Significance
Traditional interpretation of animal phylogeny suggests traits, such as mesoderm, muscles, and neurons, evolved only once given the assumed placement of sponges as sister to all other animals. In contrast, placement of ctenophores as the first branching animal lineage raises the possibility of multiple origins of many complex traits considered important for animal diversification and success. We consider sources of potential error and increase taxon sampling to find a single, statistically robust placement of ctenophores as our most distant animal relatives, contrary to the traditional understanding of animal phylogeny. Furthermore, ribosomal protein genes are identified as creating conflict in signal that caused some past studies to recover a sister relationship between ctenophores and cnidarians.
Keywords: phylogenomics, Metazoa, Ctenophora, Porifera, Cnidaria
Abstract
Elucidating relationships among early animal lineages has been difficult, and recent phylogenomic analyses place Ctenophora sister to all other extant animals, contrary to the traditional view of Porifera as the earliest-branching animal lineage. To date, phylogenetic support for either ctenophores or sponges as sister to other animals has been limited and inconsistent among studies. Lack of agreement among phylogenomic analyses using different data and methods obscures how complex traits, such as epithelia, neurons, and muscles evolved. A consensus view of animal evolution will not be accepted until datasets and methods converge on a single hypothesis of early metazoan relationships and putative sources of systematic error (e.g., long-branch attraction, compositional bias, poor model choice) are assessed. Here, we investigate possible causes of systematic error by expanding taxon sampling with eight novel transcriptomes, strictly enforcing orthology inference criteria, and progressively examining potential causes of systematic error while using both maximum-likelihood with robust data partitioning and Bayesian inference with a site-heterogeneous model. We identified ribosomal protein genes as possessing a conflicting signal compared with other genes, which caused some past studies to infer ctenophores and cnidarians as sister. Importantly, biases resulting from elevated compositional heterogeneity or elevated substitution rates are ruled out. Placement of ctenophores as sister to all other animals, and sponge monophyly, are strongly supported under multiple analyses, herein.
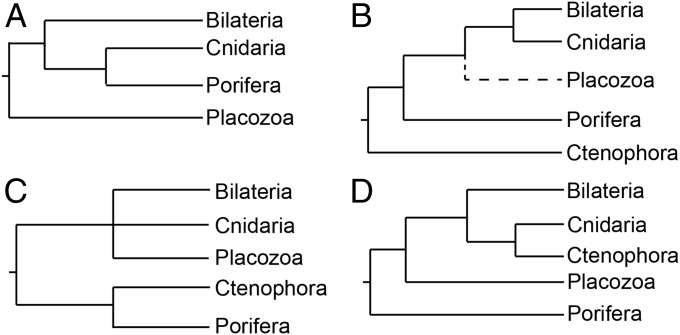
Resolving relationships among extant lineages at the base of the metazoan tree is integral to understanding evolution of complex animal traits, including nervous systems and gastrulation. Historically, sponges and placozoans, both of which have relatively simple body plans and lack neurons, have been considered to diverge from other animals earlier than ctenophores, cnidarians, and bilaterians (1). Phylogenomic studies have resulted in controversial hypotheses placing either Placozoa (Fig. 1A) (2), ctenophores (ctenophore-sister hypothesis) (Fig. 1B) (3–7), or a clade of ctenophores and sponges (Fig. 1C) (6) as sister to all remaining animals. Others (7–10) have claimed nontraditional findings resulted from systematic error and argued for traditional placement of sponges as sister to all remaining animals (Eumetazoa, or Porifera-sister hypothesis) (Fig. 1D) and a sister relationship between ctenophores and cnidarians (Coelenterata) (Fig. 1D). Limited statistical support for various hypotheses and conflict among, and even within studies, has undermined confidence in our understanding of early animal evolution. Basal metazoan relationships must be resolved with greater consistency before a consensus viewpoint is widely accepted.
Fig. 1.
Phylogenetic hypotheses from previous molecular studies. (A) Placozoa-sister hypothesis (2). (B) Ctenophora-sister hypothesis (3–7). Placozoa was not included in some studies that found support for this topology. (C) Ctenophora + Porifera-sister hypothesis (6). (D) Traditional Porifera-sister hypothesis (7–10).
Long-branch attraction (LBA) (11), which occurs when two divergent lineages are artificially inferred as related because of substitutional saturation (11), is perhaps the most often evoked explanation for controversial or spurious phylogenetic results (7–10, 12, 13). Additional sources of systematic error include poor taxon or character sampling (10, 14, 15), large amounts of missing data (16), and model misspecification (16, 17). Such errors have been implicated as influencing the position of ctenophores in metazoan phylogeny studies (5–8). For example, Ryan et al. (6) recovered a sister relationship between sponges and ctenophores in analyses where taxa with high amounts of missing data were excluded, and support for this was highest in Bayesian inference with the CAT (17) substitution model. However, ctenophores were recovered as sister to all other extant animals in maximum-likelihood analyses with greater taxon sampling (Bayesian inference never converged for datasets with more than 19 taxa). The CAT model is a site-heterogeneous model that may handle LBA artifacts better than site-homogeneous substitution models like GTR (18). Therefore, LBA plausibly influenced the phylogenetic position of ctenophores in analyses of Ryan et al. (6) that recovered ctenophores-sister. In Moroz et al. (5), strong nodal support for ctenophores sister to all other extant animals disappeared when both the strictest orthology criteria were enforced and ctenophore taxon sampling increased. Thus, further consideration of systematic error influencing phylogeny reconstruction at the base of the animal tree is desirable.
The ctenophore-sister hypothesis has challenged our understanding of early metazoan evolution, but given conflicting results (2–10), this and other hypotheses must be carefully scrutinized. Ideally, if robust datasets are assembled and causes of systematic error are accounted for, different datasets and analytical methods will converge on a single phylogenetic hypothesis (19). However, practical barriers exist in assembling robust datasets free of systematic error. For example, phylogenomic datasets are prone to missing data given the incomplete nature of transcriptome and even genome sequences, and orthology determination among distantly related species can be difficult (20, 21). Computational limitations of complex phylogenetic methods can also prevent using what may be the best theoretical phylogenetic method. Nevertheless, both data quality and appropriate methods should be emphasized if deep relationships of any organismal group are to be robustly resolved.
