Significance
Hepatitis B virus (HBV) causes substantial morbidity and mortality. A large proportion of infected individuals controls infection but does not completely eradicate HBV DNA from the liver, and flares in hepatitis can be precipitated by immunosuppression. A proportion of individuals never controls infection, and these people are at substantial risk of developing liver failure and liver cancer. Current therapies are not effective at eliminating virus, and there is a major interest in developing functional cures for HBV infection. We identified host cell signaling molecules that can restrict the ability to eradicate infected cells. These molecules can be therapeutically targeted, and drugs that interfere with the function of these host cell proteins may be useful therapies to promote clearance of HBV infection.
Keywords: hepatitis B virus, cellular inhibitor of apoptosis proteins, cIAP1, cIAP2, TNF
Abstract
Hepatitis B virus (HBV) infection can result in a spectrum of outcomes from immune-mediated control to disease progression, cirrhosis, and liver cancer. The host molecular pathways that influence and contribute to these outcomes need to be defined. Using an immunocompetent mouse model of chronic HBV infection, we identified some of the host cellular and molecular factors that impact on infection outcomes. Here, we show that cellular inhibitor of apoptosis proteins (cIAPs) attenuate TNF signaling during hepatitis B infection, and they restrict the death of infected hepatocytes, thus allowing viral persistence. Animals with a liver-specific cIAP1 and total cIAP2 deficiency efficiently control HBV infection compared with WT mice. This phenotype was partly recapitulated in mice that were deficient in cIAP2 alone. These results indicate that antagonizing the function of cIAPs may promote the clearance of HBV infection.
It is estimated that 2 billion people currently living in the world have been infected with hepatitis B virus (HBV), and among these, 360 million people are chronic carriers (1). HBV causes 780,000 deaths each year and is responsible for 50% and 33% of deaths attributable to liver cancer and cirrhosis, respectively (2). The host factors and molecular pathways that impact on HBV disease and clinical outcomes are not well-understood (3). What is becoming clear is that immunosuppressive agents and particularly, biological agents, including anti-TNF therapy, can cause major flares in HBV-related disease, leading to morbidity and mortality (4, 5). Animal models and particularly, immunocompetent mouse models of persistent HBV infection have been used to dissect host–pathogen interactions that influence infection outcomes (6–8). These animal models can be used to define host cell signaling and cell death pathways that contribute to the persistence or control of HBV infection.
We induced HBV infection in two mouse models to examine the relevance of host factors in controlling infection. In a model that mimics partial control of infection, we were able to determine the importance of host cell signaling pathways through the use of gene-targeted mice. By identifying the relevant host cell signaling molecules that impact on HBV clinical outcomes, it may be possible to develop therapeutics that target host cell pathways and alter the course of HBV-related disease.
Results
Chronic HBV Infection Can Be Mimicked in a Mouse Model.
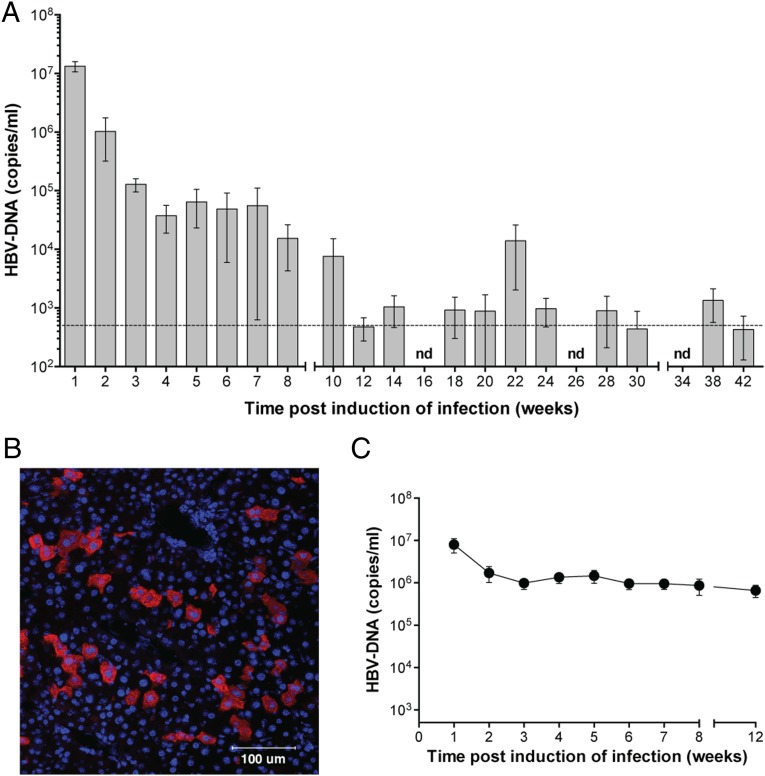
We used a previously described method to induce HBV persistence in immunocompetent mice (6). A plasmid containing a 1.2 over length sequence of HBV genotype A was hydrodynamically injected into mice, but in contrast to the previously published protocol, we did not anesthetize animals. Using this modified technique, we did not observe any injection-associated mortality, and C57BL/6 mice showed persistently high serum HBV DNA levels over 8–12 wk (Fig. 1A). Eventually, HBV DNA levels fell in all animals along with the levels of serum HBV surface antigen (HBsAg) (Fig. S1A). The decline in HBV DNA and HBsAg levels coincided with the appearance of anti-HBV antibodies in the serum (Fig. S1B). The initial control of HBV was followed by relapsing and remitting periods of HBV DNA viremia over many months (Fig. 1A). The plasmid backbone used to deliver HBV could not be detected in hydrodynamically injected animals beyond 5 wk after injection (Fig. S1C). These results suggest that the HBV genome persisted as an integrated or episomal form in hepatocytes after the plasmid that was used to induce infection had been cleared. Viral and subviral particles along with HBV e-antigen were detected in the serum, and HBV pregenomic and subgenomic RNAs were present in the livers of hydrodynamically injected animals (Fig. S1 D–F). Mice in which HBV was introduced by hydrodynamic injection began to control HBV DNA levels over a 12-wk period. The control of HBV DNA was associated with a modest, intermittent increase in levels of serum aspartate aminotransferase and alanine aminotransferase that may indicate hepatocyte dysfunction (Fig. S1 G and H). HBV core antigen (HBcAg) was detected in 4–15% of hepatocytes in infected mice using immunofluorescence staining (Fig. 1B). In contrast to C57BL/6 mice, C3H mice (endotoxin-sensitive strain) could not control serum HBV DNA levels and showed persistent viremia for at least 20 wk after induction of infection (Fig. 1C). The cause of the difference in HBV control between the two strains of mice is not clear and warrants additional investigation. Our C57BL/6 mouse model of HBV recapitulated many aspects of human HBV infection, enabling us using gene-targeted mice to dissect host factors that contributed to the initial control of HBV.
