Significance
Endosymbiotic organelles are a defining feature of eukaryotes—the last common ancestor and all extant eukaryotes possess at least a mitochondrial derivative. Although mitochondria and plastids are identified with aerobic ATP synthesis and photosynthesis, respectively, their retention by their host cells requires the merging and integration of many, often redundant, metabolic pathways. As a result, complex metabolic interdependencies arise between these formerly independent cells. Complete loss of endosymbiotic organelles, even where aerobic respiration or photosynthesis is lost, is exceedingly difficult, as demonstrated by persistence of organelles throughout secondary anaerobes and parasites. Here, we identify a rare but clear case of plastid loss in a parasitic alga and detail the metabolic disentanglement that was required to achieve this exceptional evolutionary event.
Keywords: organelle loss, plastid loss, endosymbiosis, plastid metabolism, diaminopimelate aminotransferase
Abstract
Organelle gain through endosymbiosis has been integral to the origin and diversification of eukaryotes, and, once gained, plastids and mitochondria seem seldom lost. Indeed, discovery of nonphotosynthetic plastids in many eukaryotes—notably, the apicoplast in apicomplexan parasites such as the malaria pathogen Plasmodium—highlights the essential metabolic functions performed by plastids beyond photosynthesis. Once a cell becomes reliant on these ancillary functions, organelle dependence is apparently difficult to overcome. Previous examples of endosymbiotic organelle loss (either mitochondria or plastids), which have been invoked to explain the origin of eukaryotic diversity, have subsequently been recognized as organelle reduction to cryptic forms, such as mitosomes and apicoplasts. Integration of these ancient symbionts with their hosts has been too well developed to reverse. Here, we provide evidence that the dinoflagellate Hematodinium sp., a marine parasite of crustaceans, represents a rare case of endosymbiotic organelle loss by the elimination of the plastid. Extensive RNA and genomic sequencing data provide no evidence for a plastid organelle, but, rather, reveal a metabolic decoupling from known plastid functions that typically impede organelle loss. This independence has been achieved through retention of ancestral anabolic pathways, enzyme relocation from the plastid to the cytosol, and metabolic scavenging from the parasite’s host. Hematodinium sp. thus represents a further dimension of endosymbiosis—life after the organelle.
Dinoflagellates belong to a plastid-bearing algal lineage that offers wide scope for studying plastid evolution. Photosynthetic dinoflagellates help power ocean carbon fixation and food webs, and symbiotic dinoflagellates drive tropical reef formation through their coral hosts. However, approximately half of dinoflagellates are heterotrophic and adapted to micropredation and parasitism (1). Even the deepest branching members of the dinoflagellate lineage, which are predominately parasites and predators (2), are secondarily nonphotosynthetic because there is compelling evidence that the common ancestor of dinoflagellates and their sister phylum, Apicomplexa, possessed a photosynthetic plastid (3). All apicomplexans, which include the malaria parasite, have parasitic lifestyles and contain a relic nonphotosynthetic plastid called the apicoplast (except Cryptosporidium; see below). Basal members of the apicomplexan lineage Chromera and Vitrella, however, maintain photosynthesis (4, 5), and their plastids share several distinctive synapomorphies with dinoflagellates (3). Whereas apicomplexan parasites are thought to stem from a single loss of photosynthesis, photosynthesis has been lost on multiple occasions independently in dinoflagellates (6). Ciliates and colponemids represent the basal members of the infrakingdom Alveolata, but they appear to lack plastid organelles (7, 8). Despite the parallel losses of photosynthesis, all heterotrophic dinoflagellates that have been closely scrutinized apparently retain reduced forms of a plastid organelle, likely because they are metabolically dependent on plastid-based de novo synthesis of compounds such as isoprenoids (Perkinsus, Oxyrrhis, Crypthecodinium), tetrapyrroles (Oxyrrhis), and Fe–S clusters (Perkinsus) (9–12). Genes for these pathways are nucleus-encoded, and targeting signals direct the enzymes into plastid organelles. Retention of the same anabolic pathways makes the apicoplast indispensable throughout almost all of Apicomplexa (13–15).
Metabolic functions that drive the retention of plastids, even when photosynthesis is lost, are typically those found within the cytosol of eukaryotes before the gain of a plastid. Endosymbiosis, therefore, leads to pathway duplication. The presence of the plastid often leads to loss(es) of these original host-cell–based pathways, making the plastid indispensible. Loss of the plastid can only occur if the cell avoids or overcomes these dependencies, by: (i) maintaining cytosolic pathways; (ii) scavenging an exogenous supply of metabolites; (iii) eliminating the requirement for the metabolite; (iv) relocating a plastid pathway; or (v) a combination of these. Only one apicomplexan group, Cryptosporidium, has solved this complex puzzle by maintaining cytosolic fatty acid synthesis and scavenging host isoprene precursors and the tetrapyrrole heme (16–18), allowing it to lose the apicoplast. Drastic reduction of heme requirements (Cryptosporidium has only one known cytochrome) (19), and limitation to a single host to complete its life cycle, might also have contributed to overcoming plastid dependency. To date, to our knowledge, Cryptosporidium is the only substantiated case of plastid loss in any eukaryote, highlighting the difficulty of this evolutionary step.
We have investigated the basal, nonphotosynthetic dinoflagellate Hematodinium sp. for evidence of a plastid. Hematodinium species are members of the Syndiniales, an exclusively parasitic basal lineage of dinoflagellates (2). Hematodinium species parasitize decapod crustaceans, have a broad host range, and cause significant impact on commercial fisheries and wild stocks (20). We generated extensive transcriptomic RNA-sequencing (RNA-seq) and genomic data and analyzed these for evidence of a plastid and plastid-associated metabolic pathways. Our data indicate that through some previously unidentified metabolic pathway contortions, Hematodinium—like Cryptosporidium—has also solved the plastid dependency puzzle and eliminated this organelle.
Results and Discussion
No plastid or candidate plastid organelle has been reported in any ultrastructural studies of Hematodinium species (21), but molecular evidence is increasingly a more sensitive and informative approach to testing for cryptic endosymbiotic organelles. We therefore generated extensive molecular data from Hematodinium sp. and interrogated these data for evidence of either an extant or past plastid.
RNA-Seq Provides Extensive Coverage of Hematodinium Genes.
