Significance
R-loop formation has been related to genome instability and human disease, yet the role of R-loops in replication priming remains to be explored in the eukaryotic genome. This investigation discloses that transcription-dependent R-loops have the potential to initiate origin-independent replication events in ribosomal DNA. Taken together, our data suggest that R-loops contribute to transcription-driven endoreplication events and alterations in genome copy number.
Keywords: RNA:DNA hybrids, RNase H, topoisomerase 1, replication, ribosomal DNA
Abstract
DNA replication initiates at defined replication origins along eukaryotic chromosomes, ensuring complete genome duplication within a single S-phase. A key feature of replication origins is their ability to control the onset of DNA synthesis mediated by DNA polymerase-α and its intrinsic RNA primase activity. Here, we describe a novel origin-independent replication process that is mediated by transcription. RNA polymerase I transcription constraints lead to persistent RNA:DNA hybrids (R-loops) that prime replication in the ribosomal DNA locus. Our results suggest that eukaryotic genomes have developed tools to prevent R-loop–mediated replication events that potentially contribute to copy number variation, particularly relevant to carcinogenesis.
During transcription, DNA acts as a template for the synthesis of the nascent RNA. RNA synthesis is accompanied by the generation of positive and negative DNA supercoiling in front of and behind the transcription machinery, respectively (1). Unwinding of the DNA double helix by negative supercoiling may allow the RNA to hybridize to its DNA template behind the elongating RNA polymerase, leading to R-loops (2). Other elements that could potentiate R-loop accumulation include RNA:DNA hybrid-facilitating DNA sequences, such as G-quadruplex structures (3) or nicks in the nontemplate DNA strand (4).
Eukaryotic cells need to control R-loop formation to avoid replication impairment, genome instability, and life span shortening mediated by such intermediates (5–10; reviewed in ref. 11). To do so, cells catalyze the relaxation of supercoiled DNA by type I topoisomerases (12–15), thus preventing replication fork reversal (16), DNA overwinding with the potential to block replication fork progression (17), DNA unwinding (18), or R-loop–mediated blocks of ribosomal RNA synthesis (19). Other enzymatic activities involved in R-loop processing include ribonuclease H (RNase H) activities, DNA-RNA helicases, such as Sen1/senataxin (20, 21), or Ataxin-2 RNA-binding protein Pbp1 (10). The ribonuclease activity of Saccharomyces cerevisiae RNases H1 and H2 specifically cleaves the RNA moiety of the RNA:DNA hybrid structure (22), whereas RNase H2 and topoisomerase 1 (Top1) can also process ribonucleotides in duplex DNA (23, 24).
Notably, R-loops are required to initiate mitochondrial DNA replication (25) and pioneering studies connected R-loops to origin-independent replication in prokaryotic systems (26, 27). For example, DnaA-dependent initiation of DNA replication at the Escherichia coli oriC replication origin can be overcome in the absence of RNase H1 (28, 29). As a consequence, rnhA mutants can survive complete inactivation of oriC by transcription-dependent activation of so-called oriK sites (30, 31), although candidate oriK sites have been identified only recently (32). Additional evidence for R-loop–primed replication was given by the observation that rnhA mutants are prone to an increase in mutation and DNA amplification events if origin activity is suppressed. These events required removal of the RNA polymerase (RNAP) to allow conversion of an R-loop into a replication fork (33). In summary, R-loops may act as the earliest known mutagenic intermediate in transcribed regions, and accelerate adaptation to genomic stress in prokaryotes. However, the possibility that R-loops mediate replication events in eukaryotic organisms still remains to be explored.
Highly transcribed ribosomal genes have been shown to favor R-loop formation in cells lacking both RNase H and Top1 activities (19). Here, by taking advantage of R-loop promoting conditions we potentiate the formation of DNA double-strand breaks (DSBs) and detect origin-independent replication intermediates (RI) within the transcribed 35S rRNA genes. A main finding in this work is the observation of “bubble-shaped” RIs by 2D agarose gels within the actively transcribed 35S rDNA when both Top1 and cellular RNases H are depleted. Importantly, in accordance with R-loop–mediated replication these “bubbles” are no longer observed when transcription by RNAPI is constrained. Our data suggest that R-loop–mediated replication contributes to stress-induced mutation, which is potentially relevant to eukaryotic genome evolution and disease formation.
Results
R-Loops Promote Genome Instability and Noncanonical Replication Events.
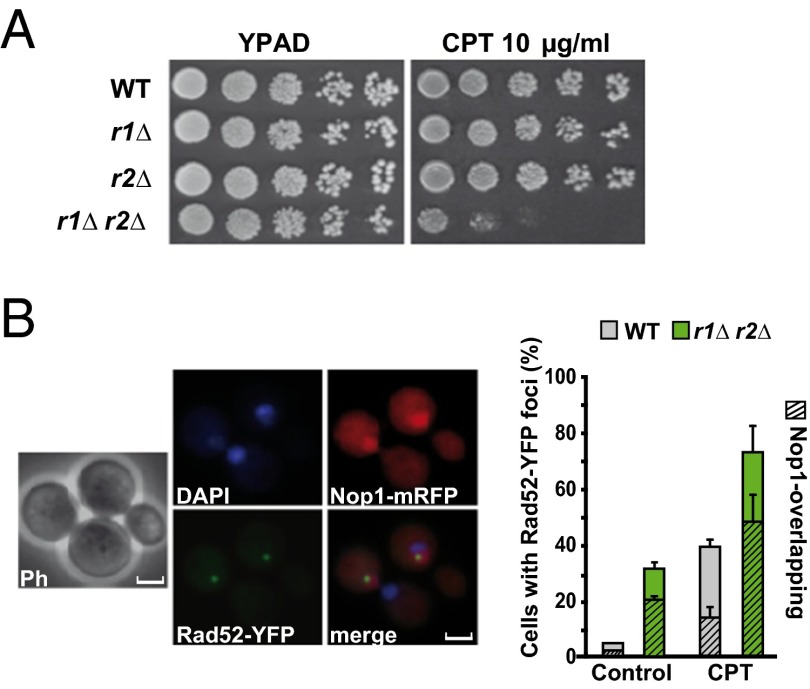
We maximized R-loop accumulation by treatment of mutants devoid of both RNase H1 and H2 (rnh1∆ rnh201∆, referred to herein as r1∆ r2∆) with the Top1 inhibitor camptothecin (CPT). CPT causes the accumulation of a covalent Top1–DNA complex that prevents religation of a nicked DNA duplex (15). Sensitivity to CPT and replicative stress generated by hydroxyurea (HU) and methyl methanesulfonate (MMS) was dependent on the removal of both RNase H activities, suggesting that both enzymes can substitute for each other (Fig. 1A and Fig. S1 A and B). Similarly, the lack of both RNase H activities was necessary to induce genomic instability. Double mutants r1∆ r2∆ showed a sixfold increase in CAN mutation rates (Fig. S1C), a 10-fold increase in loss of heterozygosity at the MAT locus (Fig. S1C), and a sevenfold increase (WT 4.5%, r1∆ r2∆ 31.6%) in S/G2 phase DNA repair centers as monitored by Rad52-YFP foci (Fig. 1B). More than half of the Rad52-YFP foci appeared to be associated with nucleolar DNA in r1∆ r2∆ cells (61% in the absence and 65% in the presence of CPT), indicating an increased susceptibility of ribosomal DNA (rDNA) to DNA damage caused by impaired processing of RNA:DNA hybrids.