Fig. 1.
Two preclinical models of chronic HBV infection. (A) Serum HBV DNA levels in C57BL/6 mice after induction of infection over 42 wk. The horizontal line denotes the limit of detection for any single sample, and bars depicted below this limit represent the average of all samples at that time point, some of which were below and some of which were above the limit of detection (if all samples were below the detection limit, no bar is depicted; n = 10). (B) Immunofluorescence staining (blue DAPI and red HBcAg) of a liver section taken from an HBV-infected C57BL/6 mouse 1 wk after induction of infection (representative of n = 10). (C) Serial measurement of serum HBV DNA levels in C3H mice after induction of HBV infection (n = 6). Graphs show means and SEMs, and data are representative of (A and B) 10 or (C) 3 independent experiments that served as controls in the remainder of the study. Experiments were blinded. nd, not detected.
Host Immune Cells Affect HBV Outcomes.
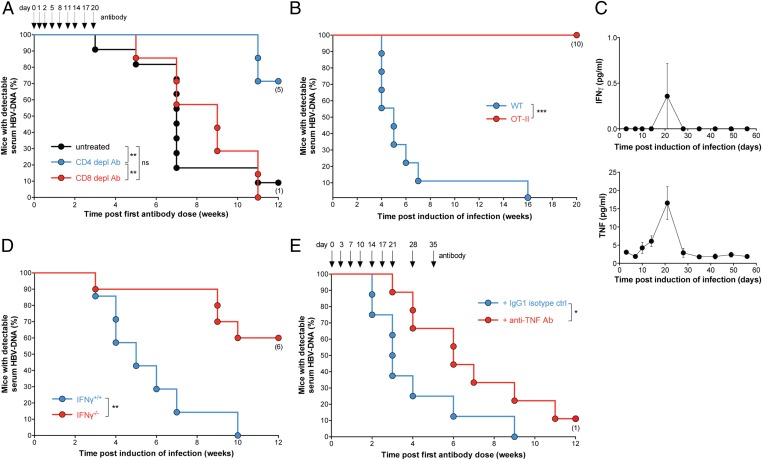
Mice that were deficient in CD3 or recombination-activating gene 1 were unable to control viral loads compared with WT mice infected with HBV (Fig. S1 I–L), implicating a role for immune cells and particularly, T cells in the control of HBV. The relative roles of CD4+ and CD8+ T cells in controlling serum HBV DNA levels were examined by depleting these cell populations using antibodies. Mice treated with CD4+ T cell-depleting antibodies 6 d after induction of infection were unable to control viremia; however, CD8+ T-cell depletion did not impair HBV control (Fig. 2A and Fig. S2A). In both instances, the extent and duration of T-cell depletion were verified by flow cytometry analysis of blood (Fig. S2B). To explore the role of CD8+ T cells using another approach, we induced infection in mice that were deficient in perforin and mice with targeted mutations in H-2Kb and H-2Db causing a loss of MHC class Ia. Both lines of gene-targeted animals have highly impaired CD8+ T-cell cytolytic function, but on infection, they behaved like WT animals (Fig. S2C). Together, these results confirmed that CD8+ T cells were not essential for the initial control of HBV in our model. Several groups have indicated that both CD4+ and CD8+ T cells assist in the control of HBV using more acute models of infection (7, 9–11). Our model may be more representative of a chronic infection in animals, where viral control is suboptimal. An effective cytotoxic CD8+ T-cell response requires MHC class I expression on target cells. To understand why CD8+ T cells play a redundant role in the initial control of HBV in our model, we examined the expression levels of MHC class I on the surface of infected hepatocytes. We found that the level of MHC class I expression on HBV-infected hepatocytes was similar to that found in uninfected hepatocytes and much lower than hepatocytes infected with lymphocytic choriomeningitis virus (LCMV), which elicits a very strong antiviral cytolytic CD8+ T-cell response (Fig. S2D). The reduced expression of MHC class I in HBV compared to LCMV infected hepatocytes may, in part, explain why CD8+ T cells play a redundant role in the control of HBV in our model.