An RNA-seq analysis of different growth stages (trophonts and dinospores) of Hematodinium sp. was undertaken and the data interrogated for evidence of either an extant or past plastid. We assembled 222,704 unique transcripts of >200 bp (Table S1). To assess what portion of the complete gene content of Hematodinium sp. these data represent, we compared them to known databases and recovered 15,510 Swiss-Prot (UniProtKB) matches (<e−5) and 9,207 Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations. Using the Core Eukaryotic Genes Mapping Approach (CEGMA) (22), we recovered 85.9% of the expected core eukaryotic genes, more than the Symbiodinium draft genome or earlier dinoflagellate transcriptomes (Table S2). The large number of unique transcripts (222,704) is likely an overestimation of the true coding capacity due to: (i) observed coding sequence redundancy where UTRs showed either divergent or unrelated sequence (the reasons for which are under investigation); and (ii) high representation of very short ORFs. Clustering of identical ORFs results in 118,749 unique predicted proteins, and exclusion of ORFs <180 amino acid length still retains 29,510 unique ORFs of mean and median length of 443 and 330 amino acids, respectively, which is comparable to known gene length means and medians (472 and 361, respectively) from a wide range of eukaryotes (23, 24). Together, these analyses suggest that we have captured a substantial fraction of Hematodinium sp. genes.
Genes Encoding Plastid Proteins Are Absent in Hematodinium.
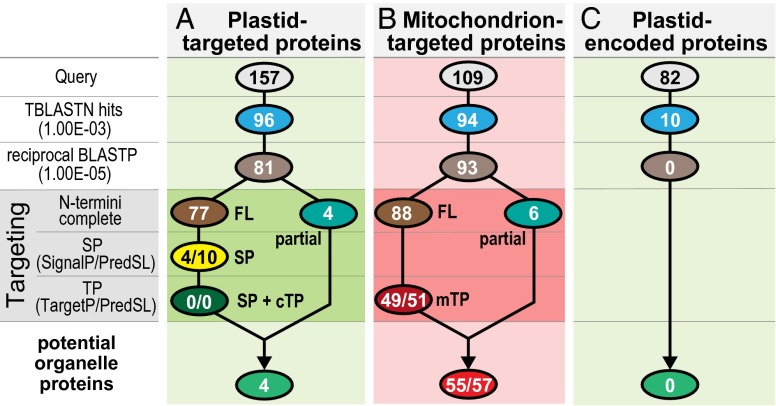
A hallmark of endosymbiosis is that the vast majority of organelle proteins are coded for in the cell nucleus and posttranslationally targeted back to the organelle. Proteins sorted to secondary plastids, including those of dinoflagellates and apicomplexans, are first directed into the endomembrane system via a canonical N-terminal signal peptide (SP) (25, 26). Plastid proteins are then directed into the plastid lumen via a chloroplast-type transit peptide (cTP) downstream of the SP, which shares conserved, recognizable features for almost all plastids (27). Most of the biochemistry of the nonphotosynthetic plastids of apicomplexans is defined by nucleus-encoded plastid proteins bearing this distinctive bipartite (SP + cTP) targeting leader (28). To search for evidence of nucleus-encoded plastid genes in Hematodinium sp., we used a curated set of 157 predicted plastid proteins from Plasmodium vivax (proteins with bipartite leaders, plastid-type functions, and apicoplast-located orthologs) (29). By using RNA-seq data, permissive TBLASTN searches (E value < e−3) were performed and scrutinized by reciprocal BLASTP searches against the National Center for Biotechnology Information (NCBI) RefSeq database (E value < e−5), and 81 proteins were recovered (Fig. 1A and Dataset S1). Many of the Plasmodium protein set represent generic cell functions (e.g., ribosomal function, tRNA ligases, transporters), and so these Hematodinium sp. matches could also represent mitochondrial or cytosolic homologs. To validate the matches as potential plastid proteins, we then analyzed them for the presence of a bipartite leader. Transcripts were first checked for the presence of the dinoflagellate 5′-splice leader (SL) and/or upstream in-frame termination codon as evidence that they are 5′-complete and that the protein N terminus is represented. Of the 81 matches, 77 contained complete ORFs. When these full-length ORFs were tested for a SP (SignalP3.0 and PredSL) followed by a cTP (TargetP1.1 and PredSL), none satisfied these criteria (Fig. 1A and Dataset S1). Of the four incomplete Hematodinium sp. sequences that could not be tested for plastid-targeting signals, two were transporters and two were unknown proteins with conserved domains, so all have credible nonplastid roles. Thus, from a test set of 157 nucleus-encoded plastid proteins, no credible transcripts were found for plastid-targeted proteins in Hematodinium sp. In contrast, the presence of recognizable bipartite targeting peptides provides evidence of reduced plastids in Perkinsus, Oxyrrhis, and Crypthecodinium (9–12).
Fig. 1.
Search strategy for plastid-encoded genes and nucleus-encoded genes for plastid-targeted (or mitochondrial-targeted) proteins. Gene query sets were from predicted apicoplast-targeted proteins common to Plasmodium (A); predicted P. falciparum mitochondrion-targeted proteins (B); and plastid-encoded genes common to apicomplexans, dinoflagellates, and chromerids (C). A list of genes and full search results are shown in Datasets S1–S3. FL, full length; mTP, mitochondrial targeting peptide.
To assess the expected likelihood of recovering nucleus-encoded organelle-targeted proteins from our dataset, an equivalent search was performed for mitochondrial proteins, with the exception of scoring for mitochondrial targeting peptides (mTP). By using a set of 109 mitochondrial proteins from P. falciparum (30), 94 putative mitochondrial proteins were identified in the Hematodinium sp. dataset. Of these proteins, 49 (TargetP1.1) and 51 (PredSL) contained credible mitochondrial targeting information—an overall recovery rate of 45 and 47%, respectively (Fig. 1B and Dataset S2). Targeting peptide predictors by their nature are not perfect and will produce some false predictions. A previous assessment of cTP prediction (ChloroP) in dinoflagellates indicated 45% success (27), similar to that shown for mTP prediction here (56–58%). Given the detection of mitochondria-targeted proteins in this system, and that of plastid proteins in other low-branching dinoflagellates, we believe the most plausible explanation is that plastid-targeted proteins are no longer present in Hematodinium sp. It is conceivable, however, that plastid-targeted proteins do exist, but are of very low abundance, too divergent, or exceptional in lacking identifiable bipartite leaders.