Here, we have assembled a more comprehensive phylogenomic dataset of metazoan lineages that branched early in animal evolution than previous studies to alleviate taxon sampling concerns. We have sequenced transcriptomes of eight additional species and used other deeply sequenced publicly available transcriptomes (including some not used in past studies). Additionally, we use a number of data-filtering steps to explore the sensitivity of these results to potential sources of error. This process includes strict orthology determination, removal of taxa and genes that may cause LBA, and removal of heterogeneous genes that may cause model misspecification. Regardless of how data were filtered, all maximum-likelihood analyses with model partitioning and all Bayesian inference analyses using a site-heterogeneous model recover ctenophores as sister to all other animals with strong support. We identify overreliance on ribosomal protein genes in some datasets (7, 9) as the source of incongruence among previous phylogenomic studies. We also find strong support for sponge monophyly in contrast to previous reports (7, 22–24).
Results
Datasets and Accounting for Biases.
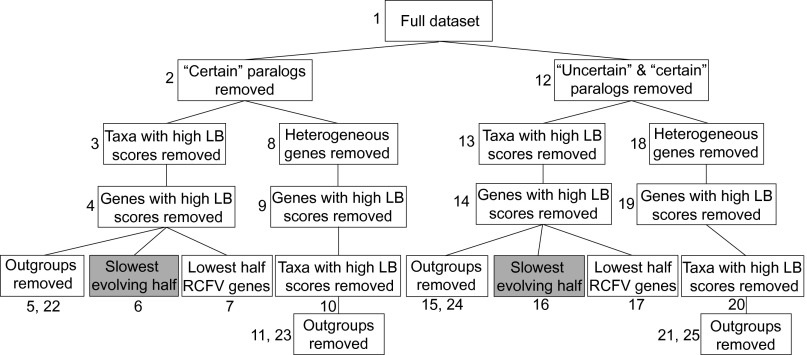
Orthology filtering of transcriptome and genome data from 76 species resulted in 251 orthologous groups (OGs) and 81,008 aligned amino acid sites (Tables S1 and S2). TreSpEx (25) further identified 83 “certain” paralogs (i.e., sequences TreSpEx classified as high-confidence paralogs) from 10 OGs. TreSpEx also identified 2,684 “uncertain” paralogs (i.e., sequences TreSpEx classified as possibly, but not definitively, paralogous) from 104 OGs. Datasets with certain and both certain and uncertain paralogs pruned were starting points for progressively filtering other causes of systematic error. Overall, 25 hierarchical datasets that had progressively fewer characters, but controlled for more potential causes of systematic errors, were assembled (Fig. 2 and Table S2) (all datasets have been deposited on figshare, doi 10.6084/m9.figshare.1334306). The percentage of gene occupancy and missing data ranged from 70–82% and 35–44%, respectively (Table S2). Other than progressive data filtering, differences between datasets analyzed here and those used in previous studies of basal metazoan relationships (3–10) are increased character sampling and less missing data compared with some studies and increased nonbilaterian taxon sampling. In contrast to Nosenko et al. (7) and Philippe et al. (9), both of which relied heavily on ribosomal proteins (i.e., 52% and 71%, respectively), our dataset did not contain a large representation of any one gene class (e.g., only 8 of 250 were ribosomal protein genes) (SI Methods).
Fig. 2.
Hierarchy of datasets with progressive data filtering. Numbers associated with each datasets are references throughout the text. Datasets 5, 11, 15, 21 have Choanoflagellates as an outgroup, and datasets 22, 23, 24, 25 have all outgroups removed. RAxML analyses were used for each dataset, and shaded datasets were also analyzed with PhyloBayes. Data matrix statistics (e.g., number of sites, percentage of missing data, and so forth) for each dataset can be found in Table S2.
Ctenophores Sister to Other Extant Animals and Monophyletic Sponges.
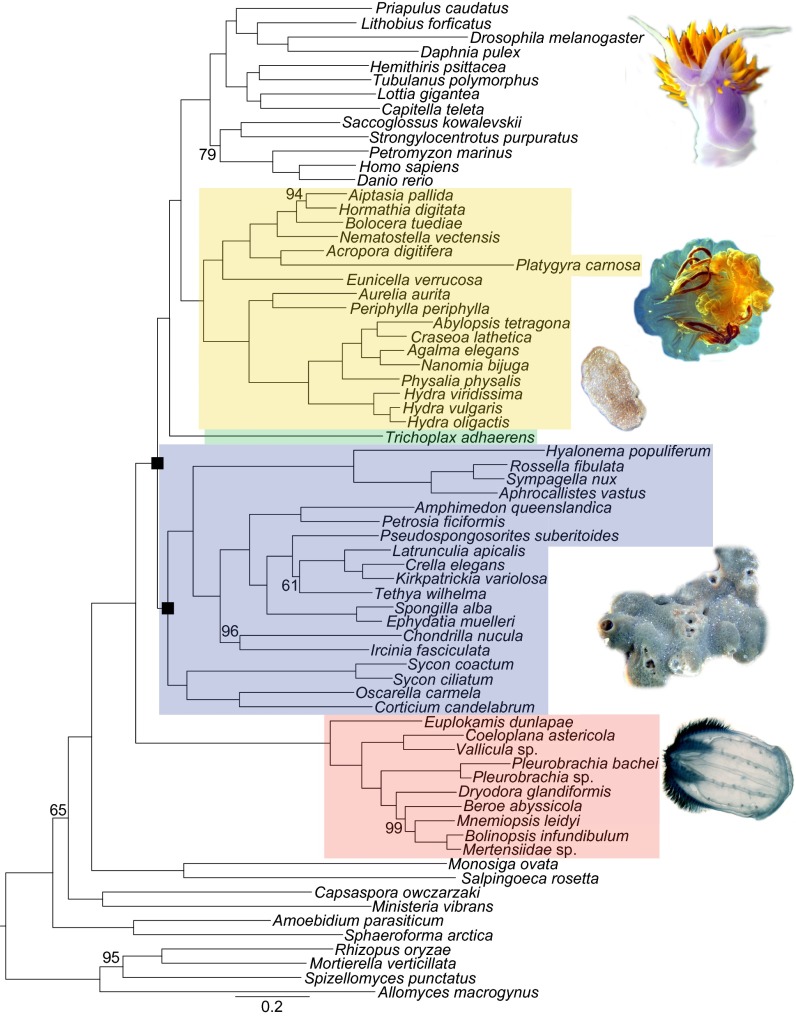
