Significance
Proton transfer is a fundamental mechanism at the core of many enzyme-catalyzed reactions. It is also exquisitely sensitive to a number of factors, including pH, electrostatics, proper active-site geometry, and chemistry. Carbonic anhydrase has evolved a fast and efficient way to conduct protons through a combination of hydrophilic amino acid side chains that coordinate a highly ordered H-bonded water network. This study uses a powerful approach, combining NMR solution studies with neutron protein crystallography, to determine the effect of pH and divalent cations on key residues involved in proton transfer in human carbonic anhydrase. The results have broad implications for our understanding of proton transfer and how subtle changes in ionization and H-bonding interactions can modulate enzyme catalysis.
Keywords: proton transfer, neutron crystallography, perturbed pKa, solution NMR
Abstract
Human carbonic anhydrase II (HCA II) uses a Zn-bound OH−/H2O mechanism to catalyze the reversible hydration of CO2. This catalysis also involves a separate proton transfer step, mediated by an ordered solvent network coordinated by hydrophilic residues. One of these residues, Tyr7, was previously shown to be deprotonated in the neutron crystal structure at pH 10. This observation indicated that Tyr7 has a perturbed pKa compared with free tyrosine. To further probe the pKa of this residue, NMR spectroscopic measurements of [13C]Tyr-labeled holo HCA II (with active-site Zn present) were preformed to titrate all Tyr residues between pH 5.4–11.0. In addition, neutron studies of apo HCA II (with Zn removed from the active site) at pH 7.5 and holo HCA II at pH 6 were conducted. This detailed interrogation of tyrosines in HCA II by NMR and neutron crystallography revealed a significantly lowered pKa of Tyr7 and how pH and Tyr proximity to Zn affect hydrogen-bonding interactions.
Carbonic anhydrases (CAs) are ubiquitous enzymes that catalyze the interconversion of CO2 and HCO3−. In humans, CAs are involved in a great variety of physiological processes, from respiration, ureagenesis, gluconeogenesis, and cerebrospinal fluid production to general acid/base homeostasis (1). The most abundant and well-characterized human CA isoform is human carbonic anhydrase II (HCA II), which displays a bell-shaped pH dependence on catalytic activity, with a maximal kcat of 106 per second at pH 7.2. In the hydration direction, the first step of catalysis is a Zn–OH−-mediated nucleophilic attack on CO2 to form HCO3− and one H+, leaving one water molecule bound to the zinc. The second, rate-limiting step occurs via proton transfer (PT) that removes one H+ from the metal-bound water to regenerate the catalytic Zn–OH− (2). This PT step is thought to adopt classic Grotthuss-type hopping and it is mediated by a well-ordered network of hydrogen-bonded (H-bonded) water molecules that spans the ∼8-Å distance between the Zn-water and proton-shuttling residue, His64 (Figs. 1–3). In previous neutron structures, the Zn-bound solvent has been observed to be an H2O (or exchanged D2O) and this may imply that H2O is the favored species over OH− in the absence of substrate (3). In chemical crystallography extended X-ray absorption fine structure can be used to determine Zn:Zn-solvent distances as a way to discriminate between H2O or OH−. For Zn–OH− the distance should be ∼1.80 Å and for Zn–H2O it should be closer to 2.0 Å (4). Despite the availability of ultra-high-resolution X-ray crystal structures of holo HCA II (no substrate present) the range of Zn:Zn-solvent distances observed has led to inconclusive results. Also, within the coordinate error it is unfeasible to determine the Zn-bound solvent species using distance with any certainty (5, 6).
Fig. 1.
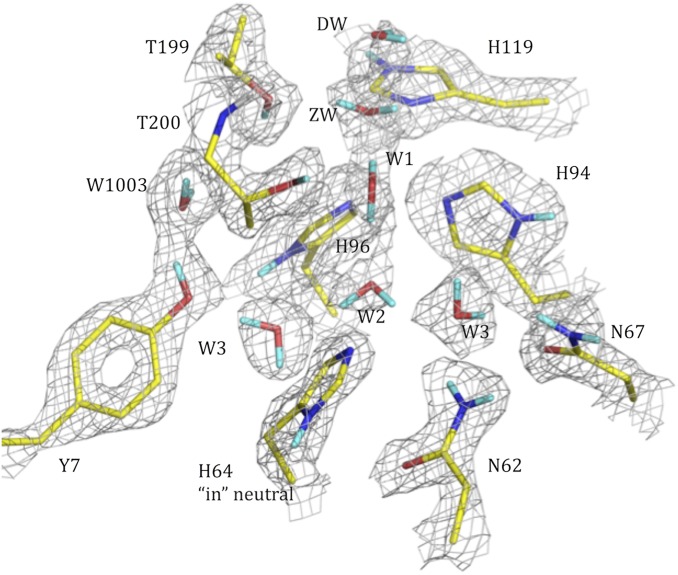
The pH 7.5 apo HCA II active site determined by neutron crystallographic studies. The 2Fo − Fc nuclear density map is shown in gray mesh (contour 1.3σ). Exchanged Ds are in cyan; unexchanged H atoms are not shown for clarity. Active-site residues and solvent molecules are labeled.
Fig. 3.