Fig. 2.
Control of HBV viremia in mice is dependent on CD4+ T cells, IFN-γ, and TNF. (A) Proportion of animals and time when C57BL/6 mice treated with the indicated antibodies or left untreated first achieved an undetectable serum HBV DNA level; CD8 and CD4 antibodies are of the same isotype and serve as controls for each other (arrows indicate timing of repeated antibody doses; n = 7–11 per group). (B) Proportion of animals and time when mice of the specified genotype first achieved an undetectable serum HBV DNA level (n = 9–10 per group). (C) Serial quantification of serum cytokine levels at the indicated time points after induction of HBV infection in C57BL/6 mice (n = 10). (D) Proportion of animals and time when mice of the indicated genotypes first achieved an undetectable serum HBV DNA level (n = 7–10). (E) Proportion of animals and time when mice treated with the indicated antibodies first achieved an undetectable serum HBV DNA level (arrows indicate timing of repeated antibody doses; n = 8–9 per group). Numbers below dots in time to event analyses represent remaining mice that have been censored. Graphs show means and SEMs, and data are representative of (A) three or (B–E) two independent experiments. Experiments were blinded. ns, not significant. *P < 0.05; **P < 0.01; ***P < 0.001 (log-rank Mantel–Cox test).
We dissected the role of CD4+ T cells in our infection model by using OT-II mice that transgenically express an α- and a β-chain of the T-cell receptor (TCR) specific for an epitope in chicken ovalbumin. The transgenically expressed receptor pairs with the CD4 coreceptor and interferes with the expression of other TCRs, thus skewing the whole CD4+ T-cell compartment and impeding responses to antigens other than ovalbumin (12). Despite possessing both CD4+ and CD8+ T cells, albeit with a highly skewed antigen recognition repertoire, these mice were unable to control HBV infection and behaved like T cell-deficient animals (Fig. 2B and Fig. S2E). These results indicate that, if the CD4+ TCR repertoire is restricted such that HBV cannot be recognized, mice were unable to control HBV infection.
TNF and IFN-γ Are Important in the Control of HBV Infection.
Immune cells, including CD4+ T cells, secrete numerous cytokines that may participate in the control of HBV infection (7). We measured the levels of several serum cytokines over the course of HBV infection in our model. We observed modest peaks in TNF, IFN-γ, IL-6, IL-12, and IL-1β 3 wk after infection was induced (Fig. 2C and Fig. S2F). To delineate the importance of some of these cytokines, we used gene-targeted animals and cytokine-neutralizing antibodies or cytokine receptor-blocking antibodies. Mice deficient in IFN-γ and mice treated with TNF-neutralizing antibody commenced 7 d after infection was induced showed impaired control of HBV infection (Fig. 2 D and E and Fig. S2G), consistent with other studies (7). Although circulating serum levels of IFN-γ and TNF were low, liver-infiltrating T cells from HBV-infected mice but not uninfected animals were capable of producing these cytokines in ex vivo restimulation tests (Fig. S2H). To investigate the role of IFN-α in limiting the initial viral load, we treated mice with a 5-d course of type I IFN receptor subunit 1 (IFNAR-1) blocking antibody (13) commenced 1 d before infection was induced. We found that mice treated with IFNAR-1 blocking antibody showed no difference in initial serum HBV DNA levels compared with mice that received isotype control antibody treatment (Fig. S2I). We verified the efficacy of our IFNAR-1 blocking antibody in mice chronically infected with the LCMV docile strain (Fig. S2J). Clear immune effects were observed in LCMV-infected mice, consistent with published reports (14, 15). The redundant role of endogenous type I IFNs in controlling viremia in our model is in contrast to results obtained in other models (7) but consistent with reports indicating that HBV does not induce a strong type I IFN response in infected cells (16, 17). Intriguingly, the abrogated type I IFN response associated with chronic HBV infection may explain why MHC class I is not highly expressed on HBV-infected hepatocytes, because previous studies have shown that type I IFN responses are required for the up-regulation of MHC class I after infection (18). Importantly, the redundancy of type I IFNs in restraining HBV DNA levels in our model indicates that the initial hydrodynamic injection of plasmid DNA did not induce a type I IFN response that artificially impacted on serum HBV DNA clearance kinetics. Our data show that TNF and IFN-γ were nonredundant cytokines mediating the initial control of HBV infection in our mouse model but that type I IFN responses were dispensable in determining the initial viral load.
Cellular Inhibitor of Apoptosis Proteins Limit TNF-Mediated HBV Clearance.