Fig. 1.
Lack of RNase H activities predominantly affects nucleolar DNA stability. (A) Tenfold serial dilutions of cells grown for 3 d on YPAD (control) or YPAD-containing 10 μg/mL CPT. (B) Rad52-YFP foci represent DNA repair centers associated to nuclear versus nucleolar DNA (rDNA; hatched). Nop1-mRFP was used as nucleolar marker. (Scale bars, 2.5 µm.) Percentage of cells containing Rad52-YFP foci counted in exponentially growing cells growing in the absence (control) or presence of CPT (10 µg/mL, 3-h treatment; Right). Data represent mean ± SD from three independent experiments. Note that the ratio of Rad52-YFP/Nop1-RFP colocalization increased from 29% (control) and 34% (+CPT) in WT cells to 61% (control) and 65% (+CPT) in r1∆ r2∆ mutants.
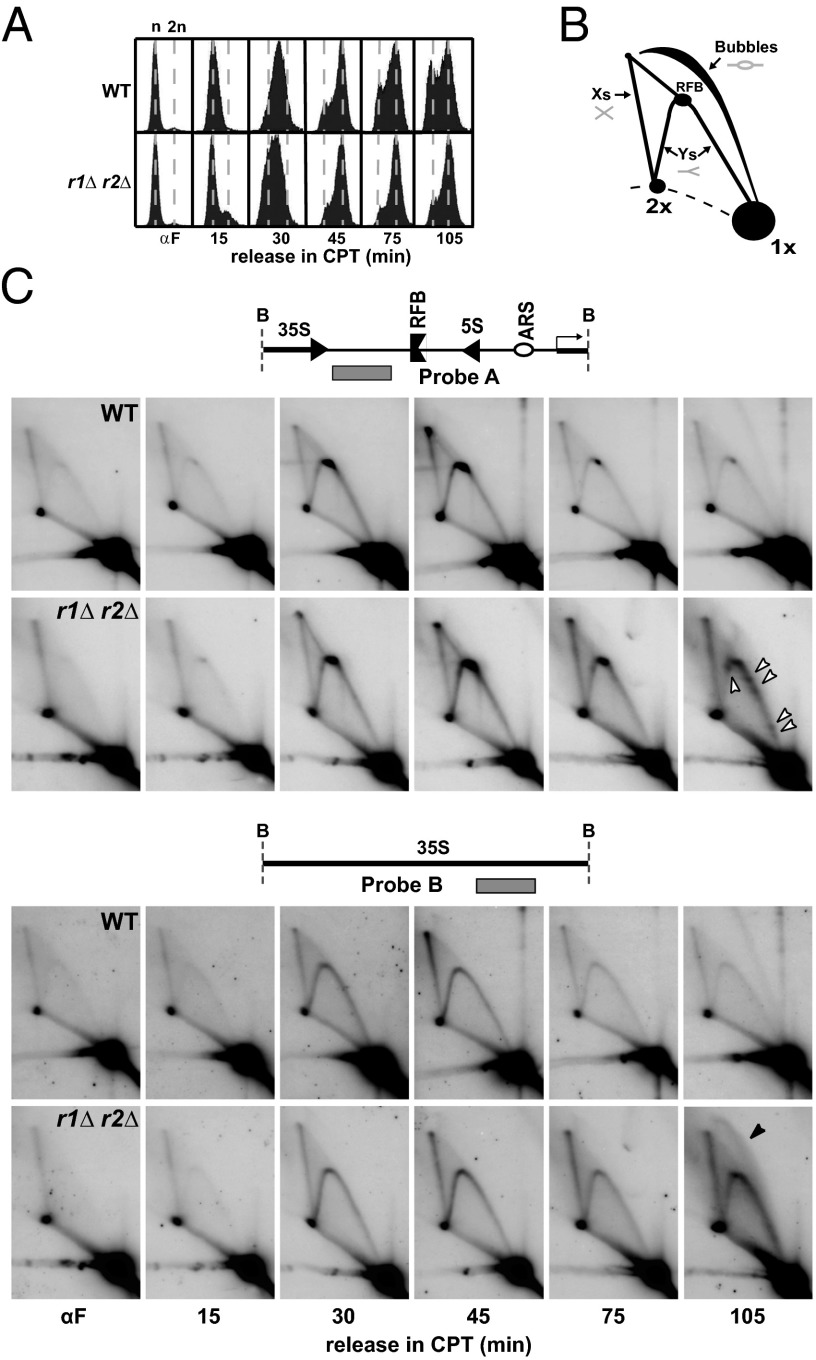
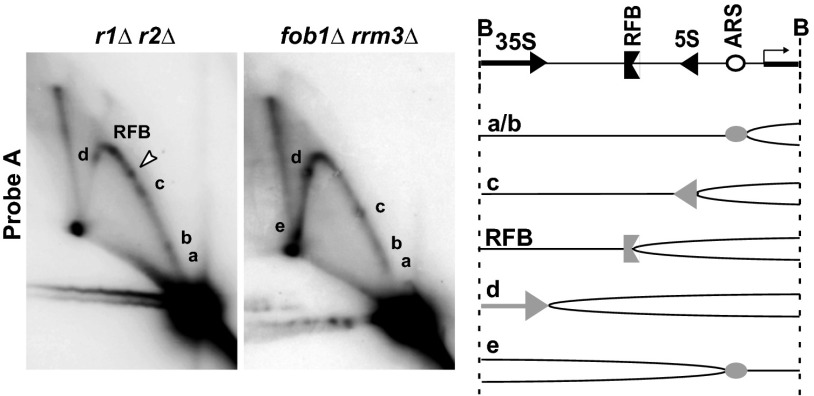
The rDNA, which is hosted in the nucleolus, is localized on chromosome XII and consists of ∼150 rDNA copies per haploid that are susceptible to stress-induced changes in repeat number and the formation of extrachromosomal rDNA circles (ERCs) caused by the excision of rDNA repeats (34, 35). Consistent with an increase in replicative stress upon CPT treatment, the number of Rad52 foci-containing cells doubled 30–90 min after release from α-factor (Fig. S2A) and CPT-treated r1∆ r2∆ mutant cells accumulated in late S or G2 phase (Fig. 2A). To further investigate the impact of CPT on rDNA maintenance, we monitored the fate of replication forks in S-phase cells. Following G1-synchronization by α-factor and release into CPT-containing medium, RIs were isolated and analyzed by 2D agarose gel electrophoresis (for interpretation of the results, see Fig. 2B). Although the S-phase specific pattern of replication (Y-arc) and recombination (X-spike) intermediates in WT and r1∆ r2∆ strains were similar, a clear difference was observed at late S/G2 phase. Whereas replication in the WT had finished, Y-arc and X-spike RIs remained in the r1∆ r2∆ mutants, and replication fork pausing sites (RFPs) (Fig. 2C, open arrowheads) appeared in the nontranscribed spacer region (Fig. 2C, probe A). These pausing sites were CPT-dependent because we could not detect them in RIs isolated from untreated r1∆ r2∆ mutants (Fig. S3A). The majority of these RFP sites overlapped with those previously described in cells lacking the Rrm3 DNA helicase (36) (Fig. 3). The observed pausing sites may be enriched at hot spots of Top1–DNA interaction sites and correspond to CPT-trapped Top1–DNA complexes, yet they correlated with potential sites of R-loop formation (37, 38), such as the 3′ end of 35S genes (d in Fig. 3) or 5S genes (c in Fig. 3), but also included sites of protein barriers such as the ribosomal autonomously replicating sequence (ARS) or 35S promoter (a/b and e in Fig. 3).