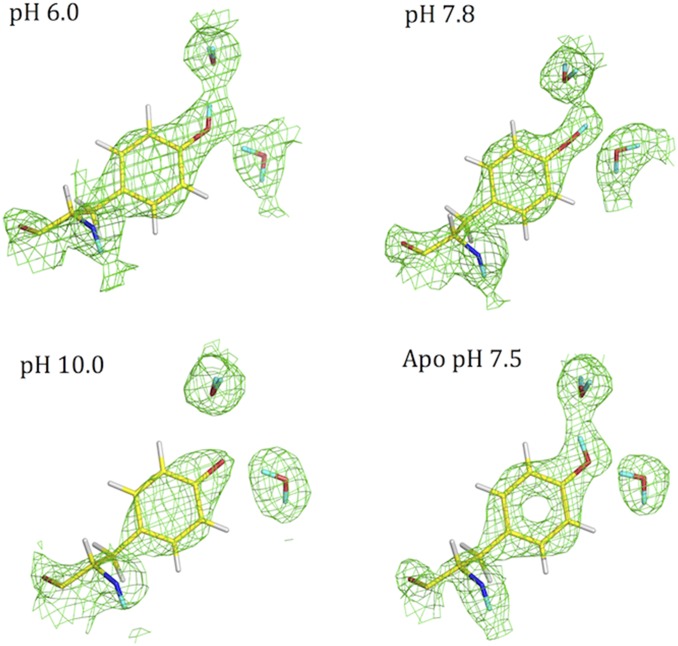
Protonation state and H-bonding interactions of Tyr7 in HCA II as a function of pH and in the absence of Zn. The 2Fo − Fc nuclear density map is shown in green mesh (contour 1.2σ). Exchanged D atoms are in cyan; unexchanged H atoms are shown in white.
His64 is thought to be one component in the delivery of excess H+ to the bulk solvent, resetting the active site for another round of catalysis (7–9). The His64Ala mutant retains 5–10% wild-type PT activity. This observation in combination with computational analysis supports the existence of alternate PT pathways (5, 8). The O atoms of the water network are observed intact in X-ray crystal structures determined from pH 5–10 without positional change (10). The water molecules are coordinated by a number of hydrophilic active-site residues, including Tyr7, Asn62, Asn67, Thr199, and Thr200 (Figs. 1 and 3). Numerous mutagenesis, computational, structural, and kinetic studies have shown the importance of these hydrophilic residues in (i) organizing and coordinating the active-site water network and (ii) maintaining an appropriate electrostatic environment for optimal PT (11–13). A previous study focused on the role of hydrophilic residues Tyr7, Asn62, and Asn67 in the active site. These residues were mutated to hydrophobic residues Phe7, Leu62, and Leu67. The mutants with Leu at positions 62 and 67 displayed slower PT kinetics than wild type but the Tyr7Phe mutant was observed to have significantly enhanced PT activity (approximately sevenfold over wild type) in the bircarbonate dehydration reaction. This unexpected behavior was ascribed to two effects: the loss of Tyr at position 7 seemed to lower the pKa of His64 from 7 to ∼6, and the solvent structure between the Zn-bound solvent and His64 was disrupted to form an unbranched network. It was believed that this and His64’s having a more acidic pKa led to the Tyr7Phe mutant’s having improved PT kinetics compared with wild type (11).
Neutron protein crystallography provides insight into enzyme mechanisms. It is the only technique that can directly, and at high resolution, visualize H and its isotope deuterium, D. Unlike X-rays, neutrons are scattered very well by H/D atoms, similarly to the other common elements found in proteins, such as C, N, and O. This allows detailed analysis of water coordination, H bonding, ligand binding, and the protonation state of certain amino acids such as His and Lys (14, 15). This information is crucial to understanding how H bonds and water mediate not only enzyme catalysis but also ligand binding. Previously, the neutron crystal structure of HCA II crystallized from a pH 10 buffer showed an incomplete H-bonded water network in the active site and a D2O molecule bound to the Zn (3). The structure also revealed that a key active-site residue, Tyr7, was deprotonated (Fig. 3). Interestingly, all of the other seven tyrosines in HCA II, including solvent-exposed surface Tyr residues that were expected to be sensitive to bulk solution pH changes, were observed to be neutral in the crystal structure (3). Although deprotonation of Tyr7 was not totally unexpected, because tyrosine in solution and in random coil polypeptides has a pKa of ∼10, this observation indicated that the pKa of this residue is significantly lowered (16, 17). In enzyme systems where tyrosines are involved with general acid–base catalysis, site-specific pKa measurements with NMR spectroscopy have revealed that some tyrosines exhibit pKas as low as 6 (18). Other examples of enzymes with catalytic tyrosines that have perturbed pKas include β-lactamase, ketosteroid isomerase, and UDP-galactose 4-epimerase (19–21). It is an important, although less understood, phenomenon that enzymes are able to fine-tune local electrostatics to support a particular function (22).
Subsequent to the characterization of the high-pH neutron structure of HCA II, an additional neutron crystal structure was determined for crystals grown at pH 7.8. Here, as expected, Tyr7 was neutral and involved in a bifurcated H bond (Fig. 3). To date, the catalytic role for Tyr7 in HCA II has not been proposed, although it is unusual for a noncatalytic tyrosine to have a perturbed pKa. Of course, the extent of the perturbation is difficult to gauge using neutron crystal structures alone because they provide only static snapshots of the residue in question at a few selected pH values. Moreover, estimating the pH inside a crystal is challenging; we are limited to measuring the pH of the crystallization solution, which gives only an approximation of the crystal’s inner pH. Furthermore, because at pH close to pKa residues are partially protonated, it is possible that one of the charged forms selectively crystallizes, shifting the observed equilibrium.
There are eight Tyr residues in HCA II with residue numbers 7, 40, 51, 88, 114, 128, 191, and 194. Tyr7 is located on the edge of the active site and is exposed to solvent. Tyr40, 114, and 128 sit on the surface and are also solvent-exposed, whereas Tyr51, 88, and 191 have their side chains buried and participate in H bonding with other parts of the protein. In this study, we probed the protonation state of all tyrosine residues in HCA II and measured their pKa in solution. We combined these studies with analysis of neutron crystallographic structures to shed light on the effect of pH on important active-site residues (such as Tyr7 and His64) and the biophysical effect of the divalent cation (Zn) in the active site.
