Significance
The B-cell–specific antibody gene-diversifying enzyme activation-induced cytidine deaminase (AID) shows high homology with apolipoprotein B mRNA-editing enzyme catalytic polypeptide 1 (APOBEC1), which edits apolipoprotein B-100 mRNA in the presence of cofactor APOBEC1 complementation factor/APOBEC complementation factor (A1CF/ACF). Here we show that the DNA cleavage and recombination functions of AID depend critically on its RNA-dependent interaction with distinct heterogeneous nuclear ribonucleoprotein (hnRNP) cofactors. Depletion of hnRNP K inhibited DNA cleavage essential to both class switch recombination (CSR) and somatic hypermutation, but depletion of hnRNP L blocked the CSR-associated recombination. Thus this is the first report, to our knowledge, identifying A1CF-like hnRNP-family editing cofactors for AID.
Keywords: class switch recombination, somatic hypermutation, activation-induced cytidine deaminase, B cell, IgH
Abstract
Activation-induced cytidine deaminase (AID) is essential for antibody class switch recombination (CSR) and somatic hypermutation (SHM). AID originally was postulated to function as an RNA-editing enzyme, based on its strong homology with apolipoprotein B mRNA-editing enzyme, catalytic polypeptide 1 (APOBEC1), the enzyme that edits apolipoprotein B-100 mRNA in the presence of the APOBEC cofactor APOBEC1 complementation factor/APOBEC complementation factor (A1CF/ACF). Because A1CF is structurally similar to heterogeneous nuclear ribonucleoproteins (hnRNPs), we investigated the involvement of several well-known hnRNPs in AID function by using siRNA knockdown and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9–mediated disruption. We found that hnRNP K deficiency inhibited DNA cleavage and thereby induced both CSR and SHM, whereas hnRNP L deficiency inhibited only CSR and somewhat enhanced SHM. Interestingly, both hnRNPs exhibited RNA-dependent interactions with AID, and mutant forms of these proteins containing deletions in the RNA-recognition motif failed to rescue CSR. Thus, our study suggests that hnRNP K and hnRNP L may serve as A1CF-like cofactors in AID-mediated CSR and SHM.
Antigen-stimulated mature B cells express activation-induced cytidine deaminase (AID), an essential enzyme for somatic hypermutation (SHM) and class switch recombination (CSR) at the Ig locus (1, 2). AID induces DNA breaks at the variable (V) and switch (S) regions during SHM and CSR, respectively. Most of the mutations produced during SHM are introduced by error-prone DNA synthesis during single-strand break (SSB) repair (3, 4). In contrast, CSR requires the conversion of SSBs to double-strand breaks (DSBs), followed by recombination between two DSB ends located in the donor and acceptor S regions (5, 6). The entire process of CSR is accomplished through elaborate DNA-repair processes involving S–S synapse formation and end-joining.
The mechanism by which AID functions differently in DNA cleavage and recombination at different loci remains unclear. Functional studies of a large number of AID mutants revealed that the N-terminal AID mutations impair SHM and CSR, indicating that the AID N terminus, which also possesses a bipartite nuclear-localization signal, is required for DNA cleavage in both SHM and CSR (7–9). On the other hand, C-terminal AID mutations suppressed the recombination activity of CSR but had no effect on SHM, indicating that the C terminus of AID, which contains a nuclear-export signal, is required for the recombination activity associated specifically with CSR (7, 9, 10). Indeed, recent studies showed that defects in the AID C terminus compromise DNA end-joining and S–S synapse formation without perturbing DNA breakage at either the V or S region (11, 12), also suggesting a specific role for the AID C terminus in the recombination step of CSR. Given AID’s small size (198 residues), we were intrigued by its diverse and compartmentalized functions.
AID is a member of the AID–apolipoprotein B mRNA-editing enzyme, catalytic polypeptide (APOBEC) cytidine deaminase family, which is related to ancestral AID-like enzymes, PmCDA1 and PmCDA2, expressed in the lamprey (13, 14). Although most of these related proteins are predicted to be involved in cytidine deamination, their targets and the molecular mechanisms are not fully elucidated (15). The best-characterized AID-like enzyme is APOBEC1, an RNA-editing enzyme that catalyzes the site-specific deamination of C to U at position 6666 of the apolipoprotein B-100 (APO B-100) mRNA, generating a premature stop codon (16–18). The edited mRNA, referred to as “APOB-48,” encodes the triglyceride carrier protein, a truncated product of the LDL carrier protein, which is encoded by APO B-100 mRNA.
The specificity of the APOBEC1 RNA-editing activity is determined by its cofactor, APOBEC1 complementation factor/APOBEC complementation factor (A1CF/ACF), a 64-kDa RNA-binding protein containing three distinct RNA-recognition motifs (RRMs) (19–23), which specifically targets a single C within a transcript of ∼14,000 bases. A conserved motif of 11 nucleotides (referred to as the “mooring sequence”), located four to six nucleotides downstream of the edited base, is critical for A1CF binding and RNA editing (24, 25). It also has been suggested that a 30-bp predicted stem–loop structure surrounding the deaminated C is involved in target-site selection, but this suggestion is controversial (26, 27). A1CF has been hypothesized to melt the secondary structure of the editing site (28), thus allowing C6666 access to APOBEC1 deamination and that A1CF is the critical factor in the APOBEC1-editing complex (21, 22, 29) that mediates the RNA-substrate recognition and base-editing specificity. Because A1CF binds to RNA and also docks APOBEC1, it is thought to function as a molecular bridge between the RNA substrate and APOBEC1.
A1CF shows strong structural similarity to the heterogeneous nuclear ribonucleoprotein (hnRNP) family members, which typically possess more than one RNA-binding module, including RRMs and K homology (KH) domains (30, 31). In particular, the RRMs of A1CF show strong homology to the RRMs found in the hnRNPs. APOBEC1 also can interact with more than one isoform of hnRNP Q, which shows almost 50% identity with A1CF (19). Members of the hnRNP protein family are involved in multiple aspects of nucleic acid metabolism and show distinct binding preferences for nucleic acids and proteins. For instance, hnRNP K possesses KH domains, which bind RNA as well as ssDNA (32), and it also contains a K-interactive region that recruits diverse proteins such as kinases and mRNA regulators. In contrast, hnRNP L contains four RRMs and preferentially binds to CA repeats in RNA (33).
