Significance
Smartphones and wearable electronics have advanced tremendously over the last several years but fall short of allowing their use for molecular diagnostics. We herein report a generic approach to enable molecular diagnostics on smartphones. The method utilizes molecular-specific microbeads to generate unique diffraction patterns of “blurry beads” which can be recorded and deconvoluted by digital processing. We applied the system to resolve individual precancerous and cancerous cells as well as to detect cancer-associated DNA targets. Because the system is compact, easy to operate, and readily integrated with the standard, portable smartphone, this approach could enable medical diagnostics in geographically and/or socioeconomically limited settings with pathology bottlenecks.
Keywords: cancer diagnostics, smartphone, molecular sensing, point-of-care diagnostics, cervical cancer
Abstract
The widespread distribution of smartphones, with their integrated sensors and communication capabilities, makes them an ideal platform for point-of-care (POC) diagnosis, especially in resource-limited settings. Molecular diagnostics, however, have been difficult to implement in smartphones. We herein report a diffraction-based approach that enables molecular and cellular diagnostics. The D3 (digital diffraction diagnosis) system uses microbeads to generate unique diffraction patterns which can be acquired by smartphones and processed by a remote server. We applied the D3 platform to screen for precancerous or cancerous cells in cervical specimens and to detect human papillomavirus (HPV) DNA. The D3 assay generated readouts within 45 min and showed excellent agreement with gold-standard pathology or HPV testing, respectively. This approach could have favorable global health applications where medical access is limited or when pathology bottlenecks challenge prompt diagnostic readouts.
The rapid dissemination of electronic communication devices such as smartphones, tablets, and wearable electronics, all with integrated sensors, creates new possibilities for inexpensive point-of-care (POC) diagnostics and care delivery. One example is detecting cancer in low- and middle-income countries where limited resources and geographical constraints often lead to missed opportunities for intervention, resulting in mortalities even with treatable cancers (1). Current efforts to control cancer thus focus on implementing population-based early screening programs; a key element for success is a cost-effective, robust diagnostic platform that can be readily deployed into POC settings (2). Whereas conventional microscopy of human samples (smears, aspirates, biopsies, blood) is the most widely used to diagnose cancer, its POC adaptation is limited by inherent drawbacks such as bulky optics, requirements for trained microscopists, and operator-dependent variability.
Recent advances in digital sensors and computational approaches have introduced new microscopy techniques. Digital holography, in particular, has emerged as one alternative to conventional bright-field microscopy. Following the initial description of lens-free holography by Kreuzer's group (3), various diffraction-based imaging systems have been developed (4–8). The majority of recent work, however, is based on identifying targets by their inherent morphology (e.g., blood cells, bacteria, Caenorhabditis elegans) (4, 9–14). We reasoned that it would be possible to impart molecular specificity to improve disease detection and phenotyping akin to other molecular profiling strategies (15, 16).
Here we describe a digital diffraction diagnostics (D3)—a computational analysis of distinct diffraction patterns generated by microbeads that bind to biological target of interest. The strategy can detect a broad range of targets (SI Appendix, Table S1): soluble proteins, nucleic acids, or cellular proteins. To provide effective POC operation at remote sites, we adopted a client-server model: the data acquired by a smartphone were digitally processed by a remote parallel-computing server. We tested the approach by exploring cancer cell profiling with immunomicrobeads. Diffraction patterns generated by microbeads were detected with a smartphone camera in a bright-field setting, and digital processing reconstructed the images of bead-bound cells to retrieve molecular information. The assay protocol enabled molecular analyses on >104 individual cells in a single still image, with the entire assay complete in 45 min. As an initial proof-of-principle in human clinical samples, we used D3 to screen for cervical cancer, the third most prevalent cancer in women worldwide, with 80% of cases occurring in resource-limited countries (16). Cellular analyses using the D3 assay reliably identified patients at high clinical risk for malignancy. To show its versatility, we further extended the D3 assay to other disease targets, including human papillomavirus (HPV) DNA and lymphoma cell detection in fine-needle aspirates.
Results
D3 Platform.
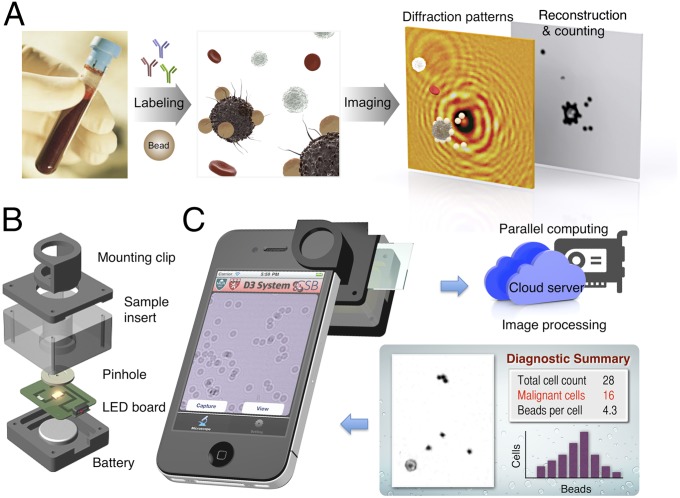
The D3 assay for cell detection is illustrated in Fig. 1A. Specimens are obtained from minimally invasive procedures (e.g., smears, brushings, fine-needle aspiration, blood draw), and cells are labeled with molecular-specific microbeads. Labeled samples are placed directly on the imaging device. Interference patterns between scattered light from the specimen and the reference light are recorded (17, 18). Digital signal processing then recovers and analyzes object images. Because individual cells are spatially resolved and only target-cell–associated beads are counted, there is no need for washing steps, leading to simplified assays. Selective microbead binding is critical to distinguishing target cells from other host cells and quantitatively profiling protein markers in individual cells.
