Significance
Phytophthora is a major threat to agriculture. However, the molecular interaction of these severe pathogens with plant hosts is poorly understood. Here, we report that the Phytophthora Suppressor of RNA Silencing 1 (PSR1) effectively promotes infection in Arabidopsis thaliana by directly targeting an essential protein containing a aspartate–glutamate–alanine–histidine-box RNA helicase domain. This PSR1-Interacting Protein 1 (PINP1) is required for the accumulation of distinct classes of endogenous small RNAs and acts as a positive regulator of plant immunity. Silencing of PINP1 impaired the assembly of microRNA-processing complexes in the nucleus, leading to defects in development and immunity. This study revealed a conserved RNA helicase as a regulator of RNA silencing and provides mechanistic insight into Phytophthora pathogenesis.
Keywords: Phytophthora pathogenesis, RxLR effector, RNA helicase, gene silencing, small RNA
Abstract
A broad range of parasites rely on the functions of effector proteins to subvert host immune response and facilitate disease development. The notorious Phytophthora pathogens evolved effectors with RNA silencing suppression activity to promote infection in plant hosts. Here we report that the Phytophthora Suppressor of RNA Silencing 1 (PSR1) can bind to an evolutionarily conserved nuclear protein containing the aspartate–glutamate–alanine–histidine-box RNA helicase domain in plants. This protein, designated PSR1-Interacting Protein 1 (PINP1), regulates the accumulation of both microRNAs and endogenous small interfering RNAs in Arabidopsis. A null mutation of PINP1 causes embryonic lethality, and silencing of PINP1 leads to developmental defects and hypersusceptibility to Phytophthora infection. These phenotypes are reminiscent of transgenic plants expressing PSR1, supporting PINP1 as a direct virulence target of PSR1. We further demonstrate that the localization of the Dicer-like 1 protein complex is impaired in the nucleus of PINP1-silenced or PSR1-expressing cells, indicating that PINP1 may facilitate small RNA processing by affecting the assembly of dicing complexes. A similar function of PINP1 homologous genes in development and immunity was also observed in Nicotiana benthamiana. These findings highlight PINP1 as a previously unidentified component of RNA silencing that regulates distinct classes of small RNAs in plants. Importantly, Phytophthora has evolved effectors to target PINP1 in order to promote infection.
Although constantly challenged by microbial parasites in the environment, plants can defend themselves from most of the attacks through innate immune systems. A basal layer of plant immunity relies on the recognition of conserved molecular signatures called microbe-associated molecular patterns (1, 2). This pattern-triggered immunity (PTI) leads to defense responses that can effectively defeat the vast majority of potential pathogens. However, successful pathogens have evolved effector proteins whose fundamental function is to subvert plant immunity (3, 4). Many effectors are delivered into the host cells and directly manipulate the functions of immune regulators (5, 6). Research on effector targets has not only revealed essential virulence strategies of the pathogens, but also helped identify novel components of plant immunity.
The genus Phytophthora contains some of the most notorious plant pathogens. For example, Phytophthora infestans is the causative agent of potato late blight that was responsible for the Great Irish Famine (7); Phytophthora ramorum is a major threat of forestry by causing the sudden oak death (8); and Phytophthora sojae is the second most destructive pathogen of soybean (9). Phytophthora spp. establish intimate associations with host plants through infection structures called haustoria, through which effectors are secreted to the extrahaustorial space; the so-called cytoplasmic effectors can then be taken up by plant cells through a host-targeting motif (10). Each Phytophthora species is predicted to encode >1,000 cytoplasmic effectors (11), and the majority of them contained the consensus RxLR motif (11, 12). This remarkably large effector repertoire reflects the high level of complexity in the Phytophthora–plant arms race and demands mechanistic analysis of effector functions to gain understanding of Phytophthora pathogenesis.
Substantial efforts have been devoted to identifying virulence targets of Phytophthora effectors, and a variety of plant processes that can be disrupted during Phytophthora infection have been revealed (13, 14). Using a functional screen, we recently identified two P. sojae RxLR effectors that can suppress the RNA-silencing process in plants (15). RNA silencing is a key mechanism of gene regulation in eukaryotes. Expression of these Phytophthora Suppressors of RNA silencing (PSRs) or a viral suppressor of RNA silencing in Nicotiana benthamiana significantly enhanced the infection of P. infestans. These findings demonstrate that RNA-silencing suppression is an important virulence strategy of Phytophthora pathogens.
