Significance
The molecular mechanisms used by noncoding RNAs to regulate gene expression are largely unknown. We have discovered a previously unidentified regulatory phenomenon underlying the transcriptional activation of the intermediate filament protein vimentin. This regulation involves the participation of a previously uncharacterized head-to-head antisense transcript that forms part of a hybrid DNA:RNA structure known as the R loop. R loops have been the focus of recent research regarding their unexpected involvement in gene expression regulation. Antisense-mediated formation of the R loop supports a local chromatin environment that ensures the optimal binding of vimentin transcriptional activators. In addition, we describe how hypermethylation of the locus in a large panel of colon cancer patients is correlated with antisense silencing and, thereby, compromises its regulatory activity.
Keywords: vimentin, antisense transcription, DNA methylation, R loop, nucleosome occupancy
Abstract
The mechanisms used by antisense transcripts to regulate their corresponding sense mRNAs are not fully understood. Herein, we have addressed this issue for the vimentin (VIM) gene, a member of the intermediate filament family involved in cell and tissue integrity that is deregulated in different types of cancer. VIM mRNA levels are positively correlated with the expression of a previously uncharacterized head-to-head antisense transcript, both transcripts being silenced in colon primary tumors concomitant with promoter hypermethylation. Furthermore, antisense transcription promotes formation of an R-loop structure that can be disfavored in vitro and in vivo by ribonuclease H1 overexpression, resulting in VIM down-regulation. Antisense knockdown and R-loop destabilization both result in chromatin compaction around the VIM promoter and a reduction in the binding of transcriptional activators of the NF-κB pathway. These results are the first examples to our knowledge of R-loop–mediated enhancement of gene expression involving head-to-head antisense transcription at a cancer-related locus.
Many well-documented instances of functional long noncoding RNAs (ncRNAs) attest to their multiple roles in regulating transcriptional programs (for a recent review, see ref. 1). The most abundant class of long ncRNAs contains natural antisense transcripts, which partially or totally overlap transcripts originating from the opposite strand. Antisense transcripts may have regulatory effects at different levels, including transcriptional regulation, epigenetic control, imprinting, alternative splicing, translation, and RNA editing (reviewed in refs. 2 and 3). Also, recent studies have addressed the role of ncRNAs as spatial regulators of 3D chromatin folding (4). However, we have a far from thorough understanding of the mechanisms underlying antisense-mediated regulation of gene expression.
VIM is a member of the group of type III intermediate filament genes whose expression increases during the epithelial-to-mesenchymal transition and that are generally associated with an enhanced ability for cell migration and invasion (5). Although the existence of antisense transcription at the VIM locus has been reported in rat and is known to influence the epigenetic status of the locus (6), it was not known whether a similar mechanism is present in humans. We present data supporting the positive regulation exerted by VIM head-to-head antisense transcript on VIM mRNA, through the formation of an RNA:DNA hybrid known as the R loop.
Results
Head-to-Head Antisense Transcription at the VIM Locus.
The region encompassing the human VIM promoter region and transcription start site (TSS) contains an additional, as yet functionally uncharacterized, transcriptional unit corresponding to the antisense strand (Fig. 1A), deposited as VIM-AS1 transcript in the University of California, Santa Cruz (UCSC) data bank (also known as BC078172 transcript). VIM-AS1 is a 1.8-kb noncoding RNA transcribed 5′ head-to-head with VIM, starting 709 bp downstream from the canonical VIM TSS. A minor (expressed at a ∼50-fold lower level) alternative VIM TSS has been described 993 bp upstream of the canonical TSS (7) (not depicted in Fig. 1A). VIM is generally scarce in epithelial cells, but it can also be expressed in epithelial cell lines as part of the adaptation to in vitro culture conditions (8, 9). VIM is expressed in normal colon mucosa, mainly in stromal cells and lymphocytes (10). We readily detected both sense and antisense transcripts in end-point PCR by using total RNA from human colon (Fig. 1 A and B) as the template. To confirm strand specificity, we carried out primer-specific reverse transcription and PCR (Fig. 1A). In addition, oligo-dT–primed and random-primed reverse transcription indicated that the VIM-AS1 antisense transcript is a polyadenylated RNA (Fig. 1B, Upper). PolyA+/− partition of total RNA and quantitative RT-PCR (RT-qPCR) confirmed that sense and antisense transcripts are both polyadenylated (Fig. 1B, Lower). However, analysis of the nuclear and the cytoplasmic fractions showed a clear enrichment of VIM-AS1 transcript in the nucleus (Fig. 1C), suggesting a possible nuclear function. RT-qPCR experiments carried out in this study indicated that the VIM mRNA is 2–3 orders of magnitude more abundant than its antisense transcript, consistent with general estimates of the abundance of noncoding antisense transcript relative to its sense partners (ref. 11; see below).
Fig. 1.

VIM-AS1 is a nuclear, polyadenylated transcript running head-to-head with VIM transcript. (A, Upper) Intronic/exonic organization of vimentin (VIM) and its antisense VIM-AS1 transcripts. Coordinates are given relative to the canonical VIM TSS and the UCSC Gene data bank (uc001iot.2) for VIM-AS1 (release hg19). (A, Lower) End-point RT-PCR from normal colon mucosa total RNA with strand-specific primers. Reverse transcription was carried out either with specific reverse primers (“R,” lanes 1–3) or with forward primers (“F,” lanes 4–6). (B, Upper) VIM-AS1 RNA transcript is polyadenylated. (B, Lower) polyA+/polyA- partition of total RNA from SW480 cells analyzed by RT-qPCR. (C) Nuclear/cytoplasmic fractionation of SW480 cells, analyzed by RT-qPCR and Western blot to assess fraction purity.
