Significance
The larval imaginal discs of the fruit fly are capable of fully regenerating mechanically damaged parts. Wound healing is initiated by the JNK signaling pathway. We followed the subsequent formation of the regenerating blastema by transcriptome profiling and identified the JAK/STAT pathway as a central regulatory node controlling local cellular and global physiological responses. This signaling cascade induces, together with the Wingless pathway, proliferation of cells forming the blastema. However, JAK/STAT also up-regulates Drosophila insulin-like peptide 8 (Dilp8), a paracrine factor involved in organismal developmental delay, thereby allowing regenerative recovery.
Keywords: Drosophila imaginal discs, epimorphic regeneration, JAK/STAT signaling
Abstract
Regeneration of fragmented Drosophila imaginal discs occurs in an epimorphic manner involving local cell proliferation at the wound site. After disc fragmentation, cells at the wound site activate a restoration program through wound healing, regenerative cell proliferation, and repatterning of the tissue. However, the interplay of signaling cascades driving these early reprogramming steps is not well-understood. Here, we profiled the transcriptome of regenerating cells in the early phase within 24 h after wounding. We found that JAK/STAT signaling becomes activated at the wound site and promotes regenerative cell proliferation in cooperation with Wingless (Wg) signaling. In addition, we showed that the expression of Drosophila insulin-like peptide 8 (dilp8), which encodes a paracrine peptide to delay the onset of pupariation, is controlled by JAK/STAT signaling in early regenerating discs. Our findings suggest that JAK/STAT signaling plays a pivotal role in coordinating regenerative disc growth with organismal developmental timing.
Epimorphic regeneration, typically represented by limb restoration in urodele amphibians (1) or fin regeneration in teleost fish (2), is an accurate posttraumatic morphogenetic process. This activity involves the formation of a blastema through the dedifferentiation and active proliferation of cells at the wound site. The redevelopment of lost limbs or fins requires considerable gene expression adjustments in the remaining cells to promote regenerative proliferation and cellular reprogramming. Although numbers of genes and signaling pathways in regeneration have been identified, the molecular mechanisms regulating the early phase of regeneration that leads to the blastema formation remain to be fully understood.
The larval imaginal discs of Drosophila melanogaster are fate-committed but undifferentiated epithelial primordia for adult appendages. The discs exhibit remarkable regeneration ability not only in vivo (3) but also, when cultured ex vivo in the adult fly abdomen (4). Of the currently used experimental systems for disc regeneration, regeneration of mechanically fragmented imaginal discs seems to replicate the natural epimorphic regeneration process most faithfully. Wound healing is the first response by which the cells at the wound edge contact and reestablish a continuous epithelium (5). Local cell proliferation at the wound site becomes pronounced as early as 24 h after cultivation and results in the blastema structure (6, 7). Both wound healing and regenerative cell proliferation are dependent on the Jun N-terminal kinase (JNK) signaling pathway (8–10). However, initiation of cell proliferation is observed even without wound closure (11, 12), indicating that wound healing and regenerative cell proliferation are controlled separately.
Several genetic approaches to dissect the early events in disc regeneration have identified prospective genes and signaling pathways, including Wingless (Wg), Myc, or Hippo signaling (7, 13–16). In addition, transcriptome analyses of regenerating disc cells have been conducted to identify genes involved in the regeneration process. Klebes et al. (17) investigated the transcriptomes of regenerating leg discs at 3–5 d after fragmentation and transdetermining blastema cells in wg-induced leg discs. The subsequent study showed that three genes, regeneration, augmenter of liver regeneration, and Matrix metalloproteinase-1 (Mmp1), which were originally described as transdetermining blastema-related genes, are also involved in the formation of the regenerated blastema in fragmented discs (7). Later, Mmp1 was shown to be required for reepithelialization during wound healing (18). Blanco et al. (19) analyzed the transcriptome of regenerating wing discs at 24 and 72 h after fragmentation. The results of Blanco et al. (19) uncovered the involvement of Notch signaling, cabut and absent, small or homeotic discs 2 in wound healing and regenerative cell proliferation.
Although these studies considerably contributed to our knowledge on early disc regeneration, our understanding on how the molecular processes therein are regulated and how regeneration is coordinately organized is, by far, not complete. In this study, we used a tracing method to isolate early regenerating disc blastemas and subjected the cells undergoing reprogramming to a time-resolved transcriptome analysis. We found that unpaired (upd) and its downstream JAK/STAT signaling are specifically activated in regenerating discs. At the wound site, activated JAK/STAT signaling regulates regenerative cell proliferation in cooperation with Wg signaling. Additionally, we found that JAK/STAT signaling also regulates the expression of Drosophila insulin-like peptide 8 (dilp8), a paracrine factor that controls timing of pupariation. Our results identify JAK/STAT signaling as a central node that coordinately controls both the interdisc regenerating process and interorgan processes.
Results
Comparison of Transcriptomes by Lineage-Tracing Regenerating Cells.
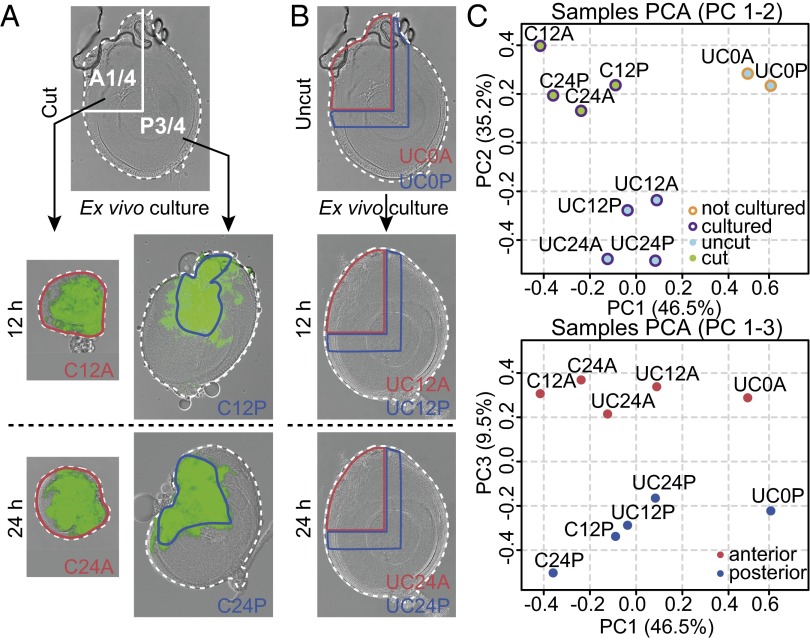
To identify regulatory networks underlying early disc regeneration, we generated transcriptome profiles of regenerating disc cells before blastema formation. When a disc fragment is ex vivo cultured in an adult fly abdomen, resident cells at the wound site activate JNK signaling and launch the regeneration program (8, 9). However, activation of wound-induced JNK signaling is transient. In the regenerating disc fragment, the expression of puckered (puc), which is one of the best characterized JNK signaling target genes encoding a JNK phosphatase (20), was detected by 3 h after ex vivo cultivation and decreased substantially by 12 h (Fig. S1 A–F). Therefore, we adapted a cell lineage-tracing system (21) to identify cells exposed to this initiating signal. We used prothoracic leg discs with a regeneration process that has been extensively studied (22–25). Because disc fragments from puc-GAL4, UAS-flp, Act-FRT-STOP-FRT-GAL4, and UAS-GFP (hereafter termed puc>>GFP) showed continuous expression of GFP in regenerating cells from as early as 6 h after ex vivo cultivation (Fig. S1 G–L), we were able to specifically isolate the regenerating cells at successive time points. For this purpose, discs were dissected into an anterior one-quarter (A1/4) fragment and a posterior three-quarters (P3/4) fragment (Fig. 1A). The early regeneration process of both fragments proceeds in a similar manner up to the formation of the blastema. However, in the subsequent reprogramming and repatterning process of the blastema, the P3/4 fragments often regrow the existing disc pattern (duplication), whereas the A1/4 fragments recreate missing regions (regeneration) (22). Sporadically, both fragments are also capable of acquiring wing fate by a transdetermination event (13). We profiled the transcriptome of GFP-marked cells within the A1/4 and P3/4 fragments at 12 and 24 h after ex vivo cultivation, two time points at which we were clearly able to observe a fluorescent signal under the stereomicroscope. As controls, the corresponding regions of uncut discs that were cultured for the same time period (12 or 24 h) or taken directly from larvae were prepared (Fig. 1B). An L-shaped region along the original rectangular cutting edge corresponding to the GFP-positive region in the regenerating P3/4 fragment was taken for comparison. In total, 30 samples (10 conditions in three biological replicates) were subjected to microarray gene expression profiling. Throughout the text, we refer to these profiles with a label encoding the condition [cut (C) and uncut (UC)], the time point [0 (not cultured), 12 (12 h), and 24 (24 h)], and the disc fragment being analyzed. Hierarchical clustering analysis of gene expression profiles revealed a good agreement among biological replicates for each condition (Fig. S2 A and B). When expression data were subjected to principal component analysis, the first two principal components defined a clear segregation of cut and cultured, uncut and cultured, and uncut and uncultured samples, whereas the third component separated the anterior from the posterior compartment (Fig. 1C). Taken together, these results support the high quality of our microarray data.