The central players of RNA silencing are small RNAs, which repress gene expression at transcriptional, posttranscriptional, and translational levels. Small RNAs are important regulators of plant immunity. Plants produce two major classes of small RNAs—microRNA (miRNA) and small interfering RNA (siRNA). miRNAs are encoded by endogenous MIR genes, whereas siRNAs are derived from invading nucleic acids, such as viruses and transgenes, and from endogenous loci, such as repeats, transposable elements, and genes (16). siRNAs play a key role in antiviral defense (17), whereas specific miRNAs have been shown to regulate PTI during bacterial, oomycete, and fungal infection (18). For example, miR393 is induced in soybean roots by P. sojae and acts as a positive regulator of soybean defense (19). Transgenic plants expressing PSR1 or PSR2 in Arabidopsis thaliana exhibit decreased abundances of small RNAs (15). In particular, PSR1 has a general impact on the accumulation of both miRNAs and siRNAs; as a result, PSR1-expressing Arabidopsis exhibits developmental defects, including serrated leaves, dwarfism, and reduced seed production. However, the host target(s) of PSR1 and the mechanism by which PSR1 suppresses small RNA accumulation in plants remains unknown.
Here, we report that PSR1 directly interacts with a nuclear protein containing the aspartate–glutamate–alanine–histidine (DEAH)-box RNA helicase domain in Arabidopsis. Silencing of this putative RNA helicase in Arabidopsis and N. benthamiana renders defects in small RNA accumulation and hypersusceptibility to Phytophthora. This study provides mechanistic insight into the suppression of host immunity by a Phytophthora suppressor of RNA silencing and highlights an evolutionarily conserved and essential protein as a regulator of RNA silencing in plants.
Results
Identification of PSR1-Associating Proteins.
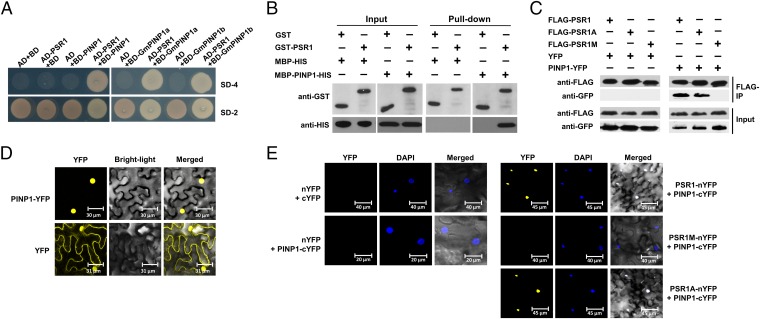
To elucidate the mechanism by which PSR1 suppresses RNA silencing in plants, we characterized PSR1-associating proteins in Arabidopsis by yeast two-hybrid screening using PSR1 without the N-terminal secretion signal (1–20 aa) as the bait. Approximately 8 × 106 yeast clones were screened in four independent experiments, and one protein (At5g13010) was repetitively identified. This protein was designated PSR1-Interacting Protein 1 (PINP1) (Fig. 1A). Because PSR1 is produced by the soybean pathogen P. sojae, we examined the interaction of PSR1 with two PINP1 homologs of soybean (Glycine max XP_003547002 and XP_003542053) and showed that both of them interact with PSR1 in yeast (Fig. 1A).
Fig. 1.
PSR1 interacts with a plant nuclear protein PINP1. (A) PSR1 interacts with PINP1 in yeast. Yeast strain AH109 was transformed with the bait plasmid pGBKT7 (BD) carrying PSR1 together with the prey plasmid pGADT7 (AD) carrying PINP1, GmPINP1a, or GmPINP1b. Transformants were selected on minimal medium lacking adenine, tryptophan, histidine, and leucine (SD-4). (B) PSR1 and PINP1 interact in vitro. GST–PSR1 and MBP–PINP1–HIS were expressed in E. coli. Coprecipitation of PINP1 with PSR1 was examined by Western blotting before (input) and after affinity purification (pull-down) using glutathione agarose beads. (C) PSR1 and PINP1 interact in planta. Total proteins were extracted from N. benthamiana leaves expressing PINP1–YFP and FLAG–PSR1. The immune complexes were pulled down by using anti-FLAG agarose gel, and the coprecipitation of PINP1 was detected by Western blotting. (D) PINP1 is exclusively located in the nucleus. PINP1–YFP was expressed in N. benthamiana through Agro-infiltration. Fluorescence was detected from epidermal cells in the infiltrated tissues by confocal microscopy at 48 h postinoculation (hpi). (E) Bimolecular fluorescence complementation analysis showing PSR1/PINP1 interaction in the nuclei of plant cells. PSR1–nYFP and PINP1–cYFP were coexpressed in N. benthamiana through Agro-infiltration. Fluorescence was detected by confocal microscopy at 48 hpi. DAPI was used to stain the nuclei. These experiments were repeated three times with similar results.