Hypermethylation in Colon Cancer Is Associated with Sense and Antisense Transcript Silencing.
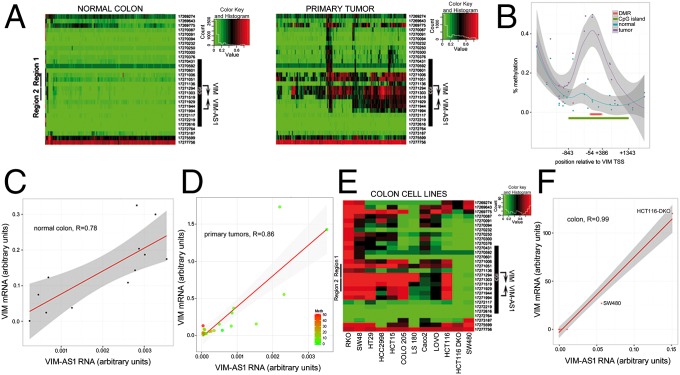
VIM promoter has been thoroughly characterized (12–18). In addition, hypermethylation of VIM promoter-associated CpG-rich island (CGI) has been reported in colon cancer (10, 19). To investigate the impact of CGI methylation on antisense transcription, we examined DNA samples from 120 normal colon and 120 primary tumors with Illumina’s HumanMethylation450 BeadChip. VIM promoter region remains largely unmethylated in normal samples, whereas primary tumors display a clearly hypermethylated CpG island (Fig. 2 A and B). Hypermethylation in primary tumors was confirmed by bisulfite sequencing in an independent subset of matched pairs of normal and tumor samples (n = 33 per type). In this independent subset, we also observed hypermethylation of tumor samples, allowing us to define a differentially methylated region (DMR, thick red line in Fig. 2B) embedded within the CpG island, indicating that hypermethylation of the VIM promoter is a hallmark of colon cancer. At the expression level, the two transcripts are positively correlated, both in normal and primary tumors (Fig. 2 C and D). Interestingly, the primary tumors with highest methylation levels display the lowest transcript abundance (Fig. 2D). Additionally, 10 of 12 colon adenocarcinoma cell lines analyzed exhibited CpG island hypermethylation (Fig. 2E). VIM and VIM-AS1 transcript levels were also positively correlated in methylated and unmethylated lines (Fig. 2F), with 100- to 100,000-fold greater levels of expression in unmethylated compared with methylated lines. Similar correlations were also observed in breast carcinomas and tumor cell lines (Fig. S1 A–C). Methylation levels inversely correlated with the quantities of VIM protein, as shown for HCT116 and HCT116-DKO (hypomorphic for the DNA methyltransferases DNMT1 and DNMT3b) cell lines (Fig. S1D). To further estimate the abundance of VIM-AS1 transcript in comparison with VIM mRNA, we performed absolute quantitation of both RNAs in methylated (HCT116) and unmethlyated (DKO, SW480, MCF10A) cell lines (Fig. S2). The results obtained indicate a difference in 2–3 orders of magnitude between VIM and VIM-AS1 levels, and an impact of methylation resulting in a reduction in expression of 2 orders of magnitude for VIM mRNA and 1 order of magnitude for VIM-AS1 transcript (Fig. S2C).
Fig. 2.
Sense/antisense transcripts are coordinately expressed in normal and tumor colon samples and inversely correlated with DNA methylation. (A) Heatmap representation of a DNA methylation microarray analysis of 120 human normal colon mucosae and 120 tumor samples. (B) Percentage methylation levels of individual CpG sites contained in the 450k array, averaged by class (normal/tumor). The position of the CGI is indicated (green line), and the differentially methylated region defined in C. (C and D) Positive Pearson's correlation coefficients between VIM (y axis) and VIM-AS1 (x axis) expression for normal and tumor colon samples. For primary tumors, the color code indicates methylation levels assessed by bisulfite sequencing. (E) Heatmap representation of a DNA methylation microarray analysis of 12 human colorectal adenocarcinoma cell lines. (F) Pearson's correlation coefficients between VIM and VIM-AS1 expression for all colon cell lines shown in E.
Antisense Knockdown Results in VIM Silencing.