All maximum-likelihood analyses with outgroups resulted in topologies with strong support [≥ 97% bootstrap support (BS)] for ctenophores sister to all other extant animal lineages (datasets 1–21 in Figs. 2 and 3 and Figs. S1–S4). Importantly, Phylobayes (26) analyses using the CAT-GTR+Γ model also resulted in phylogenies with Ctenophora sister to all other animals with 100% posterior probability (PP), and 100% PP for sponge monophyly (datasets 6 and 16 in Figs. 2 and 3 and Fig. S5 B and C). Inferred relationships among major sponge lineages (i.e., Demospongiae + Hexactinellida sister to Calcarea + Homoscleromorpha) were consistent with morphology (27, 28) and most other molecular analyses that have recovered monophyletic sponges (datasets 1–27 in Figs. 2 and 3 and Figs. S1–S5) (4, 5, 8, 9, 28–30). Alternative hypotheses of basal animal relationships (Fig. 1) were rejected by every phylogenetic analysis as measured by the approximately unbiased (AU) (31) test (P ≤ 0.001) (Table 1). Overall, our results overwhelmingly reject alternative hypotheses to ctenophores sister to all other extant animals (Table 1).
Fig. 3.
Reconstructed maximum-likelihood topology of metazoan relationships inferred with dataset 10. Maximum likelihood and Bayesian topologies inferred with other datasets (Fig. 2) have identical basal branching patterns (Figs. S1–S5). Nodes are supported with 100% bootstrap support unless otherwise noted. Support, as inferred from each dataset (Fig. 2), for nodes covered by black boxes are in Table 1.
Table 1.
AU test P values for alternative hypotheses of animal relationships and support for ctenophora-sister and sponge monophyly
| Dataset (no.) | Porifera-sister | Coelenterata | Placozoa-sister | Porifera + Ctenophora | Ctenophora-sister | Porifera monophyly |
| Full dataset (1) | 2.E-04 | 5.E-06 | 1.E-76 | 2.E-05 | 100 BS | 95 BS |
| “Certain” paralogs removed (2) | 2.E-04 | 1.E-05 | 3.E-03 | 5.E-79 | 100 BS | 90 BS |
| Taxa with high LB scores removed (3) | 6.E-06 | 2.E-06 | 1.E-62 | 2.E-07 | 100 BS | 94 BS |
| Genes with high LB scores removed (4) | 1.E-02 | 2.E-04 | 2.E-32 | 3.E-47 | 100 BS | 94 BS |
| Choanoflagellate-only outgroup (5) | 4.E-45 | 1.E-04 | 1.E-05 | 6.E-104 | 100 BS | 68 BS |
| All outgroups removed (22) | N/A | 3.E-55 | N/A | N/A | N/A | 84 BS |
| Slowest evolving half of genes (6) | 2.E-78 | 2.E-11 | 2.E-04 | 1.E-05 | 100 BS/100 PP | 95 BS/100 PP |
| Genes with lowest half of RCFV values (7) | 3.E-06 | 2.E-57 | 9.E-47 | 2.E-03 | 100 BS | 53 BS |
| Heterogeneous genes removed (8) | 7.E-52 | 2.E-49 | 9.E-05 | 3.E-05 | 100 BS | 90 BS |
| Genes with high LB scores removed (9) | 9.E-41 | 9.E-05 | 2.E-04 | 2.E-66 | 99 BS | 90 BS |
| Taxa with high LB scores removed (10) | 7.E-22 | 5.E-61 | 9.E-06 | 7.E-06 | 100 BS | 88 BS |
| Choanoflagellate-only outgroup (11) | 1.E-03 | 3.E-36 | 1.E-02 | 6.E-104 | 92 BS | 82 BS |
| All outgroups removed (23) | N/A | 2.E-04 | N/A | N/A | N/A | 92 BS |
| “Certain” and “uncertain” paralogs (12) | 6.E-39 | 1.E-05 | 4.E-44 | 4.E-05 | 100 BS | 99 BS |
| Taxa with high LB scores removed (13) | 6.E-05 | 8.E-06 | 6.E-30 | 8.E-09 | 100 BS | 99 BS |
| Genes with high LB scores removed (14) | 8.E-105 | 9.E-05 | 1.E-45 | 5.E-102 | 99 BS | 97 BS |
| Choanoflagellate-only outgroup (15) | 5.E-59 | 5.E-39 | 7.E-05 | 1.E-72 | 100 BS | 66 BS |
| All outgroups removed (24) | N/A | 2.E-07 | N/A | N/A | N/A | 92 BS |
| Slowest evolving half of genes (16) | 1.E-04 | 1.E-52 | 1.E-11 | 1.E-07 | 100 BS/100 PP | 94 BS/100 PP |
| Genes with lowest half of RCFV values (17) | 5.E-09 | 1.E-68 | 5.E-07 | 3.E-72 | 100 BS | 93 BS |
| Heterogeneous genes removed (18) | 7.E-07 | 4.E-10 | 6.E-77 | 8.E-09 | 99 BS | 100 BS |
| Genes with high LB scores removed (19) | 2.E-46 | 1.E-08 | 2.E-04 | 1.E-57 | 100 BS | 96 BS |
| Taxa with high LB scores removed (20) | 1.E-03 | 3.E-92 | 1.E-66 | 3.E-113 | 100 BS | 100 BS |
| Choanoflagellate-only outgroup (21) | 2.E-61 | 1.E-31 | 2.E-04 | 4.E-40 | 100 BS | 77 BS |
| All outgroups removed (25) | N/A | 9.E-05 | N/A | N/A | N/A | 96 BS |
| Philippe et al. (7) Maximum likelihood | 0.001 | 0.010 | 0.008 | 0.075 | 93 BS | 42 BS |
| Philippe et al. (7) Bayesian inference | — | — | — | — | N/A | 99 PP |
Dataset numbers are as in Fig. 2.
