Significance
Respiratory complex I is the largest membrane protein complex in mitochondria and has a central function in energy metabolism. Numerous human diseases are linked with complex I dysfunction or assembly defects. The concerted assembly of more than 40 subunits and the insertion of cofactors is aided by specific chaperones. In addition to eight FeS clusters, complex I comprises a Zn-binding site of unknown function. Combining X-ray structural analysis of complex I crystals with quantum chemical modeling and proteomic and spectroscopic analysis of a purified assembly intermediate, we show that accessory subunit NUMM (human ortholog NDUFS6) binds Zn at the interface of two functional modules of the enzyme complex and is required for a specific step of complex I biogenesis.
Keywords: assembly, metal protein, FeS cluster, NDUFAF2, NDUFA12
Abstract
Mitochondrial proton-pumping NADH:ubiquinone oxidoreductase (respiratory complex I) comprises more than 40 polypeptides and contains eight canonical FeS clusters. The integration of subunits and insertion of cofactors into the nascent complex is a complicated multistep process that is aided by assembly factors. We show that the accessory NUMM subunit of complex I (human NDUFS6) harbors a Zn-binding site and resolve its position by X-ray crystallography. Chromosomal deletion of the NUMM gene or mutation of Zn-binding residues blocked a late step of complex I assembly. An accumulating assembly intermediate lacked accessory subunit N7BM (NDUFA12), whereas a paralog of this subunit, the assembly factor N7BML (NDUFAF2), was found firmly bound instead. EPR spectroscopic analysis and metal content determination after chromatographic purification of the assembly intermediate showed that NUMM is required for insertion or stabilization of FeS cluster N4.
Proton-pumping NADH:ubiquinone oxidoreductase (respiratory complex I) is a multisubunit membrane protein complex with a central function in aerobic energy metabolism (1, 2). Fourteen central subunits that harbor the bioenergetic core functions are conserved from bacteria to humans. In addition, eukaryotic complex I comprises around 30 accessory subunits of largely unknown function. The structures of bacterial and mitochondrial complex I were analyzed by X-ray crystallography and electron microscopy (3–6). The central subunits can be assigned to functional modules for NADH oxidation (N-module), ubiquinone reduction (Q-module), and proton pumping (P-module). The subunits forming the N- and Q- module harbor a chain of FeS clusters that connects the NADH oxidation site with the ubiquinone reduction site where the redox energy is released to drive proton translocation.
The assembly of complex I subunits is a multistep process that proceeds via defined intermediates and is aided by a number of assembly factors (7). It also requires the concerted insertion of preformed FeS clusters into several subunits of the N- and Q-module (8). Dysfunction of complex I is the most frequent cause of mitochondrial disorders (9). Pathogenic mutations were identified not only in central subunits, encoded by either nuclear or mitochondrial DNA, but also in accessory subunits and assembly factors. The aerobic yeast Yarrowia lipolytica has been established as a yeast genetic model system to study structure and function of eukaryotic complex I, as well as complex I linked diseases (10).
In this study, we focused on the accessory subunit NUMM of Y. lipolytica complex I. NUMM belongs to a limited subset of accessory subunits that is already found in α-proteobacteria and harbors a conserved putative Zn-binding motif, comprising three cysteines and one histidine in its C-terminal part (11). Zn binding to complex I was previously reported for bovine complex I but its functional relevance and the position of the Zn site remained elusive (12, 13). A poly-alanine model of the orthologous 13-kDa subunit was tentatively fitted into the electron microscopic structure of bovine complex I but Zn binding was not shown (5). In human, mutations leading to the loss of the orthologous NDUFS6 subunit or exchange of one of the three conserved cysteine residues were shown to cause fatal diseases (14, 15). A mouse model to study complex I-linked diseases was generated by knock down of the corresponding gene (16).
We analyzed the functional significance of the Zn-binding site of the NUMM subunit in Y. lipolytica complex I and determined its position in the X-ray structure of the enzyme complex by Zn anomalous scattering. We found that deletion of the NUMM gene or site-directed mutagenesis of Zn ligands compromised a late step of complex I biogenesis that is required for stable insertion of FeS cluster N4.
Results
Deletion of the NUMM Subunit Impairs Complex I Biogenesis.
To investigate the function of the NUMM subunit and its proposed Zn-binding site (11) (Fig. S1A) in respiratory complex I we generated a chromosomal deletion of the entire reading frame of the corresponding NUMM gene by homologous recombination. Complex I content of mitochondrial membranes from the deletion strain was decreased to 41% as assessed by NADH:hexammineruthenium (HAR) oxidoreductase activity and normalized dNADH:decylubiquinone (DBQ) oxidoreductase activity dropped to 44% of the parental strain (Table 1). Complementation with variants carrying mutations in the putative Zn ligating residues C97, H110, C125, C128 did not restore complex I content and activity to the level of the parental strain (Table 1). Mutation of C128 to alanine resulted in loss of activity comparable to that found in the numm∆ strain. Change of the two other conserved cysteine residues into alanine reduced the normalized dNADH:DBQ oxidoreductase activity of complex I to 63%, whereas mutation H110A exhibited the mildest effect in reaching 76% of parental strain activity.
Table 1.
