collection. The cells were then trypsinized and plated on to fibronectin-coated glass-bottomed dishes for micromanipulation. Live nuclei were visualized after treating the cells with 3 µM SYTO 59 red fluorescent dye (Invitrogen) for 15 min.
Inducible Cell Lines.
To generate the doxycycline-inducible NIH 3T3 cells, we started with NIH 3T3 Tet-ON 3G cells (Clontech) that were retrovirally transduced with pRetroX-Tight.puro before selection with 0.5 µg/mL puromycin. Within pRetroX.Tight.puro were either the SS-HA-Sun1L-KDEL (Sun1L-KDEL) or SS-GFP-KDEL (22), the latter in which GFP replaced the HA-Sun1L protein to serve as a control. After puromycin selection, cells were screened by immunofluorescence postinduction with or without 1 µg/mL doxycycline for 18 h.
Immunostaining.
Cells were fixed in 4% paraformaldehyde for 15 min, washed with PBS, and then permeabilized and blocked with 0.1% triton X-100 in 1% BSA solution for 45 min. To immunostain for vimentin filaments the cells were incubated with vimentin primary mouse monoclonal antibody (Abcam) at 1:1,000 dilution, overnight at 4 °C. The samples were then washed with PBS and incubated with 488-nm fluorescent goat anti-mouse secondary antibody (Invitrogen) for 1 h at room temperature. To immunostain microtubules, the cells were first treated with microtubule extraction buffer containing 0.5% (mol/vol) glutaraldehyde, 0.8% formaldehyde, and 0.5% Triton X-100 in PBS for 10 min minutes and fixed with 1% (mol/vol) paraformaldehyde for another 10 min. The cells blocked with 1% BSA for 45 min were incubated with alpha tubulin rabbit polyclonal antibody (Abcam) at 1:1,000 dilution in 1% BSA solution overnight at 4 °C. The cells washed with PBS were then incubated for 1 h in goat anti-rabbit antibody (Invitrogen) at 1:500 dilution. Fixed cells were stained for F-actin with Alexa Fluor 488-nm phalloidin at 1:200 dilution and nucleus was stained with Hoechst 33342 (Life Technologies/Invitrogen) at 1:100 dilution for 1 h at room temperature.
The SS-GFP-KDEL and SS-HA-SUN1L-KDEL cells cultured on glass coverslips were fixed with 3% (mol/vol) PFA/PBS and permeabilized in 0.4% Triton X-100/PBS. Nesprin-2 was detected with rabbit anti-Nesprin 2/4 antibody (peptide antigen KKAELEWDPAGDIGGLGPLGQ; YenZyme) that cross-reacts with both human and mouse Nesprins 2 and 4. Mouse anti-HA (12CA5, 1:1,000; Covance) was used to detect SS-HA-Sun1L-KDEL fusion protein. Alexa Fluor-labeled goat anti-rabbit and goat anti-mouse (1:1,000; Invitrogen) were used to visualize the proteins and Hoechst dye 33258 (1:1,000) was used to label DNA.
Drug Treatment.
Nocodazole (Sigma-Aldrich) was used to disrupt microtubules at a concentration of 1 µM for 3 h. For F-actin disruption, cells were treated with 90 nM Cytochalasin D (Biomol) for 30 min.
siRNA Knockdown of Vimentin.
Cells were transfected with 250 nM siRNA (Sigma-Aldrich) using Lipofectamine 2000 in OPTI-MEM buffer to knock down vimentin. The siRNA oligonucleotide target sequence was CAAGAUCUGCUAAUGUUA. Nontargeting siRNA served as a control.
Cell Membrane Permeability and Cell Viability Assay.
Live/Dead reduced biohazard cell viability kit (Invitrogen) was used to test the viability of the cells undergoing nuclear manipulation. Cells were incubated with 4 µM SYTO 10 green fluorescent nucleic acid stain and 4 µM ethidium homodimer-2 (which stains the nucleus of dead cells red) for 15 min. Nuclear manipulation was performed and the cells were imaged with differential interference contrast (DIC) as well as in the green and red channels.