Ribosomal protein genes can have conflicting signal with most other genes (32). The datasets of Philippe et al. (9) and Nosenko et al. (7), which recovered cnidarians and ctenophores sister, had high proportions of ribosomal protein genes (67 of 128 and 87 of 122 genes, respectively). Nosenko et al. (7) analyzed a dataset without ribosomal protein genes and recovered ctenophores sister to all other animals, but a similar analysis has not been done for the original Philippe et al. (9) dataset. If certain topologies are recovered only with the use of a high proportion of one group of genes (e.g., ribosomal protein genes), this may indicate a phylogenetic signal that conflicts with the true evolutionary history. As such, we analyzed the Philippe et al. (9) dataset with ribosomal proteins removed (67 of 128) using maximum-likelihood and Bayesian inference, and neither reconstruction placed ctenophores and cnidarians sister as in the original study (Fig. S5 D and E). The maximum-likelihood analysis recovered strong support for ctenophores as sister to all other metazoan lineages (BS = 93) (Fig. S5D). However, Bayesian inference (Fig. S5E) recovered sponges as sister to all other metazoans, but support for this and other deep nodes were low (PP ≤ 90).
Systematic Biases and Their Effect on Phylogenetic Inference.
Long-branch (LB) scores (28), a measurement for identifying taxa and OGs that could cause LBA, were calculated for each species and OG with TreSpEx (25). In total, we identified six “long-branched” taxa, all nonmetazoans (Fig. S6A and Table S2), and 28 OGs with high LB scores compared with other OGs (Fig. S6 B and C).We found complete congruence in relationships among basal metazoan phyla in trees inferred with (datasets 1, 2, 8, 12, and 18 in Fig. 2) and without (datasets 3–7, 9–11, 13–17, 19–21, and 22–25 in Fig. 2) taxa and genes that had high LB scores, and nodal support for critical nodes showed little variation among analyses (Fig. 3 and Figs. S1–S5). Removing OGs with high amino acid compositional heterogeneity (datasets 7–11, 17–21, 23, and 25 in Fig. 2) also had no effect on branching order (Fig. 3 and Figs. S2 A–E, S3 E and F, S4 A–E, and S5A). Topologies inferred with only the slowest evolving half of OGs assembled here (datasets 6 and 16 in Fig. 2) (i.e., least saturated and least prone to homoplasy; see Fig. S7 for saturation plots) recovered high support for ctenophores sister to all other animals and sponge monophyly with both maximum-likelihood (BS = 100) (Fig. 3 and Figs. S1F and S3D) and Bayesian inference using the CAT-GTR model (PP = 1) (Fig. 3 and Fig. S5 B and C). Importantly, our datasets of the slowest evolving half of OGs were of a broad range of protein classes (SI Methods; figshare), rather than consisting of a majority of ribosomal proteins (7, 9).
Inaccurate orthology assignment can also introduce systematic error into phylogenomic analyses. Although relationships among basal lineages were unaffected, removal of paralogs as identified by TreSpEx appeared to have the greatest effect on support for some critical nodes. For example, most topologies with both certain and uncertain paralogs removed had strong support for sponge monophyly (i.e., ≥ 95% BS) (datasets 12–14 and 18–20 in Figs. 2 and 3 and Figs. S2F, S3 A, B, and F, and S4 A and B), but four analyses with only certain paralogs removed recovered low support (< 90% BS) for sponge monophyly (datasets 5, 7, 9, and 10 in Figs. 2 and 3 and Figs. S1E and S2 A, D, and E).
Because outgroup sampling has the potential to influence rooting of the animal tree, we explored outgroup sampling as well. When all outgroups except two choanoflagellates were removed (datasets 5, 11, 15, and 21 in Fig. 2), inferred nonbilaterian relationships were identical as in analyses we performed with full outgroup sampling (datasets 5, 11, 15, and 21 in Figs. 2 and 3 and Figs. S1E, S2E, S3C, and S4C), but support for sponge monophyly decreased. In these analyses the leaf-stability indices for homoscleromorph and calcareous sponges were less than 0.94, but in all other analyses they were greater than 0.97 (Fig. S5 F and G). Regardless, when choanoflagellates were the only outgroup, ctenophores were still recovered as the deepest split within the animal tree with 100% BS support. Analyses with all outgroup taxa removed (datasets 22–25 in Fig. 2) recovered identical relationships among major metazoan lineages as other analyses (Figs. S4 D–F and S5A). However, we observed low support for relationships among ctenophores, sponges, and placozoans in these analyses. This resulted from the long placozoan branch being attracted to ctenophores in the absence of outgroup taxa as indicated by bootstrap tree topologies and leaf-stability index for Trichoplax of less than 0.92, whereas leaf-stability indices were greater than 0.99 in all other analyses (Fig. S5 F and G).
Discussion
Placement of Ctenophores Sister to all Remaining Animals Is Not Sensitive to Systematic Errors.
Every analysis conducted herein strongly supported the ctenophore-sister hypothesis (Fig. 3 and Table 1). A major hurdle to wide acceptance of ctenophores as sister to other animals has been that different analyses have yielded conflicting hypotheses of early animal phylogeny (2–9). Sensitivity to the selected model of molecular evolution has been especially problematic (2–9). In contrast, both maximum-likelihood analyses using data partitioning and Bayesian analyses using the CAT-GTR model of our datasets resulted in identical branching patterns among ctenophores, sponges, placozoans, cnidarians, and bilaterians. Past critiques of studies that found ctenophores to be sister to all other animals have emphasized the CAT model as the most appropriate model for deep phylogenomics because it is an infinite mixture model that accounts for site-heterogeneity (7, 8, 29). Notably, when the CAT-GTR model was used here (datasets 6 and 16 in Fig. 2), we recovered ctenophores-sister to all other metazoans (Fig. 3 and Fig. S5 B and C).
