Safeguarding the integrity of the genome should not be left to chance. Indeed, all organisms have highly effective mechanisms to detect and remove errors and lesions in DNA. DNA mismatch repair (MMR) serves as the final safeguard in assuring the fidelity of DNA replication from bacteria to humans and backstops the nucleotide selection and exonuclease proofreading activities of replicative polymerases (Fig. 1) (1–3). The loss of MMR results in greatly elevated rates of spontaneous mutation, is the underlying cause of Lynch syndrome colon cancer, and is implicated in a subset of sporadic tumors. In its postreplication capacity, MMR functions during S phase in close association with the replication fork. This process is mediated by proliferating cell nuclear antigen protein (PCNA), a lynchpin in regulating access and activity of DNA polymerases and other proteins involved in replication, repair, and recombination (4). EGFR, a member of the HER family of receptor tyrosine kinases (RTKs) that span the plasma membrane, regulates cell proliferation, differentiation, and motility in response to extracellular ligands (e.g., EGF) (5, 6). Ligand binding promotes dimerization and allosteric activation of cytoplasmic kinase domains. Autophosphorylation initiates signaling cascades involving RAS/MAPK, PI(3)K/Akt, Jak/STAT, and others. EGFR also translocates to the nucleus where it influences replication, repair, and transcription and is linked to poor prognoses in several malignancies, but molecular mechanisms are largely unknown (7, 8). In the nucleus, EGFR phosphorylates PCNA, increasing its stability (9). This would be expected to have important consequences, but the functional significance was unclear until now. In PNAS, Ortega et al. reveal that phosphorylation of PCNA by EGFR inhibits MMR in cells and switches DNA synthesis in vitro from high-fidelity to error-prone (Fig. 2) (10). This intersection between MMR and EGFR underscores the importance of understanding the nuclear roles of EGFR and other RTKs, and highlights the relationship between the control of cell proliferation and the maintenance of genome stability, two central aspects of tumorigenesis.
Fig. 1.
Scheme for repair of replication errors. (A) Misincorporation of dT opposite dG by DNA polymerase leads to G:T mispair. (B) Template slippage at A5 repeat leads to +2 IDL (T7). MMR targets the newly synthesized strand (blue) to restore the parental sequence.
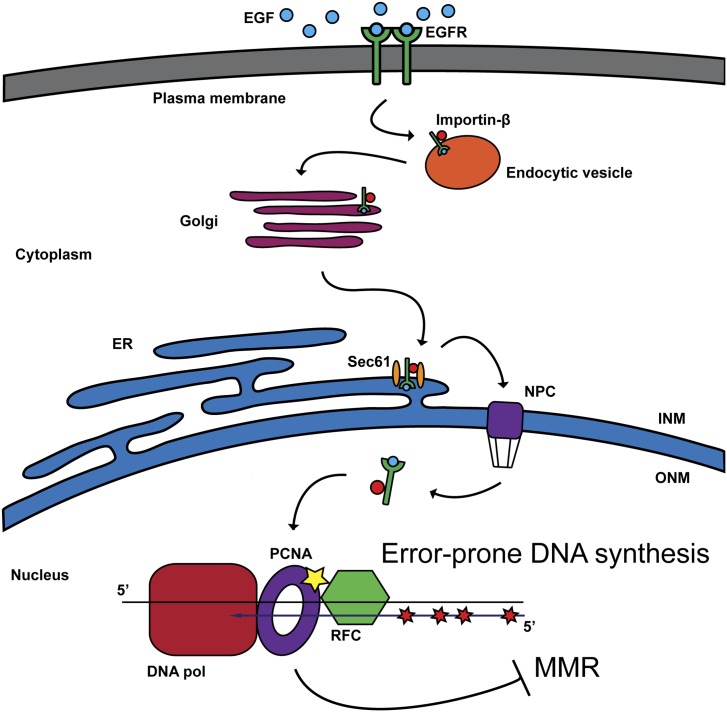
Fig. 2.
A model for nuclear transport of ligand-bound EGFR and phosphorylation of PCNA. Retrograde trafficking of EGFR delivers EGFR to the nucleus where it phosphorylates PCNA at Tyr211. Ortega et al. reveal that PCNA-pY211 inhibits MMR in colorectal cancer cell lines and promotes mutagenic DNA synthesis in vitro (10). ER, endoplasmic reticulum; INM, inner nuclear membrane; NPC, nuclear pore complex; ONM, outer nuclear membrane; RFC, replication factor C.