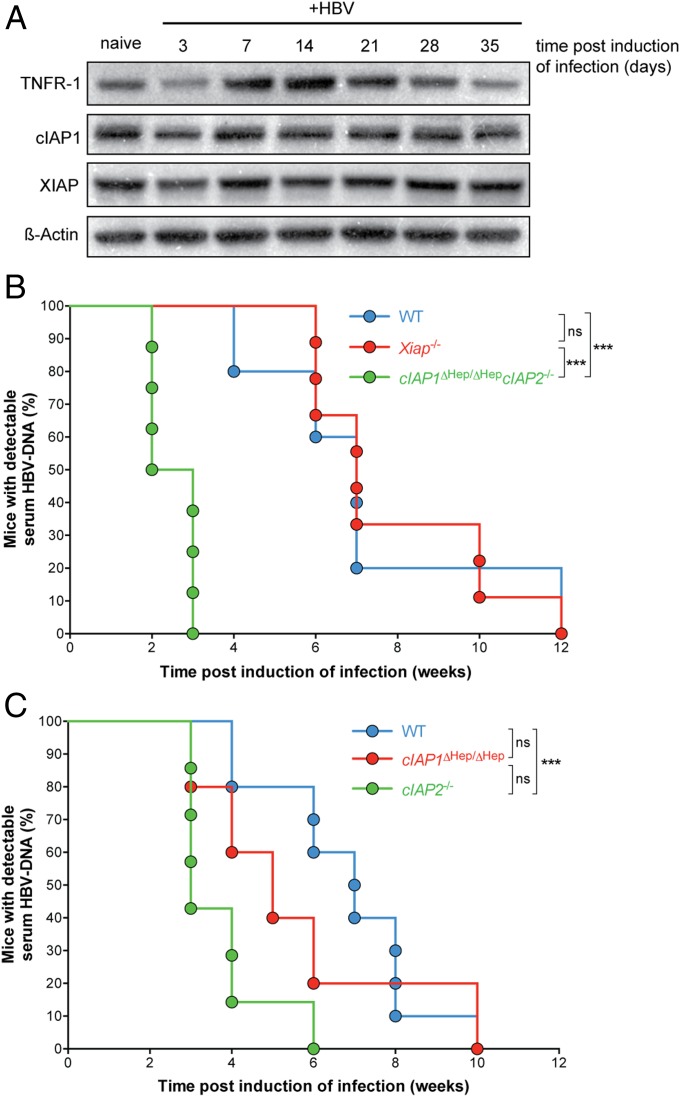
Given the importance of TNF in establishing control of HBV in our mouse model, we next investigated whether TNF signaling could be modulated to enhance HBV control and clearance. Elevated plasma levels of TNF and its receptors are present in patients with chronic HBV infection (19), and we hypothesized that this presents an opportunity to harness the efficacy of these endogenous molecules to promote clearance of HBV. In our model, we similarly found elevated levels of TNF in the serum of infected mice and increased expression of TNF receptor 1 (TNFR1) in infected livers (Figs. 2C and 3A). When TNF binds TNFR1, several outcomes are possible. TNFR1 ligation usually results in NF-κB activation and enhanced cell survival, but a second fate is possible under certain conditions, which results in cell death (20). Numerous studies have defined a role for cellular inhibitor of apoptosis proteins (cIAPs) in promoting the activation of NF-κB downstream of TNFRs (21–24), and interfering with cIAP function results in the activation of cell death pathways downstream of TNFR and perhaps, other death receptors (25). There are three major mammalian inhibitor of apoptosis proteins: X-linked inhibitor of apoptosis protein (XIAP), cIAP1, and cIAP2. We did not observe dramatic changes in cIAP1 and XIAP levels in the liver during the first weeks after induction of HBV infection in our model (Fig. 3A). The protein levels of cIAP2 could not be assessed because of a lack of reliable antibodies to mouse cIAP2. We reasoned that, if TNF and TNFR1 levels were up-regulated in the liver during HBV infection (Figs. 2C and 3A), interfering with cIAP function could result in the death of infected cells and clearance of HBV. To examine this possibility, we induced infection in inhibitor of apoptosis-deficient animals.
Fig. 3.
cIAPs restrict clearance of HBV. (A) Western blot analysis of liver protein levels after HBV infection in C57BL/6 mice at the indicated time points posthydrodynamic injection. (B and C) Proportion of animals and time when mice of the indicated genotypes first achieved an undetectable serum HBV DNA level (n = 5–9 for each group). Data are representative of three independent experiments. Experiments were blinded. ns, not significant. ***P < 0.001 (log-rank Mantel–Cox test).
XIAP-deficient (Xiap−/−) mice behaved like WT mice after infection was induced (Fig. 3B). In contrast, animals with compound mutations causing loss of cIAP1 specifically in the liver (Fig. S3A) and total loss of cIAP2 (cIAP1ΔHep/ΔHepcIAP2−/−) controlled HBV infection more rapidly compared with WT animals (Fig. 3B and Fig. S3B). Deficiency of cIAP2 was critical in promoting the early clearance of HBV, because liver-specific loss of cIAP1 alone did not recapitulate the HBV clearance kinetics seen in compound mutant mice (Fig. 3C). Our cIAP2-deficient animals lacked cIAP2 in all body compartments; therefore, to address which compartment was contributing to the control of HBV infection, we made chimeric animals. We found that liver but not hemopoietic deficiency of cIAP2 promoted clearance of HBV (Fig. S3C).