Fig. 2.
Two-dimensional gel analysis detects exceptional replication events in the rDNA of CPT-treated r1∆ r2∆ mutants. (A) FACS analysis of S-phase progression upon α-factor release of cells (WT, r1∆ r2∆) in the presence of 10 μg/mL CPT. G1 (n) and G2/M phase (2n) are indicated. (B) Schematic representation of major 2D gel signals including Y-, X-, and bubble-shaped molecules are displayed. The accumulation of replication intermediates at the RFB site as well as nonreplicating DNA (1×) are indicated. (C) Two-dimensional gel analysis of the BglII (B) digested rDNA locus. Probe A (Upper) detects the intergenic spacer region (IGS) containing the ribosomal origin of replication (ARS); the 5S gene, the RFB, and the 5′ end of the 35S gene. Replication pausing sites are indicated (open arrowheads). Probe B hybridizes to the RNAPI transcribed 35S gene. Replication intermediates indicative of bubble-shaped molecules are indicated (black arrowhead). The images are reduced by 5×. Quantification is shown in Fig. S2B.
Fig. 3.
Characterization of unscheduled replication intermediates in the rDNA. (Left) Comparison of r1∆ r2∆ and rrm3∆-dependent RFPs (indicated by lowercase letters a to d) detected by probe A. The open arrow marks a r1∆ r2∆ specific pausing site. (Right) A low-resolution mapping of the pausing sites based on their Y-arc location is shown. The images are reduced by 3.5×.
Next, we monitored replication intermediates present within the 35S gene (Fig. 2C, probe B). Although the 35S gene lacks common characteristics of replication origins that allow the binding of the prereplication complex (39), intermediates migrating as expected for bubble-shaped molecules were detected (indicated by a black arrowhead at the 105-min timepoint in Fig. 2C; see ref. 40 for a more detailed explanation of RI characteristics), indicative of replication initiation events within the 35S gene. We analyzed the bubble-shaped RIs by several means to determine the structure of these molecules (Fig. S3B). Previously it had been shown that in vitro RNase H treatment of mtDNA replication intermediates removes RNA:DNA hybrids and leads to the appearance of simple-Y structures in 2D agarose gels (41, 42). In accordance with a replicon-like structure, the bubble-shaped molecules were resistant to in vitro RNase H treatment and heat-induced branch migration. Furthermore, as expected for replicating molecules having an extendable 3′ end, in vitro DNA synthesis by Klenow/gp32 treatment could destroy the bubble-shaped molecules by strand displacement (43). These analyses rule out the presence of long stretches of R-loops, which could have a bubble-like appearance, and suggest a rapid conversion of R-loops into noncanonical replicons.
Depletion of Top1 in the Absence of RNase H Leads to Unscheduled Replication Within the 35S rDNA.
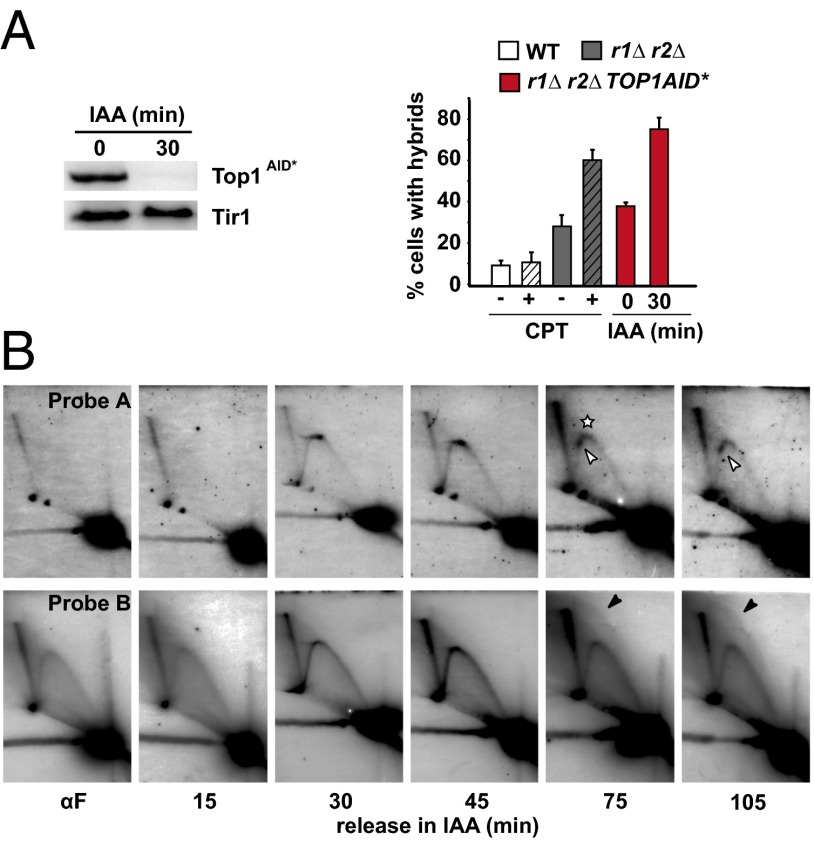
Next we generated an indole acetic acid (IAA)-inducible Top1-degron mutant (TOP1AID*) and assayed the impact of Top1 degradation on RNA:DNA hybrid formation. Top1 levels rapidly decreased upon IAA addition (Fig. 4A, Left and Fig. S4A), and consistent with previous observations (19), growth of r1∆ r2∆ mutants was inhibited in the absence of Top1 (Fig. S4A). Although 30% of r1∆ r2∆ mutant cells cross-reacted with the R-loop specific S9.6 antibody, 61% of cells were S9.6+ upon CPT addition, and similar values (39% vs. 76%) were obtained for the IAA-stimulated Top1 degradation in r1∆ r2∆ mutants, respectively (Fig. 4A, Right and Fig. S4B). This result supports the idea that the concomitant loss of RNase H and Top1 activities has an additive effect on RNA:DNA hybrid formation (13, 33).