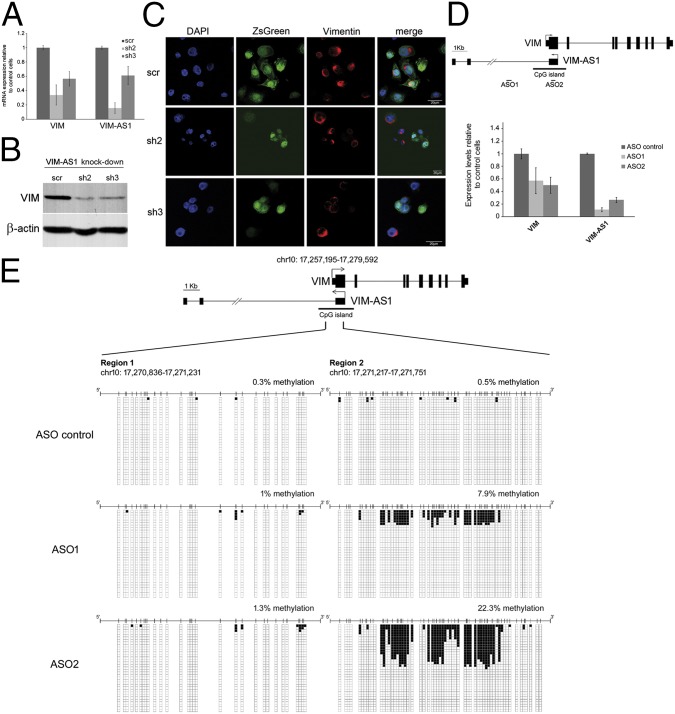
Many antisense transcripts correlate positively and are known to act in cis to regulate their sense partners (3, 20, 21). To investigate this, we used RNAi to deplete VIM-AS1 RNA in SW480 cells, which have an unmethylated VIM promoter and display high basal levels of VIM and VIM-AS1 transcripts. The shRNAs used target the last exon in VIM-AS1, in the region that does not overlap with VIM (SI Materials and Methods). Two distinct shRNAs were capable of efficiently down-regulating VIM-AS1 levels, concomitant with a two- to threefold decrease in VIM mRNA levels (Fig. 3A). This reduction was also detected at the protein level by Western blot and immunofluorescence (Fig. 3B and 3C; ZsGreen is an indicator of transduced cells). To confirm the specificity of the down-regulation, we used two locked nucleic acid (LNA)-based antisense oligonucleotide (ASO) gapmers that target, in a strand-specific manner, the 5′ region of VIM-AS1 transcript (Fig. 3D). Remarkably, both ASOs induced a marked decrease in VIM-AS1 and VIM transcripts, confirming the results obtained with the shRNAs. It is of note that ASO1 was directed against the first intron of VIM-AS, suggesting an active functional involvement for this intronic region. We next investigated whether VIM-AS1 RNA knockdown causes promoter hypermethylation that could account for VIM silencing. As seen in Fig. 3E, we observed a moderate increase in DNA methylation levels across regions 1 and 2 of the VIM promoter, which are the most highly methylated regions in colon tumors and cell lines, therefore indicating that antisense reduction results in a degree of CGI hypermethylation. The same analysis with shRNAs also shows a slight increase in methylation across region 2 of the CpG island (Fig. S3).
Fig. 3.
VIM-AS1 transcript knockdown results in VIM silencing with an effect on promoter CGI methylation. SW480 cells were transduced with lentiviral plasmids overexpressing control shRNA (scr) or shRNAs against VIM-AS1 RNA (sh2, sh3). (A) RT-qPCR analysis of VIM and VIM-AS1 RNAs. (B) Western blot to measure vimentin protein levels in the same transduced cells. (C) Immunofluorescence detection of endogenous vimentin. (D) LNA-based antisense oligonucleotides gapmers (ASOs) targeting intron 1 (ASO1) or exon 1 (ASO2) of VIM-AS1 transcript were transfected into SW480 cells and expression levels measured by RT-qPCR. (E) Bisulfite sequencing of regions 1 and 2 within VIM promoter CGI.
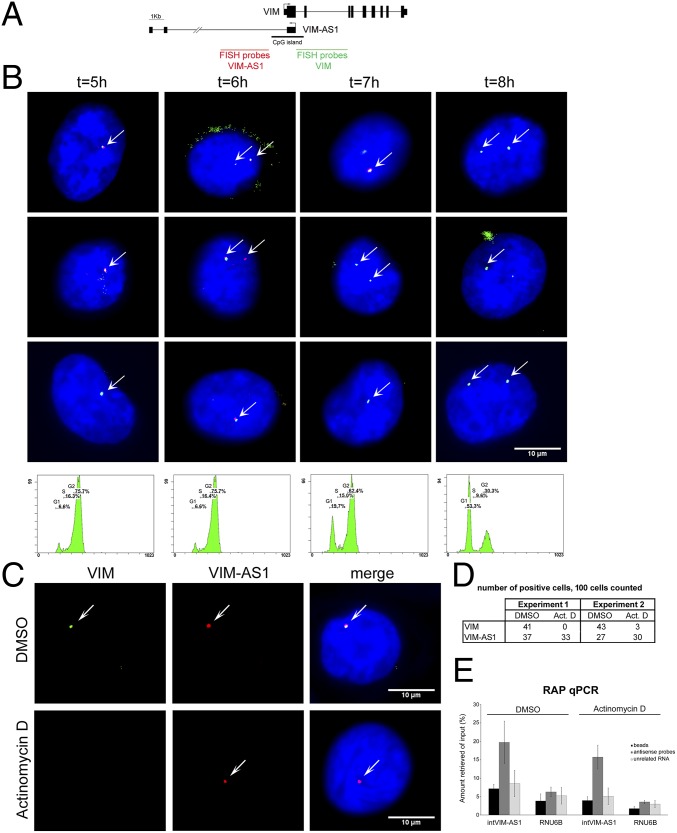
Given the suggested involvement of VIM-AS1 first intron in the regulation, we next designed specific probes to detect by RNA FISH either intron 1 of VIM or intron 1 of VIM-AS1 (Fig. 4A). As expected for intronic regions, both probes colocalize at the site of nascent transcription (Fig. 4B), with an enrichment of the VIM-AS1 probes in G2 phase. Remarkably, blocking transcription by treatment with actinomycin D resulted in the loss of VIM intron 1 signal, whereas the antisense intron remained localized near the genomic locus (Fig. 4 C and D and Fig. S4 A–C, where wider microscope fields with more cells are shown). This FISH signal could be indicative of a special stabilization of the region, possibly due to inefficient splicing. To further analyze the interaction of this intronic RNA region with the local chromatin we used the RNA antisense purification (RAP) method (22), in which a pool of 124-nt-long antisense probes designed against the first intron of VIM-AS1 was able to specifically retrieve the endogenous transcript (Fig. S4D) together with the homologous DNA region (Fig. 4E), suggesting that there is a stable RNA:DNA association in this region. Furthermore, RAP signal was maintained even when transcription was arrested, in accordance with RNA-FISH experiments.
Fig. 4.