Plastid genes can also be encoded directly in the organelle, although dinoflagellates are unusual in having reduced the number of plastid-encoded genes to only 14 (3). We searched for all known protein genes found in alveolates plastids [82 genes in total, found in dinoflagellates, apicomplexans, or chromerids (3)] and found no matches in our RNA-seq data (Fig. 1C and Dataset S3). In contrast, all five known alveolate mtDNA genes were readily detected (31), and we note that 11 of 14 dinoflagellate plastid-coding genes are represented in equivalent Lingulodinium poly-A–derived RNA-Seq data (32). Furthermore, we have now generated a draft genomic sequence for Hematodinium sp. The estimated genome size is ∼4,800 Mbp (33), consistent with the very large genomes of other dinoflagellates (ref. 34 and references therein). We currently have 4,769 Mbp of data assembled (30× coverage, N50 = 17,235 bp, 118,385 scaffolds >10 kbp, 653 scaffolds >100 kbp). Preliminary analysis of these data revealed that genes are densely populated with very large introns and are, therefore, spread over tens to hundreds of kilobase pairs. Thus, many nucleus-encoded gene sequences are currently incomplete, even on relatively large scaffolds, and sequencing and assembly of this genome is ongoing. Organelle genomes, however, are typically gene-dense, contain few or no introns, and are well represented in genomic sequence data. We searched these genomic data for the 82 alveolate plastid-encoded genes and, similar to the RNA-seq data, found none. Thus, there is a conspicuous absence of any genes for plastid proteins in Hematodinium sp., consistent with loss of this organelle.
Hematodinium Encodes Biochemical Alternatives to Plastid Functions.
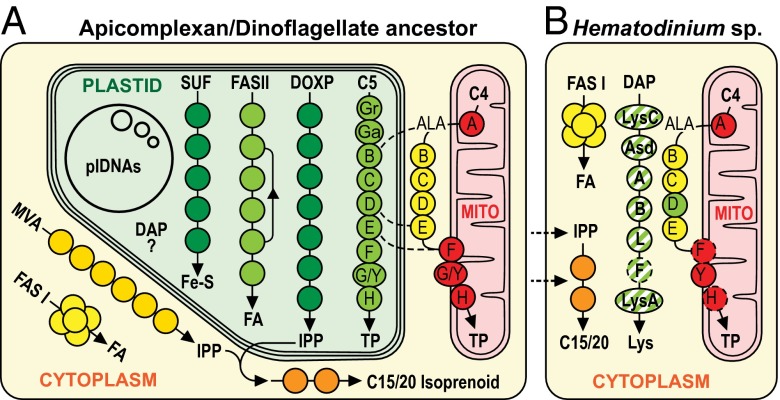
In apicomplexans, ablation of plastid biochemical pathways by gene knockouts and chemical inhibition identified several anabolic pathways that are essential for the completion of the life cycle—including fatty acid biosynthesis, tetrapyrrole synthesis, and isopentenyl pyrophosphate (IPP) (isoprene precursor) synthesis (13, 15, 35–37). The indispensability of these pathways is likely the major explanation for plastid retention. In the absence of either molecular or morphological evidence of a plastid in Hematodinium, we have analyzed our data for evidence of nonplastid alternative pathways that could alleviate the need for these typically essential plastid functions (Fig. 2).
Fig. 2.
Reconstructed metabolic pathways in the common ancestor of apicomplexans and dinoflagellates at the time of plastid gain (A) and in Hematodinium sp. (B), from transcriptomic data. Cytosolic MVA pathway is not present in any apicomplexan or dinoflagellate, but is present in ciliates and is inferred to be present at the time of plastid gain. Dashed lines indicate apicomplexan hybrid tetrapyrrole pathways; dashed circles indicate enzymes currently unidentified in transcriptomes. Enzyme color represents typical location and origin as follows: green, plastid; yellow, cytosol; red, mitochondrion. Hatched (green/white) DAP pathway indicates uncertain origin of this typically plastid-located pathway in Hematodinium. MVA, mevalonate IPP pathway; DOXP, 1-deoxy-d-xylulose-5-phosphate IPP pathway; C15/20, isoprene chains 15 and 20 carbons long derived from IPP (an external source of IPP/isoprenoids for Hematodinium sp. is predicted); SUF, plastid-type iron-sulfur cluster pathway; DAP, diaminopimelate lysine pathway; C4/C5 pathways for tetrapyrrole (TP) synthesis differ only by the reactions to δ-aminolevulinic acid (ALA) and their location.
Fatty acids are generated by using cytosolic type I, and not plastid type II, fatty acid synthase.
Fatty acids are assembled de novo in cells by iterative additions of two carbons, donated from malonyl–CoA, by fatty acid synthase (FAS) complexes. Prokarytotic FAS systems typically consists of seven separate monofunctional proteins, and this multiprotein complex, called type II FAS, occurs in plastids. Evolution of FAS in the eukaryotic cytosol has entailed fusion of most or all of the necessary enzyme modules to form the multienzyme type I FAS complex (Fig. 2). Plastid-containing organisms can, therefore, contain both type I and II FAS capabilities, or they might lose one or the other. In apicomplexans, Toxoplasma and Eimeria have both, whereas Plasmodium retains only the plastid type II form, and Cryptosporidium retains only cytosolic type I (14, 38). Thus, the common ancestor of apicomplexans and dinoflagellates must have had both forms. Although FAS in dinoflagellates is less well studied, these organisms can synthesize fatty acids de novo (39, 40). Gene disruption of type II FAS in Plasmodium shows that this plastid pathway is essential during the liver stage and, in a human strain, the insect stage of the life cycle, implying that FAS might be a function mandating plastid retention (13, 35, 41).
We searched the Hematodinium sp. transcript data and found coding sequence for all necessary FAS enzymes, but never as separate genes. Instead, they were always present on transcripts of large, multienzyme complexes, consistent with the presence of a fused type I FAS and the absence of a type II FAS complex (Fig. 2B). Our RNA-seq data recovered three large FAS-enzyme-containing transcripts (GenBank accession nos. KP739886–KP739888), whose full-length ORFs all lack a bipartite leader and which are predicted to be cytosolic, as well as two further incomplete transcripts with multiple FAS-like enzyme domains (GenBank accession nos. KP739889–KP739890; Fig. S1A). Polyketide syntases (PKS) are a related family of proteins that are considered degenerate FASs in which one or more modules have been lost, and PKS are known to occur in dinoflagellates (40). Although two of the transcripts contain all modules expected of complete type I FAS proteins, it is difficult to reliably discern FAS from PKS without biochemical validation. Therefore, we tested for de novo fatty acid synthesis by measuring incorporation of [13C]-U-glucose into Hematodinium sp. fatty acids. 13C-glucose is catabolized to pyruvate and acetyl-CoA, providing the precursors for the FAS complexes. Label was uniformly incorporated into saturated C14:0 and C16:0 fatty acids in 13C-glucose–fed Hematodinium sp., generating a series of fatty-acid isotopomers that increased by +2 atomic mass units, consistent with de novo synthesis of fatty acids from a malonyl–CoA donor (Fig. S1 B and C). These analyses suggest that Hematodinium sp. express a large type I FAS complex that would allow loss of a plastid-located type II complex.