Although replication is remarkably faithful, replicative polymerases incorporate an incorrect nucleotide leading to premutagenic, non-Watson–Crick base pairs, such as G:T, about once every 104 to 105 times. In addition, during replication through mono- and dinucleotide repeats, the template strand can misalign, forming insertion/deletion loops (IDLs) that yield insertion or deletion mutations. How do cells mitigate replication errors? A proofreading exonuclease activity that is an integral component of replicative polymerases removes misincorporated nucleotides, but it’s not ironclad. More problematic are IDLs that are likely to escape proofreading altogether. MMR serves as the final gatekeeper, excising errors in new daughter strands and giving replicative polymerases a second chance in a gap-filling step. The loss of MMR by mutation or epigenetic silencing increases spontaneous mutation by 50- to 1,000-fold and is a driver of malignancy in Lynch syndrome colon cancer and a subset of sporadic cancers (11, 12).
PCNA is a processivity factor for polymerases and likely functions in MMR at mismatch recognition, excision on the newly synthesized strand, and DNA synthesis by replicative polymerases (1, 3). In eukaryotes, mismatch repair is initiated by conserved mismatch binding proteins related to bacterial MutS: MutSα, a heterodimer of MSH2-MSH6, and MutSβ, a heterodimer of MSH2-MSH3. MutSα targets base-base mispairs and IDLs of 1–2 nucleotides, whereas MutSβ preferentially targets IDLs. MutSα and MutSβ scan the helix until a mismatch is encountered. This search occurs in the context of the advancing replication fork and PCNA, using the PCNA-interacting protein boxes of MSH6 and MSH3. MutSα and MutSβ bind to mismatches and bend the DNA using distinct molecular interactions. Differential binding and hydrolysis at two nonequivalent nucleotide binding sites license conformational changes that turn MutS proteins into sliding clamps that diffuse along the DNA. Nucleotide binding also mediates interaction with a second conserved MMR protein, MutLα, a heterodimer of two MutL homologs, MLH1 and PMS2, and the formation of a poorly defined ternary complex of MutSα or MutSβ, MutLα, and DNA.
Structural studies reveal that MLH1 and PMS2 dimerize through their C termini and upon ATP binding at an N-terminal ATP-binding domain form a DNA clamp (13). Notably, PMS2 harbors a PCNA-activated endonuclease activity that contributes to DNA excision by providing additional sites of entry for EXO1, a strict 5′ to 3′ exonuclease (14, 15). In this way MMR is bidirectional and can be initiated from a nick on either side of the mismatch. The net result of excision is a single-strand gap that is filled in with the aid of PCNA by a high-fidelity replicative polymerase, Polδ or Polε, and ligated to restore an error-free duplex.
Essential to MMR is excision targeted to the new strand containing the error and not the template strand. PCNA has a critical role. PCNA is a trimer of identical subunits arranged head-to-tail in a ring that encircles DNA (4). It loads near single-strand and double-strand DNA junctions with the help of replication factor C. PCNA is always oriented on DNA such that the same side of the trimer faces the 3′ terminus. Thus, interactions of MMR proteins with PCNA fix their orientation on DNA, and the catalytic activity of PMS2 is directed to the newly synthesized strand.
Ortega et al. use in vitro assays to dissect the effects of PCNA-pY211 on MMR in several MMR-deficient colorectal cancer cell lines with elevated levels of EGFR, and observe a correlation between levels of EGFR, PCNA-pY211, and MMR loss (10). Restoration of MMR in these colorectal cancer cells requires the addition of MMR proteins and unphosphorylated PCNA. PCNA-pY211 binds poorly to MutSα and MutSβ, suggesting that their recruitment to DNA by PCNA might be compromised. PCNA isoforms mimicking constitutively phosphorylated PCNA-Y211, -Y211D and -Y211E, inhibit MutLα endonuclease activity on mismatched DNA, and PCNA-pY211 appears to direct error-prone synthesis in vitro. These findings raise interesting questions for: (i) the interaction of MutSα and PCNA on DNA; (ii) the nature of the elusive MutSα/MutLα/DNA ternary complex; (iii) the mechanism by which PCNA activates PMS2 endonuclease; and (iv) the fidelity of DNA synthesis at gaps. However, there are broader implications as well. Tyr211 is positioned in close proximity to a critical loop and hydrophobic pocket that mediate binding to PCNA-interacting protein box-containing proteins, and modeling suggests that phosphorylation might disrupt local geometry, threatening many PCNA interactions (10). Is DNA synthesis affected in replication, translesion synthesis repair, or recombination? Additional in vitro and in vivo studies are required.
What is the spectrum of nuclear EGFR targets? Histone H4 phosphorylation at Tyr72 by EGFR modulates DNA synthesis and repair and is correlated with increased levels of Ki-67 proliferation marker in human breast tumors (16). DNA-PK and TIP60, an acetylase and coactivator of ataxia telangiectasia mutated (ATM), may also be targets (17). Because EGFR-directed signaling pathways are broad in scope and ubiquitous, differentiating direct vs. indirect responses is challenging; the identification of specific residues phosphorylated by EGFR is critical.