Fig. 4.
Top1AID*-depletion stimulates R-loop formation and unscheduled replication events in r1∆ r2∆ mutants. (A, Left) Western blot analysis of IAA (1 mM) induced Top1AID* protein degradation. Tir1 serves as loading control. Note that both proteins contain multiple Myc epitopes for detection. (Right) Quantification of RNA:DNA hybrids by S9.6 antibody detection in WT, r1∆ r2∆, and r1∆ r2∆ TOP1AID* mutants in the absence or presence of CPT or IAA, respectively (Right). (B) Two-dimensional gel analysis of RIs isolated from the r1∆ r2∆ TOP1AID* mutant cells grown in the presence of 1 mM IAA following α-factor release. The disappearance of the RFB signal is indicated (white asterisk). The images are reduced by 5.5×. Description as in Fig. 2.
Top1 depletion in r1∆ r2∆ mutants was accompanied by an increase in Rad52 foci, indicative of an increase in DSB accumulation in the absence of both Top1 and RNase H activities (Fig. S4C) and a cell cycle arrest in S/G2 (Fig. S4D). Some of the CPT-mediated pausing sites characterized within the intergenic space region in r1∆ r2∆ mutants (Fig. 3) were barely detectable in the triple mutant (Fig. 4B, probe A). However, a strong pausing site at the 3′ end of the 35S gene (Fig. 4B, open arrowhead) may indicate a failure of the replication machinery to bypass torsional stress generated ahead of the transcribing RNAPI. Interestingly, the appearance of this strong RFP correlates with a decrease of RF accumulation at the Fob1-dependent replication fork barrier (RFB) (Fig. 4B, asterisk). A cease in ribosomal ARS firing, increased torsional stress, or weakened Fob1 binding in the absence of Top1 (44) could contribute to the loss of the Fob1-dependent RFB signal. Nevertheless, the presence of replicon-like structures within the 35S gene (Fig. 4B; probe B, black arrowhead) upon Top1 depletion in cells lacking RNase H strengthens the idea that R-loops could mediate origin-independent replication initiation events.
Unscheduled Replication Events Are Transcription-Dependent.
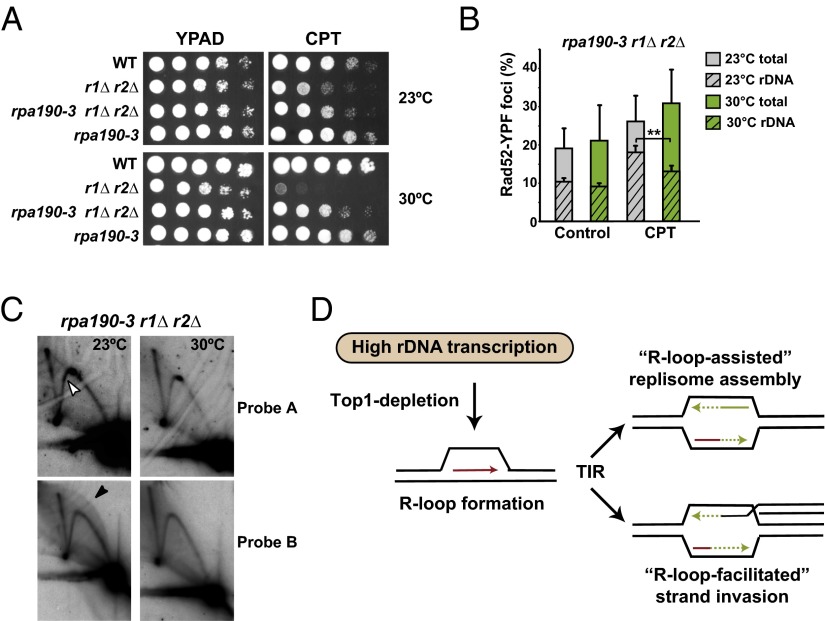
Our model predicts that RNAPI transcription would be a prerequisite for R-loop–initiated replication. To slow down rDNA transcription rates, we made use of the rpa190-3 mutant of the largest RNAPI subunit Rpa190 and the rrn3-8 mutant of Rnr3, which recruits RNAPI to the promoter of 35S rRNA genes (45). Strikingly, both rpa190-3 and rrn3-8 mutants alleviated the CPT-sensitivity of r1∆ r2∆ mutants at semipermissive temperature (Fig. 5A and Fig. S5A), reduced the formation of nucleolus-associated Rad52-foci formation and suppressed S/G2-phase cell cycle arrest (Fig. 5B and Fig. S5B). These observations suggest that the majority of the CPT-induced Top1-dependent DNA lesions are linked to rDNA transcription, as well as a potential link between rDNA damage, checkpoint activation, and growth rate. Next, we examined the fate of replication in rpa190-3 r1∆ r2∆ mutants by 2D gel electrophoresis. Cells were grown at 26 °C, before α-factor synchronization and released in CPT-containing media at permissive (23 °C) or semipermissive (30 °C) temperature. The absence of bubble-shaped replication intermediates in cells released from α-factor synchronization at semipermissive temperature (Fig. 5C and Fig. S5C) confirms a mechanism where transcription-mediated R-loops initiate replication at late S/G2 phase within the 35S rDNA.
Fig. 5.
RNAPI transcription is a prerequisite for the induction of unscheduled replication events. (A) Tenfold serial dilutions of indicated strains grown on YPAD (control) with or without 5 μg/mL CPT for 3 d at different temperatures allowing the permissive (23 °C) and semipermissive (30 °C) growth of rpa190-3 mutants (Rpa190 is the largest subunit of RNAPI). (B) Quantification of nuclear and nucleolar-associated Rad52-YFP foci in rpa190-3 r1∆ r2∆ mutants grown at indicated conditions. Data represent the mean ± SD from three independent experiments (**P < 0.001). (C) Two-dimensional gel analysis rpa190-3 r1Δ r2Δ triple mutant RIs isolated at 105 min after α-factor release in the presence of CPT at different temperatures. The presence of bubble-shaped molecules RI-derived from cells grown at 23 °C is indicated (black arrowhead). Description as in Fig. 2. (D) Model for TIR in yeast rDNA. RNA (red) and newly synthesized DNA (green) are indicated. The images are reduced by 5×. See text for explanation.