The argument for LBA (7–10) or saturated datasets (7, 8) as the reason past studies found ctenophores to be sister to all other animals seems to have been overstated. The recovered position of ctenophores was identical in analyses with (datasets 1, 2, 8, 12, and 18 in Fig. 2 and Figs. S1 A and B, S2 B and F, and S3F) and without (datasets 3–7, 9–11, 13–17, and 19–25 in Fig. 2, and Figs. S1 C–F, S2 A and C–E, S3 A–E, S4, and S5 A–C) taxa and genes with high LB scores, and analyses with the slowest evolving genes (datasets 6 and 16 in Fig. 2 and Fig. S7) also recovered ctenophores sister to all other animals (Fig. 3 and Figs. S1F, S3D, and S5 B and C). Furthermore, despite the long internal branch leading to the ctenophore clade, the position of this lineage did not change in any analysis including those when outgroups were removed (datasets 5, 11, 15, 21, and 22–25 in Fig. 2 and Figs. S1E, S2E, S3C, and S4 C–F). If this branch was being artificially attracted toward outgroups, then employment of different outgroup schemes would be expected to result in different ctenophore placement. Maximum-likelihood and Bayesian inference using the CAT-GTR model of the least saturated datasets (datasets 6 and 16 in Fig. 2 and Fig. S7) recovered identical basal relationships as our other analyses (Fig. 3 and Figs. S1F, S3D, and S5 B and C), also indicating homoplasy and model choice did not bias results. Given the consistency among our analyses that were designed to have different levels of potential biases, we conclude that the ctenophore-sister hypothesis is robust to systematic errors.
Rather than focusing on long branches, fast evolving genes, or model misspecification as influencing the position of ctenophores, the individual genes underlying datasets that resulted in a sister relationship between ctenophores and cnidarians (7, 9) should be the focus of identifying problems with phylogenetic reconstruction. A benefit of phylogenomic datasets is that multiple gene classes and many parts of the genome are analyzed. As such, phylogenomic datasets should not rely too heavily on a single gene class. Past molecular studies that found support for Coelenterata and the Porifera-sister (7, 9) hypotheses appear to have been strongly affected by a disproportionate reliance (i.e., > 50%) on ribosomal protein genes. Nosenko et al. (7) and Philipe et al. (9) found support for ctenophores sister to cnidarians, but Nosenko et al. (7) recovered ctenophores sister to all other animals when ribosomal proteins were excluded. Ribosomal protein datasets from these studies are less saturated than datasets assembled here based on linear regression of patristic distance versus uncorrected genetic distance (Fig. S7) (7–9, 25). This lower mutational saturation has been the primary rational for emphasizing ribosomal genes when inferring deep animal relationship (7). However, standard measurements of sequence saturation (7, 8, 25) average across the length of the sequence. Thus, a sequence with a few variable, highly saturated sites may appear less saturated than a sequence with numerous variable sites but less saturation per site. Furthermore, extremely low mutation rates can indicate selection and result in too little phylogenetic information, both of which could lead to the inference of incorrect relationships (33). Our maximum-likelihood analysis of Philipe et al.’s (9) dataset with ribosomal genes removed recovered support for ctenophores as sister to all other animals (Fig. S5D). Basal relationships were poorly resolved in the Bayesian analysis, which may be a result of too few characters, but ctenophores and cnidarians were not recovered as sister (Fig. S5E). Notably, a study focused only on myxozoan cnidarians (34) used the same matrix as Philippe et al. (9), but added two highly divergent cnidarians and recovered ctenophores sister to all other animals. Ribosomal protein genes have previously been identified as a potential source of phylogenetic error (32), and the above indicates that ctenophores sister to cnidarians as in Philippe et al. (9) was caused by either limited cnidarian taxon sampling, misleading signal in ribosomal genes, or both. More work is needed to assess saturation, possible convergent evolution, and selective pressures of ribosomal proteins in ctenophores, sponges, placozoans, and cnidarians. However, differences in topologies when ribosomal proteins are included or excluded strongly imply a misleading signal in ribosomal protein genes. Put simply, it appears highly improbable that all genes other than ribosomal protein genes could be recovering an incorrect phylogeny.
Sponges Are Monophyletic.
Sponge monophyly, although less controversial than the phylogenetic positions of ctenophores, cnidarians, and placozoans remains an important question as several studies have supported sponge paraphyly (7, 22–24). In regards to inferring the characteristics of the metazoan ancestor, sponge paraphyly, coupled with sponges being at the base of the metazoan phylogeny, is an attractive hypothesis that implies the metazoan ancestor was sponge-like. However, sponge monophyly was recovered in all of our analyses and best supported when the strictest orthology criteria were applied with TreSpEx (Fig. 3 and Table 1), which also removes sequences resulting from sample contamination (e.g., endosymbionts).This observation suggests that spurious paralogs or sequence contamination may have been a source of error when sponges were found paraphyletic, but datasets that have recovered sponge paraphyly were also much smaller than those analyzed here (e.g., refs. 7, 22–24). Sponge monophyly and the well-supported ctenophore-sister hypothesis complicates inferring the ancestral condition of metazoan and other major metazoan groups (e.g., Placozoa + Cnidaria + Bilateria) because many sponge characteristics are likely apomorphic traits. Our robust support of sponge monophyly agrees with morphology (35) and most other large molecular datasets (5, 6, 8–10).