Fig. 1.
Preparation of specific disc tissue for microarray analysis and sample clustering. (A) Discs were cut into A1/4 and P3/4 fragments and individually ex vivo cultured for either 12 or 24 h. The GFP-positive regions (surrounded by blue lines) were isolated for posterior samples (C12P or C24P), whereas the entire fragments (surrounded by red lines) were used for anterior samples (C12A or C24A). (B) As indicated by blue or red lines, the corresponding regions in uncut and cultured or uncut and uncultured discs, respectively, were isolated as control samples. (C) Principal component analysis (PCA) plots showing the differences and clustering of microarray samples. The first principal component (PC1; 46.5%) captured the significant variance in expression profiles between cultured and not cultured samples. (Upper) The second principal component (PC2; 35.2%) segregated the regenerating cut and cultured samples from the control uncut and cultured samples. (Lower) The third principal component (PC3; 9.5%) represented the difference between anterior and posterior samples. Plots are colored as indicated.
Clustering Analysis Discriminates Expression Changes Induced by Regeneration and Culturing Conditions.
To identify genes involved in the early regeneration process, we performed differential expression analysis (Materials and Methods) and considered only those genes that were significantly differentially expressed in at least one of four core comparisons (adjusted P value < 0.001) that test for the effect of cutting given a time point and a disc compartment (e.g., C24P–UC24P). Of the resulting 1,425 genes, 726 genes showed up-regulation, and 699 genes were down-regulated (Table 1 and Dataset S1). Expression levels of these 1,425 genes across our core comparisons as well as the four contrasts assessing the influence of cultivation (e.g., UC24P–UC0P) were subjected to k-means clustering to identify the group of genes exhibiting a common response across conditions (Fig. 2). Our analysis indicates that considerable gene expression changes were induced in the uncut discs simply by ex vivo cultivation. Nevertheless, four gene clusters exhibiting different response to cutting were identified (Fig. 2B and Fig. S2C). This analysis allowed additional partitioning of up- and down-regulated genes in core comparisons while taking into account their response to cultivation. Cluster I (151 genes) contained genes up-regulated in both core comparison and cultivation, whereas genes in cluster II (575 genes) were down-regulated by cultivation (Fig. 2 A and B). Furthermore, genes in cluster I are further up-regulated after cultivation (Fig. 2 C and D) as opposed to genes within cluster II, for which gene expression remains nearly constant across time points. These data indicate that differences in gene activity can be explained by a reduced expression in the control uncut and cultured discs.
Table 1.
Differentially expressed genes in cut and cultured discs
| Rank | Genes | Adjusted P value | Fold change* | k-Means clustering | |||
| C12A–UC12A | C24A–UC24A | C12P–UC12P | C24P–UC24P | ||||
| 1 | dilp8 | 7.56E-15 | 8.918 | 9.601 | 3.916 | 15.214 | I |
| 2 | upd | 1.60E-13 | 6.142 | 5.269 | 3.337 | 6.916 | I |
| 3 | Ama | 1.44E-12 | 2.065 | 2.585 | 1.779 | 2.528 | II |
| 4 | Pent | 5.27E-12 | −5.467 | −1.388 | −2.662 | −3.162 | III |
| 5 | Cyt-c-p | 5.57E-12 | 1.631 | 2.576 | 1.837 | 2.248 | II |
| 6 | dpr17 | 4.20E-11 | 2.197 | 2.682 | 1.677 | 1.933 | II |
| 7 | PGRP-SA | 1.07E-10 | 4.289 | 1.569 | 1.340 | 6.609 | I |
| 8 | CG17219 | 2.44E-10 | 2.012 | 2.481 | 2.101 | 2.275 | II |
| 9 | bip1 | 3.74E-10 | −2.623 | −1.261 | −1.508 | −2.099 | III |
| 10 | CG7054 | 1.01E-09 | 2.137 | 1.682 | 1.664 | 2.161 | II |
| 11 | bt | 1.02E-09 | 5.456 | 1.913 | 3.501 | 1.381 | I |
| 12 | RpS9 | 1.34E-09 | −1.974 | −1.756 | −2.758 | −1.473 | IV |
| 13 | CG17124 | 1.93E-09 | 5.851 | 2.095 | 2.013 | 5.828 | I |
| 14 | CG18641 | 1.99E-09 | 2.678 | 1.194 | 1.470 | 1.847 | II |
| 15 | br | 1.99E-09 | −2.507 | 1.058 | −1.209 | −1.697 | III |
| 16 | hoip | 2.83E-09 | 1.449 | 2.054 | 1.990 | 1.654 | II |
| 17 | CG10126 | 2.83E-09 | 2.258 | 1.535 | 1.156 | 2.236 | I |
| 18 | CG9689 | 3.34E-09 | 2.118 | 1.853 | 1.349 | 2.018 | II |
| 19 | CG11843 | 3.46E-09 | 3.029 | 1.519 | 1.636 | 2.352 | II |
| 20 | CoVIII | 3.46E-09 | 1.219 | 1.998 | 1.594 | 1.691 | II |
Top 20 genes ranked by the adjusted P values are shown.
Negative values indicate the fold down-regulation.
Fig. 2.
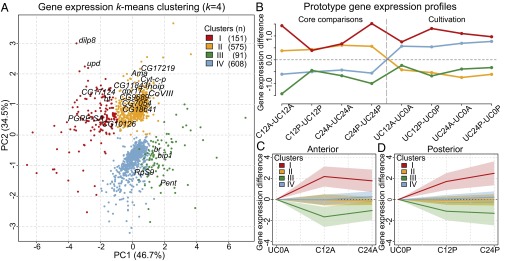
Clustering of differentially expressed genes by k-means clustering. (A) Principal component analysis plots and k-means clustering (k = 4) of 1,425 differentially expressed genes. The 20 genes from Table 1 are indicated. (B) The four clusters were created by taking into account the expression changes in both core comparisons and cultivation. (C and D) Gene expression differences between cut and cultured samples and uncut and uncultured samples are presented. The average value and variance of each cluster are shown by corresponding colored lines and pale-colored zones. PC, principal component.
Down-regulated genes in core comparisons were also partitioned in clusters III (91 genes) and IV (608 genes). The former was down-regulated in regenerating discs, whereas gene expression of the latter was mostly unchanged in regenerating cells but increased on ex vivo cultivation (Fig. 2 B–D). Therefore, our clustering approach discriminated expression changes induced by regeneration from those mediated by culturing. Genes in clusters I and III were considered to be representing the up- and down-regulated genes, respectively, in regenerating cells in cut and cultured discs. In contrast, clusters II and IV mostly reflected changes in uncut and cultured discs. Next, we assessed the biological processes represented by genes within each cluster by performing a gene ontology (GO) analysis (Table S1). The GO terms enriched in clusters I and III indicate the activation of the immune system (cluster I) and intriguingly, suppression of imaginal disc development (cluster III) in the regenerating cells of cut and cultured discs. In contrast, GO terms enriched in clusters II and IV are primarily associated with energy balancing of the uncut and cultured disc cells.
upd And dilp8 Are Up-Regulated in Early Regenerating Discs.