Discussion
Decades ago it became evident that R-loops take part in replication initiation of prokaryotic cells (46, 47). Here we present evidence that this is also the case for eukaryotic cells based on the observation that persistent R-loops can mediate unscheduled, origin-independent replication initiation in yeast chromosomal DNA. These replication events were observed in the highly transcribed 35S rRNA gene, and occurred spatially and temporally outside of the regular replication schedule. Unscheduled replication was not linked to a defined replication origin, and it was observed in late S/G2 phase of the cell cycle where replication termination and completion is expected to take place.
Which Factors and Mechanism Would Participate in Transcription-Initiated Replication Events?
Various, nonexclusive mechanisms could cooperate to trigger such transcription-initiated replication (TIR) (Fig. 5D). At present we do not know whether the presence of single-stranded DNA within an R-loop may permit strand invasion-dependent replication events, favored by the repetitive nature of the rDNA array. DSBs seem to be particularly frequent in the rDNA locus. Thus, R-loops and DSBs could stimulate recombination-driven replication events as observed in Candiada albicans mtDNA (42) or break-induced replication (48, 49) and involve the transient formation of simple Y-like replication intermediates.
The synthetic lethality observed in the absence of Top1 and RNase H activities (19, 50, and present work) complement previous notions that R-loops have an evolutionary conserved impact on transcription. E. coli Top1 mutants suffer from impaired growth and rDNA transcription (51–53), and that RNase H1 overexpression can partially compensate for the absence of Top1 (54). Other factors that possibly contribute to synthetic lethality include the accumulation of positive supercoiling generated ahead of RNAPI and in front of an advancing replication fork in convergent orientation. Such supercoiling can promote DNA extrusions and secondary structures that can be substrates for specific DNA nucleases (55).
Consistent with the observation that R-loops block replication fork progression (6), replication fork collapse at the site of nicked DNA may result in the physical presence of a “truncated” replication machinery in close vicinity to the R-loop and also explain the observed increase in DSB formation during S-phase (Fig. S2A). A truncated replication machinery potentially restarts replication from an R-loop, given that elegant in vitro experiments demonstrated that replication can restart from a purified E. coli replisome–RNAP complex, and that the replisome uses mRNA as a primer to reinitiate leading-strand synthesis after displacing a codirectional RNAP from DNA (56). One can speculate that RNAPI is no longer associated with the R-loop, a scenario that facilitates TIR without the need for factors that drive the displacement of RNAPs being head-on to a replisome (57).
In contrast to replication restart from a colliding replisome–RNAP complex, TIR events may be driven by the de novo replisome assembly at an R-loop. Assembly of the replication factor A protein complex to single-stranded DNA opposite a RNA:DNA hybrid could promote interaction with DNA replicases that are available at the end of chromosomal DNA synthesis at late S/G2 (58). There is evidence that replicases remain replication competent at S/G2 (58), and by doing so they may be able to initiate DNA synthesis within an R-loop. The DNA polα-primase subunit Pol12 is essential for replication initiation and has been suggested to act as a molecular tether during DNA replication (59). Pol12 is an essential but stable protein and its phosphorylated form appears to be required for the initial stages of DNA synthesis before the HU-sensitive elongation step (60). Pol12 remains in active and phosphorylated form in S/G2 and its inactivation by dephosphorylation only occurs while cells exit mitosis (61). Our observations suggest that RNAPI-associated R-loops and replication-competent DNA polα-primase complexes could drive S/G2-dependent TIR events.
Are TIR Events Limited to rDNA?
At a much lower frequency, TIR events may happen throughout the whole genome, and hot-spots for R-loop formation, such as highly transcribed genes, might be more prone to TIR events. Numerous enzymatic activities linked to the suppression of RNA:DNA hybrids including the THO-complex (37), the mRNA polyadenylation factor Pbp1, G4-quadruplex binding proteins Stem1 and Pif1 (10, 62), or the Sen1 subunit of the Nrd1 complex (20, 21, 63) may also be required to avoid TIR events. Mutations in the yeast Sen1 ortholog senataxin have been shown to be associated with human AOA2/ALS4 neurodegenerative disorders (64, 65). Indeed, Senataxin is needed to maintain genome integrity because of its function in the coordination of transcription, DNA replication, and the DNA damage response (reviewed in ref. 66). Thus, it would be interesting to see if TIR events are stimulated by the absence of Sen1 or other factors involved in mRNA biogenesis.
What Could be the Consequences of TIR Events?
R-loop primed, unlicensed genome replication would provide a new threat to eukaryotic genome stability. TIR carried out by a noncanonical replisome is likely to be inaccurate, and hence may have serious repercussions for genome instability. Unscheduled replication, particularly in regions of repetitive sequences, such as the rDNA array, could lead to the deletion of repetitive sequences or gene-amplification events, respectively. The rDNA repeat is located on chromosome XII and we find by pulse-field gel-electrophoresis (PFGE) analysis that the migration of chromosome XII is retarded (Fig. S6A). These initial findings indicate that loss of RNase H activity leads to rDNA expansion or intermediate structures that hamper electrophoretic separation as shown for replicating chromosomes (67). The observation that ERC formation is enhanced in r1∆ r2∆ mutants (Fig. S6B) confirms that the loss of RNase H activities causes genetic instability (Fig. S1). It would be particularly interesting to determine to which extent impaired RNA:DNA hybrid processing contributes to rereplication, gene amplification, and alterations in chromosome copy number in human cells. These events would have disastrous consequences for eukaryotic genome function and are particularly relevant to carcinogenesis (68).
Selective gene amplification is frequently observed in differentiating eukaryotic cells and results in transcript number and gene product increases in a dosage-dependent manner (69). The mechanisms by which gene amplification is achieved include DSB-dependent sister chromatid fusion and repeated breakage-fusion-bridge cycles evident in the dihydrofolate reductase locus of CHO cells (70), endoreplication of diptera chorion genes by multiple activation of replication origins within the same S-phase (71), RNA-template derived nanochromosome amplification in Stylonychia lemnae (72), or rolling circle amplification of extrachromosomal rDNA circles in Xenopus oocytes (73). Although in our study we could not distinguish if ERCs are more prone to TIR events, our results provide the possibility that ERC replication can be driven by impaired RNA transcript-processing suggesting that R-loops could have a physiological role in the control of gene amplification linked to nuclear differentiation events.
Experimental Procedures
Yeast Strains and Growth Conditions.
Yeast strains used in this study are listed in Table S1. Gene deletions were constructed by PCR-based methods (74). If not generated by a direct knock-out in the YKL background, mutant strains were backcrossed at least twice to the YKL83 strain background. Yeast strains were grown in YPAD, or synthetic complete (SC) minimal medium supplemented with 2% (wt/vol) glucose and amino acids. IAA (Sigma) was added at 500 µM for solid and 1 mM for liquid YPAD media.