Fig. 1.
D3 platform. (A) Assay schematic for cellular detection. Target cells in patient samples (e.g., blood or biopsy) are immunolabeled with microbeads, and their diffraction patterns are recorded. The diffraction images are then digitally reconstructed into object images wherein bead-labeled target cells are identified. For the detection of other types of targets, see SI Appendix, Table S1. (B) The snap-on module for a smartphone consists of an LED powered by a coin battery, a pinhole for uniform illumination with partial coherence, and a sample mount. (C) The D3-mounted smartphone’s embedded phone camera is used to record the diffraction images of the specimen. The recorded images are transferred to a server via the cloud service for real-time image reconstruction and analyses, which can be returned to the smartphone in less than 2 min.
To perform the D3 assay in POC settings, we implemented a portable sensing terminal that uses the embedded optics and communication functions of a smartphone (Fig. 1B). We constructed a snap-on module, containing both a light source and sample insert, to mount over the phone camera (SI Appendix, Fig. S1). The acquired diffraction patterns were transferred to a dedicated server for postprocessing (SI Appendix, Fig. S2). Following this step, the analytical readouts (e.g., target cell counts, bead counts per cell) and reconstructed images were sent back to the smartphone for display. This scheme frees the sensing terminals from heavy computation load, thereby making their implementation simple and cost-effective. All data were communicated through a secure cloud service, and we programmed a user-friendly interface to streamline the process (SI Appendix, Movie S1). The system had a wide field-of-view (FOV), whereas microscopic resolution could be achieved through numerical reconstruction. The FOV and the spatial resolution for iPhone 4S were 14 mm2 and 2 µm, respectively. Alternatively, a miniaturized imager connected to a local computer can be used (e.g., MT9P031, Aptina Imaging Inc.; SI Appendix, Fig. S3), which had an FOV and spatial resolution of 24 mm2 and 2 µm, respectively.
D3 Processing.
To accurately detect bead-bound target cells, we formulated a processing algorithm for image reconstruction and postanalysis (Fig. 2A). The reconstruction was based on the Rayleigh–Sommerfeld diffraction principle but was extended to digitally retrieve both transmittance and phase shift of objects through an iterative optimization (19–21). In each iteration, the routine applied physical constraints (i.e., light transmittance and object supports) to a reconstructed object image and updated the corresponding diffraction patterns with retrieved phase information (see Materials and Methods for details). The method provided high phase contrast between cells and microbeads (Fig. 2B). Each type could be easily distinguished, even when the size was similar (SI Appendix, Fig. S4).
Fig. 2.
Real-time reconstruction and counting process. (A) Diagram of reconstruction and counting algorithm. A diffraction image, normalized by a reference image obtained in the absence of specimens, is reconstructed through an iterative process. Following the image reconstruction, cells and beads are detected using a counting algorithm, which scanned a reference image of a microbead over the reconstructed image. (B) Examples of the image reconstruction. Raw diffraction patterns of cancer cells and 7-µm microbeads show undecipherable patterns. The reconstruction algorithm recovers both transmittance and phase information. Cells and microbeads can be differentiated from their high phase contrast. The bead-bound cells are automatically identified, and the bead numbers are counted. The transmittance (green) and phase contrast (red) images are pseudocolored to better visualize optical properties of cells and beads. (C) Computational time for image reconstruction and cell/bead counting (1-MB pixel image) with and without GPU-implemented systems.
Following reconstruction, images were processed by the detection routine. The algorithm generated transmittance and phase-correlation maps by scanning a microbead reference image over the reconstructed images (SI Appendix, Fig. S5). Cells and microbeads could be differentiated from the transmittance and phase correlations, respectively. Subsequently, cells labeled with microbeads were automatically identified, and their individual bead counts were recorded.
To accelerate imaging analyses, we used a server equipped with a graphics processing unit (GPU). The code executed massively parallel image processing threads (>5,000) using its multicore processors, which permitted near-instantaneous image reconstruction and postanalyses (SI Appendix, Table S2). For instance, using a 448-core GPU (Tesla C-2070), a 16-bit 1024 × 1024 diffraction pattern could be analyzed within 90 ms, ∼3,000-fold faster than relying on a conventional central processing unit (4 cores 2.4 GHz; Fig. 2C).
We adopted an encrypted cloud storage for data transfer. The scheme provides a buffer for asynchronous communication between smartphones and the server, and is readily scalable to global networks. To minimize data transfer time, especially from smartphones to a cloud storage, we converted original uncompressed color images (24 MB) into lossless gray-scale images (2.9 MB, Portable Network Graphics/PNG format). When microbeads with the diameter of >5 µm were used, the file could be further compressed (0.4 MB, Joint Photographic Experts Group/JPEG format) without affecting bead counts (SI Appendix, Fig. S6). These final images can be transferred to the cloud storage within 3 min, even through legacy 2G cellular network (transfer speed, 20 kbps).
High-Throughput D3 Assay for Cell Detection.