Complex I content and ubiquinone reductase activities in mitochondrial membranes
| Strain | Complex I content*, % | Complex I activity†, % |
| nummΔ | 41 | 44 |
| n7bmlΔ | 30 | 83 |
| n7bmΔ | 26 | 35 |
| C97ANUMM | 54 | 63 |
| H110ANUMM | 60 | 76 |
| C125ANUMM | 55 | 63 |
| C128ANUMM | 54 | 37 |
Complex I content was assessed as nonphysiological NADH:hexaammineruthenium (III) oxidoreductase activity and compared with parental strain GB20 for chromosomal deletions (100% = 1.41 µmol∙min−1∙mg−1) and with plasmid complemented nummΔ deletion strain for NUMM point mutants (100% = 1.14 µmol∙min−1∙mg−1).
Inhibitor sensitive dNADH:decylubiquinone oxidoreductase activity was normalized to complex I content (100% = 0.55 µmol∙min−1 CI−1 for GB20 to compare with chromosomal deletions and 100% = 0.46 µmol∙min−1 CI−1 for plasmid complemented nummΔ deletion to compare with NUMM point mutants).
We subsequently used complexome profiling (17) of intact mitochondria to analyze complex I assembly (Fig. S2). Of the 42 known subunits of Y. lipolytica complex I (18) we confirmed assembly of 38 subunits in the parental strain. They can be assigned to the peripheral arm and the two pump modules, PP and PD, in the membrane arm. Subunit ST1 is known to be detached from complex I under BN-PAGE conditions (19); subunits ND2, ND4L and especially ND6 of the PP module are difficult to detect by mass spectrometry because they are highly hydrophobic and lack suitable cleavage sites (18). Complex I was mainly found in monomeric form but also assembled into higher molecular mass supercomplexes (20); membrane arm subcomplexes, very likely representing assembly intermediates, were detected at lower molecular mass (Fig. S2).
In strain nummΔ we detected assembly of nearly all expected subunits (Fig. S2). However, we also found a higher fraction of unassembled subunits and subcomplexes of the peripheral arm. Notably, accessory subunit N7BM did not assemble into complex I and was almost exclusively found in free form at the migration front in the Blue-Native gel (Fig. 1 and Fig. S2). On the other hand, an additional protein was found to be associated with the nearly complete complex I subassembly in nummΔ. The 27.6-kDa protein, mainly detected in free form in the parental strain, was identified as YALI0D27082p in the Y. lipolytica genome (21) and has sequence homology to a known complex I assembly factor called NDUFAF2 in Homo sapiens, B17.2L in Bos taurus, and 13.4L in Neurospora crassa (Fig. S1B) (22, 23). Because its N-terminal half was also clearly homologous to Y. lipolytica complex I subunit N7BM (Fig. S1D), we named the protein N7BML for N7BM like. Indeed, N7BM is also homologous to subunits NDUFA12, B17.2, and nuo13.4 from H. sapiens, B. taurus, and N. crassa, respectively (Fig. S1D) that are paralogs of the respective assembly factors indicated above (22–24).
Fig. 1.
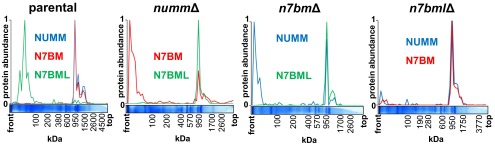
Migration profiles of complex I subunits NUMM, N7BM, and assembly factor N7BML. The migration profiles were derived from the complexome profiling analysis of intact mitochondria from the parental strain and the deletion strains for NUMM, N7BM, and N7BML shown in Fig. S2. Each panel shows the relative abundance of the indicated proteins in relation to the migration distance in the BN-gel; mass scale calibrated based on complex I, dimeric complex IV and monomeric and dimeric ATP synthase.
A Functional Link Between NUMM, N7BM, and N7BML in Complex I Assembly.
To investigate the role of N7BML in the complex I assembly process and its functional relation to its paralog, complex I subunit N7BM, we deleted the corresponding genes from the Y. lipolytica genome. In strain n7bmlΔ the complex I content of mitochondrial membranes was reduced to 30%, but the normalized ubiquinone reductase activity was preserved at a level of 83% of the parental strain. In strain n7bmΔ complex I content and normalized activity both dropped significantly to 26% and 35%, respectively. Consistent with a reduced content due to impaired assembly and/or stability, a significant amount of unassembled subunits and subassemblies was observed by complexome profiling for both deletions (Fig. S2). In strain n7bmlΔ, complete complex I was assembled. However, we also detected a prominent subassembly or fragment forming part of the Q-module (49-kDa, 30-kDa, PSST) together with accessory subunit NUFM (human ortholog NDUFA5) permitting its assignment to this structural element, in line with recent modeling of the orthologous B13 subunit of the bovine enzyme (5). Complex I of strain n7bmΔ did not lack any additional subunits (Fig. S2) but it was interesting to note that, like in strain nummΔ, N7BML was found associated with the subassembly (Fig. 1).
We attempted to purify complex I from detergent solubilized mitochondrial membranes of all three deletion strains taking advantage of the His–tag attached to the 30-kDa subunit. We succeeded to obtain pure preparations for strains nummΔ and n7bmlΔ and analyzed their subunit composition by gradient and dSDS PAGE (Fig. 2 and Fig. S3). For nummΔ, we confirmed the absence of subunit N7BM and the presence of N7BML. The loosely attached subunit ST1 was also absent. In n7bmlΔ we detected all subunits including N7BM, NUMM, and ST1. No complex I could be isolated from membranes of strain n7bmΔ, probably because of fragmentation during chromatography.
Fig. 2.