The evolutionary conservation between AID and APOBEC1 led us to postulate that AID may use different hnRNP proteins as cofactors for its roles in promoting DNA cleavage and recombination (15, 34). Because both AID-induced DNA cleavage and recombination are dependent on AID’s cytidine deaminase activity, the dual functions could involve differential target specificities, which could be determined by cofactors rather than by AID itself. Thus, the use of hnRNPs as specific cofactors for DNA cleavage and recombination may contribute to the mechanism by which AID mediates both processes.
To investigate the involvement of hnRNP family proteins in CSR, we performed siRNA screening of various hnRNPs and identified hnRNP K and hnRNP L as AID cofactors required for CSR. In addition, our finding that both hnRNPs interact with AID in an RNA-dependent manner suggests AID’s role in CSR and SHM could involve RNA editing.
Results
Both hnRNP K and hnRNP L Are Required for Efficient CSR.
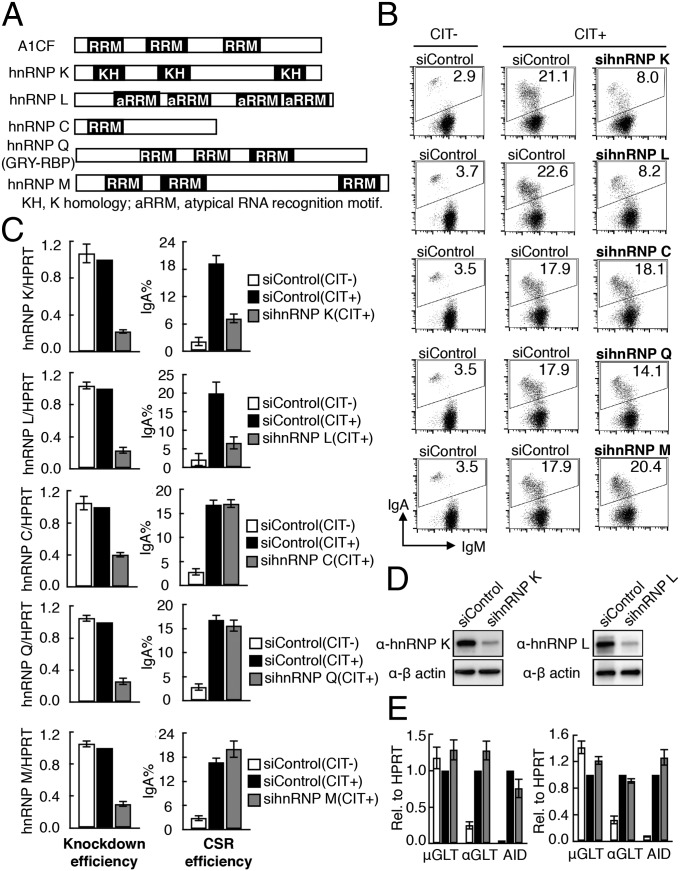
To examine whether AID function requires a cofactor similar to APOBEC1’s requirement for A1CF, we used siRNA-mediated knockdown to study the effects of candidate hnRNP depletion on CSR. Because A1CF is not expressed, we knocked down other hnRNPs individually in a mouse B-cell line, CH12F3-2A, which undergoes high-efficiency CSR from IgM to IgA in response to stimulation with CD40L, IL-4, and TGF-β (CIT). Among the hnRNPs screened, the knockdown of hnRNP K and hnRNP L (Fig. 1A) showed substantially reduced (60–70%) IgA switching (Fig. 1 B and C). The hnRNP K and hnRNP L siRNAs effectively depleted their target mRNAs, and the protein expression was significantly reduced (Fig. 1 C and D). However, no adverse effects on cell survival or proliferation were observed over the 48-h treatment period (Fig. S1 A and B). In contrast, the knockdown of hnRNP Q, hnRNP M, or hnRNP C had no effect on CSR (Fig. 1 A–C), suggesting that hnRNP K and hnRNP L are specifically required in AID’s function. We also confirmed that hnRNP K and hnRNP L knockdown did not alter the expression of the germline transcripts μGLT and αGLT or of AID, all of which are essential for CSR (Fig. 1E). This finding excluded the involvement of hnRNP K and hnRNP L in the transcriptional regulation of known CSR-associated genes. These results suggest that both hnRNP K and hnRNP L are directly required for CSR.
Fig. 1.
Knockdown of hnRNP K or hnRNP L inhibits CSR. (A) Structure of ACF and selected hnRNPs. (B) FACS profiles of IgA switching in CH12F3-2A cells transfected with the indicated hnRNP and control siRNAs. (C) Quantitative RT-PCR (qRT-PCR) analysis of hnRNP mRNA expression and CSR assay results from three independent experiments, as shown in B. The error bars represent the SD; (+) and (−) represent the present and absence of CIT stimulation. (D) Western blot analysis showing the knockdown efficiency of hnRNP K and hnRNP L. (E) qRT-PCR analysis of μGLT, αGLT, and AID transcripts. Values in C and E represent the mean of three independent experiments.
Generation of Cells Defective in hnRNP K and hnRNP L Expression.
To demonstrate unequivocally that hnRNP K and hnRNP L are required for CSR, we used clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) technology (35) in CH12F3-2A cells and generated hnRNP K- and hnRNP L-expression–defective clones, K2-20 and L11, respectively (see Materials and Methods and Fig. S2 for details). Although both alleles were disrupted, a truncated hnRNP K transcript was expressed in the K2-20 clone, which was depleted further by hnRNP K siRNA (Fig. S2D). In the L11 clone only one of the hnRNP L alleles was disrupted, but complete depletion of hnRNP L protein was achieved by introducing sihnRNP L (Fig. S2F). Because treatment of K2-20 and L11 cells with the respective siRNAs resulted in drastic inhibition of CSR (Fig. S2 D and G), this combined blocking system was used in most of the subsequent experiments.
Depletion of hnRNP K but Not of hnRNP L Reduces SHM.