RNA FISH detection shows enrichment of antisense transcription during G2 phase and intronic stability following actinomycin D treatment. (A) RNA FISH probe design. (B) MCF10A cells were synchronized and released, fixed at the indicated times and stained for RNA FISH (VIM intron 1 is in green and VIM-AS1 intron 1 is in red) or analyzed for DNA content by fluorescence-activated cell sorting (FACS) (Upper and Lower). (C) RNA FISH in control (DMSO-treated) or actinomycin D-treated MCF10A cells. (D) RNA FISH signal was counted in 100 randomly selected cells. (E) Quantitative PCR (qPCR) of the DNA captured in crosslinked MCF10A cells treated as in C, using streptavidin beads alone (beads), with antisense probes to VIM-AS1 intron 1 (antisense probes) or against the LINC00085 RNA (unrelated RNA). Enrichments represent means from two replicate experiments and are relative to the input amount used per pulldown. RNU6B is used as negative control to assess binding specificity.
Antisense Transcript Forms Part of an R-Loop Structure and Its Disruption Represses VIM Transcription.
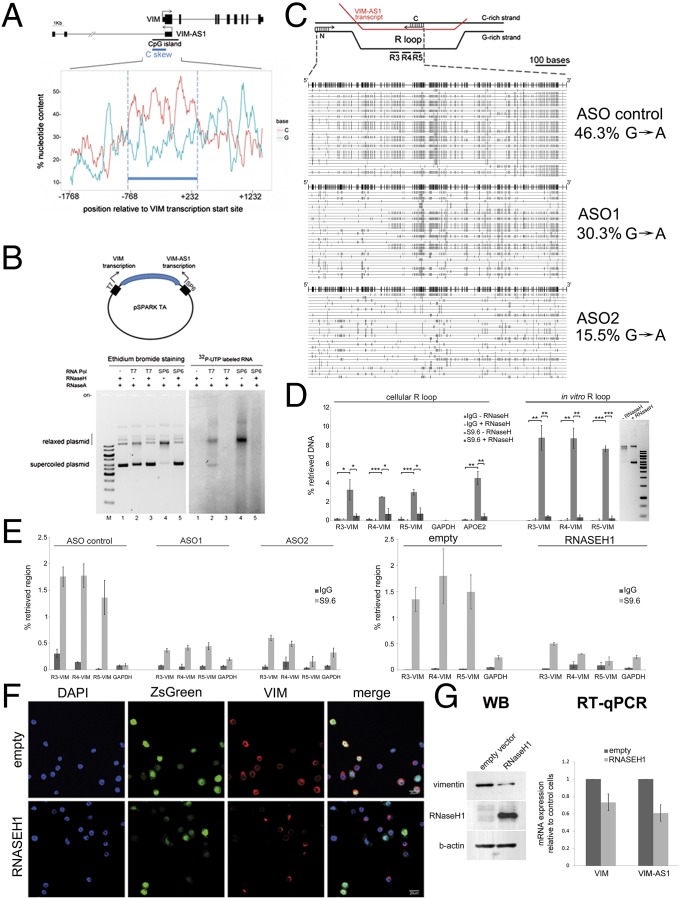
Further exploration of the genomic region between the two transcription start sites revealed an asymmetric distribution of C and G nucleotides (known as a GC skew) on the plus strand of the DNA along the first half of the CGI and coinciding with the first intron of VIM-AS1. As seen in Fig. 5A, a fragment of approximately 1 kb between VIM and VIM-AS1 TSS was particularly enriched in C nucleotides in the plus strand (C skew), whereas no enrichment was observed for A or T nucleotides (Fig. S5A). This observation points to the potential formation of R loops throughout this region. R loops are special three-stranded nucleic acid structures that form in vivo as G-rich RNA transcripts invade the DNA duplex and anneal to the template strand to generate an RNA-DNA hybrid (23), leaving the nontemplate, G-rich DNA strand in a largely single-stranded conformation. To explore this possibility, we cloned the DNA region comprising the C skew between two opposing promoters and tested R-loop formation in vitro. Transcription from T7 promoter gives rise to an RNA molecule in the direction of VIM mRNA, whereas transcription from SP6 promoter originates the antisense VIM-AS1 RNA (Fig. 5B). The formation of extended RNA:DNA hybrid structures results in topological change in the plasmid DNA that can be detected as a lower electrophoretic mobility. Transcription of the region containing the C skew led to a strong shift in DNA migration only when the template strand was the C-rich DNA strand (that is, the RNA produced is VIM-AS1). Transcription in the other direction (VIM physiological orientation) did not result in such a migration pattern (Fig. 5B, Left). To confirm the involvement of RNA, the reaction was carried out in the presence of radiolabeled [α-32P]-rUTP (Fig. 5B, Right). The DNA migration shift and radioactive signal are both abolished upon incubation with recombinant RNaseH, which digests RNA:DNA hybrids. Taken together, these properties (slower migration, orientation dependence, and sensitivity to RNaseH) indicate the presence of an R-loop structure with involvement of VIM-AS1 RNA in the vicinity of VIM TSS.
Fig. 5.