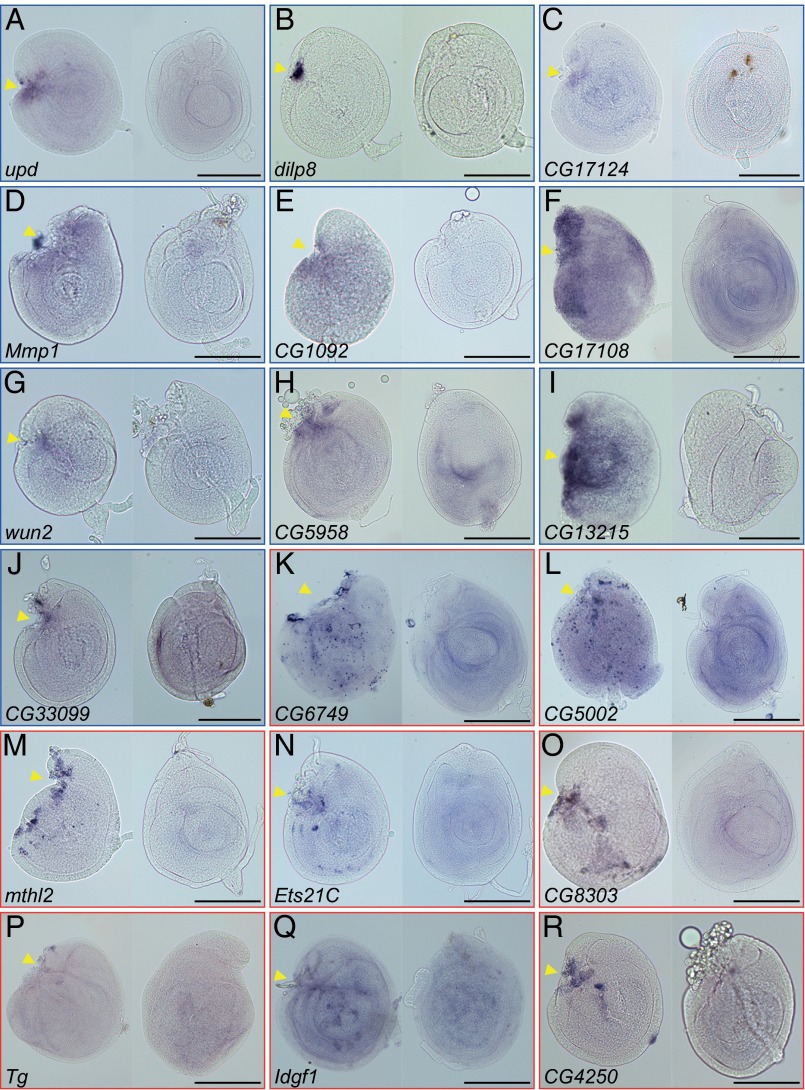
Next, we focused on the genes in cluster I and validated the up-regulation of candidate genes by in situ hybridization (Fig. 3). Cluster I contains not only many known immune-responsive genes but also, genes that were enriched in hemocytes or increased on immune challenge (26–31). Consistent with these results, the in situ staining of these transcripts often showed an irregular scattered pattern, indicating their expression in the attached hemocytes rather than the regenerating disc cells (Fig. 3 K–R). However, we recently showed that damaged discs still successfully promote regenerative cell proliferation to form blastemas under immune-deficient conditions (32). Therefore, we considered the up-regulated immune response genes in our candidate list to be unrelated to the actual process of disc regeneration.
Fig. 3.
In situ hybridization of differentially expressed transcripts in regenerating discs. (Left) The regenerating C24P fragment and (Right) the uncut and cultured disc (UC24) were hybridized with digoxigenin (DIG)-labeled RNA probes against the genes of cluster I: (A) upd, (B) dilp8, (C) CG17124, (D) Mmp1, (E) CG1092, (F) CG17108, (G) wun2, (H) CG5958, (I) CG13215, (J) CG33099, (K) CG6749, (L) CG5002, (M) mthl2, (N) Ets21C, (O) CG8303, (P) Tg, (Q) Idgf1, and (R) CG4250. The (A–J) blue or (K–R) red rectangular frame indicates the expression of mRNA signals in disc cells or attached hemocytes, respectively. In all images, anterior is to the left, and dorsal is up. Arrowheads indicate the fragmented positions. (Scale bar: 100 μm.)
We found that two genes, dilp8 and upd, were highly elevated in all four core comparisons and ranked at the top-most position not only in cluster I but also, among 1,425 differentially expressed genes (Table 1). Interestingly, recent studies revealed that both Dilp8 and Upd were involved in interorgan communication from damaged imaginal discs to other tissues or organs; Dilp8 retards the onset of pupariation (33, 34), whereas Upd act as a cytokine, activating the proliferation of circulating immune blood cells (35). In addition, the cluster I genes prominently exposed the known regeneration or wound healing-related genes: Mmp1, which is required for basement membrane disassembly (7, 18), and Dopa decarboxylase, which is involved in the epidermal wound-healing process in embryos (36). These results indicate that our assay both confirmed known regeneration factors and uncovered components previously unrelated to the regenerative process.
Upd Activates JAK/STAT Signaling in Early Regenerating Discs to Promote Regenerative Cell Proliferation.
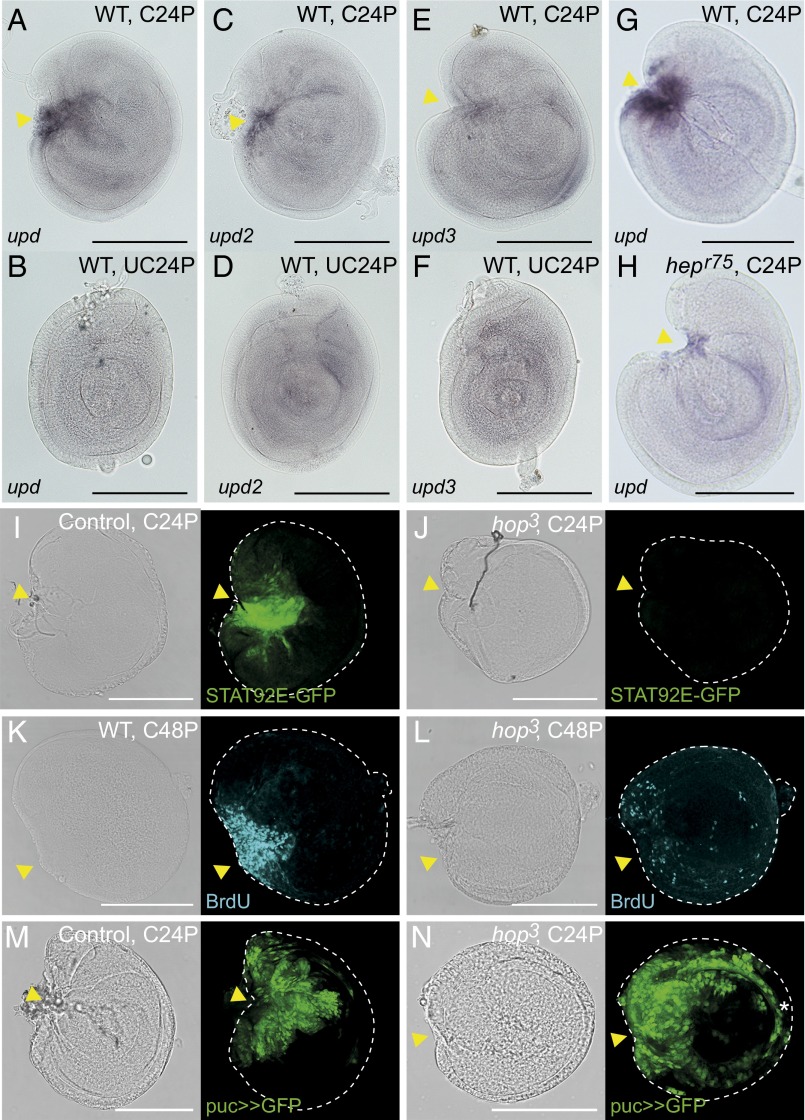
The Drosophila upd gene encodes a cytokine-like ligand for the JAK/STAT signaling pathway. There are three Upd family genes (upd, upd2, and upd3). JNK signaling-dependent activation of Upd cytokines from damaged disc cells was shown previously, and the paracrine actions of secreted Upds on hemocyte proliferation were shown (35). However, the effect of the Upds on regenerative cell proliferation in the damaged discs has not been addressed so far. In our analysis, among three Upd family genes, only upd was significantly up-regulated in regenerating cells, whereas only a moderate up-regulation of upd2 and upd3 in regenerating disc samples was observed (Fig. S3). In agreement with these results, locally elevated expression of upd at the wound site of regenerating discs was detected by in situ hybridization (Figs. 3A and 4 A and G). Conversely, upd2 and upd3 were specifically but only weakly detected at wound sites. (Fig. 4 C–F). Moreover, we noticed that upd expression disappeared by around 72 h after wounding (Fig. S4), suggesting a specific role in the early regeneration phase. Consistent with a previous observation using upd reporters (35), a hypomorphic allele of JNK-kinase Hemipterous (hepr75) diminished the ectopic up-regulation of upd at the wound site (Fig. 4H compared with Fig. 4G), thus confirming that upd expression in regenerating disc is downstream of JNK signaling.