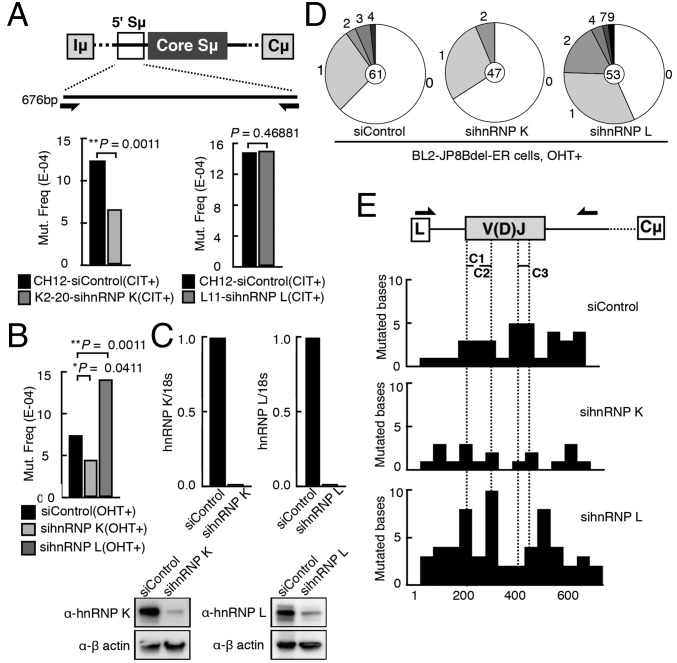
To determine whether CSR inhibition mediated by hnRNP K or hnRNP L deficiency is caused by defective AID-induced DNA breaks, we first examined the postbreak mutation signature in the Sμ regions of K2-20 and L11 cells treated with the hnRNP K and hnRNP L siRNA, respectively. Because the flanking regions of the core Sμ sequence are frequent targets of AID-induced DNA breaks, we sequenced a 676-bp sequence immediately upstream of the core Sμ sequence (Fig. 2A and Fig. S3 A and B). We observed reduction of mutation frequency in hnRNP K-depleted cells but not in hnRNP L-depleted cells, suggesting that the DNA cleavage step may be dependent specifically on hnRNP K.
Fig. 2.
Depletion of hnRNP K, but not of hnRNP L, impairs the generation of mutations in the S and V regions. (A) Sequenced region of 5′ Sµ and the total mutational frequency in the K2-20 and L11 clones treated with siRNAs targeting hnRNP K and L, respectively. (B) Mutation analysis of the V region (V4-39/JH5) in BL2-JP8Bdel-ER cells treated with the indicated siRNAs. (C, Upper) qRT-PCR of hnRNP K and hnRNP L mRNA expression in the BL2-JP8Bdel-ER knockdown cells. (Lower) Corresponding Western blot analysis. (D) Pie charts depicting the proportion of clones that contained the indicated number of mutations. The total number of clones sequenced is indicated at the center. (E) Diagram representing the rearranged IgH V region. Primers used for sequencing the region are indicated above the scheme (arrows). The distribution of mutations is represented as the percentage of mutated bases per 50 bp sequenced. C1, CDR1; C2, CDR2; C3, CDR3; L, leader.
Next, we asked whether AID-induced SHM also is dependent on hnRNP K. The SHM-proficient BL2 cell line expressing a C-terminal AID deletion mutant fused to the estrogen receptor (JP8Bdel-ER) induces SHM at a high frequency upon tamoxifen (4-OHT)-mediated activation (36). Therefore, we examined the SHM frequency in the V(D)J region of the Ig heavy-chain (IgH) locus in BL2-JP8Bdel-ER cells after either hnRNP K or hnRNP L knockdown. As expected, a significant reduction (P < 0.05) in AID-induced mutations was observed upon hnRNP K depletion (Fig. 2B and Fig. S3C), and the mutation frequency was even higher (P < 0.01) in the hnRNP L-depleted cells. The siRNA-mediated depletion of both hnRNP mRNAs was very efficient and correlated well with the reduced protein expression level (Fig. 2C). The numbers of both mutated clones and mutations per clone decreased in hnRNP K-depleted cells but increased in hnRNP L-depleted cells compared with the control BL2 cells (Fig. 2D). There was no bias in the SHM distribution profile in the rearranged V region, regardless of SHM reduction or augmentation by depleting hnRNP K or hnRNP L (Fig. 2E). These results indicate that hnRNP K, but not hnRNP L, is required for the DNA cleavage involved in SHM.
hnRNP K, but Not L, Is Required for AID-Induced DNA Breaks.
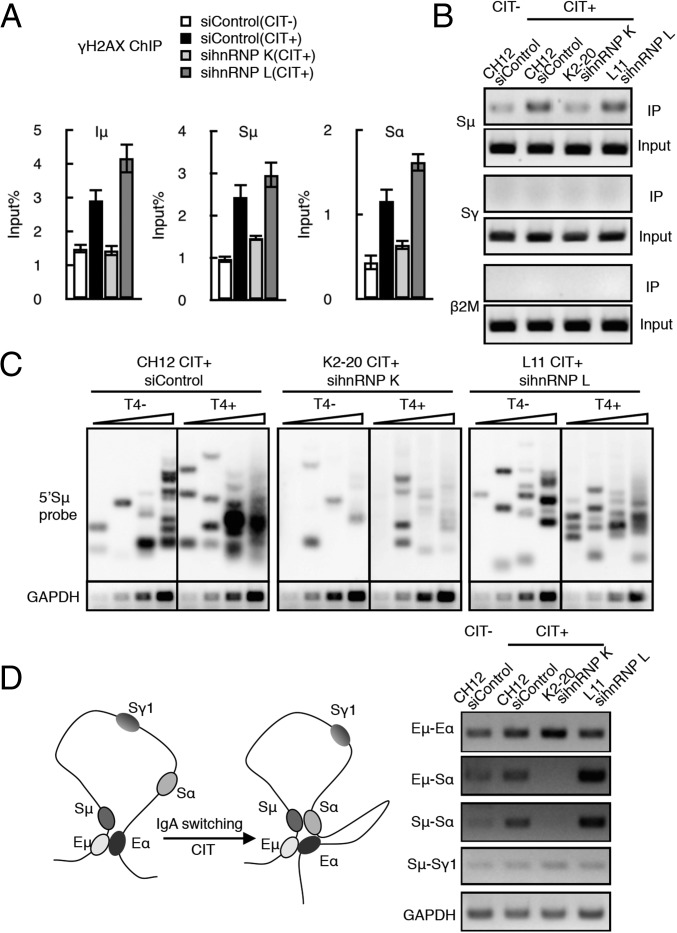
To examine the requirement for hnRNP K in S-region DNA cleavage directly, we performed the histone gamma-H2AX (γH2AX) ChIP assay, which detects DSB-induced γH2AX focus formation at DNA regions flanking DSBs (37, 38). The depletion of hnRNP K, but not of hnRNP L, significantly reduced the γH2AX signal in the Iμ, Sμ, and Sα sequences (Fig. 3A and Fig. S4A).