Imaging and Micromanipulation.
For microscopy, cells were transferred onto 35-mm glass-bottom dishes (World Precision Instruments) treated with 5 µg/mL fibronectin (BD Biosciences) overnight. The nuclei were manipulated with a micropipette with a 0.5-µm-diameter tip (Femtotip; Eppendorf) connected to an Eppendorf InjectMan micromanipulator system. A known suction pressure was applied using the Eppendorf Femtojet and the micropipette was translated with the micromanipulator joystick. The suction force is defined as the applied suction pressure multiplied by the micropipette tip cross-sectional area, which are both known. The suction force is expected to be very close to the actual force applied to the nuclear surface at the point of nucleus release (SI Text). Time-lapse fluorescence and DIC imaging was performed on a Nikon TE2000 microscope equipped with a 40× oil immersion objective and CCD camera (CoolSNAP; Photometrics). During microscopy, cells were maintained in an environmental chamber in which the temperature was kept at 37 °C, the CO2 level at 5%, and the relative humidity controlled at 100%.
Analysis.
Images captured were processed for brightness and contrast and cropped with ImageJ software (raw images are available upon request). They were imported into MATLAB to quantify outlines of the nucleus from which the length strain and translation of the back edge of the nucleus were calculated using a previously published method (31). In MATLAB, the initial shape of the nucleus and the maximum pulled shape were overlapped after correcting for the translation of the nucleus. Maximum nuclear deformation and recovery of nuclear shape were quantified as described in Fig. S10A. Fitting of time-dependent curves in Fig. 2 B and C was done with a least-squares method in MATLAB. All data are presented as mean ± SEM. All of the statistical comparisons were made with the Student’s t test.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation Grant CMMI 0954302 (to T.P.L.) and NIH Grants R01 EB014869 (to T.P.L. and J.A.N.) and R01 GM102486 (to T.P.L. and R.B.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1502111112/-/DCSupplemental.
References
- 1.Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: Mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol. 2009;10(1):75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- 2.Kutscheidt S, et al. FHOD1 interaction with nesprin-2G mediates TAN line formation and nuclear movement. Nat Cell Biol. 2014;16(7):708–715. doi: 10.1038/ncb2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Théry M, et al. Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity. Proc Natl Acad Sci USA. 2006;103(52):19771–19776. doi: 10.1073/pnas.0609267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russell RJ, et al. Sarcomere length fluctuations and flow in capillary endothelial cells. Cytoskeleton (Hoboken) 2011;68(3):150–156. doi: 10.1002/cm.20501. [DOI] [PubMed] [Google Scholar]
- 5.Fridolfsson HN, Starr DA. Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. J Cell Biol. 2010;191(1):115–128. doi: 10.1083/jcb.201004118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Starr DA. Watching nuclei move: Insights into how kinesin-1 and dynein function together. BioArchitecture. 2011;1(1):9–13. doi: 10.4161/bioa.1.1.14629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson MH, Holzbaur EL. Opposing microtubule motors drive robust nuclear dynamics in developing muscle cells. J Cell Sci. 2012;125(Pt 17):4158–4169. doi: 10.1242/jcs.108688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petrie RJ, Koo H, Yamada KM. Generation of compartmentalized pressure by a nuclear piston governs cell motility in a 3D matrix. Science. 2014;345(6200):1062–1065. doi: 10.1126/science.1256965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu J, et al. Actomyosin pulls to advance the nucleus in a migrating tissue cell. Biophys J. 2014;106(1):7–15. doi: 10.1016/j.bpj.2013.11.4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci USA. 1997;94(3):849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chancellor TJ, Lee J, Thodeti CK, Lele T. Actomyosin tension exerted on the nucleus through nesprin-1 connections influences endothelial cell adhesion, migration, and cyclic strain-induced reorientation. Biophys J. 2010;99(1):115–123. doi: 10.1016/j.bpj.