Viability Assays.
To test for sensitivity to genotoxic agents, 10-fold serial dilutions of cells were grown for 3 d on YPAD plates or YPAD-containing HU (50 mM; USBiological), MMS (10 mM; Fluka), or CPT (5 µg/mL; Santa Cruz Biotechnology), unless otherwise specified.
Quantification and Colocalization of Rad52-YFP Foci.
For colocalization of Rad52-YFP foci with the nucleolar marker Nop1, cells were cotransformed with the RAD52-YFP expressing plasmid (F. Prado, Sevilla, Spain) and a plasmid expressing NOP1-mRFP (75). Transformants were grown in exponential phase in SC with plasmid selection, in the presence or absence of 10 µg/mL CPT for 3 h, and fixed using 2.5% (vol/vol) formaldehyde. YFP fluorescence (480-nm excitation/527-nm emission) and RFP fluorescence (584-nm excitation/607-nm emission) were detected by wide-field fluorescence microscopy (DM-6000B, Leica) at 100× magnification. Images were taken using LAS AF software (Leica). For each sample, 600 cells were counted from three independent experiments. For time-course analysis of Rad52-YFP foci, cells expressing the RAD52-YFP plasmid were synchronized with α-factor and released in the presence or absence of 10 µg/mL CPT. Samples were retrieved at the specified time points following release and fixed with 2.5% (vol/vol) formaldehyde. Approximately 600 cells derived from three independent colonies were analyzed for each condition.
Cell Cycle Progression Analysis and DNA Isolation.
For G1 synchronization, MATa cells were grown to an OD600 of 0.4 in YPAD medium before α-factor (1 μg/mL; Biomedal) synchronization for 180 up to 240 min. Cells were released from α-factor treatment by washing three times in prewarmed, fresh YPAD medium containing 0.1 mg/mL Pronase E (Sigma). For FACS analysis, 1-mL samples were taken at indicated times and fixed in 70% ethanol. Samples were resuspended in PBS containing 5 μg/mL propidium iodide, following RNase A overnight treatment, and were analyzed by flow cytometry using a FACSCalibur (Becton Dickinson) and CellQuest software. For DNA analysis, cells were released in fresh media containing 10 µg/mL CPT. 100 mL samples were retrieved at the specified time points following release and arrested by adding sodium azide (to a final concentration of 0.1%). DNA was extracted according to ref. 6.
Two Dimensional Agarose Gel Electrophoresis.
In a total volume of 100 µL, about 5 µg of genomic DNA was digested with 40 units BglII for 6 h, isopropanol precipitated, and resuspended in 20 µL loading buffer. First, dimension electrophoresis was performed in 0.4% TBE-buffered agarose gels at 40 V for 20 h. A gel slice containing DNA fragments between 3 and 12 kb was cut out for second dimension resolution in 1.1% TBE-buffered agarose at 140 V for 16 h. Denatured DNA was transferred to a Hybond XL membrane (Amersham) by standard procedures. Replication intermediates were detected by hybridization with specific 32P-labeled DNA probes, matching to nucleotides 452691–453344 (probe A) and 453834–454699 (probe B) on chromosome XII. Signals were quantified using a PhosphorImager with ImageGauge software (Fuji). The relative intensity of replication intermediates was normalized to the signal intensity obtained in the 1×-spot (nonsaturating exposure).
TOP1AID* Degron Construct.
The Top1 protein was tagged at the C terminus with the IAA17 auxin-binding domain (amplified by PCR from the plasmid pKan–AID* (76) in a r1∆ r2∆ strain, which expresses TIR1 under control of the constitutive ADH promoter. Note that both proteins contain multiple Myc epitopes that can be detected by a c-Myc primary antibody. The functionality of the Top1AID* degron was confirmed by Western blot analysis against the Myc tag to confirm degradation of the Top1 protein in response to 1 mM IAA (60). Protein extracts for immunoblotting were separated by SDS/PAGE using 8% (wt/vol) polyacrylamide (37.5:1), and protein bound membranes were incubated overnight with a mouse anti–c-Myc primary antibody (Abcam) at 1:1,000 and 1:20,000 in PBS-5% (wt/vol) milk, respectively. For FACS and 2D-gel analysis of the r1∆ r2∆ TOP1AID* strain, 1 mM IAA was added to YPAD medium for the last 30 min of α-factor incubation, and cells were released in the presence of IAA.
Statistics.
Values are expressed as the mean ± SD. Significance values (P) were obtained following Student’s t test for pairwise comparisons of data and considered statistically significant for P values < 0.05.
Supplementary Material
Acknowledgments
We thank K. Labib, H. Ulrich, F. Prado, J. Baßler, and J. de la Cruz for kindly providing strains and plasmids; H. Gaillard, A. El Hage, and D. Tollervey for suggestions and critical reading of the document; and E. Fernandéz-García and H. Heluani for technical assistance. This work was supported in part by Spanish Ministry of Science and Innovation Grants BIO2006-08051 and BFU2010-21339 (to R.E.W.); Junta de Andalucía Grants BIO-026, P08-CTS-04297, and P11-CTS-7962 (to R.E.W.); a European Union FEDER grant (to R.E.W.); a Junta para la Ampliación de Estudios predoctoral training grant from the Spanish National Research Council (CSIC) (to R.S.); a predoctoral training grant from the University of Seville/El Monte Foundation (to N.G.-R.); the Spanish Ministry of Science and Innovation Grant BFU2010-16372 and Consolider Ingenio 2010 CSD2007-0015 (to A.A.); Junta de Andalucía Grants BIO-102 and P09/CVI4567 (to A.A.); and the European Union (FEDER) (A.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1501769112/-/DCSupplemental.