Fig. 4.
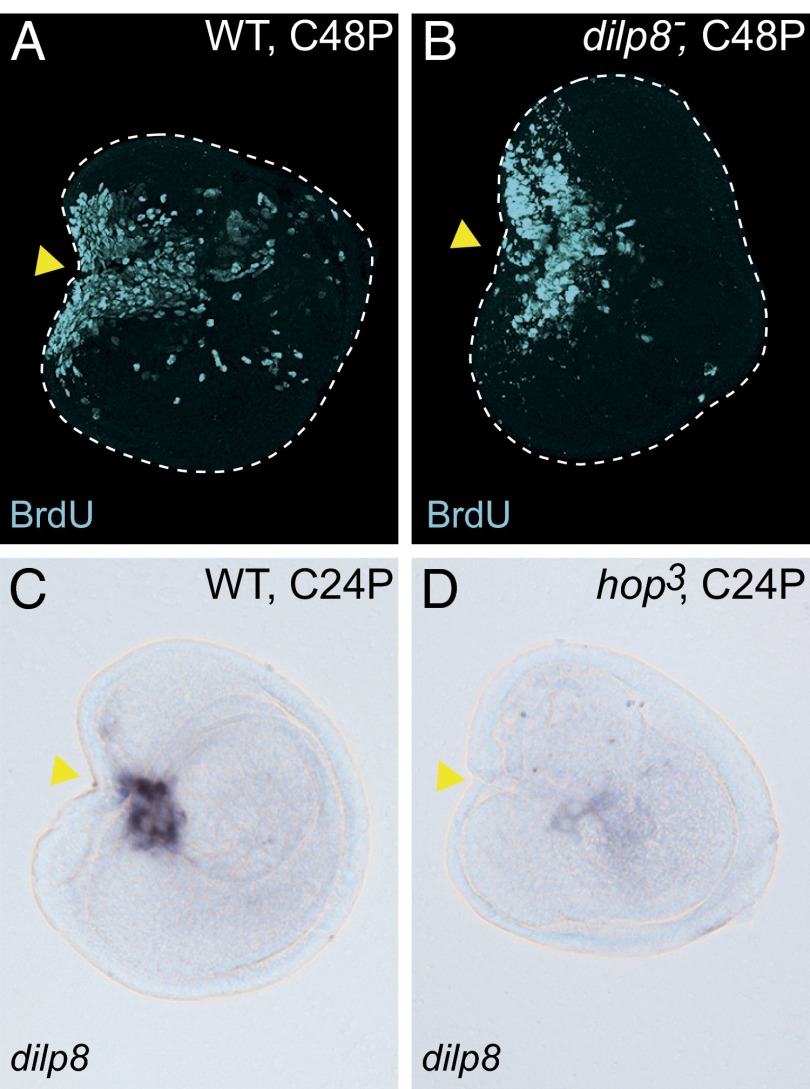
Defects of regenerative growth in hop3 JAK/STAT signaling mutant disc fragments. (A–F) The wound site-specific induction of upd, upd2, and upd3 in C24P and UC24P disc samples. (G and H) JNK signaling-dependent upd expression at the wound site. The discs of hepr75 males (hemizygote) were used for H. The in situ hybridizations for A–F and G and H were performed in the same experiment. (I) The 10xSTAT92E-GFP reporter was activated at the wound site of the WT control C24P fragment. (J) The 10xSTAT92E-GFP in the C24P fragment of the hop3 hemizygote male. (K and L) The cell proliferations of (K) regenerating WT or (L) hop3 C48P fragments were monitored by BrdU incorporation. (M and N) JNK activity (puc>>GFP cell lineage) in (M) regenerating control or (N) hop3 C24P fragments. In all images, anterior is to the left, and dorsal is up. Yellow arrowheads indicate the fragmented positions. *Abnormal activity of JNK signaling in hop3 discs (Fig. S5C). (Scale bar: 100 μm.)
We next examined whether the up-regulation of upd activates JAK/STAT signaling in regenerating disc cells using a 10xSTAT92E-GFP reporter (37). Discs, in which the GFP expression is under the control of STAT-binding enhancer, were fragmented and cultivated in a non-GFP host fly abdomen; 24 h after cultivation, a GFP signal was observed at the wound site (Fig. 4I). Notice that the cells located by the closed cutting edge (indicated by an arrowhead in Fig. 4I) have a stronger GFP signal than those in distant regions, which might reflect a graded-like activity of Upd over the wound site.
To determine the function of JAK/STAT signaling in early regenerating cells, we investigated the restoration capacities of disc mutants in this pathway. In Drosophila, the JAK protein is encoded by hopscotch (hop). Among the several hop mutant alleles that we tested, we found that the hop3 allele, which contains a 4-nt deletion resulting in a frameshift at the C-terminal end of the kinase domain (38), develops until early pupal stages. The leg discs grew to almost normal size in hop3 third instar larvae, but the morphologies were not exactly the same as those of the WT (Fig. S5 A–D). We first tested whether the mutation has an impact on the JAK/STAT signaling cascade. The activation of 10xSTAT92E-GFP in the fragmented discs was virtually blocked in the mutant background (Fig. 4J). We noticed, however, that the rectangular cutting edge of the P3/4 disc fragment was normally closing at 24 h after cultivation (arrowhead in Fig. 4J), suggesting that JAK/STAT signaling is not required for the wound-healing process. Next, we examined the proliferative capacity of the hop3 mutant cells in disc regeneration. Because the active cell proliferation at the wound site begins at 24 h after fragmentation/cultivation (Fig. S1 K″ and L″), the proliferative cells that entered S phase in the subsequent 24-h window (i.e., 24–48 h after cultivation) were labeled by BrdU incorporation. Whereas the WT disc fragments showed massive cell proliferation at the wound site (Fig. 4K), the hop3 disc fragments exhibited a substantial impairment in cell proliferation (Fig. 4L). Furthermore, the impaired JAK/STAT signaling in the hop3 mutation does not influence the activity of JNK signaling (puc>>GFP) in the disc regeneration process (Fig. 4 M and N and Fig. S5 D and E). Taken together, these results suggest that JAK/STAT signaling plays a role in regenerative cell proliferation downstream of JNK signaling.
To exclude the possibility that the observed defect in regenerative growth was caused by the underlying imperfect development of hop3 discs, we examined the regenerative cell proliferation using discs expressing a dominant negative form of the JAK/STAT signaling receptor Domeless (Dome∆CYT). Dome∆CYT and GFP were coexpressed in the en-GAL4–positive posterior domains. Correspondingly, a nick appeared in the posterior compartment of leg discs (Fig. S6A). The expression was temporally controlled with a temperature-sensitive GAL80 (GAL80ts) to not disturb the disc development (Fig. S6B). In the control disc, where only GFP was expressed, the cell proliferation for regeneration was locally increased next to the wound site in the posterior compartment (Fig. S6 C, C′, and C″). In contrast, we did not observe the BrdU incorporation in the Dome∆CYT-expressing posterior compartment cells, whereas cell proliferation was detected in the anterior compartment (Fig. S6 D, D′, and D″), showing a clear depletion of the proliferative capacity in the Dome∆CYT-expressing cells. These results strongly suggest that the JAK/STAT signaling is required for the regenerative cell proliferation to form the regenerated blastema.
Interrelation Between JAK/STAT Signaling and Wg Signaling Pathways for Blastema Formation.