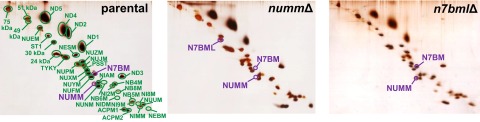
Subunit composition of purified complex I from parental, numm∆ and n7bml∆ strains. Complex I was chromatographically purified and resolved by dSDS PAGE; protein spots were stained with silver. The assignment of subunits NUMM, N7BM and complex I assembly factor N7BML labeled in violet was confirmed by LC-ESI MS/MS; assignment of all other subunits labeled in green is based on (35).
The lipid reactivated preparation of complex I from strain n7bmlΔ showed a specific ubiquinone reductase activity close to the parental strain level (Table 2). The specific activity of purified complex I from strain nummΔ was 25% comparing to 44% in mitochondrial membranes.
Table 2.
Catalytic activity and metal stoichiometry of purified complex I
| Strain | Complex I activity, % parental | Fe, atoms per Cl | Zn, atoms per Cl |
| Parental | 100 | 30 ± 2 | 1 ± 0 |
| nummΔ | 25 | 24 ± 2 | 0 |
| n7bmlΔ | 88 | n.d. | n.d. |
dNADH:decylubiquinone oxidoreductase activity of purified and lipid-reactivated parental complex I was 6.5 µmol∙min−1∙mg−1. Metal content was determined by TXRF (n = 3). n.d., not determined.
Effect of Mutations in the Zn-Binding Site on Complex I Assembly.
To assess the role of the proposed Zn-binding residues in subunit NUMM we purified small amounts of mutant complex I (Fig. S4). Interestingly, we found two forms (P1 and P2) of complex I in strains carrying mutations H110A or C125A. Form P1 eluted from the gel filtration column at the same peak position as parental complex I. P1 dominated in mutant H110A and, as judged by dSDS-PAGE, it contained only traces of N7BML. In contrast, in P2 N7BML was present in amounts comparable to the preparation from strain nummΔ. Subunit NUMM is difficult to detect in dSDS PAGE because of its diffuse migration behavior and staining. However, it appeared that NUMM was present in P1 but not in P2. Both forms of the enzyme were found in approximately equal amounts in the preparation of mutant C125A making it more difficult to separate them and differentiate their composition. Only the P2 form was found in mutant C128A suggesting a complete loss of subunit NUMM in the preparation from this strain. Overall these results indicate that integrity of the Zn-binding site is needed for proper assembly and stability of complex I and that exchange of H110 is much better tolerated compared with an exchange of C128 as is also evident from complex I content and activity in mitochondrial membranes (Table 1).
Accessory Subunit NUMM Binds Zn at the Junction of the N-Module and the Q-Module of Complex I.
The metal to protein ratio of purified complex I was determined using Total Reflection X-ray Fluorescence (TXRF). For the parental strain preparation we found a stoichiometry of one Zn atom per complex I (Table 2). No Zn was detected in three independent preparations from the nummΔ strain confirming that the CX8HX14CX2C motif of NUMM is the only Zn-binding site of complex I.
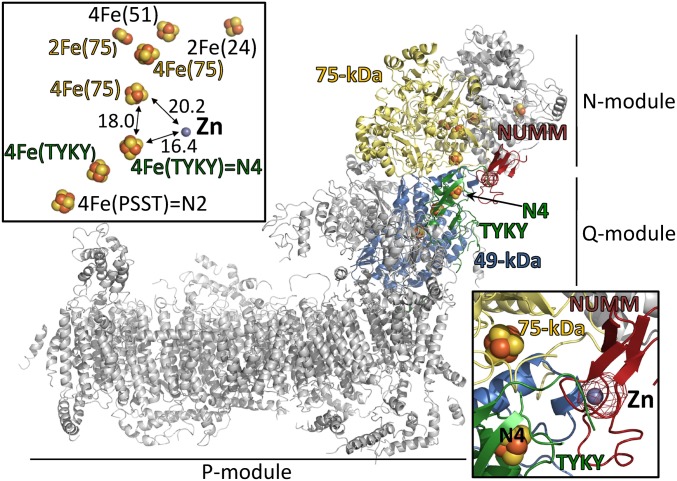
Complex I was crystallized and X-ray diffraction data were collected at the K-edge of Zn. After data processing, we obtained an anomalous Fourier map showing a density peak of 13σ, which could be unambiguously assigned to Zn (Fig. S5). Combining the anomalous data with experimental electron density maps and the X-ray structure of the Y. lipolytica complex (4, 6) positioned the Zn at the interface between the N- and Q-modules of the peripheral arm (Fig. 3 and Table S1). However, lower local order in the upper half of the peripheral arm precluded us from unambiguously building the NUMM subunit based on our experimental X-ray electron density maps. Instead we prepared a homology model for the C-terminal part of the subunit based on the structure of a bacterial NUMM ortholog determined in a structural genomics project (PDB ID code 2JVM). As the metal binding site was not occupied in the reference structure, we added Zn in silico and optimized the model of the binding site using hybrid quantum mechanics/molecular mechanics (QM/MM) calculations (Fig. S6 and Table S2). The model was fitted into the X-ray structure of Y. lipolytica complex I guided by the Zn peak of the anomalous Fourier map, experimental electron density maps, and the orientation of the β-strands of the superimposed 5-Å resolution electron-microscopy structure of bovine complex I (5) (Fig. 3). The C-terminal domain of NUMM consists of a two-stranded β-sheet involving strands β2 and β3 and a three-stranded β-sheet composed of strands β1, β4 and β5 (Fig. S6). The single Zn(II) ion is coordinated by three cysteine residues and one histidine; C97 resides at the tip of the β2 strand, whereas the other two cysteine residues, C125 and C128, are located in the loop connecting strands β4 and β5. The QM/MM calculations suggest that the coordination of the Zn is nearly tetrahedral, with the Zn ligands located at 2.3–2.4 Å (Table S2).The single histidine ligand H110 resides in the long β2-β3 loop, and is stabilized by a hydrogen bond with the backbone of P111 (Fig. S6). The C-terminal NUMM domain interacts with the 75-kDa, 49-kDa, and TYKY subunits (Fig. 3). Notably, the Zn is in close vicinity of two FeS clusters involved in electron transfer and distances could be accurately determined from the peaks in the respective iron and Zn anomalous Fourier maps. It is situated 20.2 Å away from one of the two 4Fe4S clusters of the 75-kDa subunit (4[Fe]75H) and 16.4 Å away from one of the two 4Fe4S clusters of the TYKY subunit (4[Fe]TY1). The latter cluster corresponds to EPR signature N4, whereas the cluster in the 75-kDa subunit is EPR silent (25). Due to limited resolution, subunit N7BM could not be identified unambiguously in the electron density maps.