With its large FOV (14 mm2 for iPhone 4S and 24 mm2 for the image sensor) and rapid image processing, the D3 platform enabled high-throughput cellular analyses. In a single image acquisition, more than 100,000 individual objects can be detected at microscopic resolution (Fig. 3A and SI Appendix, Fig. S7). The dynamic range of detection spanned over 4 orders of magnitude (Fig. 3B), with the object concentration reaching up to ∼108 microbeads or cells per milliliter of sample (SI Appendix, Figs. S7–S9). The capacity for high-density imaging, combined with the recognition of bead-bound cells, made it possible to detect target cells in the presence of abundant host cells (e.g., leukocytes) and unbound microbeads. Importantly, this eliminated the need for external washing and purification steps. The assay throughput could be further increased by flowing target cells through a microfluidic channel and imaging in real-time (FOV, 2.5 × 2.5 mm2; 10 frames per s) (Fig. 3C and SI Appendix, Fig. S10 and Movie S2). This is akin to a flow cytometer with imaging capabilities for quantitative analysis (22), but in a miniaturized system.
Fig. 3.
High-throughput cell and bead identification. (A) Diffraction and reconstructed images of 7-µm microbeads at a concentration of 5 × 107 beads per mL. More than 105 objects can be detected at microscopic resolution from a single image (also see SI Appendix, Fig. S7). (B) Comparison between the D3-counted microbeads and leukocytes and their expected counts. Note the linearity (R2 > 0.97) even at very high bead or cell concentrations (4 × 108 beads per mL and 1 × 108 cells per mL). (C) Temporal image reconstruction of cells in flow. Diffraction images were recorded at 4 frames per s and reconstructed in real time by the D3 server. A bead-labeled cancer cell (black arrow), a nontargeted cell (red), and a free-floating microbead (blue) are automatically identified. (Inset) High-resolution details of the bead-labeled cell undergoing rotational motion in the flow stream.
Quantitative Profiling.
We next applied the D3 assay to cell profiling (Fig. 4A). Human cancer cell lines were immunolabeled with microbeads to generate specific diffraction signatures. Whereas it is possible to use antibody-coated microbeads directly on cells, we found that a two-step approach increased bead binding (23). We thus opted for highly efficient bioorthogonal [e.g., between transcyclooctene and tetrazine (24)] or streptavidin/biotin approaches. For example, using the latter, cells were first targeted by biotinylated antibodies and subsequently incubated with streptavidin-coated microbeads. We further tested commercially available microbeads in different sizes (diameter 3–22 µm). A bead diameter between 5 and 7 µm was found to optimize accurate bead counting while minimizing cell clustering (SI Appendix, Fig. S11).
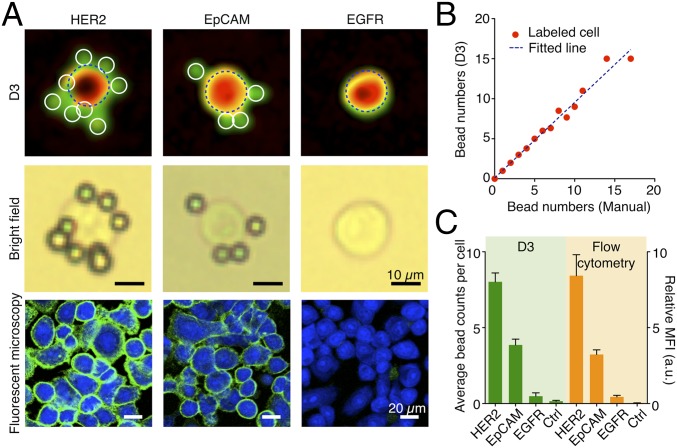
Fig. 4.
Detecting cancer cell markers with immunobead labeling. (A) Cancer cells (SkBr3, human breast carcinoma) were immunobead-labeled for HER2, EpCAM, and EGFR. (Top Row) Representative reconstructed images in pseudocolor (green, transmittance; red, phase). (Middle Row) Corresponding bright-field micrographs. (Bottom Row) Cells labeled with fluorescent antibodies for comparison. HER2, human epidermal growth factor receptor 2; EpCAM, epithelial cell adhesion molecule; EGFR, epidermal growth factor receptor. (B) The bead numbers on labeled cells, determined automatically by D3, were in good agreement (R2 = 0.97) with those counted manually from microscope images. (C) The average bead count per cell correlated with the expression level of a target marker as determined by flow cytometry (R2 = 0.99). MFI, mean fluorescence intensity. a.u., arbitrary unit.
To validate D3-based cellular profiling, we measured the expression of three protein markers, human epidermal growth factor receptor 2 (HER2)/neu, epithelial cell adhesion molecule (EpCAM), and epidermal growth factor receptor (EGFR), on human cells (SkBr3, human breast carcinoma). Samples were immunolabeled with 7-µm microbeads and analyzed using the D3 system (Fig. 4A and SI Appendix, Fig. S12). The average bead count per cell was highest for HER2/neu targeting (8.0 beads per cell), followed by EpCAM (3.9 beads per cell) and EGFR (0.5 beads per cell); these results were consistent with immunofluorescence microscopy. Unlike microscopy, however, the D3 assay enabled simultaneous analyses of far greater numbers of cells (>10,000 cells) because of its wide FOV. The cellular bead counts generated with automated D3 analyses were statistically identical with those from manual microscopy enumeration (P = 0.43; paired t test; Fig. 4B and SI Appendix, Fig. S13). Further comparison with flow cytometry validated D3 assay’s analytically capacity (Fig. 4C). The number of beads per cell correlated linearly with levels of marker expression (R2 = 0.99). Similar D3 profiling on a different cell line (A431, human epidermoid carcinoma) also matched well with immunofluorescence microscopy and flow cytometry results (SI Appendix, Fig. S14).