References
- 1.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci USA. 1987;84(20):7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomas M, White RL, Davis RW. Hybridization of RNA to double-stranded DNA: Formation of R-loops. Proc Natl Acad Sci USA. 1976;73(7):2294–2298. doi: 10.1073/pnas.73.7.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18(13):1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roy D, Zhang Z, Lu Z, Hsieh CL, Lieber MR. Competition between the RNA transcript and the nontemplate DNA strand during R-loop formation in vitro: A nick can serve as a strong R-loop initiation site. Mol Cell Biol. 2010;30(1):146–159. doi: 10.1128/MCB.00897-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell. 2003;12(3):711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Wellinger RE, Prado F, Aguilera A. Replication fork progression is impaired by transcription in hyperrecombinant yeast cells lacking a functional THO complex. Mol Cell Biol. 2006;26(8):3327–3334. doi: 10.1128/MCB.26.8.3327-3334.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gan W, et al. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 2011;25(19):2041–2056. doi: 10.1101/gad.17010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helmrich A, Ballarino M, Tora L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol Cell. 2011;44(6):966–977. doi: 10.1016/j.molcel.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 9.Castellano-Pozo M, García-Muse T, Aguilera A. R-loops cause replication impairment and genome instability during meiosis. EMBO Rep. 2012;13(10):923–929. doi: 10.1038/embor.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salvi JS, et al. Roles for Pbp1 and caloric restriction in genome and lifespan maintenance via suppression of RNA-DNA hybrids. Dev Cell. 2014;30(2):177–191. doi: 10.1016/j.devcel.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 11.Aguilera A, García-Muse T. R loops: From transcription byproducts to threats to genome stability. Mol Cell. 2012;46(2):115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Wang JC. Interaction between DNA and an Escherichia coli protein omega. J Mol Biol. 1971;55(3):523–533. doi: 10.1016/0022-2836(71)90334-2. [DOI] [PubMed] [Google Scholar]
- 13.Wang JC. Cellular roles of DNA topoisomerases: A molecular perspective. Nat Rev Mol Cell Biol. 2002;3(6):430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 14.Leppard JB, Champoux JJ. Human DNA topoisomerase I: Relaxation, roles, and damage control. Chromosoma. 2005;114(2):75–85. doi: 10.1007/s00412-005-0345-5. [DOI] [PubMed] [Google Scholar]
- 15.Pommier Y. DNA topoisomerase I inhibitors: Chemistry, biology, and interfacial inhibition. Chem Rev. 2009;109(7):2894–2902. doi: 10.1021/cr900097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sogo JM, Lopes M, Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002;297(5581):599–602. doi: 10.1126/science.1074023. [DOI] [PubMed] [Google Scholar]
- 17.Tuduri S, et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol. 2009;11(11):1315–1324. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.French SL, et al. Distinguishing the roles of topoisomerases I and II in relief of transcription-induced torsional stress in yeast rRNA genes. Mol Cell Biol. 2011;31(3):482–494. doi: 10.1128/MCB.00589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El Hage A, French SL, Beyer AL, Tollervey D. Loss of topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010;24(14):1546–1558. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell. 2011;42(6):794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mischo HE, et al. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell. 2011;41(1):21–32. doi: 10.1016/j.molcel.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerritelli SM, Crouch RJ. Ribonuclease H: The enzymes in eukaryotes. FEBS J. 2009;276(6):1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazzaro F, et al. RNase H and postreplication repair protect cells from ribonucleotides incorporated in DNA. Mol Cell. 2012;45(1):99–110. doi: 10.1016/j.molcel.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim N, et al. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332(6037):1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu B, Clayton DA. RNA-DNA hybrid formation at the human mitochondrial heavy-strand origin ceases at replication start sites: An implication for RNA-DNA hybrids serving as primers. EMBO J. 1996;15(12):3135–3143. [PMC free article] [PubMed] [Google Scholar]
- 26.Kogoma T. A novel Escherichia coli mutant capable of DNA replication in the absence of protein synthesis. J Mol Biol. 1978;121(1):55–69. doi: 10.1016/0022-2836(78)90262-0. [DOI] [PubMed] [Google Scholar]
- 27.Kogoma T, Maldonado RR. DNA polymerase I in constitutive stable DNA replication in Escherichia coli. J Bacteriol. 1997;179(7):2109–2115. doi: 10.1128/jb.179.7.2109-2115.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horiuchi T, Maki H, Sekiguchi M. RNase H-defective mutants of Escherichia coli: A possible discriminatory role of RNase H in initiation of DNA replication. Mol Gen Genet. 1984;195(1-2):17–22. doi: 10.1007/BF00332717. [DOI] [PubMed] [Google Scholar]
- 29.Kogoma T. Absence of RNase H allows replication of pBR322 in Escherichia coli mutants lacking DNA polymerase I. Proc Natl Acad Sci USA. 1984;81(24):7845–7849. doi: 10.1073/pnas.81.24.7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kogoma T, von Meyenburg K. The origin of replication, oriC, and the dnaA protein are dispensable in stable DNA replication (sdrA) mutants of Escherichia coli K-12. EMBO J. 1983;2(3):463–468. doi: 10.1002/j.1460-2075.1983.tb01445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Massy B, Fayet O, Kogoma T. Multiple origin usage for DNA replication in sdrA(rnh) mutants of Escherichia coli K-12. Initiation in the absence of oriC. J Mol Biol. 1984;178(2):227–236. doi: 10.1016/0022-2836(84)90141-4. [DOI] [PubMed] [Google Scholar]
- 32.Maduike NZ, Tehranchi AK, Wang JD, Kreuzer KN. Replication of the Escherichia coli chromosome in RNase HI-deficient cells: Multiple initiation regions and fork dynamics. Mol Microbiol. 2014;91(1):39–56. doi: 10.1111/mmi.12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wimberly H, et al. R-loops and nicks initiate DNA breakage and genome instability in non-growing Escherichia coli. Nat Commun. 2013;4:2115. doi: 10.1038/ncomms3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinclair DA, Guarente L. Extrachromosomal rDNA circles—A cause of aging in yeast. Cell. 1997;91(7):1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 35.Takeuchi Y, Horiuchi T, Kobayashi T. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 2003;17(12):1497–1506. doi: 10.1101/gad.1085403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivessa AS, Zhou JQ, Zakian VA. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell. 2000;100(4):479–489. doi: 10.1016/s0092-8674(00)80683-2. [DOI] [PubMed] [Google Scholar]
- 37.Chan YA, et al. Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLoS Genet. 2014;10(4):e1004288. doi: 10.1371/journal.pgen.1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.El Hage A, Webb S, Kerr A, Tollervey D. Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria. PLoS Genet. 2014;10(10):e1004716. doi: 10.1371/journal.pgen.1004716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raghuraman MK, et al. Replication dynamics of the yeast genome. Science. 2001;294(5540):115–121. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- 40.Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51(3):463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 41.Holt IJ, Lorimer HE, Jacobs HT. Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell. 2000;100(5):515–524. doi: 10.1016/s0092-8674(00)80688-1. [DOI] [PubMed] [Google Scholar]