Tetrapyrrole biosynthesis occurs in the cytosol.
Tetrapyrrole biosynthesis is required for the synthesis of heme and chlorophylls, and it occurs via two related pathways (19) (Fig. 2A). The C4 pathway, which occurs in heterotrophic eukaryotes, commences with the synthesis of δ-aminolevulinic acid (ALA) from glycine and succinyl-CoA by ALA synthase (ALAS) in the mitochondrion. ALA is exported to the cytosol, where the enzymes porphobilinogen synthase (HemB), porphobilinogen deaminase (HemC), uroporphyrinogen III synthase (HemD), and uroporphyrinogen III decarboxylase (HemE) catalyze the production of coproporphyrinogen, which is then transported back into the mitochondrion to be converted into heme by coproporphyrinogen oxidase (HemF), protoporphyrinogen oxidase (HemG/Y), and ferrochelatase (HemH). In contrast, most photosynthetic eukaryotes produce tetrapyrroles through the plastid-located C5 pathway. Here, ALA is synthesized from glutamyl–tRNA by glutamyl–tRNA reductase (GTR) followed by glutamate 1-semialdehyde aminotransferase (GSA-AT). ALA is then converted into tetrapyrroles via the same enzymes found in the C4 pathway, but these enzymes are mostly derived from the cyanobacterium that gave rise to plastids. In most photosynthetic eukaryotes, the plastid C5 pathway has been retained in place of the ancestral C4 pathway, and the bulk of tetrapyrroles are incorporated into chlorophylls.
Tetrapyrrole synthesis in apicomplexans and their photosynthetic relative Chromera velia is unusual in that elements of both the C4 and C5 pathways have been used in a pathway of hybrid origin (19, 42) (Fig. 2A, dashed lines). Classic C4 synthesis of ALA occurs in the mitochondrion (with plastid enzymes GTR and GSA-AT being lost) and is transported into the plastid. Here, either all seven of the remaining reactions of the C5 pathway either occur in the plastid (Chromera), or only three or four reactions occur in the plastid (other apicomplexans) before protoporphyrinogen or coproporphyrinogen is exported back to the mitochondrion for final heme synthesis. In either case, a linear pathway is envisaged, and both taxonomic groups rely on their plastid for parts of this anabolic process.
Photosynthetic dinoflagellates appear to have retained a C5 plastid pathway, including GTR and GSA-AT (10, 42, 43). Although later-branching dinoflagellates lack ALAS of the C4 pathway, we have previously reported this mitochondrial C4 gene in Hematodinium sp. (44), and it is also in basal lineages Perkinsus and Oxyrrhis. Hematodinium sp. ALAS lacks an obvious mTP; however, when we expressed it as a GFP fusion in Toxoplasma, mitochondrial localization was seen, suggesting that it has a cryptic mitochondrial-targeting signal (Fig. S2). Neither plastid-type GTR nor GSA-AT are found in Hematodinium sp., so although the two alternative pathways for ALA synthesis, C4 and C5, coexisted during the early radiation of dinoflagellates, this redundancy was differentially lost in early and later-branching lineages.
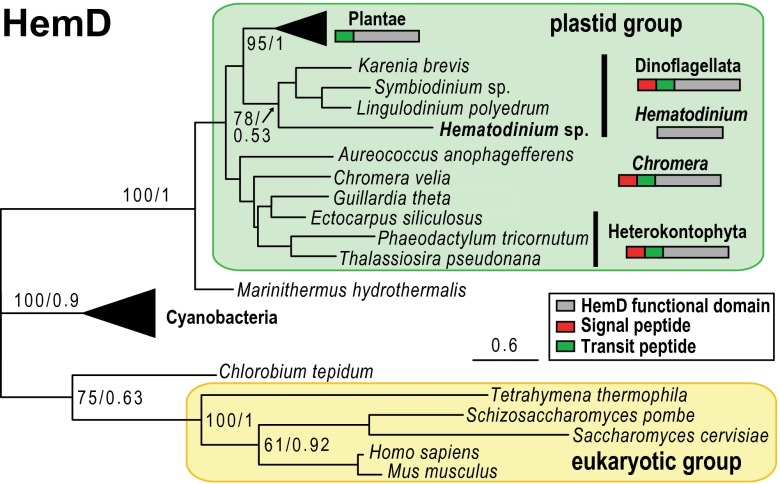
If Hematodinium contains a mosaic pathway, such as in apicomplexans (19, 42), HemB–E could still reside in the plastid. We indeed find Hematodinium sp. does contain genes for these enzymes (GenBank accession nos. KP739891–KP739896). However, they all lack plastid-targeting leaders and are predicted to be cytosolic. Further, HemB, HemC, and HemE enzymes are phylogenetically placed with the cytosolic enzymes found in other heterotrophic eukaryotes (Fig. S3). These data provide further evidence that after the gain of a plastid, duplicated elements of this pathway persisted. Hematodinium sp. HemD is the exception, in that it groups with plastids in phylogenies, and specifically with photosynthetic dinoflagellates (Fig. 3). A plastid-derived HemD provides direct evidence that a plastid was present in the Hematodinium ancestor. Relocation of plastidial HemD to the cytosol has seemingly replaced the former cytosolic enzyme (Fig. 2B). The remainder of the tetrapyrrole pathway apparently takes place in the mitochondrion, because Hematodinium sp. HemY contains a predicted mTP (Fig. 2B). This location is consistent with the mitochondrion being the major site of utilization of tetrapyrroles in the absence of chlorophyll synthesis in these parasites. Only HemF and HemH were not found in the Hematodinium sp. transcriptome; however, the ferrochelatase (HemH) has not been identified in any dinoflagellate, suggesting that a divergent enzyme is present. In summary, although the replacement of cytosolic HemD with the plastid homolog is previously unidentified, tetrapyrrole synthesis in Hematodinium sp. is otherwise typical of the classic heterotrophic cytosol/mitochondrial C4 pathway and is consistent with nonreliance on a plastid.