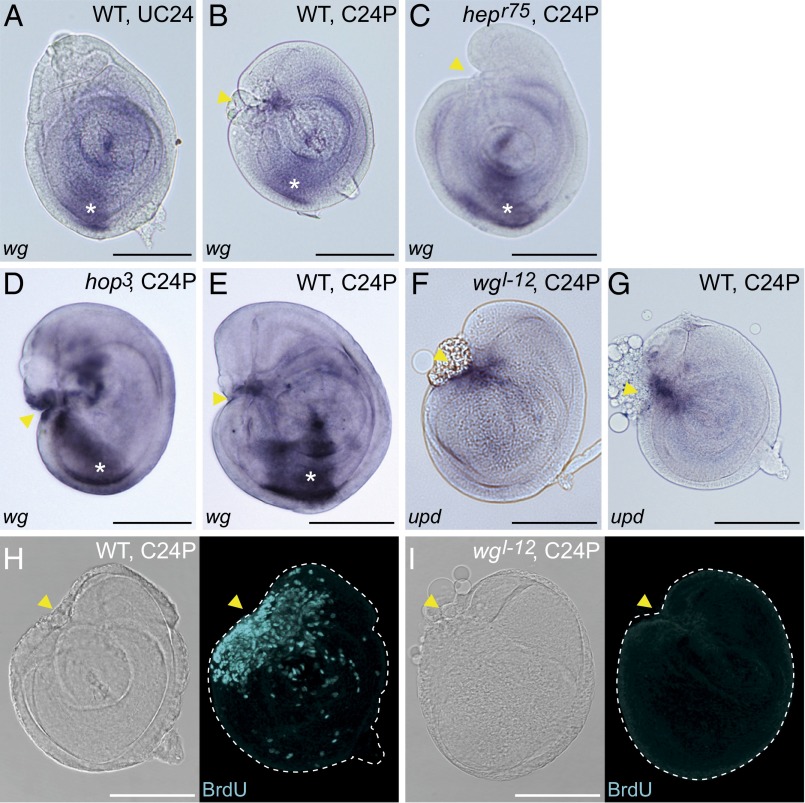
Wg is ectopically expressed at the wound site of fragmented discs before blastema formation (7). Although our microarray analysis did not detect a significant expression change of wg, we confirmed an ectopic wg expression at wound sites by in situ hybridization (Fig. 5 A and B). Wound site-associated wg was not detected after regeneration of hepr75 discs, whereas the developmental expression in the ventral region was still visible (Fig. 5C), suggesting that the activation of wg for the early regeneration process is downstream of JNK signaling.
Fig. 5.
Cooperative functions of JAK/STAT and Wg signaling pathways for blastema proliferation. (A–E) In situ hybridization with a wg probe. (A) wg Expression in WT UC24 leg discs. (B and C) wg Was ectopically up-regulated at the wound site in the WT C24P fragment (arrowhead in B) but not in the hepr75 C24P fragment (arrowhead in C). (D and E) Ectopic wg was still detected in the hop3 C24P fragment (arrowhead in D). E is the WT C24P staining control for D. *Developmental wg expression in the anterior ventral sector. (F and G) Wound site upd expression was detected in the wgl-12 C24P fragment. G is the WT C24P staining control for F. Both were cultured at 29 °C. (H and I) The C48P of either (H) WT or (I) wgl-12 disc was stained with anti-BrdU antibody. BrdU signal was not detected in regenerating wgl-12 discs. In all images, anterior is to the left, and dorsal is up. Yellow arrowheads indicate the fragmented positions. (Scale bar: 100 μm.)
Given the functional similarities between the JAK/STAT signaling in regenerative cell proliferation and that reported for Wg signaling, we investigated a potential epistatic relationship. To this end, we analyzed wg expression in fragmented hop3 mutant discs. The ectopic wg expression at the wound site in the hop3 disc was comparable with that of WT discs (Fig. 5 D and E), indicating that wg expression is independent from JAK/STAT in this process. To examine the wound site upd expression in the Wg signaling mutant discs, we used the wgl-12 mutant, which is characterized as a heat-sensitive amorphic allele (39). When the fragmented wgl-12 discs were cultured at restrictive temperatures, upd up-regulation was similar to control (Fig. 5 F and G), suggesting that the upd expression is not regulated by Wg signaling. Hence, we did not observe an epistatic relation between the two pathways. Interestingly, however, the regenerative proliferation was impaired in wgl-12 disc fragments (Fig. 5 H and I). Taken together, these results suggest that the JAK/STAT signaling and the Wg signaling are individually activated by JNK signaling in the early regeneration phase. Cooperative functions of both signaling pathways are required for the regenerative cell proliferation.
JAK/STAT Signaling Regulates dilp8 Expression in Damaged Discs.
The role of Dilp8 in retarding the onset of pupation in response to abnormalities or damage to tissues is of vital importance for fly development. We first tested if Dilp8 functions as a growth factor in disc regeneration. However, the fragmented discs of dilp8MI00727 homozygote, a strong loss-of-function allele of dilp8 (34), did not impair regenerative cell proliferation (Fig. 6 A and B), suggesting that Dilp8 does not have a direct role in blastema formation. Interestingly, the concomitant up-regulation of dilp8 and upd was observed in not only our study (Table 1) but also, previously reported transcriptome datasets from discs with growth abnormalities: the proliferative and transdetermining cells of wg-induced leg discs (17) and the genetically impaired wing discs that eventually become neoplastic (33). Because Upd-mediated JAK/STAT signaling promoted disc cell proliferation in regeneration, we asked if dilp8 expression requires JAK/STAT signaling activity. Indeed, dilp8 up-regulation was abolished in a cultivated hop3 disc fragment (Fig. 6 C and D). Therefore, JAK/STAT plays a central role not only in controlling local cell proliferation but also, by staging a physiological response, which allows the regenerative process to be accomplished by delaying development.
Fig. 6.
JAK/STAT signaling is required for dilp8 expression in wounded discs. (A and B) Anti-BrdU staining in C48P fragments of (A) WT or (B) dilp8MI00727 homozygote. (C and D) In situ hybridization with dilp8 probe. (C) The up-regulation of dilp8 in WT C24P. (D) The dilp8 up-regulation was not observed in the hop3 C24P fragment. In all images, anterior is to the left, and dorsal is up. Yellow arrowheads indicate the fragmented positions. (Scale bar: 100 μm.)
Discussion
Transcriptome Changes in the Early Phase of Disc Regeneration.
In this study, we focused on the early phase of disc regeneration and performed a time-resolved transcriptome analysis to gain a better understanding of the underlying molecular mechanisms. We used classical disc fragmentation and subsequent ex vivo culture through transplantation, which allows a precise control of tissue excision and focus on the reprogramming processes in the remaining cells. To not miss modest but significant gene expression changes in regenerating cells, we further isolated the local area involved in the regeneration process. This accurate sampling procedure made it possible to produce highly pure samples and a detailed comparison of gene expression changes in the early regenerating process.
Several previous studies published transcriptome data from regenerating discs (17, 19, 33, 34). However, comparing clusters I and III with these datasets, we found that rather few genes were shared with these studies. Differences in disc identities and regeneration timing between individual comparisons might explain this limited overlap. However, the observed dissimilarities could also be caused by the relatively heterogeneous samples of regenerating disc tissues used in previous studies. Because only part of the disc is regenerating in the neighborhood of normal tissue, it is important to restrict gene expression profiling to those cells that are directly involved in this process. Interestingly, our measured gene expression changes in early regenerating cells showed some commonalities with seemingly unrelated datasets. Indeed, up-regulation of dilp8 and upd was also observed in the transdetermining blastema of wg-induced leg discs (17) and the genetically perturbed wing discs that eventually exhibit tumorous growth (33). Surprisingly, we found considerable overlap with the dataset by Colombani et al. (33). Particularly, a strong overlap was observed with expression profiles from a specific developmental time point [116 h after egg laying (AEL)] when the cells of the impaired region activates JNK signaling without yet becoming neoplastic (Fig. S7). This result suggests that rather similar gene regulatory networks are activated in both early regenerating and early tumorigenic cells.
The sensitivity of detection of differential gene expression is, however, intrinsically limited by the disc fractionation procedure. Expression levels of the genes that originally exhibit region-specific expression patterns in the leg discs tended to have a higher expression variance across samples than more broadly expressed or not expressed genes. Although we carefully partitioned the disc regions for control samples, a slight deviation influences the expression values of these genes. For instance, in our microarray analysis, we could not detect the expression change of wg, which developmentally expresses in the anterior ventral sector (Fig. 5A), whereas its wound site-specific expression was confirmed by in situ hybridization. ANOVA of wg expression within replicates for distinct biological conditions indicated that, in A1/4 disc samples, which ideally, do not overlay with the endogenous wg domain, wg expression was highly variable (Fig. S8). In addition, the region of P3/4 samples is, indeed, overlaid with developmental wg expression; therefore, we assume that the expression of wound-induced wg would be masked in our microarray analysis. A recent study showed that the genes associated with pattern formation are temporally disrupted in damaged discs before initiating regenerative proliferation (40). In our data, cluster III comprised a significant number of developmental genes that are involved in the regional specification of leg disc development. However, the observed down-regulation of patterning genes in our microarray analysis needs to be further validated by histochemical experiments.