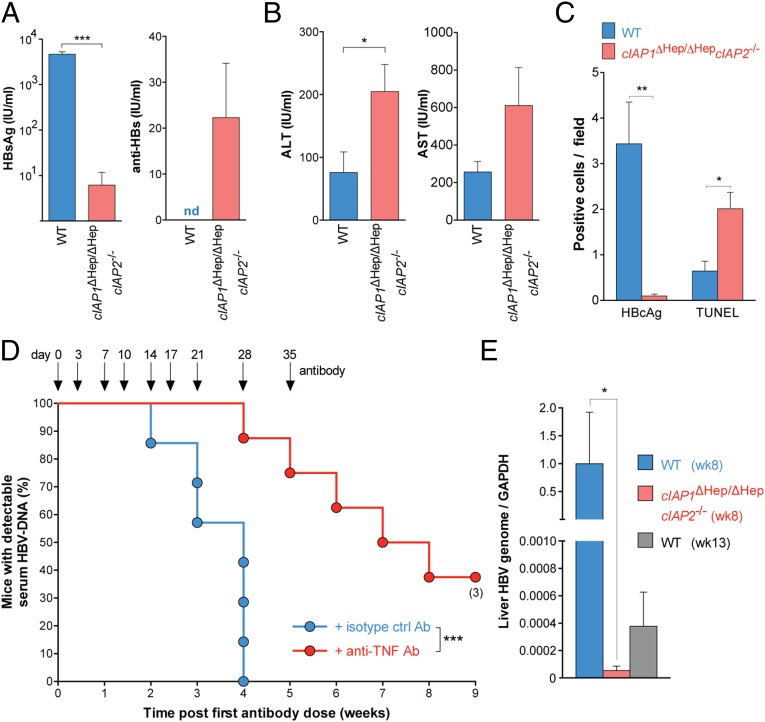
All cIAP1ΔHep/ΔHepcIAP2−/− mice had levels of serum HBV DNA below our detection limit (500 copies/mL) at 3 wk postinfection, well before any WT animals begin to control HBV (Fig. 3B and Fig. S3B). These results were not caused by inherent differences that affect the ability to induce infection through hydrodynamic injection, because cIAP1ΔHep/ΔHepcIAP2−/− and control mice had the same number of hepatocytes that expressed HBcAg and the same levels of serum HBV DNA 3 d postinduction of infection (Fig. S3D). Two weeks postinduction of infection, cIAP1ΔHep/ΔHepcIAP2−/− mice showed reduced levels of HBsAg compared with controls, and this reduction was associated with the early presence of anti-HBV antibody in the serum of cIAP1ΔHep/ΔHepcIAP2−/− mice but not controls (Fig. 4A). Coinciding with the rapid decline in serum HBV DNA and HBsAg, cIAP1ΔHep/ΔHepcIAP2−/−-infected mice showed an increase in serum transaminase levels and a reduced number of hepatocytes expressing HBcAg 2 wk after infection compared with controls (Fig. 4 B and C). The reduction in HBcAg-expressing hepatocytes was associated with a twofold increase in the number of TUNEL-positive cells (Fig. 4C), indicative of hepatocyte death, and a two- to sixfold increase in the number of activated (CD44+CD62Llo, CD69+, or PD1hi) CD4+ and CD8+ T cells in the liver of cIAP1ΔHep/ΔHepcIAP2−/−-infected animals compared with controls (Fig. S4A). The increased numbers of T cells present in the livers of cIAP-deficient mice and the increased number of TUNEL-positive cells were only observed after HBV infection and were not seen in uninfected animals (Fig. S4 A and B). Collectively, these results indicated that the loss of cIAP1 and cIAP2 resulted in the death of HBV-infected hepatocytes, and this death seemed specific, because random hepatocyte death could not explain the preferential loss of HBcAg-expressing cells in the absence of substantial collateral damage and liver destruction. The preferential or specific loss of HBcAg-expressing cells in cIAP-deficient mice may be promoted by the up-regulation of TNFR1 on hepatocytes after HBV infection (Fig. 3A) (19). Additionally, immune infiltrates may preferentially accumulate around infected hepatocytes that express HBcAg and promote their death through the local production of cytokines. We did not detect differences in systemic serum cytokine levels between WT and cIAP-deficient mice during the course of infection (Fig. S4C). To determine if the death of HBcAg-expressing hepatocytes in cIAP-deficient animals was at least, in part, mediated by TNF, which may be localized to the liver, we treated cIAP1ΔHep/ΔHepcIAP2−/−-infected mice with TNF-neutralizing antibodies. When cIAP1ΔHep/ΔHepcIAP2−/− mice received anti-TNF antibodies immediately after induction of infection and for a duration of 5 wk, they were no longer able to rapidly clear HBV (Fig. 4D). These results indicated that a major mechanism responsible for the rapid clearance of HBV in cIAP-deficient mice was through TNF-mediated death of HBcAg-expressing hepatocytes.
Fig. 4.
Deficiency of cIAPs promotes TNF-mediated clearance of HBV infection. (A and B) Serological HBV assays and serum transaminase levels performed 2 wk after induction of infection in mice of the indicated genotypes (n = 5 in each group). (C) Quantification of TUNEL staining and HBcAg expression in liver sections taken from HBV-infected mice 2 wk after induction of infection (n = 3–5 in each group). (D) Proportion of animals and time when cIAP1ΔHep/ΔHepcIAP2−/− mice treated with the indicated antibodies first achieved an undetectable serum HBV DNA level (n = 7–8 for each group). (E) RT-PCR of HBV DNA relative to GAPDH in the liver of infected mice with the indicated genotypes and at the indicated times (n = 4 in each group). Numbers below dots in time to event analyses represent remaining mice that have been censored. Graphs show means and SEMs, and data are representative of two independent experiments. (A, B, D, and E) Experiments were blinded. ALT, alanine aminotransferase; AST, aspartate aminotransferase; nd, not detected. *P < 0.05; **P < 0.01; ***P < 0.001 (A and B, unpaired two-tailed t test; C, unpaired two-tailed t test with Holm–Sidak correction; D, log-rank Mantel–Cox test; and E, Mann–Whitney test).