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isermann P, Lammerding J. Nuclear mechanics and mechanotransduction in health and disease. Curr Biol. 2013;23(24):R1113–R1121. doi: 10.1016/j.cub.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gundersen GG, Worman HJ. Nuclear positioning. Cell. 2013;152(6):1376–1389. doi: 10.1016/j.cell.2013.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dahl KN, Kahn SM, Wilson KL, Discher DE. The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J Cell Sci. 2004;117(Pt 20):4779–4786. doi: 10.1242/jcs.01357. [DOI] [PubMed] [Google Scholar]
- 15.Rowat AC, Lammerding J, Ipsen JH. Mechanical properties of the cell nucleus and the effect of emerin deficiency. Biophys J. 2006;91(12):4649–4664. doi: 10.1529/biophysj.106.086454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durand RE, Olive PL. Cytotoxicity, mutagenicity and DNA damage by Hoechst 33342. J Histochem Cytochem. 1982;30(2):111–116. doi: 10.1177/30.2.7061816. [DOI] [PubMed] [Google Scholar]
- 17.Wojcik K, Dobrucki JW. Interaction of a DNA intercalator DRAQ5, and a minor groove binder SYTO17, with chromatin in live cells—influence on chromatin organization and histone-DNA interactions. Cytometry A. 2008;73(6):555–562. doi: 10.1002/cyto.a.20573. [DOI] [PubMed] [Google Scholar]
- 18.Luxton GW, Gomes ER, Folker ES, Worman HJ, Gundersen GG. TAN lines: A novel nuclear envelope structure involved in nuclear positioning. Nucleus. 2011;2(3):173–181. doi: 10.4161/nucl.2.3.16243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mendez MG, Kojima S, Goldman RD. Vimentin induces changes in cell shape, motility, and adhesion during the epithelial to mesenchymal transition. FASEB J. 2010;24(6):1838–1851. doi: 10.1096/fj.09-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. Physical plasticity of the nucleus in stem cell differentiation. Proc Natl Acad Sci USA. 2007;104(40):15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lombardi ML, et al. The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J Biol Chem. 2011;286(30):26743–26753. doi: 10.1074/jbc.M111.233700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crisp M, et al. Coupling of the nucleus and cytoplasm: Role of the LINC complex. J Cell Biol. 2006;172(1):41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luxton GW, Starr DA. KASHing up with the nucleus: Novel functional roles of KASH proteins at the cytoplasmic surface of the nucleus. Curr Opin Cell Biol. 2014;28:69–75. doi: 10.1016/j.ceb.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roux KJ, et al. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc Natl Acad Sci USA. 2009;106(7):2194–2199. doi: 10.1073/pnas.0808602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luxton GW, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 2010;329(5994):956–959. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Worman HJ, Gundersen GG. Here come the SUNs: A nucleocytoskeletal missing link. Trends Cell Biol. 2006;16(2):67–69. doi: 10.1016/j.tcb.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 27.Levy JR, Holzbaur EL. Dynein drives nuclear rotation during forward progression of motile fibroblasts. J Cell Sci. 2008;121(Pt 19):3187–3195. doi: 10.1242/jcs.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomes ER, Jani S, Gundersen GG. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 2005;121(3):451–463. doi: 10.1016/j.cell.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 29.Lammerding J, et al. Lamins A and C but not lamin B1 regulate nuclear mechanics. J Biol Chem. 2006;281(35):25768–25780. doi: 10.1074/jbc.M513511200. [DOI] [PubMed] [Google Scholar]
- 30.Sullivan T, et al. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147(5):913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arce SH, Wu PH, Tseng Y. Fast and accurate automated cell boundary determination for fluorescence microscopy. Sci Rep. 2013;3:2266. doi: 10.1038/srep02266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.