Among differentially expressed genes in the four core comparisons, the genes in cluster II or IV were significantly down- or up-regulated in uncut and cultured disc cells, respectively, but actually, not changed in regenerating cells; hence, they were apparently up- or down-regulated in the core comparisons, respectively. The GO terms enriched in clusters II and IV indicated that ex vivo cultivation influences the energy balance in the uncut and cultured disc cells (Table S1). For example, the cluster II genes indicated a reduction of both ATP synthesis (oxidative phosphorylation) and ATP-consuming process (ribosomal biogenesis). In contrast, the GO term associated with cluster IV genes suggested that the energy reserve metabolic processes, including glycogen catabolism, become activated. Notice that clusters II and IV genes are a part of the genes influenced by cultivation. Actually, considerable gene expression changes were induced solely by the ex vivo cultivation conditions (e.g., UC12A–UC0A; the gene list from four cultivation comparisons has been deposited in the Gene Expression Omnibus database with the accession no. GSE54868). These results suggest that the transplanted disc cells require a fine-tuned balance between energy production and consumption in the exogenous environment of the host fly abdomen. In addition, hormonal conditions in the female abdomen could contribute to the observed gene expression changes.
Roles for the JAK/STAT Signaling in Early Regenerating Discs.
Drosophila studies on JAK/STAT signaling unveiled a variety of conserved roles in a wide spectrum of biological processes, including immune response, cell proliferation, and stem cell maintenance (reviewed in ref. 41). Recently, the Upd cytokine families were shown to be secreted from damaged tissue to activate the downstream JAK/STAT signaling in the immune-responsive cells, such as hemocytes and fat body cells, suggesting a role in the recruitment of immune cells (35). In this study, we reveal an additional function of the JAK/STAT signaling in the actual process of disc regeneration. Using disc mutants in JAK/STAT components, we show that this signaling cascade is required for local cell proliferation for blastema formation but not for the healing of the cut edge (Fig. 4 and Fig. S6). The involvement of JAK/STAT signaling has been also reported in cricket leg regeneration (42), suggesting a conserved role in regeneration processes of holometabolous and hemimetabolous insects.
A requirement of different signaling pathways, including Wg, Myc, and Hippo signaling, in the disc regeneration has been implicated (7, 13–16). Here, we showed that the JAK/STAT signaling regulates the regenerative cell proliferation cooperatively with Wg signaling. Our results suggest that the activities of both signaling pathways are required for the initiation of regenerative cell proliferation. This early cooperative role may also account for an old observation in regenerating leg P3/4 disc fragments; the cells along the horizontal cut edge become more proliferative than those along the vertical edge (Fig. S1L″) (6). Other than the ectopically up-regulated Wg at the wound site (7), the cells at the horizontal cut edge may receive the endogenous developmental Wg from the anterior–ventral sector. Therefore, we speculate that the horizontal cut edge cells could exhibit active cell proliferation compared with those along the ventral cut edge.
Although we have not addressed the interrelation of JAK/STAT with Myc or Hippo signaling, a previous study using clonal analyses in developing imaginal discs suggested an independence of the JAK/STAT-mediated growth regulation with not only Wg signaling but also, Myc or Hippo signaling pathways (43). Indeed, although both Myc and Yorkie (the Hippo signaling effector) regulate Cyclin E (CycE) expression (44, 45), JAK/STAT signaling leads to active CycD (46). Therefore, our results suggest that the regenerative cell proliferation requires the cooperative parallel inputs for cell cycle activation by several signaling pathways.
Smith-Bolton et al. (14) noted in a previous study that the coexpression of upd or CycD in genetically ablated (eiger-expressing) cells does not show a positive effect on recovery of wing size, unlike wg or Myc coexpression. This seemingly contradictory result could be explained by the difference between classical fragmentation/ex vivo cultivation and recently established in situ ablation methods observing compensatory proliferation induced by apoptotic cell loss. In the former, a disc loses a part instantly, and the remaining cells at the wound site have to launch the regeneration programs without large-scale apoptosis occurring. In the latter, in contrast, it takes time (40 h in the system described in ref. 14) to severely damage parts of a disc region (wing pouch). This system creates a condition of dying/undead cells flanked by apoptosis-induced cell proliferation of normal cells. Although JNK is activated in apoptotic as well as adjacent surviving cells, it has been shown that the identification of upstream and downstream signaling pathways is much dependent on the apoptosis-inducing system used (47). Hence, the complex tissue heterogeneity created by local cell death seems to obscure a straightforward dissection of the required signaling cascades during normal regeneration. Alternatively, ablation induced by local apoptosis might also trigger other signaling cascades to induce compensatory cell proliferation than the mechanical disruption. Therefore, we do not consider the previous report contradictory evidence but believe the observed role of JAK/STAT to be fundamental for the regenerative process, certainly after mechanical tissue damage.
Indeed, the aberrant activation of JAK/STAT signaling has also been implicated in a variety of malignant tumors in Drosophila as well as in mammals (48–50). In imaginal discs, the tumorous overgrowths by mutations of Polycomb group (PcG) genes or inter- or intracellular cooperation of oncogenic RasV12 and scrib− mutations resulted in abnormal activation of JAK/STAT signaling (51, 52). The study with the latter tumor model further showed that JNK signaling contributes to activate JAK/STAT signaling for developing RasV12 and scrib– tumors (52). Hence, strict control of JAK/STAT signaling is essential not only in development but also, for a correct regeneration process. Indeed, upd up-regulation in the regenerating fragmented discs is observed only in the early phase and fades by around 72 h after wounding (Fig. S4). This result suggests that the transcriptional activation of upd is temporally tightly controlled. Therefore, we speculate that the transcription of upd in early regenerating disc cells may not be maintained by PcG/trithorax group memory mechanisms but rather, specifically controlled by the transient activity of JNK signaling at the wound site. Consistent with this idea, the wound site upd expression was neither enhanced nor prolonged by reducing the genomic dose of the Pc gene (Fig. S4). However, a previous study showed that ectopic Wg expression is still strongly detected at least until 72 h after fragmentation/cultivation (7). However, it remains unclear if Wg is continuously regulating cell proliferation or whether it is involved in other processes, such as the respecification of blastema cells. It was shown that the ubiquitous wg expression in leg discs could confer the regeneration ability to the disc fragments that are otherwise unable to restore the lost part (13), arguing for the latter explanation.
The coordination of tissue growth and developmental timing is important for not only normal development but also, regenerative processes. Here, we found that JAK/STAT signaling activity is required for the expression of dilp8 (Fig. 6). This factor was recently identified as an important messenger for interorgan communication; it is secreted from the damaged imaginal discs and delays the onset of pupariation for recovery (33, 34). The Dilp8 induction was observed at the wound site of transplanted discs, indicating that the disc cells still respond to the damage as if they are in a normal physiological environment, although Dilp8 unlikely serves a function in the abdomen of the host fly. One of the previous studies showed that dilp8 induction in the damaged discs is dependent on JNK signaling (33). Here, we identified that the JAK/STAT signaling, which is activated by JNK signaling, is required for controlling dilp8 expression. Because JAK/STAT regulates regenerative cell proliferation, we propose that this signaling plays a role in the coordination between disc regeneration and developmental arrest for recovery. The expression of dilp8 is also observed in normal development; the transcription levels peak from the late second until the early third larval instar and decrease by the midthird instar stage, which is seemingly in preparation for the onset of pupariation (33). Intriguingly, the period with high dilp8 levels coincides with the detection of a high and broad JAK/STAT signaling activity in imaginal discs (37). Specific roles of the JAK/STAT pathway in growth control have been well-described in Drosophila (49). Therefore, these results suggest that the JAK/STAT signaling may also play a role in the coordination of developmental disc growth with the Dilp8-mediated control of the larval to pupal transition.