We used primers that bind all forms of HBV DNA to quantify residual HBV genome in the liver of animals and found that the initial rapid clearance of HBcAg-expressing cells was accompanied by a reduction in the amount of HBV genome in the liver of cIAP-deficient animals compared with controls (Fig. 4 C and E). Despite the reduction in HBcAg and HBV genome, most animals relapsed at later time points; 10 of 13 cIAP-deficient animals showed relapses in serum HBV DNA levels, whereas 11 of 15 WT animals showed relapses (Fig. S3B). All animals had low detectable levels of HBV genetic material that persisted in livers after initial clearance of HBcAg-expressing hepatocytes, serum HBV DNA, and serum HBsAg (Fig. 4 A, C, and E and Fig. S3B). These data may indicate that, in our model, some hepatocytes contain HBV genome that is transcriptionally silent but retains capacity to become transcriptionally active. Critically, however, the amount of residual HBV genome in the livers of cIAP-deficient animals was much lower compared with control mice (Fig. 4E). On producing HBV DNA, formerly quiescent cells were rapidly removed in infected animals (Fig. 1A and Fig. S3B). It is not clear if an analogous population of HBV-infected and transcriptionally quiescent hepatocytes is clinically relevant in maintaining long-term viral turnover in humans. Nonetheless, our data indicate that cIAPs impair the clearance of hepatocytes that express HBV antigens and/or contain HBV genome and that the loss of cIAPs promoted a rapid initial clearance of HBV in our mouse infection model.
Discussion
We induced HBV infection in a C57BL/6 mouse model and showed that the loss of host cell cIAPs promotes a rapid initial clearance of HBV in our mouse infection model. The early HBV clearance observed in cIAP-deficient mice is dependent on TNF and correlates with the loss of HBcAg-expressing hepatocytes and the appearance of TUNEL-positive dying hepatocytes. The promotion of cell death, caused by the loss of cIAPs, seems to preferentially target HBcAg-expressing hepatocytes, because mice do not develop hepatic failure or substantial collateral damage during the course of infection. Importantly, we were able to induce the same level of initial HBV infection in mice that were deficient in cIAPs; therefore, the improved HBV elimination kinetics seen in these animals were not caused by differences in the ability to induce HBV infection.
In our models, HBV produced in the liver enters the circulation, but circulating virions cannot reinfect hepatocytes or infect naïve hepatocytes, because these cells lack the cognate HBV receptor. In this respect, our models mimic patients chronically infected with HBV that are treated with HBV polymerase inhibitors, such as entecavir and tenofovir, which are now standard of care, because these patients usually have undetectable viral loads, and hence, reinfection or infection of naïve hepatocytes is not clinically relevant.
Although hydrodynamically injected plasmid containing HBV was used to induce infection in our animal models, the plasmid did not persist as the transcriptional template for HBV. Possible persistent transcriptional templates in our models include the formation of episomal HBV DNA promoted by the introduction of inverted terminal repeats flanking the HBV sequence in the constructs that we used to induce infection or alternatively, integration of HBV into the mouse genome. The nature of the transcriptional template is unimportant, because our results indicate that the loss of host cIAPs results in the death of cells containing HBV DNA, regardless of its nature.
Potent cIAP small-molecule antagonists are now being used in clinical cancer trials (26) to promote TNF-mediated killing of tumors. Our data indicate that they may have therapeutic efficacy in the treatment of chronic HBV infection and perhaps, other persistent intracellular infections.
Materials and Methods
Mice and Induction of HBV Infection.
The Walter and Eliza Hall Institute of Medical Research Animal Ethics Committee reviewed and approved all animal experiments. Gene-targeted animals used in our experiments have been described elsewhere (12, 27–34). We crossed the relevant single-mutant animals to generate compound mutant cIAP1ΔHep/ΔHepcIAP2−/− mice. The HBV DNA plasmid used to induce infection has been described previously (6). In our modification of the procedure, we did not anesthetize animals. This modification prevented any morbidity or mortality related to the procedure that might otherwise have introduced a selection bias. All mice used in experiments were between 6 and 10 wk old. Additional details, including procedures for LCMV infection and generation of chimeric mice, are provided in SI Materials and Methods.
Biochemistry, HBV serology, and genome quantification.
Sample preparation and analysis are described in SI Materials and Methods.
Antibody Treatment.
CD4+ and CD8+ T cell-depleting antibodies (rat IgG2b GK1.5–1 and rat IgG2b YTS-169, respectively) were injected at a dose of 100 μg. Animals treated with TNF-neutralizing antibodies (rat IgG1 XT22) received 200 μg i.p. Injection of a rat IgG isotype was used as a control. IFNAR-1 blocking antibody (mouse IgG1 MAR1-5A3) or isotype control (GIR-208) was used at a dose of 200 μg i.p. Additional information is detailed in SI Materials and Methods.
Histology, Immunofluorescence, and TUNEL Staining.
Preparation of tissues and staining are described in SI Materials and Methods.
Cytokine assays.
Serum cytokines were determined using the Cytometric Bead Assay Flex System Kit (BD Bioscience) according to the manufacturer’s instructions.