To address these questions, we determined the neutron structure of apo HCA II (with active-site Zn removed) crystallized at pH 7.5 to detect any influence of the divalent cation on the nearby residues and those farther away from the active site. Another neutron crystal structure of holo HCA II (with active-site Zn present) crystallized at pH 6.0 was determined to observe pH effects on all Tyr residues and the proton shuttle, His64. [13C]Tyr-labeled CA was prepared for NMR analysis and the spectra were measured from pH 5.4–11.0. It was possible to assign all eight Tyr in HCA II, providing the opportunity to interrogate their individual behaviors in solution and to compare the results to the crystal structures. To our knowledge, this is the first report of the use of neutron crystallography and NMR spectroscopy together to map tyrosine protonation states as a function of pH and the divalent cation. The observation of a perturbed pKa for Tyr at physiological pH has implications for our understanding of the PT mechanism and the overall enzyme-catalyzed reaction trajectory.
Results and Discussion
Apo HCA II at pH 7.5.
The neutron structure of apo HCA II was determined to 1.8-Å resolution with a refined Rcryst/Rfree of 20.3/21.7. Detailed analyses of Fo − Fc and 2Fo − Fc nuclear maps show that the active-site water network has the same H bonding and coordination as holo HCA II crystallized at pH 7.8 (Fig. 1; PDB ID code 3tmj). In the absence of an active-site metal, there is still a complete H-bonded network of active-site water molecules stretching to the bulk solvent. Also, as expected, Asn62, Asn67, Thr199, and Thr200 remain unchanged (Fig. 1). Although the overall water pattern and most of the active-site residues seem to be similar to holo structures, there are notable differences in the catalytically significant residues. The proton shuttling residue, His64, is neutral and well-ordered in the “inward” conformation. This is in contrast to other studies in which His64 was observed in two conformations, “inward” and “outward,” relative to the active-site opening (Fig. 1) (8, 23, 24). In the holo pH 7.8 neutron structure, there are three discernable states of His64 that differ in charge and imidazole ring orientation (PDB ID code 3tmj).
Holo HCA II pH 6.
Because the protein does not crystallize below pH 7, it was challenging to obtain the low-pH crystal form of HCA II. To obtain crystals at this low pH, we subjected them to vapor acidification, which may have led to lower-quality neutron data observed. The neutron structure of holo HCA II crystallized at pH 6 was determined to 2.0-Å resolution and the final model was refined to Rcryst/Rfree 26.3/29.1. Despite the lower quality of this dataset with regard to resolution and completeness, we could observe several readily apparent structural features.
Overall, the active-site residues seem to be very similar to the apo structure with some subtle changes in the water network. Most notably, the deep water (DW) has moved compared with its position in the substrate-free pH 7.8 and 10 neutron crystal structures (Fig. 2). A report of the holo HCA II in a complex with CO2 substrate also identified this water molecule in a shifted position (25). In the low-pH neutron structure we analyzed, the DW oxygen (O) atom is shifted by about 1.6 Å, similar to the O atom of the DW in the HCA II:CO2 complex structure.
Fig. 2.
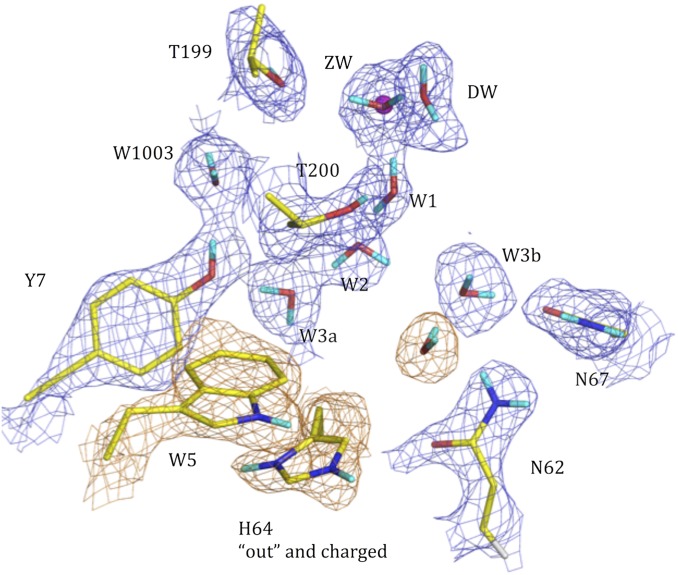
The pH 6 holo HCA II active site determined by neutron crystallographic studies. The 2Fo − Fc nuclear density map is shown in blue mesh (contour 1.3σ). Exchanged Ds are in cyan; unexchanged H atoms are not shown for clarity. Active-site residues and solvent molecules are labeled. The nuclear density for Trp5 and His64 is shown in orange.
The most interesting structural feature is His64 in the “outward” conformation while protonated and π stacking with Trp5 (Fig. 2; see close-up view in Fig. S1). This outward position has been observed in numerous X-ray crystal structures and His64 was speculated to be protonated in this position (24). Our low-pH neutron crystal structure offers the first direct evidence to our knowledge of the protonation of His64 in the “outward” conformation. Also, we observed an additional water molecule occupying the former “inward” His64 position with its O atom acting as an H-bond acceptor (HBA) from an imidazole H. The two main features of the low-pH HCA II neutron structure, namely DW moving outward and His64 in the “outward” conformation, are the same as those seen in the high-resolution X-ray crystal structure of the HCA II:CO2 complex structure (25).
Tyrosine Residues.