Fig. 3.
Position of the Zn-binding site in complex I. Structure of Yarrowia lipolytica complex I showing the position of the C-terminal NUMM domain and its bound Zn. Complex I subunits are represented in gray except for subunits TYKY (green) 49-kDa (blue), 75-kDa (yellow), and the fitted model of the C-terminal domain of NUMM (red). The Zn-specific anomalous Fourier map is contoured at 8σ and is represented in red. (Upper Left Inset) The position of the Zn is represented in context of the iron–sulfur cluster chain involved in electron transfer. Distances are measured in Å. (Lower Right Inset) Detailed view of the Zn-binding site is shown; NUMM is adjacent to elements from TYKY, 49-kDa and 75-kDa subunits.
The structural model of NUMM was used to search the Brookhaven Protein Data Bank for structurally similar proteins. The best hit was the NMR structure of the Zn-binding domain of COX4 a subunit of cytochrome c oxidase from yeast (26) (Figs. S1E and S6).
FeS Cluster N4 Is Missing from CI of Strain nummΔ and in Mutant C128A.
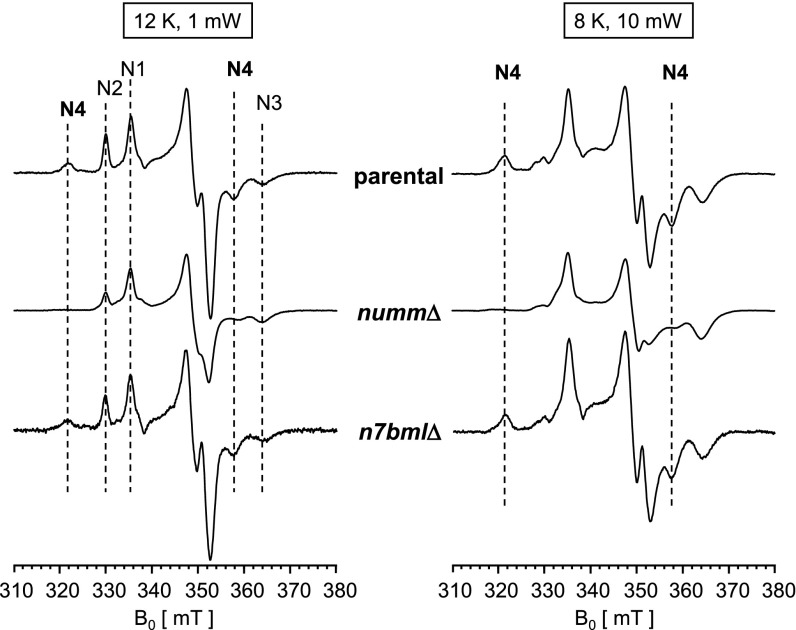
Even though complex I from the nummΔ strain contained all central subunits, it showed severe impairment of the inhibitor sensitive dNADH:DBQ oxidoreductase activity. Therefore, we used EPR spectroscopy to assess the status of its FeS clusters that function in electron transfer (Fig. 4). In complex I from the parental strain and from strain n7bmlΔ signals of clusters N1-N4 could be distinguished at 12 K and 1 mW microwave power. The characteristic signal for cluster N4 assigned to one of the two 4Fe4S clusters (4[Fe]TY1) in subunit TYKY (25) was undetectable in spectra of the preparation from strain nummΔ. The signal of cluster N2, the immediate electron donor to ubiquinone, was reduced to about half of the parental strain, whereas the signals of clusters N1 was diminished by 25% and N3 appeared unchanged. Spectra recorded at conditions optimized for detection of N4 (8 K and 10 mW microwave power) confirmed absence of the N4 signal in the preparation from strain nummΔ (Fig. 4). Signal N5 is difficult to detect but was clearly identified in wild type and nummΔ complex I at 5 K. Under these conditions the lack of N4 signal became most obvious (Fig. S7A). EPR spectra of complex I with a mutated Zn-binding site showed a clear tendency for presence of signal N4 in form P1 and reduction of signal intensity in form P2 (Fig. S7 B and C). For mutants H110A and C125A incomplete separation of P1 and P2 must be taken into account; however, a severe loss of signal N4 was evident for mutant C128A in which only form P2 was present.
Fig. 4.