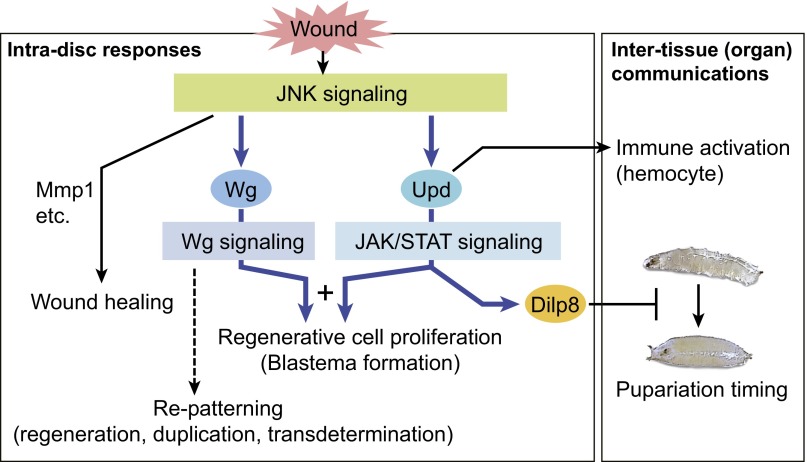
Taken together, we propose a model of signaling regulatory network and summarize the functions of Upd-JAK/STAT signaling in early disc regeneration (Fig. 7). On fragmentation, mechanical signals or other upstream signals that remain unknown activate JNK and launch the regeneration program at the wound site. For regenerative cell proliferation, the wound signal is transmitted from JNK signaling to at least two downstream signaling pathways: JAK/STAT signaling and Wg signaling through the transcriptional controls of the ligand coding genes upd and wg, respectively. A cooperative input from both JAK/STAT and Wg signaling is required to induce the local disc cells to reenter the cell cycle. Activated JAK/STAT also induces the paracrine factor Dilp8 to prolong the larval period for recovery (33, 34). In conclusion, by analyzing the early steps of disc regeneration, we identified the pivotal roles of JAK/STAT signaling in coordinating local tissue proliferation with the requirements of physiological recovery at the organismal level.
Fig. 7.
A schematic model for the roles of the JAK/STAT signaling in early disc regeneration. On wounding, upd, a ligand-coding gene for JAK/STAT signaling, is up-regulated. In a damaged imaginal disc, the activated JAK/STAT signaling has two functions. First, the JAK/STAT signaling acts for promoting regenerative cell proliferation. In this process, the JAK/STAT cooperates with the Wg signaling, which is independently activated through JNK signaling. Second, JAK/STAT signaling also induces paracrine factor Dilp8 to control developmental timing. Upd is also secreted to promote proliferation of circulating hemocytes (35). Thus, the JAK/STAT signaling has essential roles in both intradisc responses and intertissue (organ) communications during early regeneration processes. Blue arrows indicate the signaling regulatory networks revealed in this study.
Materials and Methods
Transplantation of Fragmented Imaginal Discs.
Transplantation of disc fragments was performed essentially as previously described (53). Male larvae at 96–100 h AEL were used as disc donors. The prothoracic leg discs were dissected to either A1/4 or P3/4 fragments and subsequently cultured in female adult fly abdomens.
Microarray Sample Preparation.
Ten regenerating or control discs were used for each sample preparation, which was carried out in three biological replicates as follows. (i) To obtain stage-synchronized cohorts of larvae, embryos were collected in 1 h after two cycles of 1-h precollections and incubated for exactly 100 h at 25 °C. (ii) To minimize the time variance among each sample preparation, disc fragmentation and transplantation were performed within 1 h. Usually, 30 disc fragments were transplanted for each replicate. Because different transplantation gauges were required for A1/4 and P3/4 fragments, the preparation of these samples was carried out in separate experiments. (iii) Recovery of GFP-labeled regions (or corresponding regions) was performed within 1 h. After cultivation for 12 or 24 h, transplanted discs were recovered, and 10 of 30 regenerating discs were selected. GFP-labeled regions were selected under a fluorescent stereomicroscope using a fine tungsten needle, and RNA was immediately extracted using the PicoPure RNA Isolation Kit (ARCTRUS/Life Technologies). Purified total RNAs from each sample were subjected to two rounds of amplification (GeneChip Two-Cycle cDNA Synthesis Kit and GeneChip IVT Labeling Kit; Affymetrix) and hybridized to GeneChip Drosophila Genome 2.0 Arrays (Affymetrix).
The microarray data produced in this study have been deposited in the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo/) with the accession number GSE54868.
The fly stocks, analysis of microarray data, and histochemistry are in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank E. Martin-Blanco for providing pucE69I-GAL4, J. Cast and G. Hombria for UAS-domeΔCYT, and the Bloomington Drosophila Stock Center at Indiana University and the Drosophila Genomic Resource Center for other fly stocks. We also thank the R.P. laboratory members for discussion and helpful comments on the manuscript. F.C. is a member of the Life Science Zürich Graduate School (PhD Program in Systems Biology). The work of T.K. and R.P. was supported by Deutsche Forschungsgemeinschaft Grant SPP1356 (“Pluripotency and Cellular Reprogramming”). Work in the R.P. laboratory is supported by Eidgenössische Technische Hochschule Zürich.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray data reported in this paper has been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE54868).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1423074112/-/DCSupplemental.
References
- 1.Nye HL, Cameron JA, Chernoff EA, Stocum DL. Regeneration of the urodele limb: A review. Dev Dyn. 2003;226(2):280–294. doi: 10.1002/dvdy.10236. [DOI] [PubMed] [Google Scholar]
- 2.Nakatani Y, Kawakami A, Kudo A. Cellular and molecular processes of regeneration, with special emphasis on fish fins. Dev Growth Differ. 2007;49(2):145–154. doi: 10.1111/j.1440-169X.2007.00917.x. [DOI] [PubMed] [Google Scholar]
- 3.Bryant PJ. Regeneration and duplication following operations in situ on the imaginal discs of Drosophila melanogaster. Dev Biol. 1971;26(4):637–651. doi: 10.1016/0012-1606(71)90146-1. [DOI] [PubMed] [Google Scholar]
- 4.Hadorn E, Hürlimann R, Mindek G, Schubiger G, Staub M. Developmental capacity of embryonal blastema in Drosophila following cultivation in an adult host. Rev Suisse Zool. 1968;75(3):557–569. [PubMed] [Google Scholar]
- 5.Reinhardt CA, Hodgkin NM, Bryant PJ. Wound healing in the imaginal discs of Drosophila. I. Scanning electron microscopy of normal and healing wing discs. Dev Biol. 1977;60(1):238–257. doi: 10.1016/0012-1606(77)90122-1. [DOI] [PubMed] [Google Scholar]
- 6.Schubiger G, Karpen GH. Blastema formation in regulating imaginal disc fragments of Drosophila melanogaster. Prog Clin Biol Res. 1983;110(Pt A):609–618. [PubMed] [Google Scholar]
- 7.McClure KD, Sustar A, Schubiger G. Three genes control the timing, the site and the size of blastema formation in Drosophila. Dev Biol. 2008;319(1):68–77. doi: 10.1016/j.ydbio.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bosch M, Serras F, Martín-Blanco E, Baguñà J. JNK signaling pathway required for wound healing in regenerating Drosophila wing imaginal discs. Dev Biol. 2005;280(1):73–86. doi: 10.1016/j.ydbio.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 9.Mattila J, Omelyanchuk L, Kyttälä S, Turunen H, Nokkala S. Role of Jun N-terminal Kinase (JNK) signaling in the wound healing and regeneration of a Drosophila melanogaster wing imaginal disc. Int J Dev Biol. 2005;49(4):391–399. doi: 10.1387/ijdb.052006jm. [DOI] [PubMed] [Google Scholar]
- 10.Bosch M, Baguñà J, Serras F. Origin and proliferation of blastema cells during regeneration of Drosophila wing imaginal discs. Int J Dev Biol. 2008;52(8):1043–1050. doi: 10.1387/ijdb.082608mb. [DOI] [PubMed] [Google Scholar]
- 11.Kiehle CP, Schubiger G. Cell proliferation changes during pattern regulation in imaginal leg discs of Drosophila melanogaster. Dev Biol. 1985;109(2):336–346. doi: 10.1016/0012-1606(85)90460-9. [DOI] [PubMed] [Google Scholar]
- 12.Dale L, Bownes M. Pattern regulation in fragments of Drosophila wing discs which show variable wound healing. J Embryol Exp Morphol. 1985;85:95–109. [PubMed] [Google Scholar]
- 13.Schubiger M, Sustar A, Schubiger G. Regeneration and transdetermination: The role of wingless and its regulation. Dev Biol. 2010;347(2):315–324. doi: 10.1016/j.ydbio.2010.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith-Bolton RK, Worley MI, Kanda H, Hariharan IK. Regenerative growth in Drosophila imaginal discs is regulated by Wingless and Myc. Dev Cell. 2009;16(6):797–809. doi: 10.1016/j.devcel.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grusche FA, Degoutin JL, Richardson HE, Harvey KF. The Salvador/Warts/Hippo pathway controls regenerative tissue growth in Drosophila melanogaster. Dev Biol. 2011;350(2):255–266. doi: 10.1016/j.ydbio.2010.11.020. [DOI] [PubMed] [Google Scholar]