Whereas the above examples used parallel profiling of separate images, it is also possible to perform multiplexing within the same sample using microbeads of different optical signatures. For instance, we tested three bead sizes and three different materials (e.g., polystyrene, silica, silica coated with silver) that can be effectively distinguished through nominal size and transmittance, respectively. Combining these sets could provide nine-channel multiplexing (SI Appendix, Fig. S15).
Clinical Applications of D3.
We evaluated the clinical utility of D3 by first focusing on cervical cancer surveillance. Cervical cancer maintains high incidence rates in resource-poor countries (16) where the availability of objective early screening systems is limited. We reasoned that the portable D3 system could be used as a first-line diagnostic tool without subjective readouts to promptly triage suspicious or high-risk cases. The strategy could potentially offset pathology bottlenecks and reduce repeated patient visits to central clinics, which are often complicated by geographical and/or socioeconomic constraints (25, 26).
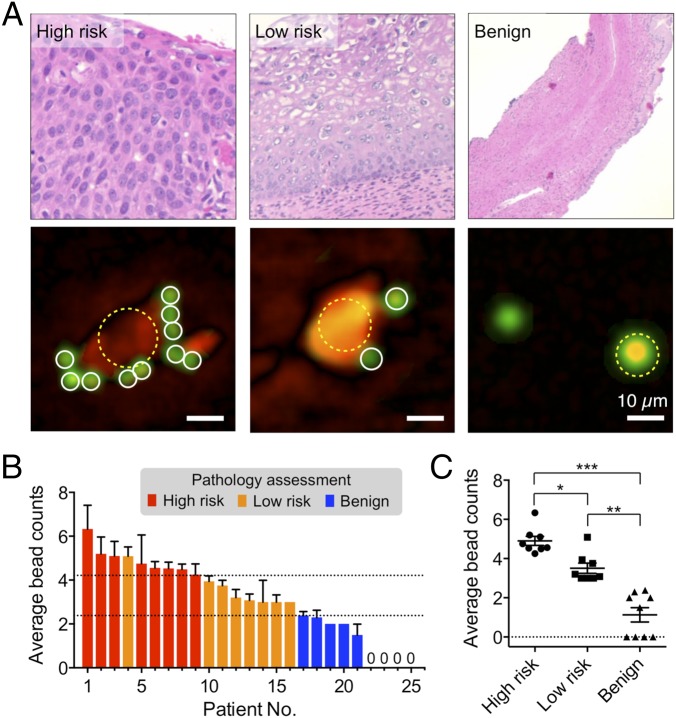
We obtained cervical specimens through brushing, loop electrosurgical excision procedure (LEEP), or biopsies during colposcopic evaluation. Twenty-five patients with previously abnormal Pap smear results were recruited. Cellular samples were labeled with a mixture of tagged antibodies against EpCAM, CD44, or tumor-associated calcium signal transducer2 (TACD2/Trop2). These cancer markers were chosen for their elevated expression in cervical cancer as described in literature (27–29). We counted the total number of bead-bound cells as well as the number of microbeads per cell. Blinded to D3 results, patient specimens were separately examined through conventional pathology, and classified into three risk classes: “high risk,” “low risk,” or “benign” (Fig. 5A) The D3 analyses showed that bead counts among targeted cells increased along the clinical risk (Fig. 5B). The mean bead counts (nbead) per targeted cell were significantly different among the clinical risk classifications (P < 0.05, Tukey multiple comparison test), indicating that nbead could serve as a single diagnostic measure (Fig. 5C). The class boundaries, determined from multiclass logistic regression, were nbead > 4.2 between high risk and low risk; and nbead > 2.4 between low risk and benign. We further dichotomized the patients into two groups, high risk versus low risk/benign, to reflect the conventional clinical process for intervention and/or follow-up. In our limited data set, the detection sensitivity was 100% and the specificity 92% (high risk vs. low risk/benign).
Fig. 5.
Cervical cancer diagnosis with the D3 platform. (A) Histology (Top) and representative reconstructed image of cells (Bottom) from high-risk (cervical intraepithelial neoplasia/CIN 2, 3), low-risk (CIN 1), and benign patient samples. Samples were targeted with a mixture of EpCAM, CD44, and Trop2 antibodies, followed by microbead labeling. More beads were bound to cells in higher risk patient samples. The transmittance (green) and the phase (red) were pseudocolored for clarification. (B and C) The profiling result of patient samples (n = 25). The average numbers of beads per labeled cell were significantly different among high-risk, low-risk, and benign groups. The dotted lines indicate the class boundaries. *P = 0.035; **P < 0.001; ***P < 0.001.
We also adopted the same D3 platform for the HPV-DNA detection (SI Appendix, Fig. S16A), given its high relevance in cervical cancer diagnostics. We applied the bead-dimer assay format: a pair of oligonucleotides, whose sequences were complementary to that of target DNA, were conjugated to silica and polystyrene microbeads, respectively. The target DNA was captured on polystyrene (PS) beads and sequentially labeled with silica beads. The hybridization yielded PS–silica bead dimers with unique diffraction signature (SI Appendix, Fig. S16B). The number of PS–silica hybrids was then counted to quantify the amount of target DNA. The D3 assay was highly sensitive; HPV 16 and 18 DNA targets could be detected down to atto-mole range without PCR amplification (SI Appendix, Fig. S16 C and D). Compared with fluorescent detection, the bead-based D3 assay benefited from simpler optics (i.e., no filter sets) and stronger light signal (bright field).