Fig. 3.
AID-induced DNA breakage is dependent on hnRNP K but not on hnRNP L. (A) DNA DSB determination by γH2AX ChIP assay using hnRNP K- or L-depleted CH12F3-2A cells. The presence or absence of CIT stimulation is indicated by (+) or (−), respectively. SD values were derived from three independent experiments. (B) Biotin-dUTP–labeled DNA break assay. PCR analysis of the S regions (Sμ and Sγ) and β2 microglobulin (β2M, control) in the K2-20 and L11 lines transfected with the indicated siRNAs. (C) LM-PCR–based DNA break assay using T4 polymerase-treated (T4+) and -untreated (T4−) DNA samples. A Southern blot analysis using an Sμ probe and the semiquantitative PCR analysis of GAPDH mRNA expression (control) are shown. DNA template samples were analyzed at threefold increasing DNA concentrations. (D, Left) Schematic view of the long-range interactions occurring at the IgH locus during CIT-induced IgA switching in CH12F3-2A cells, which brings Sμ and Sα into close proximity. (Right) 3C PCR analysis is shown under four different conditions, as indicated. Each PCR panel represents a pair of long-range interactions at the IgH locus (indicated at left). GAPDH PCR analysis of the cross-linked DNA sample served as a loading control.
We also performed a semiquantitative detection of DNA cleavage by the direct labeling of DSB ends with biotin-dUTP, followed by pull-down of the biotinylated DNA fragments and target-site–specific PCR. The Sμ region-specific signal was reduced in hnRNP K-depleted cells compared with the control and hnRNP L-depleted cells (Fig. 3B and Fig. S4B). Regardless of hnRNP depletion, no background signal was detected at two control loci, β2M and Sγ, which were transcriptionally active and inactive, respectively, in the CIT-treated CH12F3-2A cells.
To confirm further the requirement of hnRNP K for DSB formation at the S region, we used a ligation-mediated (LM)-PCR assay, which amplifies DSB ends ligated to a linker, followed by Southern blot analysis using Sμ-specific probes (Fig. 3C and Fig. S4B). To convert overhanging ends to blunt ends, the isolated DNA samples were treated with T4 polymerase. We found that hnRNP K-depleted CH2F3-2A cells exhibited greatly reduced LM-PCR–generated DSB signals in either the presence or absence of T4 polymerase. In contrast, hnRNP L-depleted cells displayed LM-PCR signals comparable to the level in WT cells.
Next, we examined whether changes in DNA cleavage frequency correlated with changes in Sμ–Sα or Eμ–Eα synapse formation, using the chromosome conformation capture (3C) assay (12, 39), which measures the relative cross-linking frequency of two distantly located loci (Fig. 3D and Fig. S4B). Although the interaction between Eμ and Eα was slightly enhanced by CIT stimulation, it was not reduced by the depletion of either hnRNP K or hnRNP L. However, the association between Sμ and Sα was reduced dramatically when hnRNP K, but not hnRNP L, was depleted. The same phenomenon was observed when the association between Eμ and Sα was examined, confirming that synapse formation at both Sμ–Sα and Eμ–Sα was disrupted by depletion of hnRNP K but not hnRNP L (Fig. 3D). Notably, the efficiency of synapse formation was elevated by hnRNP L depletion, which also correlated with the elevated γH2AX signal in the absence of hnRNP L (Fig. 3A). It is possible that hnRNP L is involved in the end-joining step after synapse formation between the cleaved ends. If so, the lack of end repair may prolong synapsis and the processing of DSBs in the absence of hnRNP L. Thus, four independent lines of evidence support the conclusion that AID-induced DNA cleavage requires hnRNP K but not hnRNP L.
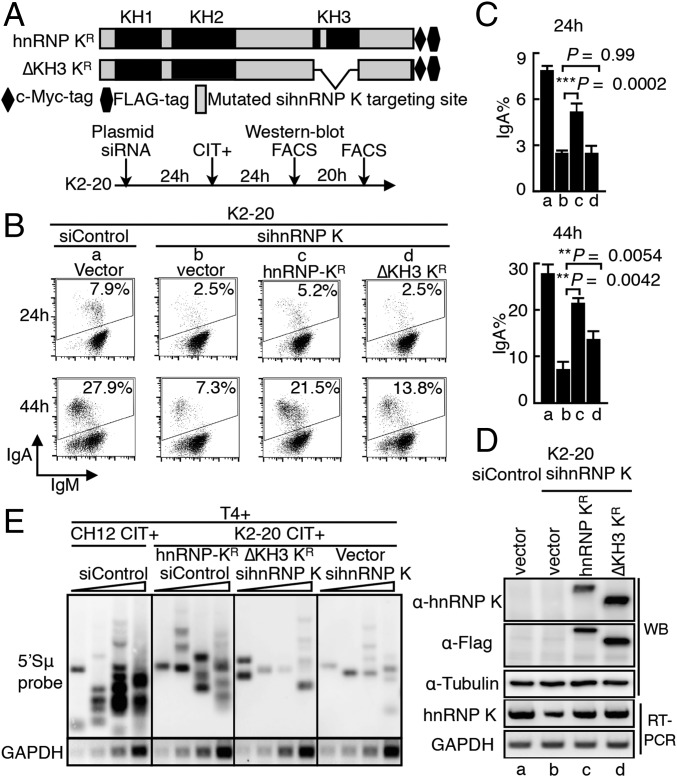
KH Domains of hnRNP K Are Required for Its Function in CSR.
The KH domains of hnRNP K are known to be responsible for its RNA-binding activity (30, 40). To evaluate the importance of these domains in CSR, we constructed KH3-domain–deleted hnRNP mutants (Fig. 4A). The mutant and WT constructs were modified so that they were resistant to the hnRNP K-targeted siRNA and were tagged at the C terminus with the c-Myc-FLAG epitope. We next examined their CSR complementation efficiencies in K2-20 cells treated with hnRNP K siRNA. The introduction of WT hnRNP K (wt-KR, where “R” denotes resistance to siRNA-mediated degradation) successfully rescued the effect of hnRNP K depletion on IgA switching (Fig. 4 B and C), whereas the KH3-domain–deleted mutant showed less CSR rescue than WT hnRNP K. We confirmed the protein expression level of each of the constructs when the endogenous hnRNP K was depleted (Fig. 4D and Fig. S5D).