PSR1 Interacts with PINP1 in Vitro and in Planta.
To validate the physical interaction of PSR1 with PINP1, we carried out in vitro pull-down assays using glutathione S-transferase (GST)-tagged PSR1 and maltose-binding protein (MBP)-HIS tagged PINP1 that were expressed in Escherichia coli. As shown in Fig. 1B, the MBP–PINP1–HIS proteins were specifically enriched in GST–PSR1-bound glutathione resins. We further examined the interaction of PSR1 with PINP1 in planta. PINP1–YFP was transiently expressed in N. benthamiana together with FLAG–PSR1 using Agro-infiltration. Total proteins were extracted from the infiltrated leaves and incubated with anti-FLAG resins. PINP1–YFP, but not YFP, was significantly enriched in the FLAG–PSR1 precipitates (Fig. 1C). These results confirmed the interaction of PSR1 with PINP1 in vitro and in plant cells.
PINP1 is homologous to MUT6 of Chlamydomonas reinhardtii, which was shown to regulate the silencing of transgenes and transposons (20). The MUT6 family proteins contain a conserved DEAH-box RNA helicase domain and are predicted to locate in the nucleus (21). When expressing PINP1–YFP in N. benthamiana using Agro-infiltration, yellow fluorescence was exclusively observed from the nuclei of epidermal cells (Fig. 1D). This localization of PINP1 is consistent with the localization of PSR1, which is also mainly in the nucleus (15). To further characterize the PSR1–PINP1 protein complex in plant cells, we conducted the bimolecular fluorescence complementation (BiFC) experiment. PSR1 and PINP1 were fused to the N- or C-terminal half of YFP (nYFP or cYFP, respectively) and coexpressed in N. benthamiana. Strong fluorescence was observed exclusively from the nucleus (Fig. 1E), suggesting that the PSR1–PINP1 complex is located in the nucleus.
PSR1 contains a putative nuclear localization signal (NLS). Previous results showed that a PSR1 mutant (PSR1M) with the putative NLS mutated lost the nuclear localization as well as the RNA silencing suppression activity in N. benthamiana (15). Consistent with this prior finding, PSR1M no longer interacts with PINP1 in plant cells (Fig. 1 C and E). On the contrary, another mutant of PSR1, PSR1A, which lacks the host-targeting motif RxLR, can still associate with PINP1 (Fig. 1 C and E). This result is expected because the RxLR motif is believed to contribute to effector entry and therefore should be dispensable after the effectors enter the host cell. We also examined the interaction of PINP1 with PSR1 fused to a nuclear export signal (NES) or mutated “nes.” A coimmunoprecipitation experiment showed that the interaction of PSR1–NES with PINP1 was abolished (Fig. S1). Together, these results strongly suggest that PSR1 associates with PINP1 in plant nuclei and that the nuclear localization of PSR1 is required for this interaction.
PINP1 Affects Small RNA Accumulation.
Expression of PSR1 in Arabidopsis resulted in an across-the-board reduction of small RNAs, including miRNAs and siRNAs (15). We therefore examined whether PINP1 also plays a role in the accumulation of small RNAs. We were unable to obtain homozygous lines of three T-DNA insertion mutants available for the PINP1 locus, indicating that loss-of-function mutation of PINP1 might be embryonic lethal (Table S1). Therefore, we generated PINP1-silenced lines of Arabidopsis using artificial miRNAs (amiRNAs).