Conclusions
For more than a century, sponges were traditionally considered sister to all other extant metazoans because unlike ctenophores, cnidarians, and bilaterians, they lack true tissues and body symmetry (36, 37). Sponges also possess choanocyte cells that are similar in morphology to choanoflagellates, the sister group to metazoans (36). However, Mah et al. (38) found that homology between choanoflagellates and sponge choanocytes is not as definitive as previously assumed. Similarly, some authors have argued that Placozoans are sister to all other animals because they lack neural and muscular systems and also share similarities in mitochondrial genome size with choanoflagellaes (2, 39). A common theme of these two hypotheses is the placement of morphologically simple animals near the base of the animal tree, but complexity is not a good proxy for metazoan evolution (40). Challenges to long-held viewpoints of morphological complexity and assumed improbability of convergent evolution must not be dismissed simply because they seem unlikely at face value, especially considering a growing body of evidence that supports convergent evolution of many animal traits including neurons (5, 6, 41–43). Furthermore, the Porifera-sister hypothesis lacks critical evaluation and homology of many characters in these taxa still need thorough analysis (44). Overall, findings presented here robustly support ctenophores as sister to all remaining animals.
Methods
Taxon and Character Sampling.
Taxon sampling included previously available data and eight new transcriptomes from two choanoflagellates, three glass sponges (Hexactinellida), two demosponges, and a deep-sea cnidarian (Scyphozoa) (Table S1). Briefly, RNA was extracted, reverse-transcribed, and amplified using the SMART kit (Clontech), and sequenced on an Illumina HiSeq (SI Methods). Raw or assembled transcriptome data for 68 additional species were retrieved from public databases (Table S1).
Raw Illumina transcriptome data were digitally normalized using normalize-by-median.py (45) with a k-mer size of 20, a desired coverage of 30, and four hash tables with a lower bound of 2.5 × 109. Normalized Illumina reads were assembled using default parameters in Trinity v20131110 (46). Raw 454 transcriptome data were assembled with Newbler (47).
Orthology Determination and Data Filtering.
Putative orthologs were determined for each species using HaMStR v13.2 (48) using the model organism core ortholog set. OGs, determined by HaMStR, were further processed using a custom pipeline that filtered OGs with too much missing data (i.e., OGs with less than 37 species), aligned sequences, and filtered potential paralogs (https://github.com/kmkocot) (SI Methods).
TreSpEx (25), which requires individual gene trees for each OG, was used to identify putative paralogs and exogenous contamination missed by our initial orthology inference approach. Gene trees were inferred with RAxML v.8.0.2 (49) with 100 rapid bootstrap replicates followed by a full maximum-likelihood inference; each tree was inferred with the LG+Γ model, which was by far the most common best-fitting model when the complete dataset was partitioned (see below). Paralogs in the initial dataset were detected with TreSpEx using the automated BLAST method and the prepackaged Capitella teleta and Helobdella robusta BLAST databases. This method identified two classes of paralogs: “certain” or sequences that are high-confidence paralogs, and “uncertain” or sequences that are potential paralogs. From the initial, 251 gene dataset, we created one dataset by removing certain paralogs and another dataset with both certain and uncertain paralogs pruned (Fig. 2); after pruning, OGs with fewer than 37 taxa were removed. LB scores (25) were calculated for each taxon and OG with TreSpEx. Following Struck (25), these values were plotted in R (Fig. S6) (50), and outliers were identified as taxa or genes that could cause LBA artifacts. After removal of taxa and genes, each OG was ranked by evolutionary rate with a custom python script (https://github.com/nathanwhelan) following Telford et al. (51). Datasets with only the slowest half of remaining genes were then generated to assess if fast evolving homoplasious genes were biasing inferences (Fig. 2).
We used BaCoCa (52) and two metrics (χ2-test of heterogeneity and relative composition frequency variability; RCFV) (53) to identify genes with amino acid compositional heterogeneity. Some datasets were further filtered by removing non-choanoflagellate outgroups and all outgroup taxa to determine if outgroup choice affects inferred relationships. Saturation of each filtered dataset with full outgroup sampling was explored with TreSpEx and plotted in R (Fig. S7) to provide a further metric to compare datasets.
Phylogenetics.
In addition to removing compositionally heterogeneous genes from some datasets, two approaches were used to handle site-heterogeneity: (i) partitioning schemes for each dataset and associated protein substitution models were determined using the relaxed clustering method in PartitonFinder (54) with 20% clustering and the corrected Akaike information criterion; (ii) a site-heterogeneous mixture model, CAT-GTR+Γ, was used in PhyloBayes (26). Maximum-likelihood topologies were inferred with RAxML using partitions as indicated by ParitionFinder, associated best-fit substitution models, and the gamma parameter to model rate-heterogeneity. Nodal support was measured with 100 fast bootstrap replicates. Phylobayes analyses were run with two chains until the maxdiff statistic between chains was below 0.3 as measured by bpcomp (26). Convergence was also assessed with tracecomp (26) to ensure each parameter had a maximum discrepancy between chains of less than 0.3 and an effective sample size of at least 50. Computational demands and convergence issues prevented us from using the CAT-GTR+Γ model for most datasets. Therefore, Bayesian phylogenies are only reported for the two analyses of the slowest evolving half of OGs. Leaf-stability indices (55) for each taxon were measured in PhyUtility (56) to identify potentially unstable taxa in each dataset.
Maximum-likelihood and Bayesian inference trees were also inferred from the Philippe et al. (9) dataset with ribosomal proteins removed (i.e., 67 of 128 genes) to determine if a single gene class-biased phylogenetic inference. Ribosomal proteins were filtered from the original dataset following data matrix annotations (9) and the matrix was split into individual genes for model testing using a custom R script (https://github.com/nathanwhelan).
The AU test (31) was used to determine if a priori hypotheses of basal metazoan relationships could be rejected (Fig. 1). Topological constraints were enforced in RAxML and the most likely tree given this constraint was inferred with the same partitioning scheme and models used for unconstrained phylogenetic inference. Per site log-likelihoods for trees were calculated in RAxML and AU tests were performed in Consel (57).