Further investigation of all eight tyrosine residues in HCA II reveals additional features (Figs. S2–S4 and Table 1). Out of the eight, Tyr7 is the only residue that is observed in three distinct states: neutral [HBA and H-bond donor (HBD) to water molecules W3a and W1003], neutral (bifurcated HBD to two water molecules), and deprotonated/charged (HBA) (Fig. 3). As expected, we observed the other seven tyrosines always as neutral (Table 1). Four tyrosine residues—Tyr40, Tyr114, Tyr128, and Tyr191—were unchanged in H-bonding interactions over the pH range studied by crystallography. These four tyrosines were also insensitive to the presence or absence of the divalent cation Zn in the active site (Table 1). The remaining three Tyr residues—Tyr51, Tyr88, and Tyr194—underwent subtle changes in their H-bonding patterns between the holo and apo forms to compensate for the absence of metal and to maintain H-bonding interactions (Figs. S2–S4). Table 1 summarizes the Tyr residues and their interactions from all four neutron structures of HCA II.
Table 1.
Tyrosine in HCA II interactions, H bonds, and distance to active-site Zn
| Tyr no. (distance from Zn, Å) | pH 6 holo (this work, 4y0j) | pH 7.8 holo (PDB ID code 3tmj) | pH 10 holo (PDB ID code 3kkx) | pH 7.5 apo (this work, 4q49) |
| 7 (7) | Neutral (HBD and HBA to 2 waters) | Neutral (bifurcated HBD to 2 waters) | Deprotonated/charged (HBA to water) | Neutral (HBD and HBA to 2 waters) |
| 40 (28) | Neutral (HBD to water) | Unchanged | Unchanged | Unchanged |
| 51 (14) | Neutral (*HBA to protonated His122) | Neutral (HBA to protonated His122) | Neutral (HBA to protonated His122) | Neutral (HBD to neutral His122) |
| 88 (17) | Neutral (HBD to –CO Asp139, HBA to 2 waters) | Neutral (HBD to –CO of Asp139, HBA to 2 waters) | Neutral (HBD to water) | Neutral (HBD and HBA to 2 waters) |
| 114 (18) | Neutral (HBD and HBA to 2 waters) | Unchanged | Unchanged | Unchanged |
| 128 (21) | Neutral (HBD to Asp139) | Unchanged | Unchanged | Unchanged |
| 191 (20) | Neutral (HBD to –CO of Lys45) | Unchanged | Unchanged | Unchanged |
| 194 (10) | Neutral (HBA to protonated His107, HBD to Ser29) | Neutral (HBA to protonated His107, HBD to Ser29) | Neutral (HBA to protonated His107, HBD to Ser29) | Neutral (HBD to neutral His107, HBA to Ser29) |
The position of the D atom shared between Tyr and His residues is challenging to assign with absolute confidence based on crystallographic data alone. However, we know from the NMR data that all Tyr residues other than Tyr7 are neutral in the pH range studied. In these cases, we also calculated a series of omit maps to help guide the placement of the D atom, while taking the surrounding H bonds into account. Interestingly, Tyr51, 88, and 194 side chains are buried and interact with other residues and water. Also, these changing tyrosine residues are all between 7 and 17 Å away from the zinc. They seem to be insensitive to pH but may be sensitive to the absence of metal. That these effects operate within a certain distance from the zinc site seems to indicate possible long-range electrostatic effects operating in HCA II. An alternate explanation is that the changes in Tyr H bonding could also be due to a series of local effects that are propagated over a distance, culminating in local changes.
NMR Spectroscopy of Tyr in HCA II.
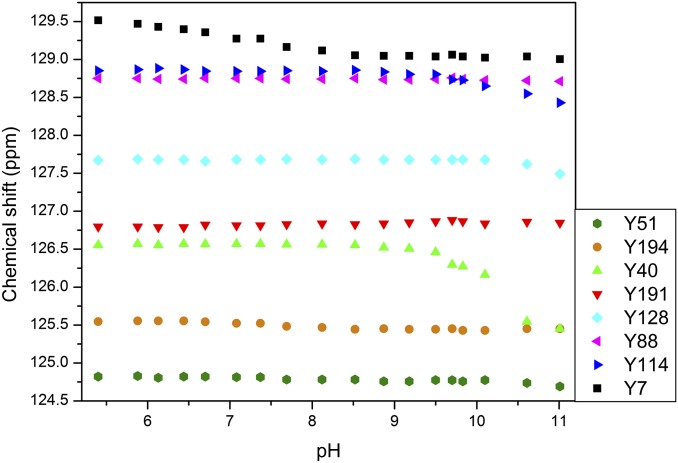
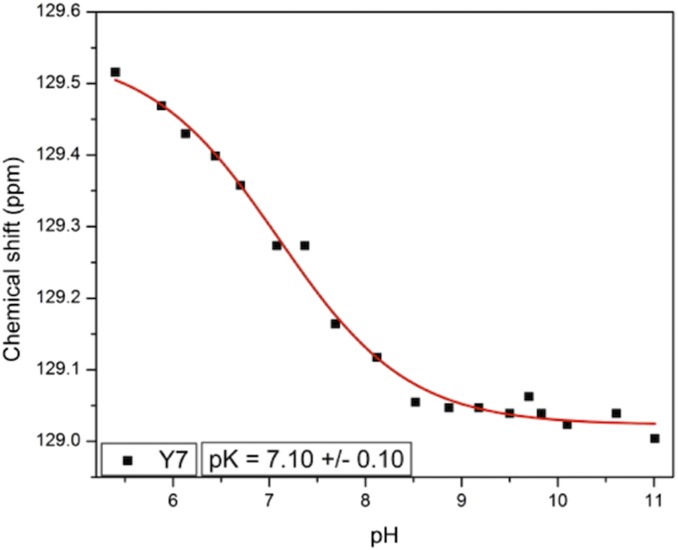
Although neutron crystal structures give explicit information about Tyr residues and their charged states, they provide discrete snapshots and do not indicate protonation changes over a broad pH range and associated pKa values. NMR spectra measured on [13C]Tyr-labeled HCA II from pH 5.4–11.0 fill in these gaps and provide details not obtainable from static crystal structures alone. Owing to specific [13C]Tyr labeling, only the Tyr residues were observed and analyzed. The assignments of Cγ carbon frequencies from HNCACB, HSQC, and CG(CB)HB spectra are shown in Fig. S5. The assignments of Cζ carbon frequencies are shown in Fig. S6. Fig. 4 shows the chemical shift changes for all Tyr residues between pH 5.4 and 11.0. The integrity of the protein was followed using 1H-15N heteronuclear single quantum coherence (HSQC) experiments, which showed well-resolved and dispersed spectra in the pH range from 5.4 to 11.0. When the pH was raised to 11.47, the HSQC spectrum showed the collapse of resonances, indicating protein unfolding. No data at higher pH could therefore be obtained. Tyr7 (black squares in Fig. 4) is the only Tyr that changes at close to neutral pH, consistent with observations of the neutron crystal structures (Table 1 and Fig. 3) (3). Close to pH 10, two additional Tyr residues, Tyr40 and 114, start to deprotonate. Tyr51, 128, and 194 begin shifting when the pH is greater than 10.5. This is not surprising because this pH is close to the expected pKa for free Tyr. Fig. 5 shows the chemical shift dependence of Tyr7 with a calculated pKa of 7.1.