Fig. 4.
Requirement for the RNA-binding domains of hnRNP K for CSR. (A) Representation of the siRNA-resistant and epitope (c-Myc-FLAG)-tagged hnRNP K constructs. WT and KH3-deleted hnRNP K are designated as hnRNP KR and ΔKH3 KR, respectively. (B) CSR complementation assay performed by cotransfecting K2-20 cells with KR constructs and hnRNP K siRNA. The percentages of IgA-switched cells are indicated in the FACS profiles. (C) Efficiency of CSR rescue calculated from three independent experiments. (D, Upper) Western blot (WB) analysis shows the protein expression of the indicated KR constructs in K2-20 cells. (Lower) RT-PCR analysis of hnRNP K mRNA expression using primers M4 and M5 (Fig. S2C). (E) Southern blot analysis of LM-PCR–based DNA break assay using T4 polymerase-treated (T4+) DNA samples as in Fig. 3. K-20 cells were cotransfected with the indicated siRNAs and the hnRNP K-expressing constructs resistant to sihnRNP K-mediated degradation. After 24 h of transfection, cells were stimulated with CIT and were harvested for DNA break assay 24 h later.
The CSR rescue abilities of the other hnRNP K KH-domain mutants also were examined in CH12F3-2A cells after siRNA-mediated knockdown of endogenous hnRNP K (Fig. S5). The results showed that none of the KH-domain–deleted mutants could rescue CSR completely, indicating that the full CSR activity of hnRNP K requires the presence of all the KH domains. However, the presence of a single KH domain was sufficient for partial CSR rescue. To verify further the importance of KH domains in AID-induced DNA break, we examined the ability of wt-KR and the mutant with KH3-domain deletion (ΔKH3R) to complement DNA breaks (Fig. 4E). As expected, cotransfection of wt-KR significantly counteracted the sihnRNP K-mediated reduction of the S region DNA break and elevated the DNA-break signal close to the WT level. In contrast, introduction of mutant ΔKH3R barely counteracted the inhibition of DNA breaking caused by hnRNP K depletion. Taking these findings together, we conclude that KH-type RRM domains of hnRNP K are critical to AID’s function in DNA breaking, an indispensable and early step for both SHM and CSR.
Atypical RRM Domains of hnRNP L Are Required for Its Function in CSR.
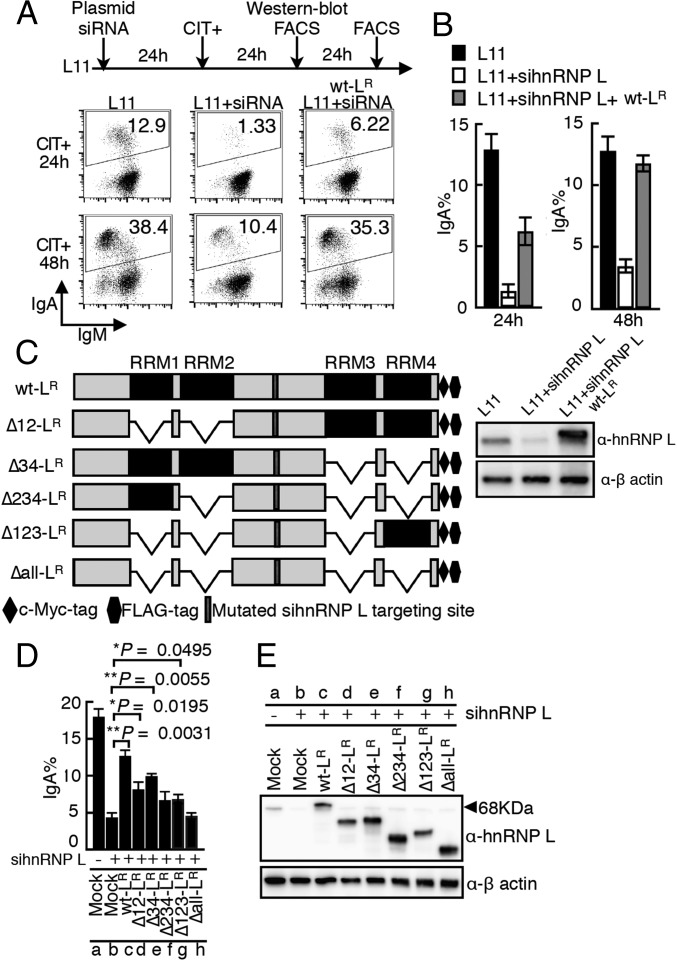
Next, we used a similar approach to determine the structural requirements of hnRNP L for CSR. As shown above, the introduction of hnRNP L siRNA into L11 cells strongly inhibits CSR. The introduction of a construct expressing siRNA-resistant c-myc-FLAG–tagged WT hnRNP L (wt-LR) into the hnRNP L-depleted cells complemented the CSR defect very efficiently, especially at 48 h after transfection (Fig. 5 A and B). Then constructs expressing siRNA-resistant hnRNP L mutants with various RRM-domain deletions (Fig. 5C) were prepared, and their protein expression levels were confirmed (Fig. 5E). Mutants lacking any one of the RRM domains were partially defective in the CSR rescue function (Fig. 5D), and the mutant devoid of all four RRMs (Δall-LR) lost all CSR-rescue ability. These finding suggest that an RNA-dependent activity is involved in hnRNP L’s CSR activity and show that the presence of only a single RRM domain in hnRNP L supports partial CSR recovery.
Fig. 5.
The RNA-binding domains of hnRNP L are required for CSR. (A) CSR complementation assay performed by cotransfecting L11 cells with the siRNA-resistant WT hnRNP L construct (wt-LR) and hnRNP L siRNA. The percentages of IgA switched cells are indicated in the FACS profiles. (B) CSR rescue efficiencies from three independent experiments. A representative Western blot analysis shows the degradation of endogenous but not exogenous hnRNP L (wt-LR). (C) Representation of the various hnRNP mutants used in CSR complementation experiments. (The “Δ” with numbers indicates the specific RRM domain deleted.) (D) Summary of CSR rescue efficiencies of the hnRNP L mutants from three independent experiments. (E) Western blot analysis shows the protein expression associated with each of the LR constructs.