Fig. 3.
HemD (uroporphyrinogen III synthase) phylogeny showing a plastid-derived protein in Hematodinium sp. Presence/absence of targeting leader sequences for the plastid clade is indicated. Support values (ML bootstraps/Bayesian posterior probabilities) are shown only for major clades. Wedges indicate collapsed clades shown in full in Fig. S3.
Lysine biosynthesis takes place in the cytosol.
Prokaryotes and plants synthesize lysine via the intermediate diaminopimelate (DAP) (45). Fungi use a different pathway with an alternative intermediate (α-aminoadipic acid), and animals are unable to synthesis lysine, relying on dietary intake (45). The canonical prokaryotic DAP pathway uses nine enzymes to convert aspartate into lysine: LysC, Asd, DapA, DapB, DapD, DapC, DapE, DapF, and LysA. Chloroplasts are the site of lysine synthesis from aspartate in plants (46), a function assumed to originate from their cyanobacterial forebears. However, plants lack identifiable dapD, dapC, and dapE genes, suggesting the presence of an alternative pathway. A novel enzyme, LL-diaminopimelate aminotransferase (DapL), was recently discovered in Arabidopsis thaliana, which replaces DapC–E, and defines a new variant of the DAP pathway (47, 48). DapL is also reported from green algae, cyanobacteria, and among other Bacteria and Archaea (48, 49). In broad genome and transcriptome surveys of diverse eukaryotes (MMETSP) (50), we identify DapL also in red algae and among secondary plastid-containing algae (heterokonts, haptophytes, cryptophytes, and basal dinoflagellates) (Fig. S4). In eukaryotes, DapL is restricted to plastid-containing lineages.
We found dapL in Hematodinium sp. (GenBank accession no. KP739901), along with the genes for the other enzymes of the DAP pathway (LysC, Asd, DapA, DapB, and LysA; only DapF was not found; GenBank accession nos. KP739897–KP739900 and KP739902). All plant, green algal, and red algal DapL proteins contain an N-terminal plastid-targeting transit peptide for plastid targeting, and heterokont proteins (e.g., diatom Phaedactylum and brown algae Ectocarpus) bear a bipartite leader (SP plus transit peptide), showing plastid retention of this pathway through secondary endosymbiosis. All of the Hematodinium sp. DAP enzymes lack obvious plastid-targeting presequences, and this pathway is therefore predicted to be cytosolic in Hematodinium sp. One explanation for these data is that a plastid-derived DAP pathway was relocated to the cytosol, similar to relocation of HemD. Support for this hypothesis, however, is equivocal. In DapL phylogenies, the position of Hematodinium sp., and other basal dinozoan taxa Perkinsus marinus and Oxyrrhis marina, was not clearly resolved, with members of the bacterial phyla Chlamydiae, Spirochaetes, and Planctomycetes interrupting the eukaryotic/plastid clade (Fig. S4). Furthermore, DapL is not found in Apicomplexa [where host lysine is used (51)], and we did not find it in photosynthetic dinoflagellate data, so a precedent and source of a plastid-located DAP pathway in this lineage is not apparent. An alternative hypothesis would be external gain of this pathway by lateral gene transfers. The current phylogenies, however, are not able to provide strong support for this scenario either (Fig. S4). The origin of the Hematodinium DAP pathway, therefore, is presently unclear. Nevertheless, location of this lysine biosynthetic pathway in the cytosol of a eukaryote is both previously unknown and provides no barrier to loss of a plastid in Hematodinium sp.
A de novo isoprenoid biosynthesis pathway is absent.
Isoprenoids are essential lipid molecules that mainly act as precursors for the synthesis of sterols, chlorophyll, and quinones. They are used as lipid anchors for some proteins and as prosthetic groups in tRNAs and proteins. Isoprenoids are built from the five-carbon precursors IPP and its isomer, dimethylallyl pyrophosphate (DMAPP). Two different pathways exist for IPP synthesis in eukaryotes (52). The canonical mevalonate (MVA) pathway consists of six enzymes and occurs in the cytosol of most heterotrophic eukaryotes, as well as some autotrophs, such as plants. A second, nonmevalonate (or DOXP) pathway, occurs in plastids, and uses seven enzymes derived from cyanobacteria. These pathways use distinct enzymes and metabolic intermediates. In particular, the MVA pathway uses acetyl-CoA and acetoacetyl-CoA and has MVA as a key intermediate, whereas the DOXP pathway uses pyruvate and glyceraldehyde-3-phosphate, resulting in production of the distinct intermediate, 1-deoxy-d-xylulose-5-phosphate (DOXP). These pathways converge with the interconversion of IPP and DMAPP by the enzyme IPP isomerase.
Within alveolates, ciliates use the MVA pathway, whereas apicomplexans exclusively use the plastid-located DOXP pathway (8, 36). Dinoflagellates also only appear to contain a plastid DOXP pathway, irrespective of whether they are photosynthetic or secondarily nonphotosynthetic (9, 10, 53, 54). Indeed, in the basal lineage P. marinus, the most substantial evidence for a plastid is presence of all seven DOXP enzyme genes (six published, plus GenBank accession no. AB445015) with bipartite plastid-targeting leaders (12). Thus, the MVA pathway appears to have been entirely abandoned in favor of the plastid pathway in the common ancestor of dinoflagellates and apicomplexans since they diverged from ciliates (Fig. 2A).
Unexpectedly, we found no evidence for the presence of either the DOXP or the MVA pathway in Hematodinium sp. Nevertheless, we readily identified pathways for assembly and utilization of IPP/DMAPP-containing molecules in Hematodinium sp., including a farnesyl–pyrophosphate synthase (GenBank accession no. KP739903), which produces farnesyl–pyrophosphate through a sequential condensation reaction of DMAPP with two units of IPP and the alpha- and beta-subunits of both farnesyltransferase and geranylgeranyltransferase (GenBank accession nos. KP739904–KP739907), which add either a C15 or C20 prenyl-group to prenylate proteins. These data strongly suggest that Hematodinium sp. can scavenge IPP and/or DMAPP or other isoprenoids (Fig. 2B). A similar salvage mechanism operates in other parasites lacking a de novo pathway, such as Cryptosporidium (17). In this context, it is noteworthy that crustaceans produce and circulate the methylated linear C15-isporenoid molecule, methyl farnesoate, as an important crustacean hormone. Methyl farnesoate is secreted from the mandibular organs into the hemolymph, where Hematodinium grows, and in crustaceans it reaches concentrations up to 0.6 µM and plays a role in morphogenesis, molt cycles, and other metabolic responses in these animals (55). Such a molecule might contribute either directly to Hematodinium isoprenoids or conceivably be metabolized back to IPP and DMAPP. Regardless of the source of scavenged molecules, the absence of a DOXP pathway is again consistent with loss of a plastid from Hematodinium sp.