- 42.Gerhold JM, Aun A, Sedman T, Jõers P, Sedman J. Strand invasion structures in the inverted repeat of Candida albicans mitochondrial DNA reveal a role for homologous recombination in replication. Mol Cell. 2010;39(6):851–861. doi: 10.1016/j.molcel.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 43.Wellinger RE, Schär P, Sogo JM. Rad52-independent accumulation of joint circular minichromosomes during S phase in Saccharomyces cerevisiae. Mol Cell Biol. 2003;23(18):6363–6372. doi: 10.1128/MCB.23.18.6363-6372.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krawczyk C, Dion V, Schär P, Fritsch O. Reversible Top1 cleavage complexes are stabilized strand-specifically at the ribosomal replication fork barrier and contribute to ribosomal DNA stability. Nucleic Acids Res. 2014;42(8):4985–4995. doi: 10.1093/nar/gku148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wittekind M, et al. Isolation and characterization of temperature-sensitive mutations in RPA190, the gene encoding the largest subunit of RNA polymerase I from Saccharomyces cerevisiae. Mol Cell Biol. 1988;8(10):3997–4008. doi: 10.1128/mcb.8.10.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marians KJ. Prokaryotic DNA replication. Annu Rev Biochem. 1992;61:673–719. doi: 10.1146/annurev.bi.61.070192.003325. [DOI] [PubMed] [Google Scholar]
- 47.Gowrishankar J. End of the beginning: Elongation and termination features of alternative modes of chromosomal replication initiation in bacteria. PLoS Genet. 2015;11(1):e1004909. doi: 10.1371/journal.pgen.1004909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saini N, et al. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature. 2013;502(7471):389–392. doi: 10.1038/nature12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kraus E, Leung WY, Haber JE. Break-induced replication: A review and an example in budding yeast. Proc Natl Acad Sci USA. 2001;98(15):8255–8262. doi: 10.1073/pnas.151008198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chon H, et al. RNase H2 roles in genome integrity revealed by unlinking its activities. Nucleic Acids Res. 2013;41(5):3130–3143. doi: 10.1093/nar/gkt027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hraiky C, Raymond MA, Drolet M. RNase H overproduction corrects a defect at the level of transcription elongation during rRNA synthesis in the absence of DNA topoisomerase I in Escherichia coli. J Biol Chem. 2000;275(15):11257–11263. doi: 10.1074/jbc.275.15.11257. [DOI] [PubMed] [Google Scholar]
- 52.Drolet M. Growth inhibition mediated by excess negative supercoiling: The interplay between transcription elongation, R-loop formation and DNA topology. Mol Microbiol. 2006;59(3):723–730. doi: 10.1111/j.1365-2958.2005.05006.x. [DOI] [PubMed] [Google Scholar]
- 53.Baaklini I, et al. Hypernegative supercoiling inhibits growth by causing RNA degradation. J Bacteriol. 2008;190(22):7346–7356. doi: 10.1128/JB.00680-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Drolet M, et al. Overexpression of RNase H partially complements the growth defect of an Escherichia coli delta topA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc Natl Acad Sci USA. 1995;92(8):3526–3530. doi: 10.1073/pnas.92.8.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lobachev KS, Gordenin DA, Resnick MA. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108(2):183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- 56.Pomerantz RT, O’Donnell M. The replisome uses mRNA as a primer after colliding with RNA polymerase. Nature. 2008;456(7223):762–766. doi: 10.1038/nature07527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pomerantz RT, O’Donnell M. Direct restart of a replication fork stalled by a head-on RNA polymerase. Science. 2010;327(5965):590–592. doi: 10.1126/science.1179595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Longhese MP, Plevani P, Lucchini G. Replication factor A is required in vivo for DNA replication, repair, and recombination. Mol Cell Biol. 1994;14(12):7884–7890. doi: 10.1128/mcb.14.12.7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Collins KL, Russo AA, Tseng BY, Kelly TJ. The role of the 70 kDa subunit of human DNA polymerase alpha in DNA replication. EMBO J. 1993;12(12):4555–4566. doi: 10.1002/j.1460-2075.1993.tb06144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Foiani M, Marini F, Gamba D, Lucchini G, Plevani P. The B subunit of the DNA polymerase alpha-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol Cell Biol. 1994;14(2):923–933. doi: 10.1128/mcb.14.2.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Foiani M, Liberi G, Lucchini G, Plevani P. Cell cycle-dependent phosphorylation and dephosphorylation of the yeast DNA polymerase alpha-primase B subunit. Mol Cell Biol. 1995;15(2):883–891. doi: 10.1128/mcb.15.2.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paeschke K, et al. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497(7450):458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tudek A, et al. Molecular basis for coordinating transcription termination with noncoding RNA degradation. Mol Cell. 2014;55(3):467–481. doi: 10.1016/j.molcel.2014.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moreira MC, et al. Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat Genet. 2004;36(3):225–227. doi: 10.1038/ng1303. [DOI] [PubMed] [Google Scholar]
- 65.Chen YZ, et al. Senataxin, the yeast Sen1p orthologue: characterization of a unique protein in which recessive mutations cause ataxia and dominant mutations cause motor neuron disease. Neurobiol Dis. 2006;23(1):97–108. doi: 10.1016/j.nbd.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 66.Skourti-Stathaki K, Proudfoot NJ. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014;28(13):1384–1396. doi: 10.1101/gad.242990.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cha RS, Kleckner N. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science. 2002;297(5581):602–606. doi: 10.1126/science.1071398. [DOI] [PubMed] [Google Scholar]
- 68.Schwab M. Amplification of oncogenes in human cancer cells. Bioessays. 1998;20(6):473–479. doi: 10.1002/(SICI)1521-1878(199806)20:6<473::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 69.Tanaka H, Yao MC. Palindromic gene amplification—An evolutionarily conserved role for DNA inverted repeats in the genome. Nat Rev Cancer. 2009;9(3):216–224. doi: 10.1038/nrc2591. [DOI] [PubMed] [Google Scholar]
- 70.Trask BJ, Hamlin JL. Early dihydrofolate reductase gene amplification events in CHO cells usually occur on the same chromosome arm as the original locus. Genes Dev. 1989;3(12A):1913–1925. doi: 10.1101/gad.3.12a.1913. [DOI] [PubMed] [Google Scholar]
- 71.Heck MM, Spradling AC. Multiple replication origins are used during Drosophila chorion gene amplification. J Cell Biol. 1990;110(4):903–914. doi: 10.1083/jcb.110.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heyse G, Jönsson F, Chang WJ, Lipps HJ. RNA-dependent control of gene amplification. Proc Natl Acad Sci USA. 2010;107(51):22134–22139. doi: 10.1073/pnas.1009284107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hourcade D, Dressler D, Wolfson J. The amplification of ribosomal RNA genes involves a rolling circle intermediate. Proc Natl Acad Sci USA. 1973;70(10):2926–2930. doi: 10.1073/pnas.70.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14(10):953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 75.Ulbrich C, et al. Mechanochemical removal of ribosome biogenesis factors from nascent 60S ribosomal subunits. Cell. 2009;138(5):911–922. doi: 10.1016/j.cell.2009.06.045. [DOI] [PubMed] [Google Scholar]
- 76.Morawska M, Ulrich HD. An expanded tool kit for the auxin-inducible degron system in budding yeast. Yeast. 2013;30(9):341–351. doi: 10.1002/yea.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.