Extending the clinical utility of D3 to other malignancies, we conducted a pilot study to detect lymphoma cells in fine-needle aspirates (FNA) of lymph nodes in patients with lymphadenopathy. POC lymphoma diagnostics would be of particular interest in sub-Saharan Africa plagued by a high prevalence of AIDS-related cancers and lymphoma (the “second wave of AIDS”) (25, 30, 31). Freshly harvested FNA samples were incubated with immunobeads specific to CD20, one lymphoma marker (SI Appendix, Fig. S17). Subsequent D3 analysis allowed the correct diagnosis of four patients with pathology confirmed lymphoma and also excluded the diagnosis in another four patients with confirmed benign lymphadenopathy.
Discussion
Global cancer rates continue to increase, and the World Health Organization predicts new cases to rise to 19.3 million by 2025 as the world's population grows, ages, and gains access to antiretroviral drugs (1, 32–35). Rapid cancer screening, however, remains an unmet clinical need. The D3 strategy reported here could address some of the diagnostic challenges in resource-limited areas. It capitalizes on the already widespread distribution of smartphones and allows fast (minutes–hours for a final answer), very low cost (compared with sectioning, microscopes, and flow cytometers), and simple diagnostics. Molecular diagnoses are achieved by integrating immunolabeling assays, cloud computing, and digital processing. The resulting system enables quantitative and operator-independent cellular analysis and reports not only cancer cell counts but also the expression levels of molecular markers. In a pilot study of cervical cancer screening, we used three molecular markers to define risk categories based on invasiveness. The D3 assay was fast (40 min for immunolabeling, most of which is “hands-off” time and 3 min for data processing; SI Appendix, Table S3) and cost-effective ($1.80 per assay; SI Appendix, Table S4). We expect that the assay costs will ultimately be much cheaper once scaled up and further optimized.
We anticipate further improvements in some analytical capabilities of D3. First, a next-generation system would incorporate multiplexed cellular detection based on different optical properties of microbeads. We have shown that microbeads can be differentiated based on their size and absorbance (SI Appendix, Fig. S15). Applying these signatures would enable multiplexed molecular profiling of the same cells to improve detection accuracy. In parallel, superresolution approaches could be used to improve the spatial resolution (6, 14, 36–38), and thereby further boost D3's multiplexing capacity. Compressive sensing is particularly appealing (39, 40), as it can numerically reconstitute high-frequency information (i.e., small features in images) with no modification in the current D3 setup. By incorporating compressive sensing, we expect to reconstruct images with submicrometer resolution (∼0.6 µm), which would allow analyses of all types of mammalian cells. Second, DNA detection should be further developed for POC operation. This would require implementing disposable cartridges (41, 42) and portable systems (43, 44) for DNA extraction and amplification. Finally, the platform should be tested for robust field operation. We plan to establish a lyophilization protocol for transport and storage of reagents (e.g., antibodies, microbeads) (45). The validation tests will be extended using larger cohorts, variably skilled operators, and diverse environment settings to obtain better statistical power (reproducibility, accuracy, diagnostic performance). In this light, the work presented here has catalyzed two larger clinical trials focused, respectively, on cervical cancer (including HPV testing) and lymphoma detection (FNAs of enlarged lymph nodes). We envision that these advances will position D3 as a versatile screening tool for various cancer types (e.g., cervix, breast, lymphoma) and infectious diseases (e.g., Ebola, HIV, tuberculosis) with applications in field work, mobile clinics, and home care settings.
Materials and Methods
D3 Imaging Terminal.
Electrical components [e.g., light-emitting diode (LED), battery socket, switch] were purchased (Digi-Key) and mounted on a custom-designed printed circuit board. The main body of the snap-on module was machined out of black acrylic plastic, and housed the light source (590 nm), a 100-µm pinhole, and a minilens. The focal length (4 mm) of the lens was similar to that of the embedded lens (4.2 mm) in the phone camera (SI Appendix, Fig. S1). By pairing these two lenses, we achieved a unit magnification to maximize the FOV, and used the phone camera without any modification. The size of the snap-on was 4 × 4 × 5.5 cm3.
D3 App.
The D3 app was programmed with user-friendly interfaces for operation in iOS 6 or higher version. The app has three main screens: (i) image capturing and processing, (ii) reconstruction parameters settings, and (iii) data communication with a cloud service. The images captured by a phone camera module were saved as uncompressed data (PNG) or compressed image (JPEG), and uploaded along with other imaging information (e.g., wavelength, diagnosis location, time, patient data) into a dedicated folder in a cloud storage (Dropbox). The uploaded images were processed by a D3 image GPU server (see below), and the results were saved into a subfolder. The D3 app subsequently downloaded reconstructed images and analysis results. All data were encrypted according to the 256-bit Advanced Encryption Standard. The application program interface library from the vendor (Dropbox) was used to implement the data transfer routine.
D3 Imaging Server.