Conclusion
Compilation of genomic evidence for plastid-type metabolic pathways that occur in apicomplexans, dinoflagellates, and ciliates provides insights into the pathways that were present in the common ancestor at the time of plastid gain (Fig. 2A). These analyses suggest that there have been many different lineage-specific losses and retentions of host and/or plastid pathways. For example: (i) loss of MVA-derived IPP from the ancestor of apicomplexans and dinoflagellates; (ii) loss of type I FAS from Plasmodium and type II FAS from piroplasms (14, 38); and (iii) multiple losses and retentions of C4 and C5 enzymes for tetrapyrrole synthesis and complex rerouting of these pathways in both dinoflagellates and apicomplexans (19, 42). Although the origin of the DAP pathway for lysine biosynthesis in Hematodinium sp. is uncertain, this pathway is present in red algal plastids and most red-derived secondary plastids, so it is likely that this pathway was originally present in the apicomplexan/dinoflagellate ancestor also but was lost once or perhaps multiple times. The metabolic roles of plastids, once gained, therefore, appear to have been highly dynamic in the evolution of these lineages, and it is thus possible that dependency upon plastid functions beyond photosynthesis occurred relatively quickly. The chaotic distribution of pathway gains and losses might also suggest rapid radiation after organelle acquisition. The rarity of plastid loss among the eukaryotic lineages that have lost photosynthesis suggests the likelihood of gaining metabolic freedom from plastids once they have been integrated is extremely low. Hematodinium sp. appears to have achieved plastid loss by retention of cytosolic pathways for synthesis of fatty acids and tetrapyrroles and supplementing loss of the DOXP pathway by scavenging intermediates for isoprenoid synthesis, likely from its crustacean hosts. The lack of plastid-type II FAS and DOXP pathways would also have removed the need for plastid Fe-S clusters that function as prosthetic groups in both pathways, thereby allowing further loss of this pathway, whereas both mitochondrial and cytosolic Fe-S cluster assembly pathways remain present. However, enzyme relocation has also been necessary, with one plastid enzyme for tetrapyrrole (HemD) relocated from the plastid to the cytosol. In comparison with Hematodinium, plastid loss from Cryptosporidium, the only other substantiated case of endosymbiotic organelle loss, appears to have been achieved primarily by extreme parasite reduction and a more complete reliance on host animal metabolites. Together, these taxa illustrate the complex machinations required to undo endosymbiosis and the reasons why it is so rarely achieved.
Materials and Methods
Transcriptome Sequencing, Assembly, and Analysis.
Hematodinium sp. was cultured as described (33). Hematodinium sp. dinospores were harvested upon sporulation from infected Nephrops norvegicus lobster (courtesy of N. Beevers, University of Glasgow). Total RNA was extracted, polyA-enriched, sequenced, and assembled by using the de novo assembler “Trinity” as described in SI Materials and Methods. Following assembly, ORFs were predicted using EMBOSS (Version 6.5.7) getorf (56). The CEGMA test for percentage of highly conserved core eukaryotic genes was performed as described (22).
Plastid Gene Searches, Targeting Sequence Prediction, and Phylogenies.
Similarity searches were performed by using NCBI BLAST. SignalP (Version 3.1) and TargetP (Version 1.1) predictions were made at standard settings as described (2007) (57). PredSL was run as described (58). For phylogenies, nearest homologs were identified from public databases including from the MMETSP (50). Alignments were generated by using MAFFT (59), manually corrected, and ambiguous sites were removed. Maximum-likelihood phylogenies were performed by using RAxML (60) using the best-fit model (LG+I+G+8) inferred by Prottest3 (61). Bayesian analyses were performed with MrBayes (Version 3.2.1) (62) by using the WAG+I+G+4 model. Markov Chain Monte Carlo runs of 1,100,000 generations were calculated with trees sampled every 200 generations and with a prior burn-in of 100,000 generations.
Genomic sequencing and metabolomics methods are outlined in SI Materials and Methods and Dataset S4.
Supplementary Material
Acknowledgments
We thank Nick Katris for assistance with Toxoplasma transformation and Ellen Nisbet for critically reading this report. This work was supported by Australian Research Council (ARC) Grants DP130100572 and DP1093395; a King Abdullah University of Science and Technology Faculty Baseline Research Fund; and Victorian Life Sciences Computation Initiative Grant VR0254. S.G.G. was supported by Science Foundation Ireland Grant 13/SIRG/2125; F. was supported by an Australia Award; A.M.C. and A.B. were supported by ARC Centre of Excellence in Plant Cell Walls Grant CE110001007; and M.J.M. was supported by the National Health and Medical Research Council as a Principal Research Fellow.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.M.A. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KP739886–KP739907).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1423400112/-/DCSupplemental.