What triggers nuclear transport of EGFR and related T-cell receptors? One model features retrograde trafficking of EGFR bound to importin-β in endocytic vesicles, transit through the Golgi apparatus and endoplasmic reticulum, and nuclear entry via the nuclear pore complex and Sec61 β (8). How does ionizing radiation signal nuclear transport? Is it via chemical/physical modification at the cell membrane: proteins or lipids? Is it a DNA damage signal from the nucleus? And, what is the relationship between nuclear transport and endocytosis-mediated recycling and degradation of RTKs vital to their regulation (8, 18)? Understanding the intersection between cell proliferation and genome stability will reveal much about what diverts EGFR from normal paths to more reckless ones.
Acknowledgments
This work was supported by the Division of Intramural Research, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
See companion article on page 5667.
References
- 1.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 2.Hsieh P, Yamane K. DNA mismatch repair: Molecular mechanism, cancer, and ageing. Mech Ageing Dev. 2008;129(7-8):391–407. doi: 10.1016/j.mad.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiricny J. Postreplicative mismatch repair. Cold Spring Harb Perspect Biol. 2013;5(4):a012633. doi: 10.1101/cshperspect.a012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mailand N, Gibbs-Seymour I, Bekker-Jensen S. Regulation of PCNA-protein interactions for genome stability. Nat Rev Mol Cell Biol. 2013;14(5):269–282. doi: 10.1038/nrm3562. [DOI] [PubMed] [Google Scholar]
- 5.Yarden Y, Pines G. The ERBB network: At last, cancer therapy meets systems biology. Nat Rev Cancer. 2012;12(8):553–563. doi: 10.1038/nrc3309. [DOI] [PubMed] [Google Scholar]
- 6.Arteaga CL, Engelman JA. ERBB receptors: From oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell. 2014;25(3):282–303. doi: 10.1016/j.ccr.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brand TM, et al. Nuclear EGFR as a molecular target in cancer. Radiother Oncol. 2013;108(3):370–377. doi: 10.1016/j.radonc.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 8.Han W, Lo HW. Landscape of EGFR signaling network in human cancers: Biology and therapeutic response in relation to receptor subcellular locations. Cancer Lett. 2012;318(2):124–134. doi: 10.1016/j.canlet.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang SC, et al. Tyrosine phosphorylation controls PCNA function through protein stability. Nat Cell Biol. 2006;8(12):1359–1368. doi: 10.1038/ncb1501. [DOI] [PubMed] [Google Scholar]
- 10.Ortega J, et al. Phosphorylation of PCNA by EGFR inhibits mismatch repair and promotes misincorporation during DNA synthesis. Proc Natl Acad Sci USA. 2015;112:5667–5672. doi: 10.1073/pnas.1417711112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hewish M, Lord CJ, Martin SA, Cunningham D, Ashworth A. Mismatch repair deficient colorectal cancer in the era of personalized treatment. Nat Rev Clin Oncol. 2010;7(4):197–208. doi: 10.1038/nrclinonc.2010.18. [DOI] [PubMed] [Google Scholar]
- 12.Peltomäki P. Epigenetic mechanisms in the pathogenesis of Lynch syndrome. Clin Genet. 2014;85(5):403–412. doi: 10.1111/cge.12349. [DOI] [PubMed] [Google Scholar]
- 13.Guarné A, Charbonnier JB. Insights from a decade of biophysical studies on MutL: Roles in strand discrimination and mismatch removal. Prog Biophys Mol Biol. 2015 doi: 10.1016/j.pbiomolbio.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 14.Pluciennik A, et al. PCNA function in the activation and strand direction of MutLα endonuclease in mismatch repair. Proc Natl Acad Sci USA. 2010;107(37):16066–16071. doi: 10.1073/pnas.1010662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goellner EM, et al. PCNA and Msh2-Msh6 activate an Mlh1-Pms1 endonuclease pathway required for Exo1-independent mismatch repair. Mol Cell. 2014;55(2):291–304. doi: 10.1016/j.molcel.2014.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chou RH, et al. EGFR modulates DNA synthesis and repair through Tyr phosphorylation of histone H4. Dev Cell. 2014;30(2):224–237. doi: 10.1016/j.devcel.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dittmann K, et al. Nuclear epidermal growth factor receptor modulates cellular radio-sensitivity by regulation of chromatin access. Radiother Oncol. 2011;99(3):317–322. doi: 10.1016/j.radonc.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Goh LK, Sorkin A. Endocytosis of receptor tyrosine kinases. Cold Spring Harb Perspect Biol. 2013;5(5):a017459. doi: 10.1101/cshperspect.a017459. [DOI] [PMC free article] [PubMed] [Google Scholar]