EPR spectra of complex I isolated from the numm∆ and n7bml∆ strains. EPR spectra of purified complexes were recorded at different temperatures and microwave power settings after reduction with 1.8 mM NADH. At 12 K, 1 mW microwave power clusters N1–N4 are detectable in the control. Although n7bmlΔ CI displayed a spectrum identical to the parental one, in complex I of nummΔ EPR signal N4 is missing and intensities of signals N2 and N1 are diminished by 50% and 25%, respectively. At 8 K, 10 mW, conditions more suitable for detection of N4, the lack of N4 EPR signal in nummΔ is evident. Under these conditions N2 is hardly detectable due to distinct power saturation. Spectra were normalized to an equal signal amplitude of N3. Characteristic contributions of individual iron–sulfur clusters to the EPR spectrum are indicated.
Because complex I harbors also two EPR invisible FeS clusters of which one is also ligated by TYKY, we analyzed iron to protein ratios by TXRF (Table 2). For the parental enzyme, the determined stoichiometry of 30 ± 2 (n = 3) Fe per complex I is consistent with the expected value of 28 Fe. In the preparation from strain nummΔ, we found a decreased Fe:complex I stoichiometry of 24 ± 2 (n = 3), consistent with the absence of cluster N4, but giving no indication for the loss of additional FeS clusters.
Discussion
Complex I is a metalloprotein comprising eight canonical redox active FeS clusters coordinated by residues of central subunits of the matrix arm (1, 2). Within the large number of accessory subunits, NUMM stands out by harboring an additional putative Zn-binding site (11). In this study we investigated its functional significance and position in the enzyme complex.
After deletion of the NUMM gene from the genome of the strictly aerobic yeast Y. lipolytica, we observed a clear and specific effect on complex I biogenesis. The nummΔ mutant assembled all central and accessory subunits of the membrane and peripheral arm with the only exception of the accessory N7BM and ST1 subunits. The association of the subassembly with the assembly factor N7BML indicated that in the nummΔ strain complex I biogenesis is stalled leading to accumulation of a specific assembly intermediate. Subunit ST1 was previously shown to have no impact on complex I assembly and fully functional enzyme was isolated after deletion of the corresponding gene (27). The blocked assembly step must therefore involve the interplay of subunits NUMM and N7BM with assembly factor N7BML. N7BML interacts strongly with the assembly intermediate as it remained firmly bound even after chromatographic purification. In line with previous studies, the deletion of the N7BML gene did not prevent assembly of complete and active complex I (23, 28). However, in Y. lipolytica mitochondria we found a marked reduction of complex I content suggesting that assembly efficiency was decreased. We found that N7BM and N7BML were never present together in any complex I (sub)assembly, strongly supporting the idea that the subunit N7BM replaces its paralog the assembly factor N7BML during complex I biogenesis (24). In any case, detachment of N7BML from the assembly intermediate required the presence of complex I subunits N7BM and NUMM in line with results recently obtained for N. crassa, where binding of the N7BML ortholog 13.4L to nascent complex I was shown immunologically (23). However, although we find a strict linkage between NUMM and N7BM insertion, the orthologous proteins were found to be incorporated independently in N. crassa.
The human ortholog of N7BML, complex I assembly factor NDUFAF2, was previously found to associate with a large subcomplex present in mitochondria of patients with mutations in subunits NDUFV1 or NDUFS4 (22). Moreover, in a patient lacking NDUFS6, the human ortholog of NUMM, formation of an 800-kDa subcomplex was reported and NDUFAF2 was found to associate with this subcomplex (29). Based on the analysis of assembly profiles of individual complex I subunits, NDUFAF2 was suggested to be responsible for attachment of the electron input module of the enzyme including central subunits NDUFV1 and NDUFV2 to preassembled complex I (29). However, complexome profiling and our thorough analysis of highly purified complexes using different electrophoresis techniques clearly showed that none of the three deletions prevented assembly of the corresponding 51-kDa and 24-kDa central subunits of Y. lipolytica complex I. The observation of smaller subcomplexes in patient material might be explained by unspecific effects, for example, decreased stability of mutant complex I and secondary loss of subunits during analysis of the human enzyme.
Comparison of metal to protein ratios of the nummΔ assembly intermediate with parental complex I clearly demonstrated that NUMM indeed binds Zn as suggested previously (11). In agreement with the 1:1 molar stoichiometry determined by TXRF, a unique strong Zn signal per complex I monomer was observed in anomalous Fourier maps positioning the Zn close to cluster N4 and the 4Fe4S cluster of the 75-kDa subunit. To elucidate the role of the Zn-binding site for complex I function and assembly, we generated a series of site-directed mutants of the putative Zn ligands. Analysis of activities and size exclusion profiles during protein purification showed that integrity of the Zn-binding site in NUMM is required to complete complex I biogenesis. Mutation of H110 was better tolerated than mutation of C125, whereas exchange of C128 had the strongest effect. Notably, mutation of C115 in human complex I subunit NDUFS6, corresponding to C128 of NUMM of Y. lipolytica, was identified to cause fatal neonatal lactic acidosis (15). The consistent lack of NUMM and presence of N7BML in form P2 found in the three investigated site-directed mutants suggests that a functional metal binding site is required for proper folding and association of the subunit with the nascent enzyme complex. Remarkably, we detected significant structural similarity of the C-terminal domains of complex I subunit NUMM and subunit COX4 of cytochrome oxidase from S. cerevisiae (26). COX4 binds Zn with the same set of ligands , whereas the orthologous subunit in mammals binds Zn with a four cysteine coordination (Fig. S1E). Interestingly, like in NUMM, mutation of the Zn-binding cysteine residues in COX4 was shown to have drastic effects on assembly, whereas mutation of the histidine residue was less detrimental (26). The results of our mutagenesis study are in agreement with a structural role of Zn binding that was also proposed to be essential for folding and functionality of COX4.