Fig. 4.
Chemical shift changes of Cγ resonances for all tyrosine residues in HCA II as a function of pH.
Fig. 5.
Chemical shift changes of Cγ resonance for Tyr7 in HCA II as a function of pH. The fit gives the apparent pKa of ∼7.1.
Although accurate pKa values for Tyr40 and Tyr114 could not be determined because the chemical shift values for these residues do not level off at the highest pH accessible, the fit of the initial part of the titration curves yields lower bounds for these pKa values. These are 10.25 for Tyr40 and 10.45 for Tyr114. Both of these residues are solvent-exposed; thus, it is not surprising that their pKa values are similar to those in model peptides. Given the initial shift change for Tyr51 above pH 10.5, we estimated its pKa to be in the 11.5–12.0 range, whereas for Tyr88, 128, 191, and 194 the estimated the pKa values will be greater than 12. This wide range of pKas, from ∼7–12 as measured for residues that are either buried or solvent-exposed indicates modulation by local environments and H bonding and how they affect the ionization of residues.
An interesting observation from the NMR data is that the total chemical shift changes for Tyr7 during the titrations seem to be very small. As determined previously, chemical shift changes upon deprotonation for 13Cγ and 13Cζ resonances in peptides are −6.2 and +10.4 ppm, respectively (26). In photoactive yellow protein, chemical shift changes for tyrosine 13Cγ were determined to be between −3.8 and −4.8 ppm (27), while Baturin et al. observed changes for tyrosine 13Cζ resonances in B. circulans xylanase between +9.0 and +9.5 ppm (19). In this study, we observed a total chemical shift change for Tyr7 13Cγ of −0.5 ppm, with an apparent pKa of 7.1, which is very low for a tyrosine hydroxyl group (Fig. 5). The data for 13Cζ of Tyr7 showed a total chemical shift change of +0.3 ppm, with similar apparent pKa (Fig. S7B).
One could argue that the small overall chemical shift changes for Tyr7 carbons reflect electrostatic changes in the neighborhood of Tyr7 rather than actual deprotonation. Tyr7 is in fact close (<5Å) to His64 with a pKa of ∼7.2 and deprotonation of this histidine could change the electrostatics around Tyr7 and lead to small chemical shift changes of Tyr7 resonances. The evidence contradicts this argument, however. First, it is the Tyr7 Cζ carbon that is closer to His64 rather than the Cγ carbon, yet the Cγ carbon experiences larger chemical shift changes. Additionally, as evidenced by the neutron crystal structures, at pH 6 Tyr7 is fully protonated, whereas at pH 10.0 it is deprotonated. Because the NMR titrations extended over a pH range from 5.4 to 11.0, the chemical shift changes of Tyr7 carbon resonances should then reflect the deprotonation of this residue.
The reason for the decreased chemical shift sensitivity of Tyr7 carbon resonances remains ambiguous. One possibility is the unique participation of Tyr7 in the proton-conducting water network extending from the Zn atom to His64. These water molecules are well ordered and seem to be conserved in all crystal structures discussed here. Tyr7 forms relatively short and strong H bonds with water molecules from this ordered network. At low pH, the O–H bond of Tyr7 is polarized by participating in the H bond as a donor. At high pH, the electron density is withdrawn from the charged O by participating in the H bond as an acceptor. If these H bonds are very stable, as suggested by the conservation of the water network and the short O…O distances, the overall change in electron density on the tyrosine ring between the neutral and charged state may be small, thus leading to small overall chemical shift changes.