Immunophenotyping and restimulation of hepatic immune cells.
Preparation of samples, staining, and analysis are described in SI Materials and Methods.
EM.
Approximately 10 μL serum was placed onto 300 mesh carbon-coated copper grids (ProSciTech) and incubated at room temperature for 1 min. Grids were washed with PBS, stained with 3% (vol/vol) phosphotungstic acid (pH 7.4), and viewed using a Tecnai 12 Electron Microscope (FEI).
Northern and Western Blots.
Procedures are detailed in SI Materials and Methods.
Statistical Analysis.
Prism 6.0d software (Graph Pad Software) was used to perform statistical tests. Groups were compared using an unpaired two-tailed t test, and Holm–Sidak correction was applied for multiple comparisons. Nonparametric data were analyzed using the Mann–Whitney test, and time to event analyses were performed using the log-rank Mantel–Cox test. Animal sample sizes were estimated based on the predetermined degree of inherent variability in the end points being measured, and animals, after age and sex matching, were randomly allocated to treatment groups by an independent animal technician.
Supplementary Material
Acknowledgments
We thank Stephen Condon and Stephen Locarnini for discussions. Pei-Jer Chen and Ding-Shinn Chen constructed the hepatitis B virus infection vector for hydrodynamic injection. David Vaux generated the cellular inhibitor of apoptosis protein 1loxP/loxP (cIAP1loxP/loxP) and cIAP2FRT/lFRT mice. Paul Hertzog provided reagents, and Lynne Waddington assisted with EM. Linda Earnest-Silveira provided laboratory support. The following mouse strain was obtained through National Institute of Allergy and Infectious Diseases Exchange Program 04215 C57BL/6-H-2Kb-H-2Db. This work was supported by Australian Research Council Future Fellowship Award (to U.N.) and Grant FT1301000166; National Health and Medical Research Council Australia Career Development Award 637350 and Grants 541901 (to J.S.), 541902 (to J.S.), 1006592 (to M.P.), 1045549 (to M.P.), and 1065626 (to M.P.); the Victorian State Government Operational Infrastructure Support; and the Independent Research Institutes Infrastructure Support Scheme of the Australian Government National Health and Medical Research Council.
Footnotes
Conflict of interest statement: The Walter and Eliza Hall Institute of Medical Research has a research license agreement with TetraLogic Pharmaceuticals Corporation, Inc., the manufacturer of the cellular inhibitor of apoptosis protein antagonist birinapant. TetraLogic Pharmaceuticals Corporation, Inc. has filed a patent cooperation treaty application on behalf of The Walter and Eliza Hall Institute of Medical Research. J.S. is on the scientific advisory board of and M.P. provides consultative advice to TetraLogic Pharmaceuticals Corporation, Inc. J.S. has options on a small number of shares in TetraLogic Pharmaceuticals Corporation, Inc. C.G.B is employed by TetraLogic Pharmaceuticals Corporation, Inc.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1502390112/-/DCSupplemental.
References
- 1.WHO Hepatitis B vaccines. Wkly Epidemiol Rec. 2009;84(40):405–419. [PubMed] [Google Scholar]
- 2.Lozano R, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ganem D, Prince AM. Hepatitis B virus infection—natural history and clinical consequences. N Engl J Med. 2004;350(11):1118–1129. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- 4.Shale MJ, et al. Review article: Chronic viral infection in the anti-tumour necrosis factor therapy era in inflammatory bowel disease. Aliment Pharmacol Ther. 2010;31(1):20–34. doi: 10.1111/j.1365-2036.2009.04112.x. [DOI] [PubMed] [Google Scholar]
- 5.Vassilopoulos D, Calabrese LH. Risks of immunosuppressive therapies including biologic agents in patients with rheumatic diseases and co-existing chronic viral infections. Curr Opin Rheumatol. 2007;19(6):619–625. doi: 10.1097/BOR.0b013e3282f05b63. [DOI] [PubMed] [Google Scholar]
- 6.Huang LR, Wu HL, Chen PJ, Chen DS. An immunocompetent mouse model for the tolerance of human chronic hepatitis B virus infection. Proc Natl Acad Sci USA. 2006;103(47):17862–17867. doi: 10.1073/pnas.0608578103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang PL, et al. Immune effectors required for hepatitis B virus clearance. Proc Natl Acad Sci USA. 2010;107(2):798–802. doi: 10.1073/pnas.0913498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tzeng HT, et al. Tumor necrosis factor-alpha induced by hepatitis B virus core mediating the immune response for hepatitis B viral clearance in mice model. PLoS ONE. 2014;9(7):e103008. doi: 10.1371/journal.pone.0103008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guidotti LG, et al. Intracellular inactivation of the hepatitis B virus by cytotoxic T lymphocytes. Immunity. 1996;4(1):25–36. doi: 10.1016/s1074-7613(00)80295-2. [DOI] [PubMed] [Google Scholar]
- 10.Nakamoto Y, Guidotti LG, Pasquetto V, Schreiber RD, Chisari FV. Differential target cell sensitivity to CTL-activated death pathways in hepatitis B virus transgenic mice. J Immunol. 1997;158(12):5692–5697. [PubMed] [Google Scholar]