Significant reduction of the N4 EPR signal in the NUMM deletion and in P2 fractions of site-directed mutants showed that the presence of NUMM is a prerequisite for stable insertion of the corresponding FeS cluster into the TYKY subunit consistent with the close distance of NUMM bound Zn to this redox-center. The iron to protein ratio is in good agreement with absence of only one 4Fe4S cluster but does not exclude partial loss of further clusters, e.g., cluster N2 that also exhibited diminished EPR signal intensity. Extensive loss of activity in the deletion and point mutants can be explained by lack of at least one cluster in the electron transfer chain from NADH to the ubiquinone reduction site. Assuming strict conservation of the structure, the loss of cluster N4 would increase the longest electron transfer distance in the deletion mutant to 22 Å, which is too long for efficient electron transfer according to the most simplified version of the electron tunneling equation (30). However, the exact electron transfer distance in the mutant is unknown and the simplified “Dutton ruler” assumes the generic values of 0.7 eV for the reorganization energy and 0 eV for ΔGo. The observed residual activity of 25% of the purified nummΔ complex I indicated that electron transfer with a rate of ∼50 s−1 was still possible in the absence of cluster N4.
The insertion of preformed FeS clusters requires interaction of the target apo-protein with an intricate assembly machinery (8). However, previous structural analysis of mitochondrial complex I showed that the central subunits of the Q-module that harbor FeS clusters, TYKY and PSST, are largely surrounded and cut off from the environment by central and accessory subunits (4–6). As this is incompatible with FeS cluster insertion after complete assembly we propose that the function of N7BML is to keep nascent complex I in an “open” state and to retain apo-proteins competent for interaction with the FeS cluster transfer machinery. Only after completion of cluster insertion NUMM and N7BM bind and release the assembly factor. The position of the Zn-binding site in the enzyme complex and its close distance to N4 suggests that it may function as a lock for the cluster entry site. In the absence of NUMM, the open state would persist and the cluster would remain exposed to solvent and fragile.
It seems remarkable that a three-cysteine/one-histidine coordination is also found in two proteins that are involved in transfer of FeS clusters to apo-proteins, IscU (31) and mitoNEET (32). Interestingly, both polypeptides were shown to bind Zn under specific conditions (33, 34). We have no indication for an active role of NUMM in cluster transfer but this possibility cannot be completely excluded at this stage.
Taken together, our study sheds light on the molecular basis of the fatal effects observed in patients carrying mutations in the human NUMM ortholog NDUFS6 (14, 15) as we demonstrated that NUMM harboring a functional Zn site is essential for stable insertion of FeS cluster N4 and thus for proper complex I assembly and function.
Materials and Methods
Deletion Strains and Site-Directed Mutagenesis of Subunit NUMM from Yarrowia lipolytica.
The entire NUMM (YALI0D19030g), N7BM (YALI0B00792g) and N7BML (YALI0D27082g) ORFs were deleted in Y. lipolytica strain GB20 (mus51Δ, nugm-Htg2, ndh2i, lys11−, leu2−, ura3−, MatB) as described (35). Point mutations in subunit NUMM were introduced by complementation of the nummΔ strain with plasmid pUB4 carrying variants of the NUMM coding sequence generated by PCR.
Preparation of Intact Mitochondria, Mitochondrial Membranes, and Purification of Complex I.
All preparations were essentially carried out as described in ref. 35. The purity of the complex I preparation from the numm∆ strain was enhanced by ion exchange chromatography on a MonoQ column (GE). Specific NADH:hexaammineruthenium and deamino-NADH:decylubiquinone oxidoreductase activities were measured in mitochondrial membranes and purified enzymes after lipid activation as described (36).
Electrophoresis.
Mitochondrial membranes or intact mitochondria from Y. lipolytica were solubilized in 1.0 g/g DDM or 4 g/g digitonin, respectively, and separated by blue native electrophoresis (BN-PAGE) with a 4–16% acrylamide gradient (37). The subunit composition of purified complex I was analyzed by dSDS-PAGE (38).
Mass Spectrometry.
Proteins from gel-spots were identified essentially as described in ref. 39. Complexome profiling analysis was done according to ref. 17 except that the raw data were analyzed using MaxQuant for protein identification and label free quantification. Mass calibration of each slice was done according to ref. 20.
EPR Spectroscopy and Metal Determination.
X-band EPR spectra were recorded as described (40) and as detailed in the figure legend. Metal to protein ratios were determined by Total-Reflection X-Ray Fluorescence (TXRF) as described (35).
Anomalous X-Ray Diffraction and Data Processing.
Purified complex I from Y. lipolytica was crystallized as described previously (4). Crystals were flash-cooled in liquid nitrogen prior data collection at the Swiss Light Source (SLS) synchrotron. Highly redundant anomalous diffraction data (Table S1) were collected at the Zn K-edge (1.2833 Å) and at a slightly lower energy (1.2861 Å), to confirm the nature of the element responsible for the signal. Data were processed with imosflm (41) and scaled with scala. Using available phases, an anomalous difference Fourier map was calculated. For both measured wavelengths, strong difference peaks were visible around the FeS clusters as iron shows still a strong signal at the Zn K-edge. A single additional peak was observed in the map obtained using the diffraction data at the Zn K-edge.
Computational Methods for QM and QM/MM Models of the Zn-Binding Site in NUMM.