The D3 server (HP xw4600 workstation, Hewlett Packard) had the following system specifications (SI Appendix, Table S2): CPU, Core2 Duo E8500 3.16 GHz (Intel); memory, 8-GB DDR2 (double data rate2); GPU, Tesla C-2070 (Nvidia); operating system, Ubuntu 12.04 64-bit. The GPU had 448 CUDA (Compute Unified Device Architecture) cores and 6 GB memory. The signal processing program was written in C++ language and used vendor-provided modules (CUDA extensions, CUDA driver 5.0, CUFFT library). The image analysis workflow is shown in SI Appendix, Fig. S2. The imaging server cyclically polled a dedicated folder in a cloud storage (Dropbox). When new images and image information were uploaded, the imaging server executed image reconstruction and postimaging analyses (cell and bead counting). The reconstructed images and counting statistics were then saved into a subfolder. When synchronized in the cloud storage, the image and data files could be accessed by users.
Image Reconstruction.
The size of an uncompressed image file on the iPhone 4S was ∼24 MB (2,448 × 3,264 pixels, 24-bit RGB). This raw image file was converted to gray-scale PNG (∼2.9 MB) or JPEG (∼0.4 MB) files (SI Appendix, Fig. S6), and normalized by a reference image recorded without samples. The normalization removed intrinsic defects and accurately calculated object transmittance (21). The normalized diffraction images were up-sampled four times through linear interpolation and used as input data for reconstruction. The reconstruction was based on phase retrieval algorithms, which can recover phase information from intensity-based diffraction patterns through iterative processes (4, 18, 19). The algorithm has four steps: (i) back-propagate an input image, (ii) apply constraints, (iii) forward-propagate updated image, and (iv) update retrieved phase information. First, the normalized diffraction image was numerically back-propagated by an optical distance between the object and the imager. We calculated the optimal optical distance by finding a focal depth with the sharpest object boundary (46). Calculating field propagation was based on the Rayleigh–Sommerfeld diffraction integral in a convolution approach, where the propagated field was calculated by the inverse Fourier transform of the multiplication between the Fourier transform of field and the transfer function (20). During the first iteration, object supports were defined using a segmentation method, where object boundaries were found by thresholding intensity variances (46). For the back-propagated image, pixels outside the object supports were regarded as background, and their transmittance values (i.e., the modulus of field) were set to unity. If a pixel inside the object support had a transmittance value larger than unity due to artificial twin-image superimposition, its transmittance value was also forced to unity. After applying the constraints, the updated image was propagated to the image plane, where the forward-propagated field had nonzero phase information. The phase information was added to the measured diffraction image as a new input. The process was usually repeated 10∼30 times until the reconstructed image with retrieved phase information converged.
Counting Algorithm.
For detecting beads and cells, a microbead reference image was scanned over the reconstructed images to generate modulus and phase correlation maps as shown in SI Appendix, Fig. S5. The reference microbead image was obtained by averaging microbead images in a pure bead solution. The correlation coefficients for modulus and phase were calculated from pixel-to-pixel comparison between reference and reconstructed images. Cells were first detected when a local maximum phase correlation coefficient was larger than the threshold value of phase correlation coefficient, which was obtained from images of pure cell population. For each cell detected, microbeads within the object support were detected from the local maxima of the modulus correlation coefficients. Microbeads within a distance of 10 pixels (or 22 µm) from a center of cells were considered bound to cells. This search radius was experimentally determined for 7-µm beads. The detection algorithm was validated by comparing bead counts with manual counting using a conventional microscopy. The microscope was equipped with a 10× objective (N.A. = 0.17) and a cooled CCD camera (7.4 × 7.4-µm2 pixel size; Spot RT3, Diagnostic Instruments Inc.), and produced images with 1.4 pixel/µm resolution.
Clinical Samples.
The clinical study was approved by the Partners Healthcare Institutional Review Board (Massachusetts General Hospital/Brigham and Women's Hospital). Informed consent was obtained from adult women who were referred to the Colposcopy Clinic for previously abnormal Pap smears. Samples were obtained by brushing, cervical biopsy, or LEEP. One clinical provider (M.A.-W.) performed all cervical procedures and provided excess or otherwise discarded ectocervical or endocervical specimens. Biopsies entailed visualizing the exocervix and bathing with 5% acetic acid using clinically standard procedures. Suspicious aceto-white epithelial changes were identified with a colposcope (Foto Optik; Leisegang Medical Inc.) followed by punch biopsies. Before the use of acetic acid, brushing samples were collected with surgical brushes (Surgipath C-E Brush, Leica Microsystems; Cytobrush Plus GT Gentle Touch, BD Surepath). Samples were suspended in sterile PBS (1 mL), and incubated (30 min, 20 °C) with a mixture of antibodies against EpCAM (clone MAB9601, R&D Systems), CD44 (clone IM7, Biolegend), and Trop2 (Clone 162–46.2, Abcam). The antibody concentration was 5 µg/mL for each. Targeted samples were then incubated with streptavidin-coated polystyrene microbeads (0.5 mg, 7-μm diameter, Spherotech) for 10 min at room temperature and analyzed using the D3 platform. Here, the sample volume examined was 10 µL; the number of microbeads per cell was automatically counted using D3 software. All experiments were conducted blind to pathology results and clinical interpretation of risk. Please see SI Appendix for cell preparation, fluorescence measurements, HPV-DNA detection, and clinical lymph node samples.
Supplementary Material
Acknowledgments
The authors thank M. Birrer, D. Dizon, and M. Del Carmen for many helpful suggestions; Melina Pectasides for assistance with clinical sample collection; and K. Joyes for reviewing the manuscript. This work was supported in part by US NIH Grants R01-HL113156 (to H.L.), R01-EB010011 (to R.W.), R01-EB00462605A1 (to R.W.), T32CA79443 (to R.W.), K12CA087723-11A1 (to C.M.C.), Massachusetts General Hospital Physician Scientist Development Award (to C.M.C.), National Heart, Lung, and Blood Institute Contract HHSN268201000044C (to R.W.), and Department of Defense Ovarian Cancer Research Program Award W81XWH-14-1-0279 (to H.L.)