- 11.Thimme R, et al. CD8(+) T cells mediate viral clearance and disease pathogenesis during acute hepatitis B virus infection. J Virol. 2003;77(1):68–76. doi: 10.1128/JVI.77.1.68-76.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based α- and β-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol. 1998;76(1):34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- 13.Sheehan KC, et al. Blocking monoclonal antibodies specific for mouse IFN-alpha/beta receptor subunit 1 (IFNAR-1) from mice immunized by in vivo hydrodynamic transfection. J Interferon Cytokine Res. 2006;26(11):804–819. doi: 10.1089/jir.2006.26.804. [DOI] [PubMed] [Google Scholar]
- 14.Teijaro JR, et al. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science. 2013;340(6129):207–211. doi: 10.1126/science.1235214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson EB, et al. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science. 2013;340(6129):202–207. doi: 10.1126/science.1235208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wieland S, Thimme R, Purcell RH, Chisari FV. Genomic analysis of the host response to hepatitis B virus infection. Proc Natl Acad Sci USA. 2004;101(17):6669–6674. doi: 10.1073/pnas.0401771101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lütgehetmann M, et al. Hepatitis B virus limits response of human hepatocytes to interferon-α in chimeric mice. Gastroenterology. 2011;140(7):2074–2083. doi: 10.1053/j.gastro.2011.02.057. [DOI] [PubMed] [Google Scholar]
- 18.Lang KS, et al. Toll-like receptor engagement converts T-cell autoreactivity into overt autoimmune disease. Nat Med. 2005;11(2):138–145. doi: 10.1038/nm1176. [DOI] [PubMed] [Google Scholar]
- 19.Marinos G, et al. Tumor necrosis factor receptors in patients with chronic hepatitis B virus infection. Gastroenterology. 1995;108(5):1453–1463. doi: 10.1016/0016-5085(95)90694-0. [DOI] [PubMed] [Google Scholar]
- 20.Silke J. The regulation of TNF signalling: What a tangled web we weave. Curr Opin Immunol. 2011;23(5):620–626. doi: 10.1016/j.coi.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 21.Bertrand MJ, et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30(6):689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 22.Feltham R, et al. Tumor necrosis factor (TNF) signaling, but not TWEAK (TNF-like weak inducer of apoptosis)-triggered cIAP1 (cellular inhibitor of apoptosis protein 1) degradation, requires cIAP1 RING dimerization and E2 binding. J Biol Chem. 2010;285(23):17525–17536. doi: 10.1074/jbc.M109.087635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahoney DJ, et al. Both cIAP1 and cIAP2 regulate TNFalpha-mediated NF-kappaB activation. Proc Natl Acad Sci USA. 2008;105(33):11778–11783. doi: 10.1073/pnas.0711122105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varfolomeev E, et al. c-IAP1 and c-IAP2 are critical mediators of tumor necrosis factor alpha (TNFalpha)-induced NF-kappaB activation. J Biol Chem. 2008;283(36):24295–24299. doi: 10.1074/jbc.C800128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vince JE, et al. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell. 2007;131(4):682–693. doi: 10.1016/j.cell.2007.10.037. [DOI] [PubMed] [Google Scholar]
- 26.Benetatos CA, et al. Birinapant (TL32711), a bivalent SMAC mimetic, targets TRAF2-associated cIAPs, abrogates TNF-induced NF-κB activation, and is active in patient-derived xenograft models. Mol Cancer Ther. 2014;13(4):867–879. doi: 10.1158/1535-7163.MCT-13-0798. [DOI] [PubMed] [Google Scholar]
- 27.Pérarnau B, et al. Single H2Kb, H2Db and double H2KbDb knockout mice: Peripheral CD8+ T cell repertoire and anti-lymphocytic choriomeningitis virus cytolytic responses. Eur J Immunol. 1999;29(4):1243–1252. doi: 10.1002/(SICI)1521-4141(199904)29:04<1243::AID-IMMU1243>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 28.Kägi D, et al. Cytotoxicity mediated by T cells and natural killer cells is greatly impaired in perforin-deficient mice. Nature. 1994;369(6475):31–37. doi: 10.1038/369031a0. [DOI] [PubMed] [Google Scholar]
- 29.Dalton DK, et al. Multiple defects of immune cell function in mice with disrupted interferon-gamma genes. Science. 1993;259(5102):1739–1742. doi: 10.1126/science.8456300. [DOI] [PubMed] [Google Scholar]
- 30.Malissen M, et al. Altered T cell development in mice with a targeted mutation of the CD3-ε gene. EMBO J. 1995;14(19):4641–4653. doi: 10.1002/j.1460-2075.1995.tb00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spanopoulou E, et al. Functional immunoglobulin transgenes guide ordered B-cell differentiation in Rag-1-deficient mice. Genes Dev. 1994;8(9):1030–1042. doi: 10.1101/gad.8.9.1030. [DOI] [PubMed] [Google Scholar]
- 32.Schile AJ, García-Fernández M, Steller H. Regulation of apoptosis by XIAP ubiquitin-ligase activity. Genes Dev. 2008;22(16):2256–2266. doi: 10.1101/gad.1663108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gardam S, et al. Deletion of cIAP1 and cIAP2 in murine B lymphocytes constitutively activates cell survival pathways and inactivates the germinal center response. Blood. 2011;117(15):4041–4051. doi: 10.1182/blood-2010-10-312793. [DOI] [PubMed] [Google Scholar]
- 34.Postic C, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274(1):305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.