Tyr7 plays a coordinating role in the proton-conducting water network, but it was previously surmised that it is mostly involved in maintaining an appropriate electrostatic environment to support PT (11–13). The pH of normal blood is 7.35–7.50, which is slightly above the pKa for Tyr7, suggesting that roughly half the population would be deprotonated (28). Given this pKa that is within the physiological pH range, there are profound implications for the role of Tyr7 in PT under normal conditions. When protonated at physiological pH, Tyr7 acts as a bifurcated HBD to W1003 and W3a in the water network (Figs. 1, 3, and 4). This “normal” configuration supports the H bond-connected water network and is seemingly ready to transport the excess proton from the Zn-bound solvent to the proton-shuttling residue, His64. However, Tyr7 was observed in the neutron crystal structure to be deprotonated at high pH, causing a rearrangement of the water molecules into a configuration where W3a is an HBD to Tyr7. Such a configuration would not support PT through the water molecules alone, and the proton moving from the Zn-bound water may be captured by the phenolate of Tyr7 before it reaches His64. In this way, Tyr7 can operate as a proton trap in wild-type HCA II, a possibility that would support the kinetic measurements of the Tyr7Phe HCA II mutant that indicated faster kinetics than in wild type (11). In the next step Tyr7 can easily donate its H to the adjacent water that in turn passes it along to His64, or His64 can occupy a rotamer that favors direct transfer from Tyr7. This direct transfer scenario would necessitate the rearrangement of two or three water molecules as well as the rotation of the His64 side chain along χ2, which should be energetically unfavorable.
The pKa of the proton donor:acceptor groups (ZW, His64) in the HCA II active site are closely matched at ∼7 and our data indicates that Tyr7 also belongs to this group. For efficient PT the pKa values of the proton donor:acceptor groups need to closely match. We propose that involvement of Tyr7 in the H-bonding network and proximity to His64 results in lowering the pKa of Tyr7 by ∼3 pH units. For His64 to switch between “in” and “out” conformations with minimal energetic cost, Tyr7 should participate in rearranging of the H-bonded network. This is possible if Tyr7 pKa is close to that of His64. This suggests several scenarios for the PT pathway in wild-type HCA II. All involve proton movement from the ZW to His64 through (i) W1–W2, (ii) W1–W2–Tyr7 and W3a, or (iii) W1–W2–Tyr7–His64. We believe that Tyr7 is also a catalytic residue. Pathways ii and iii are longer and/or may involve significant solvent rearrangement that can be expected to be kinetically and energetically less favorable. Tyr7 as a PT partner directly with His64 or a proton trap further supports the observation that the Tyr7Phe HCA II mutant has even higher PT activity than does the wild-type HCA II (11). From the Tyr7Phe kinetic results it was shown that Tyr7 has a significant role in modulating the pKa of His64 at near-neutral values and that removing Tyr7 causes the His64 to become more acidic, lowering His64 pKa to ∼6. This makes His64 a better proton donor in the bicarbonate dehydration reaction, causing an increase in PT activity in the Tyr7Phe mutant (11–13). These earlier measurements support our proposal that Tyr7 is a catalytic residue involved in stabilizing the PT network, thus enhancing PT.
The presence of a strongly electronegative Tyr in a finely balanced electrostatic environment will have a major impact on PT, electrostatics, and the overall energetics of PT. Tyrosinate could also modulate alternative, less-efficient PT routes out of the active site under certain nonideal pH conditions, such as during blood alkalosis. It is also possible that Tyr7 can act as a proton hole when it is deprotonated. The mechanistic determinants of a proton hole in this environment require that OH− is generated first by protonation of the acceptor with subsequent transfer of the OH− to the donor via the active-site water network (29). The explicit protonation information on active-site residues determined by static and dynamic (crystallographic and NMR spectroscopy) methods supports mechanisms involving the effects of electrostatics on proton holes and catalysis in HCA II (22, 29). Such a mechanism can operate in conjunction with the classic Grotthuss mechanism, or it can represent an alternate route under certain active-site conditions (30).
Conclusion
To our knowledge, this is the first example of using NMR spectroscopy and neutron crystallography complementarily to gain insight into the electrostatics of a protein active site and how such electrostatics affect the charged states and H bonding of amino acid residues. In HCA II, the behavior of Tyr7 has implications for how PT can proceed under different conditions, especially at physiological pH. Similar effects may operate in other enzyme systems that use PT during catalysis. This is a relatively unexplored area and the results are useful for computational biology where experimental knowledge of the positions of H atoms is important. The results show that the protonation states of His and Tyr residues are sensitive to both pH and the presence of metal cofactor. Although this observation seems intuitive, this is the first study to our knowledge that explores these effects in a systematic way. These factors change how His and Tyr residues H-bond to water and other amino acids and we have shown that it is possible to study these directly and at high resolution. It is noteworthy that the combination of neutron crystallography and NMR spectroscopy allows for detailed interpretation of experimental data, creating a powerful combined approach.
Experimental Procedures
Crystal Preparation for Neutron Crystallography.
Several batches of wild-type HCA II (∼400 mg total) were prepared and the expression and purification protocols are described in detail in Supporting Information. Both apo and holo HCA II were crystallized at room temperature for the neutron experiments by mixing HCA II in a 1:1 ratio with 1.15 M sodium citrate and 100 mM Tris (pH 8.5) in a 500-μL drop. The drops were allowed to undergo vapor diffusion against a 25-mL reservoir consisting of 1.4 M sodium citrate and 100 mM Tris at pH 8.5. To produce the low-pH form, crystals were grown at pH 8.5 using the same protocol for the apo, but before mounting the pH in the drop was lowered to ∼pH 6 by adding 5 mL glacial acetic acid directly to the reservoir. The sandwich box was closed and the acetic acid vapor acidified the drop. After ∼10 min the pH in the drop was checked with a microprobe pH meter and it was measured to be below pH 6. Suitable HCA II crystals were selected and mounted in thick-walled quartz capillaries with a liquid plug of perdeuterated reservoir solution. The crystals were left for ∼4 wk to undergo vapor H/D exchange before neutron data collection.
Room-Temperature X-Ray and Neutron Data Collection, Structure Determination, and Refinement.