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1501815112/-/DCSupplemental.
References
- 1.Varmus H, Trimble EL. Integrating cancer control into global health. Sci Transl Med. 2011;3(101):01cm28. doi: 10.1126/scitranslmed.3002321. [DOI] [PubMed] [Google Scholar]
- 2.Chin CD, Linder V, Sia SK. Commercialization of microfluidic point-of-care diagnostic devices. Lab Chip. 2012;12(12):2118–2134. doi: 10.1039/c2lc21204h. [DOI] [PubMed] [Google Scholar]
- 3.Xu W, Jericho MH, Meinertzhagen IA, Kreuzer HJ. Digital in-line holography for biological applications. Proc Natl Acad Sci USA. 2001;98(20):11301–11305. doi: 10.1073/pnas.191361398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia-Sucerquia J, et al. Digital in-line holographic microscopy. Appl Opt. 2006;45(5):836–850. doi: 10.1364/ao.45.000836. [DOI] [PubMed] [Google Scholar]
- 5.Gurkan UA, et al. Miniaturized lensless imaging systems for cell and microorganism visualization in point-of-care testing. Biotechnol J. 2011;6(2):138–149. doi: 10.1002/biot.201000427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng G, Lee SA, Antebi Y, Elowitz MB, Yang C. The ePetri dish, an on-chip cell imaging platform based on subpixel perspective sweeping microscopy (SPSM) Proc Natl Acad Sci USA. 2011;108(41):16889–16894. doi: 10.1073/pnas.1110681108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim SB, et al. A cell-based biosensor for real-time detection of cardiotoxicity using lensfree imaging. Lab Chip. 2011;11(10):1801–1807. doi: 10.1039/c1lc20098d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenbaum A, et al. Imaging without lenses: Achievements and remaining challenges of wide-field on-chip microscopy. Nat Methods. 2012;9(9):889–895. doi: 10.1038/nmeth.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seo S, et al. High-throughput lens-free blood analysis on a chip. Anal Chem. 2010;82(11):4621–4627. doi: 10.1021/ac1007915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mudanyali O, Bishara W, Ozcan A. Lensfree super-resolution holographic microscopy using wetting films on a chip. Opt Express. 2011;19(18):17378–17389. doi: 10.1364/OE.19.017378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SA, et al. Color capable sub-pixel resolving optofluidic microscope and its application to blood cell imaging for malaria diagnosis. PLoS ONE. 2011;6(10):e26127. doi: 10.1371/journal.pone.0026127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenbaum A, Akbari N, Feizi A, Luo W, Ozcan A. Field-portable pixel super-resolution colour microscope. PLoS ONE. 2013;8(9):e76475. doi: 10.1371/journal.pone.0076475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su T-W, Choi I, Feng J, Huang K, McLeod E, Ozcan A. 2013. Sperm trajectories form chiral ribbons. Sci Rep 3:1664. [DOI] [PMC free article] [PubMed]
- 14.Zheng G, Horstmeyer R, Yang C. Wide-field, high-resolution Fourier ptychographic microscopy. Nat Photonics. 2013;7(9):739–745. doi: 10.1038/nphoton.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ojesina AI, et al. Landscape of genomic alterations in cervical carcinomas. Nature. 2014;506(7488):371–375. doi: 10.1038/nature12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schiffman M, Solomon D. Clinical practice. Cervical-cancer screening with human papillomavirus and cytologic cotesting. N Engl J Med. 2013;369(24):2324–2331. doi: 10.1056/NEJMcp1210379. [DOI] [PubMed] [Google Scholar]
- 17.Fung J, et al. Measuring translational, rotational, and vibrational dynamics in colloids with digital holographic microscopy. Opt Express. 2011;19(9):8051–8065. doi: 10.1364/OE.19.008051. [DOI] [PubMed] [Google Scholar]
- 18.Mudanyali O, et al. Compact, light-weight and cost-effective microscope based on lensless incoherent holography for telemedicine applications. Lab Chip. 2010;10(11):1417–1428. doi: 10.1039/c000453g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fienup JR. Phase retrieval algorithms: A comparison. Appl Opt. 1982;21(15):2758–2769. doi: 10.1364/AO.21.002758. [DOI] [PubMed] [Google Scholar]
- 20.Kreis TM. Frequency analysis of digital holography with reconstruction by convolution. Opt Eng. 2002;41(8):1829–1839. [Google Scholar]
- 21.Latychevskaia T, Fink H-W. Solution to the twin image problem in holography. Phys Rev Lett. 2007;98(23):233901. doi: 10.1103/PhysRevLett.98.233901. [DOI] [PubMed] [Google Scholar]
- 22.Basiji DA, Ortyn WE, Liang L, Venkatachalam V, Morrissey P. Cellular image analysis and imaging by flow cytometry. Clin Lab Med. 2007;27(3):653–670, viii. doi: 10.1016/j.cll.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agasti SS, et al. Supramolecular host-guest interaction for labeling and detection of cellular biomarkers. Angew Chem Int Ed Engl. 2012;51(2):450–454. doi: 10.1002/anie.201105670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haun JB, Devaraj NK, Hilderbrand SA, Lee H, Weissleder R. Bioorthogonal chemistry amplifies nanoparticle binding and enhances the sensitivity of cell detection. Nat Nanotechnol. 2010;5(9):660–665. doi: 10.1038/nnano.2010.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chabner BA, Efstathiou J, Dryden-Peterson S. 2013. Cancer in Botswana: The second wave of AIDS in Sub-Saharan Africa. Oncologist 18(7):777–778. [DOI] [PMC free article] [PubMed]