VIM-AS1 RNA forms an R-loop structure whose disruption represses VIM transcription in SW480 cells. (A) Percentage of C and G nucleotides in the VIM promoter reveals the presence of a C skew region (thick blue line). For each position on the DNA plus strand, the percentage abundance of each nucleotide within the surrounding 100 nt is counted, with a sliding window of 1 nt. (B) In vitro R loop formation assay indicates participation of the VIM-AS1 transcript. (C) In vivo detection of R loop formation within the C skew region. (Upper) The diagram depicts the RNA:DNA hybrid and the displaced, single-stranded, minus DNA strand. (Lower) PCR amplification and sequencing of 30 clones corresponding to the first half of the C skew-containing region under different ASO treatment. The upper reference line depicts every G position (vertical lines), and every G-to-A change on the plus strand of the sequenced clones is indicated in light gray by a vertical line. Of the 30 clones represented, 23, 29, and 26 (for ASO control, ASO1 and ASO2, respectively) correspond to unique patterns. (D) DRIP with the S9.6 antibody. Signal intensity is presented relative to the input DNA. Three different amplicons (R3, R4, R5, shown in C) were measured. GAPDH and APOE promoters were analyzed as negative and positive controls, respectively. *P < 0.05; **P < 0.01; ***P < 0.001. (E) DRIP experiments under ASO treatment (Left), or overexpression of RNASEH1 (Right). The same genomic regions as in D were analyzed. (F) Immunofluorescence detection of endogenous vimentin (red) in cells overexpressing RNASEH1 (green). (G, Left) Western blot of total protein extracts from RNASEH1-positive cells (enriched by FACS). (Right) RT-qPCR of total RNA extracted from the pool of transfected cells.
To confirm the formation of the R loop in vivo, we used a native bisulfite treatment of SW480 genomic DNA (which converts only accessible cytosines in any DNA template), followed by PCR with primers specific to the first half of the predicted R loop-forming region, ligation, and sequencing of the resulting clones (24) (Fig. 5C). This method reveals single-strandedness either of the G-rich or C-rich strand, depending on the type of conversion: C-to-T changes in the sequence of the plus strand are indicative of single-strandedness in the C-rich strand (plus strand), whereas G-to-A changes in the plus strand indicate single-strandedness in the G-rich strand (minus strand). In control-transfected cells, all clones sequenced featured long stretches (>100 bp) of uninterrupted G-to-A conversions (Fig. 5C, Upper). These changes are qualitative indications of the existence of an unprotected, single-stranded minus strand, suggesting that R-loop formation occurs endogenously at the VIM promoter with the participation of the antisense transcript. Interestingly, ASO gapmers designed against VIM-AS1 made G-to-A changes less frequent, suggesting the involvement of the antisense VIM-AS1 transcript in R-loop formation in vivo (Fig. 5C, Lower).
As further proof of the existence of an R-loop structure near VIM TSS in vivo, we performed DNA:RNA immunoprecipitation (DRIP) experiments with the S9.6 antibody (25). Consistent with the previous data, we were able to detect specific R-loop formation by DRIP in an RNaseH-sensitive manner along the C skew region (Fig. 5D, Left). For comparison and pulldown efficiency estimations, a known amount of in vitro generated R loop was subject to parallel DRIP experiments (Fig. 5D, Right). Importantly, DRIP signal was decreased in ASO-treated cells (Fig. 5E, Left), and in cells transduced with a lentiviral vector encoding human ribonuclease H1 (RNASEH1) (Fig. 5E, Right), indicating that knockdown of VIM-AS1 transcripts and overexpression of RNASEH1 both result in R-loop resolution. In addition, immunofluorescence analysis indicate that overexpression of the protein reduced vimentin protein levels in cells expressing the transfected protein (Fig. 5F). This result was confirmed by Western blotting after sorting of RNASEH1-overexpressing cells by FACS (Fig. 5G, Left). Accordingly, both VIM-AS1 and VIM mRNA levels were down-regulated under conditions of RNASEH1 overexpression, as detected by RT-qPCR (Fig. 5G, Right). A further reduction in VIM levels was observed in Caco2 cells at the mRNA and protein levels (Fig. S5B). Similarly, in the two breast cell lines, MCF7 and MCF10A, VIM levels were also sensitive to RNASEH1 overexpression (Fig. S5C), indicating that R-loop formation has a generally positive effect on its expression. It is worth noting that VIM-AS1 RNA levels were also significantly diminished in all cell lines in which RNASEH1 was overexpressed, possibly implying that most of its transcripts are R loop-associated and direct targets of RNASEH1 digestion. Because R-loop formation has been associated with DNA methylation protection (25), we performed bisulfite sequencing to assess changes in methylation in VIM CGI under conditions where R-loop formation is disfavored. A slight increase was observed in Caco2 cell line when we overexpressed RNaseH1 (Fig. S5D), whereas no change was seen in SW480 cells (Fig. S5E). This difference is probably due to the fact that antisense levels in Caco2 cells are much lower than in SW480 and it might be easier to achieve a more complete resolution of R loops. Finally, the effect of RNASEH1 overexpression on VIM levels does not result from general changes in expression (Fig. S5F).
Antisense Transcription and R-Loop Structure Support Local Chromatin Decondensation.