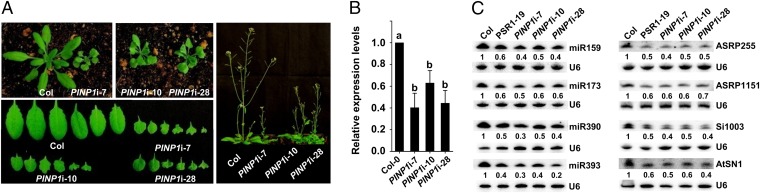
Expressing an amiRNA in Arabidopsis eco. Col-0 allowed us to obtain >30 transgenic lines from independent transformation events. In general, these transgenic plants exhibit severe developmental defects, including serrated leaves, dwarfism, late flowering, and reduced seed production (Fig. 2A). The severity of the developmental defects of individual lines is mostly correlated to the silencing efficiencies of PINP1, indicating that these phenotypes are likely caused by the reduced expression of PINP1 (Fig. 2B). Remarkably, these phenotypes are reminiscent of PSR1-expressing plants (15) and similar to miRNA biogenesis mutants. Indeed, all of the miRNAs that we examined showed reduced accumulation in PINP1-silenced lines (Fig. 2C and Fig. S2A), suggesting that PINP1 affects miRNA levels in Arabidopsis.
Fig. 2.
PINP1 plays a role in small RNA accumulation in Arabidopsis. (A) Silencing of PINP1 leads to developmental defects. Photos of wild-type (Col-0) and three independent PINP1-silenced lines (PINP1i-7, PINP1i-10, and PINP1i-28) were taken after 4 wk (Left) and 8 wk (Right) of growth. (B) Transcript abundances of PINP1 in the silenced lines compared with wild-type (Col-0) were determined by qRT-PCR. AtUBQ10 was used as the internal standard. Values are means ± SDs (as error bars) from three independent replicates. Letters represent differences with statistical significance (P < 0.01) as determined by Duncan’s multiple test. (C) Northern blots showing endogenous small RNA abundances in PINP1-silenced plants and the PSR1 transgenic line PSR1-19. Results from four representative miRNAs, two ta-siRNAs (ASRP255 and ASRP1151), and two heterochromatic siRNAs (Si1003 and AtSN1) are presented. U6 serves as the loading control. Numbers below each blot indicate relative abundances of the small RNA. Data from additional miRNAs and siRNAs are shown in Fig. S2A. These experiments were repeated twice with similar results.
In addition to miRNAs, we found that the abundances of representative endogenous siRNAs, including trans-acting siRNAs (ta-siRNAs) and heterochromatic siRNAs, were reduced in the PINP1-silenced lines (Fig. 2C and Fig. S2A). Corresponding to the decreased small RNA levels, transcripts of a few miRNA and siRNA target genes accumulated to higher levels in PINP1-silenced plants (Fig. S2B), confirming that PINP1 has a general role in regulating small RNA levels in Arabidopsis. These results are also consistent with the previously demonstrated function of PSR1 (15).
We further determined the specific step(s) during miRNA biogenesis that involves PINP1. Mature miRNAs are processed from primary miRNA (pri-miRNA) precursors, which are transcripts of the MIR genes, by the RNase III-like enzyme known as Dicer-like 1 (DCL1) (16). Quantitative RT-PCR (qRT-PCR) showed that the abundances of pri-miRNAs were either unaffected or slightly higher in PINP1-silenced plants (Fig. S3A). On the contrary, the pri-miRNA levels were significantly reduced in another Arabidopsis mutant, not2a-1 2b-1 (Fig. S3A), which is known to have reduced transcription of MIR genes (22). This result suggests that PINP1 is not required for MIR gene transcription or the stability of pri-miRNAs, but may facilitate the processing of pri-miRNAs to produce mature miRNAs. Consistent with this hypothesis, the abundances of precursor miRNAs (pre-miRNAs), the processing products of pri-miRNAs and precursors of mature miRNAs, were significantly reduced in PINP1-silenced plants (Fig. S3B). These results are consistent with the activity of PSR1, which also affects the levels of mature miRNAs and pre-miRNAs, but has no effect on pri-miRNAs (15).
PINP1-Silenced Plants Are Hypersusceptible to Phytophthora capsici.