Supplementary Material
Acknowledgments
We thank members of the Molette Biology Laboratory for Environmental and Climate Change Studies at Auburn University for help with bioinformatics and data collection, especially Damien Waits. This work was made possible in part by a grant of high-performance computing resources and technical support from the Alabama Supercomputer Authority and was supported by the US National Aeronautics and Space Administration (Grant NASA-NNX13AJ31G) and in part by National Science Foundation (Grant 1146575). This is Molette Biology Laboratory Contribution 36 and Auburn University Marine Biology Program Contribution 128.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the NCBI Sequence Read Archive, www.ncbi.nlm.nih.gov/sra (accession no. PRJNA278284). Transcriptome assemblies, phylogenetic datasets, and an annotation file were deposited to figshare, figshare.com (doi: 10.6084/m9.figshare.1334306).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1503453112/-/DCSupplemental.
References
- 1.Dohrmann M, Wörheide G. Novel scenarios of early animal evolution—Is it time to rewrite textbooks? Integr Comp Biol. 2013;53(3):503–511. doi: 10.1093/icb/ict008. [DOI] [PubMed] [Google Scholar]
- 2.Dellaporta SL, et al. Mitochondrial genome of Trichoplax adhaerens supports placozoa as the basal lower metazoan phylum. Proc Natl Acad Sci USA. 2006;103(23):8751–8756. doi: 10.1073/pnas.0602076103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunn CW, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452(7188):745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- 4.Hejnol A, et al. Assessing the root of bilaterian animals with scalable phylogenomic models. Proc Biol Sci. 2009;276(1802):4261–4270. doi: 10.1098/rspb.2009.0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moroz LL, et al. The ctenophore genome and the evolutionary origins of neural systems. Nature. 2014;510(7503):109–114. doi: 10.1038/nature13400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryan JF, et al. NISC Comparative Sequencing Program The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science. 2013;342(6164):1242592. doi: 10.1126/science.1242592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nosenko T, et al. Deep metazoan phylogeny: When different genes tell different stories. Mol Phylogenet Evol. 2013;67(1):223–233. doi: 10.1016/j.ympev.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 8.Philippe H, et al. Resolving difficult phylogenetic questions: Why more sequences are not enough. PLoS Biol. 2011;9(3):e1000602. doi: 10.1371/journal.pbio.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Philippe H, et al. Phylogenomics revives traditional views on deep animal relationships. Curr Biol. 2009;19(8):706–712. doi: 10.1016/j.cub.2009.02.052. [DOI] [PubMed] [Google Scholar]
- 10.Pick KS, et al. Improved phylogenomic taxon sampling noticeably affects nonbilaterian relationships. Mol Biol Evol. 2010;27(9):1983–1987. doi: 10.1093/molbev/msq089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felsenstein J. Cases in which parsimony and compatability methods will be positively misleading. Syst Zool. 1978;27(4):401–410. [Google Scholar]
- 12.Boussau B, et al. Strepsiptera, phylogenomics and the long branch attraction problem. PLoS ONE. 2014;9(10):e107709. doi: 10.1371/journal.pone.0107709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Straub SCK, et al. Phylogenetic signal detection from an ancient rapid radiation: Effects of noise reduction, long-branch attraction, and model selection in crown clade Apocynaceae. Mol Phylogenet Evol. 2014;80:169–185. doi: 10.1016/j.ympev.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 14.Heath TA, Hedtke SM, Hillis DM. Taxon sampling and the accuracy of phylogenetic analyses. J Syst Evol. 2008;46(3):239–257. [Google Scholar]
- 15.Jeffroy O, Brinkmann H, Delsuc F, Philippe H. Phylogenomics: The beginning of incongruence? Trends Genet. 2006;22(4):225–231. doi: 10.1016/j.tig.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Roure B, Baurain D, Philippe H. Impact of missing data on phylogenies inferred from empirical phylogenomic data sets. Mol Biol Evol. 2013;30(1):197–214. doi: 10.1093/molbev/mss208. [DOI] [PubMed] [Google Scholar]
- 17.Lartillot N, Brinkmann H, Philippe H. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol Biol. 2007;7(Suppl 1):S4. doi: 10.1186/1471-2148-7-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tavaré S. Some probabilistic and statistical problems in the analysis of DNA sequences. Lect Math Life Sci. 1986;17:57–86. [Google Scholar]
- 19.Philippe H, Delsuc F, Brinkmann H, Lartillot N. Phylogenomics. Annu Rev Ecol Evol Syst. 2005;36:541–562. [Google Scholar]
- 20.Altenhoff AM, Dessimoz C. Phylogenetic and functional assessment of orthologs inference projects and methods. PLOS Comput Biol. 2009;5(1):e1000262. doi: 10.1371/journal.pcbi.1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gabaldón T. Large-scale assignment of orthology: Back to phylogenetics? Genome Biol. 2008;9(10):235. doi: 10.1186/gb-2008-9-10-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borchiellini C, et al. Sponge paraphyly and the origin of Metazoa. J Evol Biol. 2001;14(1):171–179. doi: 10.1046/j.1420-9101.2001.00244.x. [DOI] [PubMed] [Google Scholar]
- 23.Sperling EA, Pisani D, Peterson KJ. Poriferan paraphyly and its implications for Precambrian paleobiology. Geol Soc Lond Spec Publ. 2007;286:355–368. [Google Scholar]
- 24.Sperling EA, Peterson KJ, Pisani D. Phylogenetic-signal dissection of nuclear housekeeping genes supports the paraphyly of sponges and the monophyly of Eumetazoa. Mol Biol Evol. 2009;26(10):2261–2274. doi: 10.1093/molbev/msp148. [DOI] [PubMed] [Google Scholar]