References
- 1.Coats DW. Parasitic life styles of marine dinoflagellates. J Eukaryot Microbiol. 1999;46:402–409. [Google Scholar]
- 2.Bachvaroff TR, et al. Dinoflagellate phylogeny revisited: Using ribosomal proteins to resolve deep branching dinoflagellate clades. Mol Phylogenet Evol. 2014;70:314–322. doi: 10.1016/j.ympev.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janouškovec J, Horák A, Oborník M, Lukes J, Keeling PJ. A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc Natl Acad Sci USA. 2010;107(24):10949–10954. doi: 10.1073/pnas.1003335107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Dooren GG, Striepen B. The algal past and parasite present of the apicoplast. Annu Rev Microbiol. 2013;67:271–289. doi: 10.1146/annurev-micro-092412-155741. [DOI] [PubMed] [Google Scholar]
- 5.Moore RB, et al. A photosynthetic alveolate closely related to apicomplexan parasites. Nature. 2008;451(7181):959–963. doi: 10.1038/nature06635. [DOI] [PubMed] [Google Scholar]
- 6.Saldarriaga JF, Taylor FJ, Keeling PJ, Cavalier-Smith T. Dinoflagellate nuclear SSU rRNA phylogeny suggests multiple plastid losses and replacements. J Mol Evol. 2001;53(3):204–213. doi: 10.1007/s002390010210. [DOI] [PubMed] [Google Scholar]
- 7.Janouškovec J, et al. Colponemids represent multiple ancient alveolate lineages. Curr Biol. 2013;23(24):2546–2552. doi: 10.1016/j.cub.2013.10.062. [DOI] [PubMed] [Google Scholar]
- 8.Eisen JA, et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4(9):e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slamovits CH, Keeling PJ. Plastid-derived genes in the nonphotosynthetic alveolate Oxyrrhis marina. Mol Biol Evol. 2008;25(7):1297–1306. doi: 10.1093/molbev/msn075. [DOI] [PubMed] [Google Scholar]
- 10.Sánchez-Puerta MV, Lippmeier JC, Apt KE, Delwiche CF. Plastid genes in a non-photosynthetic dinoflagellate. Protist. 2007;158(1):105–117. doi: 10.1016/j.protis.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 11.Fernández Robledo JA, et al. The search for the missing link: A relic plastid in Perkinsus? Int J Parasitol. 2011;41(12):1217–1229. doi: 10.1016/j.ijpara.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuzaki M, Kuroiwa H, Kuroiwa T, Kita K, Nozaki H. A cryptic algal group unveiled: A plastid biosynthesis pathway in the oyster parasite Perkinsus marinus. Mol Biol Evol. 2008;25(6):1167–1179. doi: 10.1093/molbev/msn064. [DOI] [PubMed] [Google Scholar]
- 13.Vaughan AM, et al. Type II fatty acid synthesis is essential only for malaria parasite late liver stage development. Cell Microbiol. 2009;11(3):506–520. doi: 10.1111/j.1462-5822.2008.01270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seeber F, Soldati-Favre D. Metabolic pathways in the apicoplast of apicomplexa. Int Rev Cell Mol Biol. 2010;281:161–228. doi: 10.1016/S1937-6448(10)81005-6. [DOI] [PubMed] [Google Scholar]
- 15.Ramakrishnan S, et al. Apicoplast and endoplasmic reticulum cooperate in fatty acid biosynthesis in apicomplexan parasite Toxoplasma gondii. J Biol Chem. 2012;287(7):4957–4971. doi: 10.1074/jbc.M111.310144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu P, et al. The genome of Cryptosporidium hominis. Nature. 2004;431(7012):1107–1112. doi: 10.1038/nature02977. [DOI] [PubMed] [Google Scholar]
- 17.Bessoff K, Sateriale A, Lee KK, Huston CD. Drug repurposing screen reveals FDA-approved inhibitors of human HMG-CoA reductase and isoprenoid synthesis that block Cryptosporidium parvum growth. Antimicrob Agents Chemother. 2013;57(4):1804–1814. doi: 10.1128/AAC.02460-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu G, Marchewka MJ, Keithly JS. Cryptosporidium parvum appears to lack a plastid genome. Microbiology. 2000;146(Pt 2):315–321. doi: 10.1099/00221287-146-2-315. [DOI] [PubMed] [Google Scholar]
- 19.van Dooren GG, Kennedy AT, McFadden GI. The use and abuse of heme in apicomplexan parasites. Antioxid Redox Signal. 2012;17(4):634–656. doi: 10.1089/ars.2012.4539. [DOI] [PubMed] [Google Scholar]
- 20.Stentiford GD, Shields JD. A review of the parasitic dinoflagellates Hematodinium species and Hematodinium-like infections in marine crustaceans. Dis Aquat Organ. 2005;66(1):47–70. doi: 10.3354/dao066047. [DOI] [PubMed] [Google Scholar]
- 21.Appleton PL. 1996. Investigations on the cytology and life-cycle of the parasitic dinoflagellate Hematodinium sp associated with mortality of Nephrops norvegicus. Dissertation (University of Glasgow, Glasgow)
- 22.Parra G, Bradnam K, Korf I. CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23(9):1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 23.Tiessen A, Pérez-Rodríguez P, Delaye-Arredondo LJ. Mathematical modeling and comparison of protein size distribution in different plant, animal, fungal and microbial species reveals a negative correlation between protein size and protein number, thus providing insight into the evolution of proteomes. BMC Res Notes. 2012;5:85. doi: 10.1186/1756-0500-5-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brocchieri L, Karlin S. Protein length in eukaryotic and prokaryotic proteomes. Nucleic Acids Res. 2005;33(10):3390–3400. doi: 10.1093/nar/gki615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nassoury N, Cappadocia M, Morse D. Plastid ultrastructure defines the protein import pathway in dinoflagellates. J Cell Sci. 2003;116(Pt 14):2867–2874. doi: 10.1242/jcs.00517. [DOI] [PubMed] [Google Scholar]
- 26.Waller RF, Reed MB, Cowman AF, McFadden GI. Protein trafficking to the plastid of Plasmodium falciparum is via the secretory pathway. EMBO J. 2000;19(8):1794–1802. doi: 10.1093/emboj/19.8.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patron NJ, Waller RF. Transit peptide diversity and divergence: A global analysis of plastid targeting signals. BioEssays. 2007;29(10):1048–1058. doi: 10.1002/bies.20638. [DOI] [PubMed] [Google Scholar]
- 28.Ralph SA, et al. Tropical infectious diseases: Metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat Rev Microbiol. 2004;2(3):203–216. doi: 10.1038/nrmicro843. [DOI] [PubMed] [Google Scholar]
- 29.Carlton JM, et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455(7214):757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Dooren GG, Stimmler LM, McFadden GI. Metabolic maps and functions of the Plasmodium mitochondrion. FEMS Microbiol Rev. 2006;30(4):596–630. doi: 10.1111/j.1574-6976.2006.00027.x. [DOI] [PubMed] [Google Scholar]