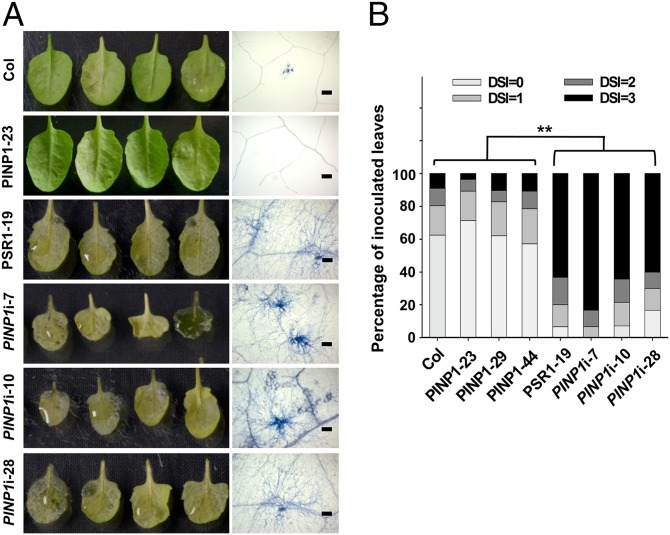
PSR1 promotes the infection of P. infestans when expressed in N. benthamiana (15). We therefore investigated the role of PINP1 in plant defense during Phytophthora infection. For this purpose, we used a pathosystem with Arabidopsis as the host and the Phytophthora capsici isolate LT263 as the pathogen (23, 24). A drastic enhancement of susceptibility was observed from PINP1-silenced plants (Fig. 3A). A similar hypersusceptibility phenotype was also observed in PSR1-expressing plants (Fig. 3A). Under our experimental conditions, ∼60–85% of the inoculated leaves in these plants showed severe water-soaked lesions at 3 d postinoculation (dpi), representing a disease index of 3 (Fig. 3B). In contrast, <10% of the inoculated leaves from wild-type plants exhibited severe disease symptoms in this category, and >50% of the inoculated leaves did not show visible symptoms. These results demonstrate that silencing of PINP1 significantly affects the resistance of Arabidopsis to P. capsici. Although we could not exclude the possibility that the compromised development of PINP1-silenced plants may contribute to the hypersusceptibility phenotype, our experiments strongly suggest a positive role of PINP1 in regulating Arabidopsis defense. We also examined the susceptibility of PINP1-overexpressing plants, but did not observe significant changes compared with wild-type plants (Fig. 3).
Fig. 3.
Silencing of PINP1 leads to hypersusceptibility of Arabidopsis to P. capsici strain LT263. Adult leaves of 4-wk-old wild-type plants (Col-0), 4-wk-old PINP1-overexpressing plants (PINP1-23), 5.5-wk-old PSR1-expressing plants (PSR1-19), and 7-wk-old PINP1-silenced plants were detached and inoculated with zoospore suspension of P. capsici strain LT263 (1 × 105 zoospores per mL). Disease symptoms were monitored at 3 d postinoculation (dpi), and the disease severity index (DSI) of each leaf was determined. Forty leaves from 15–20 plants were inoculated and analyzed in each line. Note that 4-wk-old PSR1-expressing or PINP1-silenced plants were too small for inoculation. (A) Photos of inoculated leaves (Left) and microscope pictures of Phytophthora hyphae extension (Right) in the leaves at 3 dpi. Trypan blue was used to stain the hyphae for visualization. (Scale bars: 250 μm.) (B) Quantitative analysis of disease severity. **P < 0.01 (as determined by the Wilcoxon rank-sum test). This experiment was repeated three times with similar results.
PINP1 Facilitates the Subnuclear Localization of pri-miRNA Processing Complex.
Because silencing of PINP1 in Arabidopsis results in reduced miRNA accumulation without interfering with the pri-miRNA levels, we suspected that PINP1 may affect the accumulation and/or function of DCL1, which is responsible for the processing of pri-miRNAs to produce pre-miRNAs and then mature miRNAs (16, 25). qRT-PCR showed similar abundances of dcl1 transcripts in wild-type and PINP1-silenced plants (Fig. S4A); Western blotting confirmed that DCL1 protein levels were unchanged or slightly enhanced in PINP1-silenced plants (Fig. S4B). These results demonstrate that the decreased miRNA accumulation in PINP1-silenced plants is not due to reduced DCL1 levels, suggesting that PINP1 may assist the function of the DCL1 complex. To test this possibility, we examined the impact of PINP1 on the localization of DCL1. DCL1 is exclusively located in the nucleus. Although a small percentage of the DCL1 proteins are diffusely distributed throughout the nucleoplasm, the majority are enriched in round nuclear speckles called dicing bodies, or D bodies (26). Because pri-miRNAs are recruited to D bodies, it is proposed that the maintenance of these defined nuclear speckles is important for the assembly, and hence the function, of pri-miRNA processing complex (26).