X-ray and neutron data collection details, the dataset, and refinement statistics can be found in Supporting Information and Table S1. Room-temperature X-ray data were collected on an in-house Rigaku HighFlux home source using a standard approach. Room-temperature neutron data to 1.8-Å resolution were collected from a H/D-exchanged apo HCA II crystal (∼2 mm3). These data were collected at the monochromatic neutron diffractometer BIODIFF located at the FRM-II reactor in Garching, Germany. Time-of-flight Laue neutron data to 2.0-Å resolution were collected from an H/D-exchanged, low-pH holo HCA II crystal (∼2 mm3) at the Protein Crystallography Station (PCS) at Los Alamos Neutron Science Center (LANSCE). Experimental data and model coordinates of the apo and low pH structures have been deposited with the Protein Data Bank (ID codes 4q49 and 4y0j, respectively).
Isotope-Labeled HCA II for NMR.
Several 13C isotopically Tyr-labeled versions of wild-type HCA II were prepared for pH titrations to ultimately determine the pKa values of the tyrosine residues. Details on expression and purification can be found in Supporting Information. The isotopically labeled Tyr used for these experiments was either [U-13C, 15N]tyrosine (CNLM-439-0.5; Cambridge Isotopes) or [13Cζ]tyrosine synthesized in-house.
NMR Spectroscopy Measurements and Assignments.
All experiments were recorded on 1.0–1.3 mM samples of labeled CA on a Bruker AVANCE III 700 MHz instrument equipped with a TCI cryoprobe and processed using TOPSPIN 3.2 software (Bruker Biospin). Several spectra at various pH were recorded on labeled HCA II to determine pH-dependent chemical shift changes for Tyr signals (Fig. 4). Specific details on the measurements and assignments can be found in Supporting Information. Specific assignments of Tyr residues were obtained from a combination of the following 2D and 3D NMR experiments: 1H-15N HSQC, HNCACB, HBCB(CGCD)HD, HBCB(CGCDCE)HE, and aromatic HMBC. Amide proton and nitrogen assignments previously obtained by Venters et al. (31) were used as a starting point. Assignments of 13Cγ resonances were obtained from a combination of HNCACB, HSQC, and CG(CB)HB experiments.
pKa Determination from NMR Data.
The chemical shifts of all assigned resonances were plotted as a function of pH using Origin 7 software. The data were then fitted to a sigmoidal Boltzmann function using a nonlinear least squares algorithm in Origin 7 and the midpoint of the transition was reported as the pKa value. The fits were performed for both Cζ and Cγ titrations. The chemical shift error was estimated as 0.01–0.02 ppm based on the resolution of the spectra and repeated experiments.
Supplementary Material
Acknowledgments
We thank Pete Silks for providing the labeled Tyr used in some of the labeling experiments and Esko Oksanen for useful discussions. R.M. and S.Z.F. thank Los Alamos National Laboratory (LANL) Principal Associate Directorate for Science, Technology, and Engineering for investing in key equipment that enabled many of the measurements reported here. We also thank Dr. Virginia A. Unkefer for help with editing the manuscript. S.Z.F., J.-P.B., and C.J.U. were partially funded through the Protein Crystallography Station from the Department of Energy Office of Biological and Environmental Research. S.Z.F. and R.M. were also funded through LANL Early Career Grant 20110535ER.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4q49 and 4y0j).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1502255112/-/DCSupplemental.
References
- 1.Frost SC. Physiological functions of the alpha class of carbonic anhydrases. Subcell Biochem. 2014;75:9–30. doi: 10.1007/978-94-007-7359-2_2. [DOI] [PubMed] [Google Scholar]
- 2.Silverman DN, Lindskog S. The catalytic mechanism of carbonic anhydrase: Implications of a rate-limiting protolysis of water. Acc Chem Res. 1988;21:30–36. [Google Scholar]
- 3.Fisher Z, et al. Neutron structure of human carbonic anhydrase II: A hydrogen-bonded water network “switch” is observed between pH 7.8 and 10.0. Biochemistry. 2011;50(44):9421–9423. doi: 10.1021/bi201487b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergquist C, Fillebeen T, Morlok MM, Parkin G. Protonation and reactivity towards carbon dioxide of the mononuclear tetrahedral zinc and cobalt hydroxide complexes, [Tp(Bu)t(,Me)]ZnOH and [Tp(Bu)t(,Me)]CoOH: Comparison of the reactivity of the metal hydroxide function in synthetic analogues of carbonic anhydrase. J Am Chem Soc. 2003;125(20):6189–6199. doi: 10.1021/ja034711j. [DOI] [PubMed] [Google Scholar]
- 5.Fisher SZ, et al. Atomic crystal and molecular dynamics simulation structures of human carbonic anhydrase II: Insights into the proton transfer mechanism. Biochemistry. 2007;46(11):2930–2937. doi: 10.1021/bi062066y. [DOI] [PubMed] [Google Scholar]
- 6.Avvaru BS, et al. A short, strong hydrogen bond in the active site of human carbonic anhydrase II. Biochemistry. 2010;49(2):249–251. doi: 10.1021/bi902007b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silverman DN, McKenna R. Solvent-mediated proton transfer in catalysis by carbonic anhydrase. Acc Chem Res. 2007;40(8):669–675. doi: 10.1021/ar7000588. [DOI] [PubMed] [Google Scholar]
- 8.Tu CK, Silverman DN, Forsman C, Jonsson BH, Lindskog S. Role of histidine 64 in the catalytic mechanism of human carbonic anhydrase II studied with a site-specific mutant. Biochemistry. 1989;28(19):7913–7918. doi: 10.1021/bi00445a054. [DOI] [PubMed] [Google Scholar]