- 25.Struck TH. TreSpEx-detection of misleading signal in phylogenetic reconstructions based on tree information. Evol Bioinform Online. 2014;10:51–67. doi: 10.4137/EBO.S14239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lartillot N, Rodrigue N, Stubbs D, Richer J. PhyloBayes MPI: Phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol. 2013;62(4):611–615. doi: 10.1093/sysbio/syt022. [DOI] [PubMed] [Google Scholar]
- 27.van Soest RWM. Deficient Merlia normani Kirkpatrick, 1908, from the Curacao reefs, with a discussion on the phylogenetic interpretation of sclerosponges. Contrib Zool. 1984;54(2):211–219. [Google Scholar]
- 28.Gazave E, et al. No longer Demospongiae: Homoscleromorpha formal nomination as a fourth class of Porifera. Hydrobiologia. 2012;687(1):3–10. [Google Scholar]
- 29.Dohrmann M, Janussen D, Reitner J, Collins AG, Wörheide G. Phylogeny and evolution of glass sponges (Porifera, Hexactinellida) Syst Biol. 2008;57(3):388–405. doi: 10.1080/10635150802161088. [DOI] [PubMed] [Google Scholar]
- 30.Voigt O, Adamski M, Sluzek K, Adamska M. Calcareous sponge genomes reveal complex evolution of α-carbonic anhydrases and two key biomineralization enzymes. BMC Evol Biol. 2014;14(1):230. doi: 10.1186/s12862-014-0230-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51(3):492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- 32.Bleidorn C, et al. On the phylogenetic position of Myzostomida: Can 77 genes get it wrong? BMC Evol Biol. 2009;9(1):150. doi: 10.1186/1471-2148-9-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edwards SV. Natural selection and phylogenetic analysis. Proc Natl Acad Sci USA. 2009;106(22):8799–8800. doi: 10.1073/pnas.0904103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nesnidal MP, Helmkampf M, Bruchhaus I, El-Matbouli M, Hausdorf B. Agent of whirling disease meets orphan worm: phylogenomic analyses firmly place Myxozoa in Cnidaria. PLoS ONE. 2013;8(1):e54576. doi: 10.1371/journal.pone.0054576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ax P. Multicellular Animals: A New Approach to the Phylogenetic Order in Nature. Springer; Berlin: 1996. [Google Scholar]
- 36.Nielsen C. Six major steps in animal evolution: Are we derived sponge larvae? Evol Dev. 2008;10(2):241–257. doi: 10.1111/j.1525-142X.2008.00231.x. [DOI] [PubMed] [Google Scholar]
- 37.Srivastava M, et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466(7307):720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mah JL, Christensen-Dalsgaard KK, Leys SP. Choanoflagellate and choanocyte collar-flagellar systems and the assumption of homology. Evol Dev. 2014;16(1):25–37. doi: 10.1111/ede.12060. [DOI] [PubMed] [Google Scholar]
- 39.Osigus H-J, Eitel M, Bernt M, Donath A, Schierwater B. Mitogenomics at the base of Metazoa. Mol Phylogenet Evol. 2013;69(2):339–351. doi: 10.1016/j.ympev.2013.07.016. [DOI] [PubMed] [Google Scholar]
- 40.Halanych KM. Metazoan phylogeny and the shifting comparative framework. In: Roubos EW, Wendelaar-Bonga SE, Vaudry H, De Loof A, editors. Recent Developments in Comparative Endocrinology and Neurobiology. Shaker; Maastrict, The Netherlands: 1999. pp. 3–7. [Google Scholar]
- 41.Dunn CW, Giribet G, Edgecombe GD, Hejnol A. Animal phylogeny and its evolutionary implications. Annu Rev Ecol Evol Syst. 2014;45:371–395. [Google Scholar]
- 42.Liebeskind BJ, Hillis DM, Zakon HH. Convergence of ion channel genome content in early animal evolution. Proc Natl Acad Sci USA. 2015;112(8):E846–E851. doi: 10.1073/pnas.1501195112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moroz LL. Convergent evolution of neural systems in ctenophores. J Exp Biol. 2015;218(Pt 4):598–611. doi: 10.1242/jeb.110692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Halanych KM. The ctenophore lineage is older than sponges? That cannot be right! Or can it? J Exp Biol. 2015;218(Pt 4):592–597. doi: 10.1242/jeb.111872. [DOI] [PubMed] [Google Scholar]
- 45.Brown T, Howe C, Zhang A, Pyrkosz Q, Brom AB. 2012. A reference-free algorithm for computational normalization of shotgun sequencing data. arXiv:1203.4802.
- 46.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437(7057):376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ebersberger I, Strauss S, von Haeseler A. HaMStR: Profile hidden markov model based search for orthologs in ESTs. BMC Evol Biol. 2009;9:157. doi: 10.1186/1471-2148-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.R Core Development Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2014. [Google Scholar]
- 51.Telford MJ, et al. Phylogenomic analysis of echinoderm class relationships supports Asterozoa. Proc Biol Sci. 2013;281(1786):20140479. doi: 10.1098/rspb.2014.0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kück P, Struck TH. BaCoCa—A heuristic software tool for the parallel assessment of sequence biases in hundreds of gene and taxon partitions. Mol Phylogenet Evol. 2014;70:94–98. doi: 10.1016/j.ympev.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 53.Zhong M, et al. Detecting the symplesiomorphy trap: A multigene phylogenetic analysis of terebelliform annelids. BMC Evol Biol. 2011;11(1):369. doi: 10.1186/1471-2148-11-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lanfear R, Calcott B, Kainer D, Mayer C, Stamatakis A. Selecting optimal partitioning schemes for phylogenomic datasets. BMC Evol Biol. 2014;14:82. doi: 10.1186/1471-2148-14-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thorley JL, Wilkinson M. Testing the phylogenetic stability of early tetrapods. J Theor Biol. 1999;200(3):343–344. doi: 10.1006/jtbi.1999.0999. [DOI] [PubMed] [Google Scholar]
- 56.Smith SA, Dunn CW. Phyutility: A phyloinformatics tool for trees, alignments and molecular data. Bioinformatics. 2008;24(5):715–716. doi: 10.1093/bioinformatics/btm619. [DOI] [PubMed] [Google Scholar]
- 57.Shimodaira H, Hasegawa M. CONSEL: For assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17(12):1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.