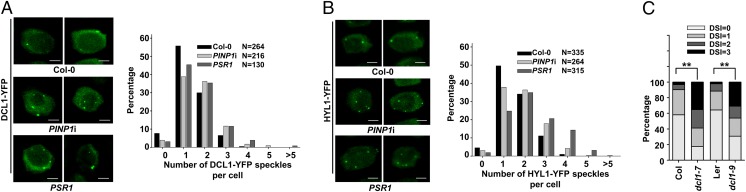
We analyzed the number of DCL1-containing nuclear speckles in wild-type and PINP1-silenced cells of Arabidopsis roots. In wild-type plants, >90% of a total of 264 cells harbor two or fewer D bodies in the nucleus and only <7% of the cells harbor three or more D bodies (Fig. 4A). Interestingly, silencing of PINP1 led to an increase on the number of DCL1-containing speckles in the nucleus. Analysis of 216 PINP1-silenced cells showed that >14% of the cells contain three or more D bodies in the nucleus. A similar observation was also made in PSR1-expressing cells (Fig. 4A).
Fig. 4.
Subnuclear localizations of DCL1 and HYL1 are altered in PINP1-silenced plants. (A and B) Localizations of DCL1–YFP (A) and HYL1–YFP (B) were examined in root cells from the meristematic zone of wild-type (Col-0) seedlings or seedlings expressing amiRPINP1 (PINP1i) or PSR1 by confocal microscopy. (Scale bars: 2.5 μm.) Percentage distributions of cells harboring different numbers of subnuclear speckles were analyzed by the Kolmogorov–Smirnov test. The distribution in wild-type cells is significantly different (P < 0.001) from those in PINP1-silenced and PSR1-expressing cells. N represents the total number of cells analyzed in each line. (C) Arabidopsis mutants dcl1-7 (in Col-0 background) and dcl1-9 [in Landsberg erecta (Ler) background] are hypersusceptible to P. capsici strain LT263. Forty detached leaves of 4-wk-old wild-type plants (Col-0 and Ler) and 6-wk-old dcl1 mutants were inoculated with zoospore suspension (1 × 105 zoospores per mL) and analyzed for DSI at 3 dpi. **P < 0.01 (as determined by the Wilcoxon rank-sum test). This experiment was repeated twice with similar results.
To further confirm that PINP1 affects the assembly of D bodies, we examined the subnuclear localization of HYL1, a double-stranded RNA-binding protein that functions and localizes together with DCL1 to process miRNAs (16, 27). Similar to what was observed in DCL1-containing nuclear speckles, a significantly higher percentage of PINP1-silenced (22.7%) and PSR1-expressing cells (38.4%) harbor three or more HYL1-containing nuclear speckles, compared with only 12% in wild-type cells (Fig. 4B). These results suggest that PINP1 is required for the correct subnuclear localization of the dicing complex for miRNA processing.
Mislocalization of DCL1 to D bodies impaired its activity. A mutated DCL1, DCL1-9, which is truncated in the C-terminal 73 amino acids, fails to localize to D bodies (26), and Arabidopsis plants carrying this mutated allele exhibit severe defects in miRNA biogenesis (25). Consistent with a role of DCL1 in anti-Phytophthora defense, dcl1-9 is hypersusceptible to P. capsici strain LT263 (Fig. 4C). Furthermore, dcl1-7, containing an amino acid substitution (P415S) within the DECH-box RNA helicase domain and an impaired function in miRNA processing (25), also showed enhanced susceptibility (Fig. 4C). These results suggest that PINP1 contributes to plant immunity, likely through its role in affecting the assembly of DCL1-containing pri-miRNA processing complexes in the nucleus.
Silencing of PINP1 Homologs in N. benthamiana Enhanced the Susceptibility to P. infestans and Affected miRNA Accumulation.
PINP1 is an evolutionarily conserved protein with homologs found from the genomes of both dicots and monocots (Fig. S5). We next investigated PINP1 homologs in N. benthamiana using Virus-Induced Gene Silencing (VIGS) to determine whether they perform a similar function on small RNA biogenesis and immunity as in Arabidopsis. Sequence analysis revealed two potential PINP1 homologous genes in the N. benthamiana, designated NbPINP1a and NbPINP1b (Fig. S5). Both homologs contain the conserved motifs that are characteristic for DEAH-box RNA helicases, similar to MUT6 in Chlamydomonas and PINP1 in Arabidopsis (Fig. S6).