- 31.Jackson CJ, Gornik SG, Waller RF. The mitochondrial genome and transcriptome of the basal dinoflagellate Hematodinium sp.: Character evolution within the highly derived mitochondrial genomes of dinoflagellates. Genome Biol Evol. 2012;4(1):59–72. doi: 10.1093/gbe/evr122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beauchemin M, et al. Dinoflagellate tandem array gene transcripts are highly conserved and not polycistronic. Proc Natl Acad Sci USA. 2012;109(39):15793–15798. doi: 10.1073/pnas.1206683109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gornik SG, et al. Loss of nucleosomal DNA condensation coincides with appearance of a novel nuclear protein in dinoflagellates. Curr Biol. 2012;22(24):2303–2312. doi: 10.1016/j.cub.2012.10.036. [DOI] [PubMed] [Google Scholar]
- 34.LaJeunesse TC, Lambert G, Andersen RA, Coffroth MA, Galbraith DW. Symbiodinium (Pyrrhophyta) genome sizes (DNA conent) are smallest among dinoflagellates. J Phycol. 2005;41:880–886. [Google Scholar]
- 35.Yu M, et al. The fatty acid biosynthesis enzyme FabI plays a key role in the development of liver-stage malarial parasites. Cell Host Microbe. 2008;4(6):567–578. doi: 10.1016/j.chom.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeh E, DeRisi JL. Chemical rescue of malaria parasites lacking an apicoplast defines organelle function in blood-stage Plasmodium falciparum. PLoS Biol. 2011;9(8):e1001138. doi: 10.1371/journal.pbio.1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Botté CY, Dubar F, McFadden GI, Maréchal E, Biot C. Plasmodium falciparum apicoplast drugs: Targets or off-targets? Chem Rev. 2012;112(3):1269–1283. doi: 10.1021/cr200258w. [DOI] [PubMed] [Google Scholar]
- 38.Zhu G. Current progress in the fatty acid metabolism in Cryptosporidium parvum. J Eukaryot Microbiol. 2004;51(4):381–388. doi: 10.1111/j.1550-7408.2004.tb00384.x. [DOI] [PubMed] [Google Scholar]
- 39.Sonnenborn U, Kunau W-H. Purification and properties of the fatty acid synthetase complex from the marine dinoflagellate, Crypthecodinium cohnii. Biochim Biophys Acta Lipids Lipid Metab. 1982;712:523–534. doi: 10.1016/0005-2760(82)90280-6. [DOI] [PubMed] [Google Scholar]
- 40.Van Dolah FM, et al. Subcellular localization of dinoflagellate polyketide synthases and fatty acid synthase activity. J Phycol. 2013;49:1118–1127. doi: 10.1111/jpy.12120. [DOI] [PubMed] [Google Scholar]
- 41.van Schaijk BCL, et al. Type II fatty acid biosynthesis is essential for Plasmodium falciparum sporozoite development in the midgut of Anopheles mosquitoes. Eukaryot Cell. 2014;13(5):550–559. doi: 10.1128/EC.00264-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koreny L, Sobotka R, Janouškovec J, Keeling PJ, Oborník M. Tetrapyrrole synthesis of photosynthetic chromerids is likely homologous to the unusual pathway of apicomplexan parasites. Plant Cell. 2011;23(9):3454–3462. doi: 10.1105/tpc.111.089102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patron NJ, Waller RF, Keeling PJ. A tertiary plastid uses genes from two endosymbionts. J Mol Biol. 2006;357(5):1373–1382. doi: 10.1016/j.jmb.2006.01.084. [DOI] [PubMed] [Google Scholar]
- 44.Danne JC, Gornik SG, Macrae JI, McConville MJ, Waller RF. Alveolate mitochondrial metabolic evolution: Dinoflagellates force reassessment of the role of parasitism as a driver of change in apicomplexans. Mol Biol Evol. 2013;30(1):123–139. doi: 10.1093/molbev/mss205. [DOI] [PubMed] [Google Scholar]
- 45.Velasco AM, Leguina JI, Lazcano A. Molecular evolution of the lysine biosynthetic pathways. J Mol Evol. 2002;55(4):445–459. doi: 10.1007/s00239-002-2340-2. [DOI] [PubMed] [Google Scholar]
- 46.Mills WR, Wilson KG. Amino acid biosynthesis in isolated pea chloroplasts: Metabolism of labeled aspartate and sulfate. FEBS Lett. 1978;92:129–132. [Google Scholar]
- 47.Hudson AO, Singh BK, Leustek T, Gilvarg C. An LL-diaminopimelate aminotransferase defines a novel variant of the lysine biosynthesis pathway in plants. Plant Physiol. 2006;140(1):292–301. doi: 10.1104/pp.105.072629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCoy AJ, et al. L,L-diaminopimelate aminotransferase, a trans-kingdom enzyme shared by Chlamydia and plants for synthesis of diaminopimelate/lysine. Proc Natl Acad Sci USA. 2006;103(47):17909–17914. doi: 10.1073/pnas.0608643103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dobson RCJ, Girón I, Hudson AO. L,L-diaminopimelate aminotransferase from Chlamydomonas reinhardtii: A target for algaecide development. PLoS ONE. 2011;6(5):e20439. doi: 10.1371/journal.pone.0020439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keeling PJ, et al. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): Illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 2014;12(6):e1001889. doi: 10.1371/journal.pbio.1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shanmugasundram A, Gonzalez-Galarza FF, Wastling JM, Vasieva O, Jones AR. Library of Apicomplexan Metabolic Pathways: A manually curated database for metabolic pathways of apicomplexan parasites. Nucleic Acids Res. 2013;41(Database issue):D706–D713. doi: 10.1093/nar/gks1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eisenreich W, Bacher A, Arigoni D, Rohdich F. Biosynthesis of isoprenoids via the non-mevalonate pathway. Cell Mol Life Sci. 2004;61(12):1401–1426. doi: 10.1007/s00018-004-3381-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waller RF, Patron NJ, Keeling PJ. Phylogenetic history of plastid-targeted proteins in the peridinin-containing dinoflagellate Heterocapsa triquetra. Int J Syst Evol Microbiol. 2006;56(Pt 6):1439–1447. doi: 10.1099/ijs.0.64061-0. [DOI] [PubMed] [Google Scholar]
- 54.Bachvaroff TR, Place AR. From stop to start: Tandem gene arrangement, copy number and trans-splicing sites in the dinoflagellate Amphidinium carterae. PLoS ONE. 2008;3(8):e2929. doi: 10.1371/journal.pone.0002929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nagaraju GPC. Is methyl farnesoate a crustacean hormone? Aquaculture. 2007;272:39–54. [Google Scholar]
- 56.Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16(6):276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 57.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2(4):953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 58.Petsalaki EI, Bagos PG, Litou ZI, Hamodrakas SJ. PredSL: A tool for the N-terminal sequence-based prediction of protein subcellular localization. Genomics Proteomics Bioinformatics. 2006;4(1):48–55. doi: 10.1016/S1672-0229(06)60016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27(8):1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.