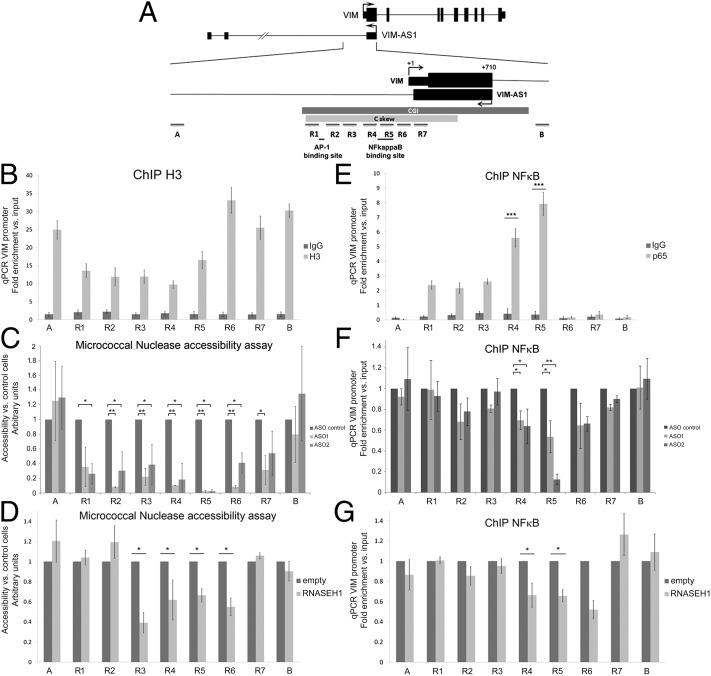
We next attempted to establish whether R-loop formation had any effect at some other level of chromatin conformation. Nucleosome occupancy is known to be lower in the vicinity of the TSSs of actively transcribed genes (26). In accordance with this premise, histone H3 becomes less prevalent in regions immediately upstream of VIM TSS, as revealed by chromatin immunoprecipitation (ChIP) experiments (Fig. 6 A and B). These results might indicate an open conformation coincident with the presence of C skew and R-loop formation. To test the effect of antisense transcription and R-loop formation on the level of chromatin compaction, we isolated native chromatin from control, VIM-AS1–depleted and RNASEH1-overexpressing SW480 cells. The graphs in Fig. 6 C and D illustrate the differences in DNA recovery during the first 10 min of micrococcal nuclease digestion, whereby higher values indicate a more thoroughly digested DNA fragment and, thus, greater accessibility to the nuclease, which is associated with more open chromatin and lower nucleosome density. Remarkably, VIM-AS1 RNA depletion resulted in a three- to fivefold less accessible chromatin conformation in regions 2–6 within the C skew, whereas locations further upstream or downstream did not change significantly (Fig. 6C). A similar effect was seen upon overexpression of RNASEH1 (Fig. 6D). Accordingly, the quantity of histone H3 increased under conditions of antisense knockdown or RNASEH1 overexpression (Fig. S6). These results indicate that R-loop formation is necessary for maintaining an open chromatin and suggest that transcription of the minus strand relaxes local chromatin and possibly keeps the VIM template strand more accessible to the transcriptional machinery.
Fig. 6.
Disruption of antisense transcription and R-loop formation results in chromatin compaction and loss of NF-κB binding in the VIM gene promoter. (A) Fragments analyzed by qPCR in the VIM promoter. (B) Chromatin immunoprecipitation experiments with histone H3 antibody (H3) and control antibody (IgG) in SW480 cells. (C) Micrococcal nuclease accessibility assay on nuclei isolated from ASO-treated SW480 cells. (D) As in C, but comparing overexpression of RNASEH1 with empty vector. (E) Chromatin immunoprecipitation experiments with p65/RelA antibody or control (IgG) antibody in SW480 cells. (F) As in E, in ASO-treated cells. Levels were calculated relative to control samples. (G) As in F, but comparing overexpression of RNASEH1 with control-transfected cells. Throughout the figure, *P < 0.05; **P < 0.01; ***P < 0.005 from Student's t test.
Antisense Transcription and R Loop Enhance NF-κB Binding to the VIM Promoter.
Enhancer binding sites and negative elements have been characterized for the VIM promoter (12, 13, 27). Specifically, binding sites for p65/RelA in the NF-κB pathway are present in regions 4 and 5 of the central region of the C skew (Fig. 6A). To determine whether R-loop formation can affect their binding, we performed ChIP experiments on cellular factors of the NF-κB pathway. In accordance with previous studies, binding of p65 following TNF-α stimulation was specifically enriched in regions 4 and 5 in SW480 cells (Fig. 6E). The same two regions displayed diminished binding following knockdown of the antisense VIM-AS1 RNA (Fig. 6F) or overexpression of RNASEH1 (Fig. 6G). Taken together, our results indicate that transcriptional activation of VIM is supported through the cotranscriptional formation of a stable R-loop structure by a head-to-head antisense transcript. This regulatory mechanism could be a general characteristic of GC-rich promoters with divergent sense/antisense transcription and asymmetrically distributed G and C nucleotides. Interestingly, the high-mobility group protein HMGA2 gene (plus strand) is transcribed head-to-head with the ribosomal protein SA pseudogene RPSAP52 from a C skew-containing locus (Fig. S7A). Similar to VIM-AS1, RPSAP52 transcription forms R-loop structures in vitro (Fig. S7B) and its depletion down-regulates the sense HMGA2 transcript (Fig. S7C) concomitantly with an increase in chromatin compaction, as measured by micrococcal nuclease accessibility assays (Fig. S7D).
Discussion
Our results imply a positive role for R-loop formation by a head-to-head antisense transcript in the regulation of sense transcript expression. Originally considered to be rare transcriptional byproducts, R loops may have a more general role as a mechanism of gene regulation (28–31). This regulatory mechanism is compatible with generally low levels of antisense RNA, because only two target molecules of DNA are present per cell. We have estimated the absolute abundance of VIM-AS1 transcripts in different cell lines (Fig. S2) to correspond to a few copies per cell. Importantly, this amount represents the spliced transcript and may not reflect the actual abundance of the functional, intron-containing species. Related to this point, our RNA-FISH data suggests that the antisense region involved in R-loop formation is present as stable RNA in approximately one-third of cells in G2 phase, and in much lower levels in other phases of the cell cycle (Fig. 4). According to our DRIP experiments and taking into account the efficiency of the technique in our hands, we can estimate that, in nonsynchronous cultures, at most 20% of VIM promoters form an R loop. Remarkably, R-loop abundance has been associated to cell cycle progression (32, 33). In this context, it is of note that VIM expression has long been known to be cell cycle dependent (34, 35). Further studies are needed to explore the detailed link between cell cycle and R-loop–mediated regulation of gene expression in the VIM locus.