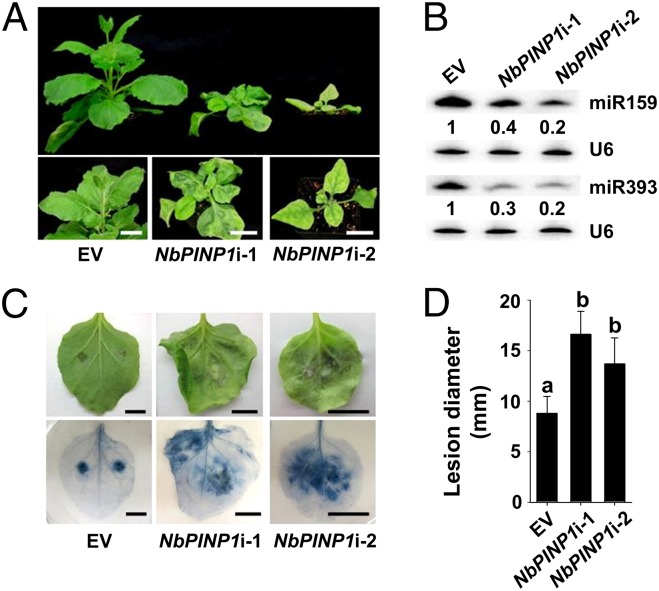
Two DNA fragments designed to target both NbPINP1a and NbPINP1b were cloned into the tobacco rattle virus (TRV)-based VIGS vector to knock down their expression in N. benthamiana. Each VIGS construct successfully silenced both genes with a higher silencing efficiency obtained from the construct NbPINP1i-2 (Fig. S7). Similar to Arabidopsis, silencing of NbPINP1a/b also led to developmental defects, including downward curling at the edges, dwarfism, and late flowering (Fig. 5A). This phenotype is more severe in plants expressing NbPINP1i-2, consistent with the lower expression of NbPINP1a/b genes (Fig. S7). Furthermore, as PINP1 was found to be required for small RNA accumulation in Arabidopsis, the abundances of miRNA159 and miRNA393 in the NbPINP1a/b-silenced N. benthamiana leaves were also greatly reduced (Fig. 5B). Importantly, when inoculated by P. infestans isolate 1306, NbPINP1a/b-silenced leaves allowed enhanced infection and showed more severe disease symptoms (Fig. 5 C and D). Together, these experiments demonstrate that PINP1 is a conserved component of small RNA biogenesis and immunity in plants.
Fig. 5.
PINP1 homologs in N. benthamiana contribute to small RNA biogenesis and immunity. (A) Silencing of NbPINP1a/b leads to developmental defects. Pictures of N. benthamiana plants expressing the empty TRV vector (EV) or the gene-silencing construct NbPINP1i-1 or NbPINP1i-2 were taken at 21 d after Agro-infiltration. (B) Northern blotting showing reduced abundances of miR159 and miR393 in NbPINP1a/b-silenced leaves. U6 serves as the loading control. Numbers below each blot indicate relative abundances of the miRNA. (C) NbPINP1a/b-silenced plants are hypersusceptible to P. infestans isolate 1306. NbPINP1a/b-silenced leaves were inoculated with 30 μL of zoospores suspension (4 × 104 zoospores per mL), and disease symptoms (Upper) were examined at 5 dpi. (C, Lower) Trypan blue staining was used to visualize lesions. (D) Sizes of lesions caused by P. infestans infection. Values are means ± SD. Letters represent differences with statistical significance (P < 0.01) as determined by Duncan’s multiple range test. These experiments were repeated three times with similar results.
Discussion
Although Phytophthora spp. are responsible for many devastating diseases of crops and forestry trees, our understanding of the molecular basis of Phytophthora pathogenicity is limited. Infection of plants by Phytophthora entails complex defense/counterdefense cross-talk, which is reflected by the hundreds to thousands of effector proteins that are predicted from each Phytophthora genome. The majority of Phytophthora effectors have a conserved N-terminal RxLR motif. Effectors with a similar host-targeting signal are also found in parasitic fungi and protozoa, indicating an evolutionarily conserved means of eukaryotic pathogens to deliver virulence proteins into host cells (12). To date, the functions of the vast majority of eukaryotic pathogen effectors remain unknown.