- 26.Ramogola-Masire D, et al. Cervical cancer prevention in HIV-infected women using the “see and treat” approach in Botswana. J Acquir Immune Defic Syndr. 2012;59(3):308–313. doi: 10.1097/QAI.0b013e3182426227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dall P, et al. Surface protein expression and messenger RNA-splicing analysis of CD44 in uterine cervical cancer and normal cervical epithelium. Cancer Res. 1994;54(13):3337–3341. [PubMed] [Google Scholar]
- 28.Varughese J, et al. Cervical carcinomas overexpress human trophoblast cell-surface marker (Trop-2) and are highly sensitive to immunotherapy with hRS7, a humanized monoclonal anti-Trop-2 antibody. Am J Obstet Gynecol. 2011;205(6):e1–e7. doi: 10.1016/j.ajog.2011.06.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Went PTH, et al. Frequent EpCam protein expression in human carcinomas. Hum Pathol. 2004;35(1):122–128. doi: 10.1016/j.humpath.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 30.Varmus H, Kumar HS. Addressing the growing international challenge of cancer: A multinational perspective. Sci Transl Med. 2013;5(175):cm2. doi: 10.1126/scitranslmed.3005899. [DOI] [PubMed] [Google Scholar]
- 31.Livingston J. Cancer in the shadow of the AIDS epidemic in southern Africa. Oncologist. 2013;18(7):783–786. doi: 10.1634/theoncologist.2013-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beaglehole R, et al. Lancet NCD Action Group UN High-Level Meeting on Non-Communicable Diseases: Addressing four questions. Lancet. 2011;378(9789):449–455. doi: 10.1016/S0140-6736(11)60879-9. [DOI] [PubMed] [Google Scholar]
- 33.Daar AS, et al. Grand challenges in chronic non-communicable diseases. Nature. 2007;450(7169):494–496. doi: 10.1038/450494a. [DOI] [PubMed] [Google Scholar]
- 34.Narayan KMV, Ali MK, del Rio C, Koplan JP, Curran J. Global noncommunicable diseases—lessons from the HIV-AIDS experience. N Engl J Med. 2011;365(10):876–878. doi: 10.1056/NEJMp1107189. [DOI] [PubMed] [Google Scholar]
- 35.Trimble EL. Global cooperation in gynecologic cancer. J Gynecol Oncol. 2010;21(1):1–2. doi: 10.3802/jgo.2010.21.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gazit S, Szameit A, Eldar YC, Segev M. Super-resolution and reconstruction of sparse sub-wavelength images. Opt Express. 2009;17(26):23920–23946. doi: 10.1364/OE.17.023920. [DOI] [PubMed] [Google Scholar]
- 37.Bishara W, et al. Holographic pixel super-resolution in portable lensless on-chip microscopy using a fiber-optic array. Lab Chip. 2011;11(7):1276–1279. doi: 10.1039/c0lc00684j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mudanyali O, et al. Wide-field optical detection of nanoparticles using on-chip microscopy and self-assembled nanolenses. Nat Photonics. 2013;7(3):247–254. doi: 10.1038/nphoton.2012.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brady DJ, Choi K, Marks DL, Horisaki R, Lim S. Compressive holography. Opt Express. 2009;17(15):13040–13049. doi: 10.1364/oe.17.013040. [DOI] [PubMed] [Google Scholar]
- 40.Rivenson Y, Stern A, Javidi B. Overview of compressive sensing techniques applied in holography [Invited] Appl Opt. 2013;52(1):A423–A432. doi: 10.1364/AO.52.00A423. [DOI] [PubMed] [Google Scholar]
- 41.Hoffmann J, Mark D, Lutz S, Zengerle R, von Stetten F. Pre-storage of liquid reagents in glass ampoules for DNA extraction on a fully integrated lab-on-a-chip cartridge. Lab Chip. 2010;10(11):1480–1484. doi: 10.1039/b926139g. [DOI] [PubMed] [Google Scholar]
- 42.Liu C, Mauk MG, Hart R, Qiu X, Bau HH. A self-heating cartridge for molecular diagnostics. Lab Chip. 2011;11(16):2686–2692. doi: 10.1039/c1lc20345b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang S, et al. Low cost extraction and isothermal amplification of DNA for infectious diarrhea diagnosis. PLoS ONE. 2013;8(3):e60059. doi: 10.1371/journal.pone.0060059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang L, Mancuso M, Lu Z, Akar G, Cesarman E, Erickson D. 2014. Solar thermal polymerase chain reaction for smartphone-assisted molecular diagnostics. Sci Rep 4:4137. [DOI] [PMC free article] [PubMed]
- 45.Bhambhani A, Blue JT. 2010. Lyophilization strategies for development of a high-concentration monoclonal antibody formulation: Benefits and pitfalls. Am Pharm Rev 13(1):31–38.
- 46.McElhinney CP, et al. Depth-independent segmentation of macroscopic three-dimensional objects encoded in single perspectives of digital holograms. Opt Lett. 2007;32(10):1229–1231. doi: 10.1364/ol.32.001229. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.