Our data indicate that conditions that favor an R loop lead to decreased nucleosome occupancy and increased binding of transcription factors of the NF-κB pathway, which are known to activate VIM expression upon mitogenic stimuli (36). Binding activity [by unknown protein factor(s)] specific to single-stranded DNA present on the minus strand immediately downstream of NF-κB binding elements has also been reported (37), although its regulatory potential is not known. Formation of the R loop would enhance such binding. Alternatively, we cannot rule out the possibility that this binding indicates the presence of some unknown factor that stabilizes R loop formation, as has been shown in Arabidopsis (38). Either way, the presence of a stable R-loop structure allows the maintenance of an open local chromatin conformation and enhances transcription factor binding to the displaced, single-stranded minus DNA strand. In summary, our results are consistent with a model (Fig. S8) in which an intact R loop with participation of VIM-AS1 transcript is essential for the optimal recognition of VIM promoter by transcriptional regulators, and specifically indicate activation by the NF-κB pathway, implicating R-loop structures in a previously unidentified positive role in gene transcription at the loci of bidirectional sense/antisense transcription.
Materials and Methods
Additional methods are described in the SI Materials and Methods.
In Vitro R-Loop Formation Assay.
R-loop formation was tested in vitro essentially as described in ref. 39. Genomic regions of the VIM or HMGA2 promoter were PCR-amplified (with oligos VIMRloop1for and VIMRloop1rev, and HMGA2Rloop1for and HMGA2Rloop1rev, respectively) and cloned into pSPARK TA vector with the antisense strand under SP6 promoter. In vitro transcription reactions were carried out in both directions for 45 min at 37 °C with either SP6 or T7 RNA polymerases in the presence of 0.15 μCi/μL of α-[32P]-UTP, and further digested with RNaseA and RNaseH as indicated, for 30 min at 37 °C. Nucleic acids were phenol-extracted, loaded onto a 1% agarose gel and run in 1× Tris/borate/EDTA. After electrophoresis, the gel was stained with SYBR Safe DNA Gel Stain (Life Technologies) and UV-visualized. Following picture acquisition, the gel was dried and exposed to an autoradiography film for radiolabel detection.
DRIP.
DRIP was performed as described in ref. 39. Genomic DNA was extracted from SW480 cells by SDS/Proteinase K treatment, phenol-chloroform extraction and ethanol precipitation. DNA was then digested with HindIII, EcoRI, XbaI, and BamHI restriction enzymes. Samples were then either mock-treated or digested with RNaseH for a further 2 h. After phenol/chloroform extraction and precipitation, samples were resuspended in IP buffer (0.05% Triton X-100 in PBS) and immunoprecipitated with the anti-DNA-RNA hybrid (S9.6) antibody. Retrieved fragments were analyzed by qPCR and compared with appropriate dilution of input DNA. An amplicon from GAPDH promoter (lacking target sites for the restriction enzymes above) was used as a negative control.
Chromatin Immunoprecipitation.
NF-κB (p65) ChIP experiments were done as described (40). See the SI Materials and Methods for further details.
Supplementary Material
Acknowledgments
We thank Dr. Verónica Dávalos for helpful discussions and A. Aguilera for the gift of the S9.6 antibody. This work was supported by Ministerio de Ciencia e Innovación (MICINN) Grants SAF2011-22895 (to S.G.) and SAF2011-22803 (to M.E.), La Marató de TV3 20131610 (to S.G.), the European Research Council Advanced Grant 268626–EPINORC project (to M.E.), Cellex Foundation, the Health and Science Departments of the Catalan Government (Generalitat de Catalunya), and Fondo de Investigación Sanitaria Grant PI08-1345 (M.E.). S.G. is funded by the Ramón y Cajal Research Program (MICINN). M.E. is an Institució Catalana de Recerca i Estudis Avançats Research Professor. R.B.-S. is a recipient of a Predoctoral Research Grant file BES-2012-058840 from Ministerio de Economía y Competitividad.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1421197112/-/DCSupplemental.
References
- 1.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338(6113):1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 2.Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15(6):423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Magistri M, Faghihi MA, St Laurent G, 3rd, Wahlestedt C. Regulation of chromatin structure by long noncoding RNAs: Focus on natural antisense transcripts. Trends Genet. 2012;28(8):389–396. doi: 10.1016/j.tig.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai F, et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494(7438):497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang Y, Massagué J. Epithelial-mesenchymal transitions: Twist in development and metastasis. Cell. 2004;118(3):277–279. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 6.Tomikawa J, et al. Single-stranded noncoding RNAs mediate local epigenetic alterations at gene promoters in rat cell lines. J Biol Chem. 2011;286(40):34788–34799. doi: 10.1074/jbc.M111.275750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Z, Kahns S, Nielsen AL. Identification of a novel vimentin promoter and mRNA isoform. Mol Biol Rep. 2010;37(5):2407–2413. doi: 10.1007/s11033-009-9751-8. [DOI] [PubMed] [Google Scholar]
- 8.Kokkinos MI, et al. Vimentin and epithelial-mesenchymal transition in human breast cancer—observations in vitro and in vivo. Cells Tissues Organs. 2007;185(1-3):191–203. doi: 10.1159/000101320. [DOI] [PubMed] [Google Scholar]
- 9.Pieper FR, et al. Regulation of vimentin expression in cultured epithelial cells. Eur J Biochem. 1992;210(2):509–519. doi: 10.1111/j.1432-1033.1992.tb17449.x. [DOI] [PubMed] [Google Scholar]
- 10.Chen WD, et al. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J Natl Cancer Inst. 2005;97(15):1124–1132. doi: 10.1093/jnci/dji204. [DOI] [PubMed] [Google Scholar]