- 9.Shimahara H, et al. Tautomerism of histidine 64 associated with proton transfer in catalysis of carbonic anhydrase. J Biol Chem. 2007;282(13):9646–9656. doi: 10.1074/jbc.M609679200. [DOI] [PubMed] [Google Scholar]
- 10.Fisher Z, et al. Structural and kinetic characterization of active-site histidine as a proton shuttle in catalysis by human carbonic anhydrase II. Biochemistry. 2005;44(4):1097–1105. doi: 10.1021/bi0480279. [DOI] [PubMed] [Google Scholar]
- 11.Fisher SZ, et al. Speeding up proton transfer in a fast enzyme: Kinetic and crystallographic studies on the effect of hydrophobic amino acid substitutions in the active site of human carbonic anhydrase II. Biochemistry. 2007;46(12):3803–3813. doi: 10.1021/bi602620k. [DOI] [PubMed] [Google Scholar]
- 12.Maupin CM, et al. Effect of active-site mutation at Asn67 on the proton transfer mechanism of human carbonic anhydrase II. Biochemistry. 2009;48(33):7996–8005. doi: 10.1021/bi901037u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mikulski R, et al. Kinetic and crystallographic studies of the role of tyrosine 7 in the active site of human carbonic anhydrase II. Arch Biochem Biophys. 2011;506(2):181–187. doi: 10.1016/j.abb.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams PD, Mustyakimov M, Afonine PV, Langan P. Generalized X-ray and neutron crystallographic analysis: More accurate and complete structures for biological macromolecules. Acta Crystallogr D Biol Crystallogr. 2009;65(Pt 6):567–573. doi: 10.1107/S0907444909011548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langan P, Chen JC. Seeing the chemistry in biology with neutron crystallography. Phys Chem Chem Phys. 2013;15(33):13705–13712. doi: 10.1039/c3cp51760h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thurlkill RL, Grimsley GR, Scholtz JM, Pace CN. pK values of the ionizable groups of proteins. Protein Sci. 2006;15(5):1214–1218. doi: 10.1110/ps.051840806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimsley GR, Scholtz JM, Pace CN. A summary of the measured pK values of the ionizable groups in folded proteins. Protein Sci. 2009;18(1):247–251. doi: 10.1002/pro.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendriks J, Hellingwerf KJ. pH Dependence of the photoactive yellow protein photocycle recovery reaction reveals a new late photocycle intermediate with a deprotonated chromophore. J Biol Chem. 2009;284(8):5277–5288. doi: 10.1074/jbc.M805904200. [DOI] [PubMed] [Google Scholar]
- 19.Baturin SJ, Okon M, McIntosh LP. Structure, dynamics, and ionization equilibria of the tyrosine residues in Bacillus circulans xylanase. J Biomol NMR. 2011;51(3):379–394. doi: 10.1007/s10858-011-9564-7. [DOI] [PubMed] [Google Scholar]
- 20.Kato-Toma Y, Iwashita T, Masuda K, Oyama Y, Ishiguro M. pKa measurements from nuclear magnetic resonance of tyrosine-150 in class C beta-lactamase. Biochem J. 2003;371(Pt 1):175–181. doi: 10.1042/BJ20021447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sigala PA, et al. Quantitative dissection of hydrogen bond-mediated proton transfer in the ketosteroid isomerase active site. Proc Natl Acad Sci USA. 2013;110(28):E2552–E2561. doi: 10.1073/pnas.1302191110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riccardi D, König P, Guo H, Cui Q. Proton transfer in carbonic anhydrase is controlled by electrostatics rather than the orientation of the acceptor. Biochemistry. 2008;47(8):2369–2378. doi: 10.1021/bi701950j. [DOI] [PubMed] [Google Scholar]
- 23.Nair SK, Christianson DW. Structural properties of human carbonic anhydrase II at pH 9.5. Biochem Biophys Res Commun. 1991;181(2):579–584. doi: 10.1016/0006-291x(91)91229-6. [DOI] [PubMed] [Google Scholar]
- 24.Maupin CM, Voth GA. Preferred orientations of His64 in human carbonic anhydrase II. Biochemistry. 2007;46(11):2938–2947. doi: 10.1021/bi062170f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Domsic JF, et al. Entrapment of carbon dioxide in the active site of carbonic anhydrase II. J Biol Chem. 2008;283(45):30766–30771. doi: 10.1074/jbc.M805353200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norton R, Bradbury J. Carbon-13 nuclear magnetic resonance study of tyrosine titrations. J Chem Soc Chem Commun. 1974;21:870–871. [Google Scholar]
- 27.Oktaviani NA, et al. Comprehensive determination of protein tyrosine pKa values for photoactive yellow protein using indirect 13C NMR spectroscopy. Biophys J. 2012;102(3):579–586. doi: 10.1016/j.bpj.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chawla S, et al. Exogenous sphingosine-1-phosphate boosts acclimatization in rats exposed to acute hypobaric hypoxia: assessment of haematological and metabolic effects. PLoS ONE. 2014;9(6):e98025. doi: 10.1371/journal.pone.0098025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riccardi D, et al. “Proton holes” in long-range proton transfer reactions in solution and enzymes: A theoretical analysis. J Am Chem Soc. 2006;128(50):16302–16311. doi: 10.1021/ja065451j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mai BK, Park K, Duong MP, Kim Y. Proton transfer dependence on hydrogen-bonding of solvent to the water wire: A theoretical study. J Phys Chem B. 2013;117(1):307–315. doi: 10.1021/jp310724g. [DOI] [PubMed] [Google Scholar]
- 31.Venters RA, Farmer BT, 2nd, Fierke CA, Spicer LD. Characterizing the use of perdeuteration in NMR studies of large proteins: 13C, 15N and 1H assignments of human carbonic anhydrase II. J Mol Biol. 1996;264(5):1101–1116. doi: 10.1006/jmbi.1996.0699. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.