The putative Zn-binding site in the NUMM subunit was modeled using quantum chemical density functional theory (DFT) calculations and hybrid quantum mechanics/classical mechanics (QM/MM) simulations. The computational methods are extensively described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the Swiss Light Source (SLS, Villigen, Switzerland) for allocation of beam time and constant excellent support, and Karin Siegmund, Andrea Duchene, Marijke Beenes, and Claudia Rittmeyer for excellent technical assistance. This work was supported by Deutsche Forschungsgemeinschaft Grant ZI 552/3-1, by the Excellence Initiative of the German Federal and State Governments (EXC 115 to V.Z. and U.B., EXC 294 BIOSS to C.H.), and by a fellowship of the Humboldt Foundation (to S.G.-C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424353112/-/DCSupplemental.
References
- 1.Brandt U. Energy converting NADH:quinone oxidoreductase (complex I) Annu Rev Biochem. 2006;75:69–92. doi: 10.1146/annurev.biochem.75.103004.142539. [DOI] [PubMed] [Google Scholar]
- 2.Hirst J. Mitochondrial complex I. Annu Rev Biochem. 2013;82:551–575. doi: 10.1146/annurev-biochem-070511-103700. [DOI] [PubMed] [Google Scholar]
- 3.Baradaran R, Berrisford JM, Minhas GS, Sazanov LA. Crystal structure of the entire respiratory complex I. Nature. 2013;494(7438):443–448. doi: 10.1038/nature11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunte C, Zickermann V, Brandt U. Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science. 2010;329(5990):448–451. doi: 10.1126/science.1191046. [DOI] [PubMed] [Google Scholar]
- 5.Vinothkumar KR, Zhu J, Hirst J. Architecture of mammalian respiratory complex I. Nature. 2014;515(7525):80–84. doi: 10.1038/nature13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zickermann V, et al. Structural biology. Mechanistic insight from the crystal structure of mitochondrial complex I. Science. 2015;347(6217):44–49. doi: 10.1126/science.1259859. [DOI] [PubMed] [Google Scholar]
- 7.Mimaki M, Wang X, McKenzie M, Thorburn DR, Ryan MT. Understanding mitochondrial complex I assembly in health and disease. Biochim Biophys Acta. 2012;1817(6):851–862. doi: 10.1016/j.bbabio.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Lill R, et al. The role of mitochondria in cellular iron-sulfur protein biogenesis and iron metabolism. Biochim Biophys Acta. 2012;1823(9):1491–1508. doi: 10.1016/j.bbamcr.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Koopman WJH, Willems PHGM, Smeitink JAM. Monogenic mitochondrial disorders. N Engl J Med. 2012;366(12):1132–1141. doi: 10.1056/NEJMra1012478. [DOI] [PubMed] [Google Scholar]
- 10.Kerscher S, Dröse S, Zwicker K, Zickermann V, Brandt U. Yarrowia lipolytica, a yeast genetic system to study mitochondrial complex I. Biochim Biophys Acta. 2002;1555(1-3):83–91. doi: 10.1016/s0005-2728(02)00259-1. [DOI] [PubMed] [Google Scholar]
- 11.Yip CY, Harbour ME, Jayawardena K, Fearnley IM, Sazanov LA. Evolution of respiratory complex I: “supernumerary” subunits are present in the alpha-proteobacterial enzyme. J Biol Chem. 2011;286(7):5023–5033. doi: 10.1074/jbc.M110.194993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giachini L, et al. EXAFS reveals a structural zinc binding site in the bovine NADH-Q oxidoreductase. FEBS Lett. 2007;581(29):5645–5648. doi: 10.1016/j.febslet.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 13.Shinzawa-Itoh K, et al. Bovine heart NADH-ubiquinone oxidoreductase contains one molecule of ubiquinone with ten isoprene units as one of the cofactors. Biochemistry. 2010;49(3):487–492. doi: 10.1021/bi9016318. [DOI] [PubMed] [Google Scholar]
- 14.Kirby DM, et al. NDUFS6 mutations are a novel cause of lethal neonatal mitochondrial complex I deficiency. J Clin Invest. 2004;114(6):837–845. doi: 10.1172/JCI20683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spiegel R, et al. Mutated NDUFS6 is the cause of fatal neonatal lactic acidemia in Caucasus Jews. Eur J Hum Genet. 2009;17(9):1200–1203. doi: 10.1038/ejhg.2009.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ke BX, et al. Tissue-specific splicing of an Ndufs6 gene-trap insertion generates a mitochondrial complex I deficiency-specific cardiomyopathy. Proc Natl Acad Sci USA. 2012;109(16):6165–6170. doi: 10.1073/pnas.1113987109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heide H, et al. Complexome profiling identifies TMEM126B as a component of the mitochondrial complex I assembly complex. Cell Metab. 2012;16(4):538–549. doi: 10.1016/j.cmet.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Angerer H, et al. A scaffold of accessory subunits links the peripheral arm and the distal proton-pumping module of mitochondrial complex I. Biochem J. 2011;437(2):279–288. doi: 10.1042/BJ20110359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgner N, et al. Subunit mass fingerprinting of mitochondrial complex I. Biochim Biophys Acta. 2008;1777(10):1384–1391. doi: 10.1016/j.bbabio.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 20.Nübel E, Wittig I, Kerscher S, Brandt U, Schägger H. Two-dimensional native electrophoretic analysis of respiratory supercomplexes from Yarrowia lipolytica. Proteomics. 2009;9(9):2408–2418. doi: 10.1002/pmic.200800632. [DOI] [PubMed] [Google Scholar]