RNA-Dependent Interactions of hnRNP K and hnRNP L with AID.
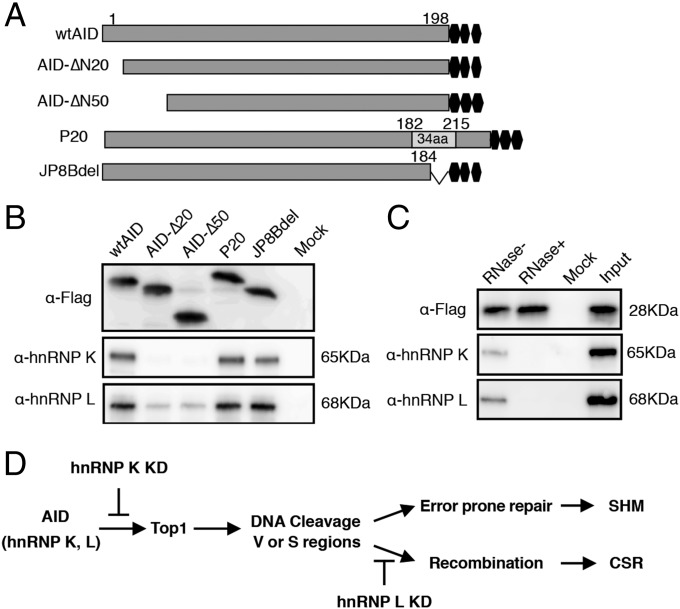
We hypothesized that AID may form specific complex(es) with hnRNP K and hnRNP L, analogous to the APOBEC1–A1CF–RNA complex (23, 41, 42). Given that AID’s function appears to be dependent on the RNA-binding domains of hnRNP K and L, it is likely that the AID–hnRNP complex contains specific target RNA and plays an important role in RNA-substrate recognition. To examine interactions among AID, hnRNP, and RNA, we used the PAR-CLIP (photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation) technology, which detects stable RNA–protein complexes formed by UV cross-linking of 4-thio uracil (4-SU)–labeled RNA and its associated proteins (43). To allow pull-down of the cross-linked RNP complexes with the anti-FLAG antibody, we generated constructs for expressing C-terminal Flag-tagged AID (Fig. 6A). The PAR-CLIP assay was performed using HEK293T cells expressing FLAG-tagged WT AID or mutant forms of AID containing C- or N-terminal deletions. All the constructs expressed well, and AID was immunoprecipitated quite efficiently (Fig. 6B). Both hnRNP K and L were easily detected in association with WT AID and with the C-terminally defective AID mutants (P20 and JP8Bdel). However, little binding of the hnRNPs was detected in association with the N-terminally truncated loss-of-function mutants, suggesting that the N terminus plays a crucial role in the formation of the AID–hnRNP complex (Fig. 6B). In addition, RNase treatment completely abolished the interactions between AID and hnRNP K or hnRNP L, showing that these interactions are completely RNA dependent (Fig. 6C). We also examined the expression of various hnRNP proteins in B cells and found that none of them were induced upon CIT stimulation (Fig. S6), suggesting that the induction of AID expression may lead to the formation of specific RNP–cofactor complexes.
Fig. 6.
RNA-dependent interactions of AID with hnRNP K and L. (A) Schematic view of the 3xFLAG-tagged WT AID and C- or N-terminal deletion mutants expressed in HEK293T cells and used in the PAR-CLIP assay. (B) Western blot analysis of the anti-Flag immunoprecipitations from 293T cells expressing WT or mutant FLAG-tagged AID and treated with 4-SU. The results show the association of the various forms of AID with hnRNP K and L and that the AID N terminus is required for their association. (C) RNase treatment of the cell lysates prevents AID association with hnRNPs. Neither hnRNP K nor hnRNP L coprecipitated with AID if the cell lysates were pretreated with RNase. (D) AID in association with an hnRNP (K or L) complex may edit specific RNAs and regulate DNA cleavage and recombination steps independently.
Discussion
Here we investigated the role of the A1CF-like hnRNP proteins in AID-mediated CSR and SHM. We found that two hnRNPs, K and L, exhibited differential regulatory effects on the two genetic events. HnRNP K deficiency reduced both CSR and SHM, whereas the absence of hnRNP L blocked only CSR without perturbing SHM at either the V or S region. Two independent methods of estimating DNA breakage (biotin-dUTP end labeling and LM-PCR), as well as the evaluation of γH2AX accumulation and break-end synapse formation at S regions, confirmed that hnRNP K is required for the formation of AID-dependent DNA breaks, resulting in the loss of CSR and SHM in hnRNP K-deficient cells. In contrast, hnRNP L depletion led to enhanced, rather than reduced, γ-H2AX accumulation and to increased S–S synapse formation. Thus, hnRNP K appears to function as an AID cofactor during the formation of DNA breaks, which are required for both CSR and SHM, whereas hnRNP L is involved primarily in regulating the recombination step of CSR.
Analysis of the role of the RNA-binding motifs in hnRNP K and L indicated that all the motifs are required for full CSR activity, suggesting that CSR/SHM regulation by hnRNP cofactors involves an RNA-dependent interaction with AID. Consistent with this theory, we also found that the interaction of AID and its hnRNP cofactors is susceptible to RNase-induced dissociation. Furthermore, the N terminus of AID appears to play a critical role in the interactions, because an N-terminal truncated loss-of-function form of AID loses its ability to bind the hnRNPs.
Recently, hnRNP Q6 was identified as an APOBEC1-interacting protein that is required for APOBEC1’s association with IL-8 mRNA (44). In that study, the complex was involved in IL-8 mRNA stabilization by an as yet unknown mechanism, suggesting that the APOBEC1–hnRNP Q6 complex may have a novel function associated with RNA processing rather than with C-to-U editing. More recently, mice lacking RBM47, an RNA-binding protein with three RRMs, were shown to be defective in APO B-100 editing mediated by APOBEC1 (45). These findings suggest that APOBEC1 RNA-editing complexes may comprise multiple RNA-binding or hnRNP cofactors that are involved in APO B-100 editing. Therefore, similar to APOBEC1, which forms a dimer in the APOBEC1–A1CF RNA editing complex (46), AID may form cofactor-specific RNP complexes to mediate its DNA cleavage and recombination functions. In addition, we cannot exclude the possibility that other hnRNP/RNA-binding proteins also are required for the editosome formation and AID’s function.