- 16.Sun G, Irvine KD. Regulation of Hippo signaling by Jun kinase signaling during compensatory cell proliferation and regeneration, and in neoplastic tumors. Dev Biol. 2011;350(1):139–151. doi: 10.1016/j.ydbio.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klebes A, et al. Regulation of cellular plasticity in Drosophila imaginal disc cells by the Polycomb group, trithorax group and lama genes. Development. 2005;132(16):3753–3765. doi: 10.1242/dev.01927. [DOI] [PubMed] [Google Scholar]
- 18.Stevens LJ, Page-McCaw A. A secreted MMP is required for reepithelialization during wound healing. Mol Biol Cell. 2012;23(6):1068–1079. doi: 10.1091/mbc.E11-09-0745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanco E, et al. Gene expression following induction of regeneration in Drosophila wing imaginal discs. Expression profile of regenerating wing discs. BMC Dev Biol. 2010;10:94. doi: 10.1186/1471-213X-10-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martín-Blanco E, et al. puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev. 1998;12(4):557–570. doi: 10.1101/gad.12.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weigmann K, Cohen SM. Lineage-tracing cells born in different domains along the PD axis of the developing Drosophila leg. Development. 1999;126(17):3823–3830. doi: 10.1242/dev.126.17.3823. [DOI] [PubMed] [Google Scholar]
- 22.Schubiger G. Regeneration, duplication and transdetermination in fragments of the leg disc of Drosophila melanogaster. Dev Biol. 1971;26(2):277–295. doi: 10.1016/0012-1606(71)90127-8. [DOI] [PubMed] [Google Scholar]
- 23.Abbott LC, Karpen GH, Schubiger G. Compartmental restrictions and blastema formation during pattern regulation in Drosophila imaginal leg discs. Dev Biol. 1981;87(1):64–75. doi: 10.1016/0012-1606(81)90061-0. [DOI] [PubMed] [Google Scholar]
- 24.Gibson MC, Schubiger G. Hedgehog is required for activation of engrailed during regeneration of fragmented Drosophila imaginal discs. Development. 1999;126(8):1591–1599. doi: 10.1242/dev.126.8.1591. [DOI] [PubMed] [Google Scholar]
- 25.Schubiger G. Anlageplan, Determinationszustand und Transdeterminationsleistungen der männlichen Vorderbeinscheibe von Drosophila melanogaster. Wilhelm Roux Arch Entwickl Mech Org. 1968;160:9–40. doi: 10.1007/BF00573645. [DOI] [PubMed] [Google Scholar]
- 26.Boutros M, Agaisse H, Perrimon N. Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev Cell. 2002;3(5):711–722. doi: 10.1016/s1534-5807(02)00325-8. [DOI] [PubMed] [Google Scholar]
- 27.Irving P, et al. New insights into Drosophila larval haemocyte functions through genome-wide analysis. Cell Microbiol. 2005;7(3):335–350. doi: 10.1111/j.1462-5822.2004.00462.x. [DOI] [PubMed] [Google Scholar]
- 28.Stramer B, et al. Gene induction following wounding of wild-type versus macrophage-deficient Drosophila embryos. EMBO Rep. 2008;9(5):465–471. doi: 10.1038/embor.2008.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kocks C, et al. Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell. 2005;123(2):335–346. doi: 10.1016/j.cell.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 30.Kurucz E, et al. Nimrod, a putative phagocytosis receptor with EGF repeats in Drosophila plasmatocytes. Curr Biol. 2007;17(7):649–654. doi: 10.1016/j.cub.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 31.Johansson KC, Metzendorf C, Söderhäll K. Microarray analysis of immune challenged Drosophila hemocytes. Exp Cell Res. 2005;305(1):145–155. doi: 10.1016/j.yexcr.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 32.Katsuyama T, Paro R. Innate immune cells are dispensable for regenerative growth of imaginal discs. Mech Dev. 2013;130(2-3):112–121. doi: 10.1016/j.mod.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 33.Colombani J, Andersen DS, Léopold P. Secreted peptide Dilp8 coordinates Drosophila tissue growth with developmental timing. Science. 2012;336(6081):582–585. doi: 10.1126/science.1216689. [DOI] [PubMed] [Google Scholar]
- 34.Garelli A, Gontijo AM, Miguela V, Caparros E, Dominguez M. Imaginal discs secrete insulin-like peptide 8 to mediate plasticity of growth and maturation. Science. 2012;336(6081):579–582. doi: 10.1126/science.1216735. [DOI] [PubMed] [Google Scholar]
- 35.Pastor-Pareja JC, Wu M, Xu T. An innate immune response of blood cells to tumors and tissue damage in Drosophila. Dis Model Mech. 2008;1(2-3):144–154. doi: 10.1242/dmm.000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mace KA, Pearson JC, McGinnis W. An epidermal barrier wound repair pathway in Drosophila is mediated by grainy head. Science. 2005;308(5720):381–385. doi: 10.1126/science.1107573. [DOI] [PubMed] [Google Scholar]
- 37.Bach EA, et al. GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr Patterns. 2007;7(3):323–331. doi: 10.1016/j.modgep.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 38.Luo H, et al. The Drosophila Jak kinase hopscotch is required for multiple developmental processes in the eye. Dev Biol. 1999;213(2):432–441. doi: 10.1006/dbio.1999.9390. [DOI] [PubMed] [Google Scholar]
- 39.Bejsovec A, Martinez Arias A. Roles of wingless in patterning the larval epidermis of Drosophila. Development. 1991;113(2):471–485. doi: 10.1242/dev.113.2.471. [DOI] [PubMed] [Google Scholar]
- 40.Díaz-García S, Baonza A. Pattern reorganization occurs independently of cell division during Drosophila wing disc regeneration in situ. Proc Natl Acad Sci USA. 2013;110(32):13032–13037. doi: 10.1073/pnas.1220543110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arbouzova NI, Zeidler MP. JAK/STAT signalling in Drosophila: Insights into conserved regulatory and cellular functions. Development. 2006;133(14):2605–2616. doi: 10.1242/dev.02411. [DOI] [PubMed] [Google Scholar]
- 42.Bando T, et al. Analysis of RNA-Seq data reveals involvement of JAK/STAT signalling during leg regeneration in the cricket Gryllus bimaculatus. Development. 2013;140(5):959–964. doi: 10.1242/dev.084590. [DOI] [PubMed] [Google Scholar]
- 43.Rodrigues AB, et al. Activated STAT regulates growth and induces competitive interactions independently of Myc, Yorkie, Wingless and ribosome biogenesis. Development. 2012;139(21):4051–4061. doi: 10.1242/dev.076760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P. Drosophila myc regulates cellular growth during development. Cell. 1999;98(6):779–790. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tapon N, et al. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110(4):467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 46.Tsai YC, Sun YH. Long-range effect of upd, a ligand for Jak/STAT pathway, on cell cycle in Drosophila eye development. Genesis. 2004;39(2):141–153. doi: 10.1002/gene.20035. [DOI] [PubMed] [Google Scholar]
- 47.Fan Y, et al. Genetic models of apoptosis-induced proliferation decipher activation of JNK and identify a requirement of EGFR signaling for tissue regenerative responses in Drosophila. PLoS Genet. 2014;10(1):e1004131. doi: 10.1371/journal.pgen.1004131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boudny V, Kovarik J. JAK/STAT signaling pathways and cancer. Janus kinases/signal transducers and activators of transcription. Neoplasma. 2002;49(6):349–355. [PubMed] [Google Scholar]
- 49.Wang YH, Huang ML. Organogenesis and tumorigenesis: Insight from the JAK/STAT pathway in the Drosophila eye. Dev Dyn. 2010;239(10):2522–2533. doi: 10.1002/dvdy.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vainchenker W, Constantinescu SN. JAK/STAT signaling in hematological malignancies. Oncogene. 2013;32(21):2601–2613. doi: 10.1038/onc.2012.347. [DOI] [PubMed] [Google Scholar]
- 51.Classen AK, Bunker BD, Harvey KF, Vaccari T, Bilder D. A tumor suppressor activity of Drosophila Polycomb genes mediated by JAK-STAT signaling. Nat Genet. 2009;41(10):1150–1155. doi: 10.1038/ng.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu M, Pastor-Pareja JC, Xu T. Interaction between Ras(V12) and scribbled clones induces tumour growth and invasion. Nature. 2010;463(7280):545–548. doi: 10.1038/nature08702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ursprung H. In vivo culture of Drosophila imaginal discs. In: Wilt FW, Wessells NK, editors. Methods in Developmental Biology. Crowell; New York: 1967. pp. 485–492. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.