- 11.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322(5909):1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rittling SR, Coutinho L, Amram T, Kolbe M. AP-1/jun binding sites mediate serum inducibility of the human vimentin promoter. Nucleic Acids Res. 1989;17(4):1619–1633. doi: 10.1093/nar/17.4.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lilienbaum A, Paulin D. Activation of the human vimentin gene by the Tax human T-cell leukemia virus. I. Mechanisms of regulation by the NF-kappa B transcription factor. J Biol Chem. 1993;268(3):2180–2188. [PubMed] [Google Scholar]
- 14.Salvetti A, Lilienbaum A, Li Z, Paulin D, Gazzolo L. Identification of a negative element in the human vimentin promoter: Modulation by the human T-cell leukemia virus type I Tax protein. Mol Cell Biol. 1993;13(1):89–97. doi: 10.1128/mcb.13.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen JH, et al. PEA3 transactivates vimentin promoter in mammary epithelial and tumor cells. Oncogene. 1996;13(8):1667–1675. [PubMed] [Google Scholar]
- 16.Izmailova ES, Zehner ZE. An antisilencer element is involved in the transcriptional regulation of the human vimentin gene. Gene. 1999;230(1):111–120. doi: 10.1016/s0378-1119(99)00046-3. [DOI] [PubMed] [Google Scholar]
- 17.Wu Y, Zhang X, Salmon M, Lin X, Zehner ZE. TGFbeta1 regulation of vimentin gene expression during differentiation of the C2C12 skeletal myogenic cell line requires Smads, AP-1 and Sp1 family members. Biochim Biophys Acta. 2007;1773(3):427–439. doi: 10.1016/j.bbamcr.2006.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Y, Zhang X, Salmon M, Zehner ZE. The zinc finger repressor, ZBP-89, recruits histone deacetylase 1 to repress vimentin gene expression. Genes Cells. 2007;12(8):905–918. doi: 10.1111/j.1365-2443.2007.01104.x. [DOI] [PubMed] [Google Scholar]
- 19.Li M, et al. Sensitive digital quantification of DNA methylation in clinical samples. Nat Biotechnol. 2009;27(9):858–863. doi: 10.1038/nbt.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engreitz JM, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341(6147):1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aguilera A, García-Muse T. R loops: From transcription byproducts to threats to genome stability. Mol Cell. 2012;46(2):115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 24.Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4(5):442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 25.Ginno PA, Lott PL, Christensen HC, Korf I, Chédin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell. 2012;45(6):814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schones DE, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132(5):887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lilienbaum A, Duc Dodon M, Alexandre C, Gazzolo L, Paulin D. Effect of human T-cell leukemia virus type I tax protein on activation of the human vimentin gene. J Virol. 1990;64(1):256–263. doi: 10.1128/jvi.64.1.256-263.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Powell WT, et al. R-loop formation at Snord116 mediates topotecan inhibition of Ube3a-antisense and allele-specific chromatin decondensation. Proc Natl Acad Sci USA. 2013;110(34):13938–13943. doi: 10.1073/pnas.1305426110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ginno PA, Lim YW, Lott PL, Korf I, Chédin F. GC skew at the 5′ and 3′ ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res. 2013;23(10):1590–1600. doi: 10.1101/gr.158436.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhatia V, et al. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014;511(7509):362–365. doi: 10.1038/nature13374. [DOI] [PubMed] [Google Scholar]
- 31.Chan YA, et al. Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLoS Genet. 2014;10(4):e1004288. doi: 10.1371/journal.pgen.1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castellano-Pozo M, et al. R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Mol Cell. 2013;52(4):583–590. doi: 10.1016/j.molcel.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 33.Herrera-Moyano E, Mergui X, García-Rubio ML, Barroso S, Aguilera A. The yeast and human FACT chromatin-reorganizing complexes solve R-loop-mediated transcription-replication conflicts. Genes Dev. 2014;28(7):735–748. doi: 10.1101/gad.234070.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giese G, Kubbies M, Traub P. Cell cycle-dependent vimentin expression in elutriator-synchronized, TPA-treated MPC-11 mouse plasmacytoma cells. Exp Cell Res. 1992;200(1):118–125. doi: 10.1016/s0014-4827(05)80079-5. [DOI] [PubMed] [Google Scholar]
- 35.Giese G, Kubbies M, Traub P. High resolution analysis of cell cycle-correlated vimentin expression in asynchronously grown, TPA-treated MPC-11 cells by the novel flow cytometric multiparameter BrdU-Hoechst/PI and immunolabeling technique. J Cell Physiol. 1994;161(2):209–216. doi: 10.1002/jcp.1041610204. [DOI] [PubMed] [Google Scholar]
- 36.Duprey P, Paulin D. What can be learned from intermediate filament gene regulation in the mouse embryo. Int J Dev Biol. 1995;39(3):443–457. [PubMed] [Google Scholar]
- 37.Benazzouz A, Duprey P. The vimentin promoter as a tool to analyze the early events of retinoic acid-induced differentiation of cultured embryonal carcinoma cells. Differentiation. 1999;65(3):171–180. doi: 10.1046/j.1432-0436.1999.6530171.x. [DOI] [PubMed] [Google Scholar]
- 38.Sun Q, Csorba T, Skourti-Stathaki K, Proudfoot NJ, Dean C. R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science. 2013;340(6132):619–621. doi: 10.1126/science.1234848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu K, Roy D, Huang FT, Lieber MR. Detection and structural analysis of R-loops. Methods Enzymol. 2006;409:316–329. doi: 10.1016/S0076-6879(05)09018-X. [DOI] [PubMed] [Google Scholar]
- 40.Nowak DE, Tian B, Brasier AR. Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. Biotechniques. 2005;39(5):715–725. doi: 10.2144/000112014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.