In conclusion, we postulate that AID may use specific hnRNPs as A1CF-like cofactors to mediate its various RNA-dependent functions. We favor the idea that the AID complex containing hnRNP K may be involved in modulating topoisomerase 1 levels, possibly through miRNA modification, resulting in the DNA cleavage associated with SHM and CSR (Fig. 6D) (47, 48). The recombination-specific editing complex comprising AID and hnRNP L may support the CSR recombination step (Fig. 6D), including the process of end-joining, through mRNA modification, followed by the formation of a new protein.
Materials and Methods
CH12F3–2A cells expressing Bcl2 and BL2 (human Burkitt’s lymphoma) cells were cultured in RPMI medium 1640 (Invitrogen) supplemented with 10% (vol/vol) FBS and penicillin-streptomycin. HEK 293T cells were cultured in DMEM (Invitrogen) supplemented with 10% (vol/vol) FBS.
Detailed materials and methods (CSR assay, siRNA oligonucleotide transfection, SHM analyses, ChIP assay, DNA break assays, 3C assay, and AID immunoprecipitation) are described in SI Materials and Methods. Primers and siRNAs, siRNA oligos, antibodies, and constructs are listed in Tables S1–S4, respectively.
Supplementary Material
Acknowledgments
We thank Dr. Jianliang Xu and Dr. Afzal Husain for assistance with experiments and discussion of results. This research was supported by Grant-in-aid for Specially Promoted Research 17002015 (to T.H.) and Grant-in-Aid for Scientific Research 24590352 (to N.A.B.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. W.H. received support for his PhD scholarship from the China Scholarship Council. S.M. received support for his postdoctoral fellowship from the Human Frontier Science Program.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1506167112/-/DCSupplemental.
References
- 1.Muramatsu M, Nagaoka H, Shinkura R, Begum NA, Honjo T. Discovery of activation-induced cytidine deaminase, the engraver of antibody memory. Adv Immunol. 2007;94:1–36. doi: 10.1016/S0065-2776(06)94001-2. [DOI] [PubMed] [Google Scholar]
- 2.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102(5):553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 3.Faili A, et al. AID-dependent somatic hypermutation occurs as a DNA single-strand event in the BL2 cell line. Nat Immunol. 2002;3(9):815–821. doi: 10.1038/ni826. [DOI] [PubMed] [Google Scholar]
- 4.Yousif AS, Stanlie A, Mondal S, Honjo T, Begum NA. Differential regulation of S-region hypermutation and class-switch recombination by noncanonical functions of uracil DNA glycosylase. Proc Natl Acad Sci USA. 2014;111(11):E1016–E1024. doi: 10.1073/pnas.1402391111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: Linkage with somatic hypermutation. Annu Rev Immunol. 2002;20:165–196. doi: 10.1146/annurev.immunol.20.090501.112049. [DOI] [PubMed] [Google Scholar]
- 6.Chaudhuri J, et al. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv Immunol. 2007;94:157–214. doi: 10.1016/S0065-2776(06)94006-1. [DOI] [PubMed] [Google Scholar]
- 7.Doi T, et al. The C-terminal region of activation-induced cytidine deaminase is responsible for a recombination function other than DNA cleavage in class switch recombination. Proc Natl Acad Sci USA. 2009;106(8):2758–2763. doi: 10.1073/pnas.0813253106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shinkura R, et al. Separate domains of AID are required for somatic hypermutation and class-switch recombination. Nat Immunol. 2004;5(7):707–712. doi: 10.1038/ni1086. [DOI] [PubMed] [Google Scholar]
- 9.Ta VT, et al. AID mutant analyses indicate requirement for class-switch-specific cofactors. Nat Immunol. 2003;4(9):843–848. doi: 10.1038/ni964. [DOI] [PubMed] [Google Scholar]
- 10.Barreto VM, Magor BG. Activation-induced cytidine deaminase structure and functions: A species comparative view. Dev Comp Immunol. 2011;35(9):991–1007. doi: 10.1016/j.dci.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 11.Zahn A, et al. Activation induced deaminase C-terminal domain links DNA breaks to end protection and repair during class switch recombination. Proc Natl Acad Sci USA. 2014;111(11):E988–E997. doi: 10.1073/pnas.1320486111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabouri S, et al. C-terminal region of activation-induced cytidine deaminase (AID) is required for efficient class switch recombination and gene conversion. Proc Natl Acad Sci USA. 2014;111(6):2253–2258. doi: 10.1073/pnas.1324057111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anant S, Yu H, Davidson NO. Evolutionary origins of the mammalian apolipoproteinB RNA editing enzyme, apobec-1: Structural homology inferred from analysis of a cloned chicken small intestinal cytidine deaminase. Biol Chem. 1998;379(8-9):1075–1081. doi: 10.1515/bchm.1998.379.8-9.1075. [DOI] [PubMed] [Google Scholar]
- 14.Rogozin IB, et al. Evolution and diversification of lamprey antigen receptors: Evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat Immunol. 2007;8(6):647–656. doi: 10.1038/ni1463. [DOI] [PubMed] [Google Scholar]
- 15.Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol. 2005;22(2):367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- 16.Powell LM, et al. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 1987;50(6):831–840. doi: 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- 17.Anant S, Davidson NO. Molecular mechanisms of apolipoprotein B mRNA editing. Curr Opin Lipidol. 2001;12(2):159–165. doi: 10.1097/00041433-200104000-00009. [DOI] [PubMed] [Google Scholar]
- 18.Prohaska KM, Bennett RP, Salter JD, Smith HC. The multifaceted roles of RNA binding in APOBEC cytidine deaminase functions. Wiley Interdiscip Rev RNA. 2014;5(4):493–508. doi: 10.1002/wrna.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quaresma AJ, Oyama S, Jr, Barbosa JA, Kobarg J. The acidic domain of hnRNPQ (NSAP1) has structural similarity to Barstar and binds to Apobec1. Biochem Biophys Res Commun. 2006;350(2):288–297. doi: 10.1016/j.bbrc.2006.09.044. [DOI] [PubMed] [Google Scholar]
- 20.Mehta A, Driscoll DM. Identification of domains in apobec-1 complementation factor required for RNA binding and apolipoprotein-B mRNA editing. RNA. 2002;8(1):69–82. doi: 10.1017/s1355838202015649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris SG, et al. Extract-specific heterogeneity in high-order complexes containing apolipoprotein B mRNA editing activity and RNA-binding proteins. J Biol Chem. 1993;268(10):7382–7392. [PubMed] [Google Scholar]