- 21.Sherman DJ, et al. Génolevures Consortium Génolevures: Protein families and synteny among complete hemiascomycetous yeast proteomes and genomes. Nucleic Acids Res. 2009;37(Database issue):D550–D554. doi: 10.1093/nar/gkn859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogilvie I, Kennaway NG, Shoubridge EA. A molecular chaperone for mitochondrial complex I assembly is mutated in a progressive encephalopathy. J Clin Invest. 2005;115(10):2784–2792. doi: 10.1172/JCI26020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pereira B, Videira A, Duarte M. Novel insights into the role of Neurospora crassa NDUFAF2, an evolutionarily conserved mitochondrial complex I assembly factor. Mol Cell Biol. 2013;33(13):2623–2634. doi: 10.1128/MCB.01476-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kensche PR, Duarte I, Huynen MA. A three-dimensional topology of complex I inferred from evolutionary correlations. BMC Struct Biol. 2012;12:19. doi: 10.1186/1472-6807-12-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roessler MM, et al. Direct assignment of EPR spectra to structurally defined iron-sulfur clusters in complex I by double electron-electron resonance. Proc Natl Acad Sci USA. 2010;107(5):1930–1935. doi: 10.1073/pnas.0908050107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coyne HJ, 3rd, et al. The characterization and role of zinc binding in yeast Cox4. J Biol Chem. 2007;282(12):8926–8934. doi: 10.1074/jbc.M610303200. [DOI] [PubMed] [Google Scholar]
- 27.Abdrakhmanova A, Dobrynin K, Zwicker K, Kerscher S, Brandt U. Functional sulfurtransferase is associated with mitochondrial complex I from Yarrowia lipolytica, but is not required for assembly of its iron-sulfur clusters. FEBS Lett. 2005;579(30):6781–6785. doi: 10.1016/j.febslet.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 28.Schlehe JS, Journel MSM, Taylor KP, Amodeo KD, LaVoie MJ. The mitochondrial disease associated protein Ndufaf2 is dispensable for Complex-1 assembly but critical for the regulation of oxidative stress. Neurobiol Dis. 2013;58:57–67. doi: 10.1016/j.nbd.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lazarou M, McKenzie M, Ohtake A, Thorburn DR, Ryan MT. Analysis of the assembly profiles for mitochondrial- and nuclear-DNA-encoded subunits into complex I. Mol Cell Biol. 2007;27(12):4228–4237. doi: 10.1128/MCB.00074-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moser CC, Farid TA, Chobot SE, Dutton PL. Electron tunneling chains of mitochondria. Biochim Biophys Acta. 2006;1757(9-10):1096–1109. doi: 10.1016/j.bbabio.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 31.Markley JL, et al. Metamorphic protein IscU alternates conformations in the course of its role as the scaffold protein for iron-sulfur cluster biosynthesis and delivery. FEBS Lett. 2013;587(8):1172–1179. doi: 10.1016/j.febslet.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zuris JA, et al. Facile transfer of [2Fe-2S] clusters from the diabetes drug target mitoNEET to an apo-acceptor protein. Proc Natl Acad Sci USA. 2011;108(32):13047–13052. doi: 10.1073/pnas.1109986108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan G, et al. Competition of zinc ion for the [2Fe-2S] cluster binding site in the diabetes drug target protein mitoNEET. Biometals. 2012;25(6):1177–1184. doi: 10.1007/s10534-012-9580-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramelot TA, et al. Solution NMR structure of the iron-sulfur cluster assembly protein U (IscU) with zinc bound at the active site. J Mol Biol. 2004;344(2):567–583. doi: 10.1016/j.jmb.2004.08.038. [DOI] [PubMed] [Google Scholar]
- 35.Angerer H, et al. The LYR protein subunit NB4M/NDUFA6 of mitochondrial complex I anchors an acyl carrier protein and is essential for catalytic activity. Proc Natl Acad Sci USA. 2014;111(14):5207–5212. doi: 10.1073/pnas.1322438111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dröse S, Zwicker K, Brandt U. Full recovery of the NADH:ubiquinone activity of complex I (NADH:ubiquinone oxidoreductase) from Yarrowia lipolytica by the addition of phospholipids. Biochim Biophys Acta. 2002;1556(1):65–72. doi: 10.1016/s0005-2728(02)00307-9. [DOI] [PubMed] [Google Scholar]
- 37.Wittig I, Braun HP, Schägger H. Blue native PAGE. Nat Protoc. 2006;1(1):418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 38.Rais I, Karas M, Schägger H. Two-dimensional electrophoresis for the isolation of integral membrane proteins and mass spectrometric identification. Proteomics. 2004;4(9):2567–2571. doi: 10.1002/pmic.200400829. [DOI] [PubMed] [Google Scholar]
- 39.Wittig I, et al. Assembly and oligomerization of human ATP synthase lacking mitochondrial subunits a and A6L. Biochim Biophys Acta. 2010;1797(6-7):1004–1011. doi: 10.1016/j.bbabio.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 40.Grgic L, Zwicker K, Kashani-Poor N, Kerscher S, Brandt U. Functional significance of conserved histidines and arginines in the 49-kDa subunit of mitochondrial complex I. J Biol Chem. 2004;279(20):21193–21199. doi: 10.1074/jbc.M313180200. [DOI] [PubMed] [Google Scholar]
- 41.Battye TGG, Kontogiannis L, Johnson O, Powell HR, Leslie AGW. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.