- 22.Sowden MP, Ballatori N, Jensen KL, Reed LH, Smith HC. The editosome for cytidine to uridine mRNA editing has a native complexity of 27S: Identification of intracellular domains containing active and inactive editing factors. J Cell Sci. 2002;115(Pt 5):1027–1039. doi: 10.1242/jcs.115.5.1027. [DOI] [PubMed] [Google Scholar]
- 23.Dance GS, et al. Two proteins essential for apolipoprotein B mRNA editing are expressed from a single gene through alternative splicing. J Biol Chem. 2002;277(15):12703–12709. doi: 10.1074/jbc.M111337200. [DOI] [PubMed] [Google Scholar]
- 24.Backus JW, Smith HC. Apolipoprotein B mRNA sequences 3′ of the editing site are necessary and sufficient for editing and editosome assembly. Nucleic Acids Res. 1991;19(24):6781–6786. doi: 10.1093/nar/19.24.6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Backus JW, Smith HC. Three distinct RNA sequence elements are required for efficient apolipoprotein B (apoB) RNA editing in vitro. Nucleic Acids Res. 1992;20(22):6007–6014. doi: 10.1093/nar/20.22.6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mehta A, Driscoll DM. A sequence-specific RNA-binding protein complements apobec-1 To edit apolipoprotein B mRNA. Mol Cell Biol. 1998;18(8):4426–4432. doi: 10.1128/mcb.18.8.4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith HC. Apolipoprotein B mRNA editing: The sequence to the event. Semin Cell Biol. 1993;4(4):267–278. doi: 10.1006/scel.1993.1032. [DOI] [PubMed] [Google Scholar]
- 28.Galloway CA, Kumar A, Krucinska J, Smith HC. APOBEC-1 complementation factor (ACF) forms RNA-dependent multimers. Biochem Biophys Res Commun. 2010;398(1):38–43. doi: 10.1016/j.bbrc.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith HC, et al. In vitro apolipoprotein B mRNA editing: Identification of a 27S editing complex. Proc Natl Acad Sci USA. 1991;88(4):1489–1493. doi: 10.1073/pnas.88.4.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valverde R, Edwards L, Regan L. Structure and function of KH domains. FEBS J. 2008;275(11):2712–2726. doi: 10.1111/j.1742-4658.2008.06411.x. [DOI] [PubMed] [Google Scholar]
- 31.Han SP, Tang YH, Smith R. Functional diversity of the hnRNPs: Past, present and perspectives. Biochem J. 2010;430(3):379–392. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
- 32.Braddock DT, Baber JL, Levens D, Clore GM. Molecular basis of sequence-specific single-stranded DNA recognition by KH domains: Solution structure of a complex between hnRNP K KH3 and single-stranded DNA. EMBO J. 2002;21(13):3476–3485. doi: 10.1093/emboj/cdf352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hui J, Reither G, Bindereif A. Novel functional role of CA repeats and hnRNP L in RNA stability. RNA. 2003;9(8):931–936. doi: 10.1261/rna.5660803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kato L, et al. An evolutionary view of the mechanism for immune and genome diversity. J Immunol. 2012;188(8):3559–3566. doi: 10.4049/jimmunol.1102397. [DOI] [PubMed] [Google Scholar]
- 35.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kato L, et al. Nonimmunoglobulin target loci of activation-induced cytidine deaminase (AID) share unique features with immunoglobulin genes. Proc Natl Acad Sci USA. 2012;109(7):2479–2484. doi: 10.1073/pnas.1120791109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146(5):905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petersen S, et al. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 2001;414(6864):660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stanlie A, Yousif AS, Akiyama H, Honjo T, Begum NA. Chromatin reader Brd4 functions in Ig class switching as a repair complex adaptor of nonhomologous end-joining. Mol Cell. 2014;55(1):97–110. doi: 10.1016/j.molcel.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 40.Thisted T, Lyakhov DL, Liebhaber SA. Optimized RNA targets of two closely related triple KH domain proteins, heterogeneous nuclear ribonucleoprotein K and alphaCP-2KL, suggest Distinct modes of RNA recognition. J Biol Chem. 2001;276(20):17484–17496. doi: 10.1074/jbc.M010594200. [DOI] [PubMed] [Google Scholar]
- 41.Lellek H, et al. Purification and molecular cloning of a novel essential component of the apolipoprotein B mRNA editing enzyme-complex. J Biol Chem. 2000;275(26):19848–19856. doi: 10.1074/jbc.M001786200. [DOI] [PubMed] [Google Scholar]
- 42.Mehta A, Kinter MT, Sherman NE, Driscoll DM. Molecular cloning of apobec-1 complementation factor, a novel RNA-binding protein involved in the editing of apolipoprotein B mRNA. Mol Cell Biol. 2000;20(5):1846–1854. doi: 10.1128/mcb.20.5.1846-1854.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141(1):129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimizu Y, et al. The RNA-editing enzyme APOBEC1 requires heterogeneous nuclear ribonucleoprotein Q isoform 6 for efficient interaction with interleukin-8 mRNA. J Biol Chem. 2014;289(38):26226–26238. doi: 10.1074/jbc.M114.563221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fossat N, et al. C to U RNA editing mediated by APOBEC1 requires RNA-binding protein RBM47. EMBO Rep. 2014;15(8):903–910. doi: 10.15252/embr.201438450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ueyama T, et al. Sequential binding of cytosolic Phox complex to phagosomes through regulated adaptor proteins: Evaluation using the novel monomeric Kusabira-Green System and live imaging of phagocytosis. J Immunol. 2008;181(1):629–640. doi: 10.4049/jimmunol.181.1.629. [DOI] [PubMed] [Google Scholar]
- 47.Kobayashi M, et al. Decrease in topoisomerase I is responsible for activation-induced cytidine deaminase (AID)-dependent somatic hypermutation. Proc Natl Acad Sci USA. 2011;108(48):19305–19310. doi: 10.1073/pnas.1114522108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kobayashi M, et al. AID-induced decrease in topoisomerase 1 induces DNA structural alteration and DNA cleavage for class switch recombination. Proc Natl Acad Sci USA. 2009;106(52):22375–22380. doi: 10.1073/pnas.0911879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.