RNA silencing is a universal gene regulation mechanism in eukaryotes and serves as an important defense mechanism against pathogen infection. Therefore, it is not surprising that viruses, bacteria, and Phytophthora have all evolved effectors to suppress this process (28). The newest members of RNA-silencing suppressors, and the only ones identified so far from eukaryotic pathogens, are PSRs produced by Phytophthora (15, 24). PSR1 has a general impact on both miRNAs and siRNAs in plants and significantly enhances Phytophthora infection. Our experiments revealed that PSR1 physically associates with PINP1, an evolutionarily conserved nuclear protein containing an RNA helicase domain. Silencing of PINP1 leads to the same defects in development and immunity as observed in Arabidopsis transgenic plants expressing PSR1, suggesting that PINP1 is likely a direct virulence target of PSR1. Previous experiments showed that the nuclear localization of PSR1 is required for its biological function; this requirement is consistent with the exclusive nuclear localization of PINP1. Importantly, mutation in the putative NLS or fusion to an NES abolished the association of PSR1 with PINP1 in plant cells. These results suggest that the interaction with PINP1 is likely responsible for PSR1-mediated suppression of small RNA accumulation and plant immunity.
PINP1 belongs to the MUT6 family of proteins, which contains the DEAH-box RNA helicase domain (21). In Chlamydomonas, MUT6 is required for silencing of transgenes and transposons, and is involved in RNA turnover (20). By characterizing the PINP1-silenced lines of Arabidopsis, we discovered that PINP1 affects the accumulation of small RNAs. Although we could not exclude a role of PINP1 on miRNA stability, it seems likely that PINP1 mainly affects the biogenesis of miRNAs. Interestingly, the subnuclear localization of the miRNA processing complex containing DCL1 and HYL1 was altered in the nucleus of PINP1-silenced cells, suggesting that PINP1 may play a role in the assembly of dicing bodies. A similar phenotype was also observed in the not2 mutant of Arabidopsis. NOT2s associate with DCL1 and promote the recruitment of DCL1 to the D bodies (22). Although silencing of PINP1 leads to a similar change in the subnuclear localization of D bodies as the not2 mutant, PINP1 does not affect the abundance of pri-miRNAs as NOT2s do (Fig. S3A; ref. 22). Therefore, PINP1 functions in the miRNA biogenesis pathway on step(s) downstream of NOT2s. This finding is consistent with the observation that pre-miRNA levels were reduced in PINP1-silenced plants.
PINP1 homologs are produced by a broad range of dicots and monocots. Silencing of the PINP1 homologs results in similar developmental defects, decreased small RNA levels, and enhanced susceptibility to P. infestans in N. benthamiana, suggesting that the PINP1 family of proteins is a conserved component of RNA silencing and regulator of immunity in plants. These results assign previously unidentified functions to this conserved protein family in plants. Consistent with the hypothesis that PINP1 positively regulates plant defense through promoting miRNA processing, dcl1 mutants of Arabidopsis exhibit enhanced susceptibility to P. capsici. dcl1-9 is also more susceptible to bacterial infection (29), suggesting that DCL1 is required for plant defense against a broad range of pathogens. Interestingly, the levels of enhanced susceptibility observed in dcl1 mutants were not to the same extent compared with the PINP1-silenced plants or the PSR1-expressing plants. This result could be due to the fact that PINP1 also affects the accumulation of endogenous ta-siRNAs and heterochromatic siRNAs, which depend on the activity of DCL4 and DCL3 respectively (16). Indeed, endogenous siRNAs have been reported to regulate defense responses (30–33). Therefore, PINP1 may facilitate the functions of multiple DCLs or common DCL cofactor(s) that are responsible for both miRNA and siRNA biogenesis. Further investigations will provide molecular details on how PINP1 contributes to the assembly of dicing complexes to promote small RNA processing.
RNA helicases are key regulators of RNA metabolism and silencing. In animals, the DEAD-box RNA helicase DDX17 binds to the stem-loop structure of pri-miRNAs and facilitates their processing (34). The SDE3 family of DEAG-box RNA helicases associates with ARGONAUTs, the major effector proteins of posttranscriptional RNA silencing, and promotes the production of secondary siRNAs in plants and animals (35). Both DDX17 and SDE3 are also required for antiviral immunity(34, 35). Here, we show that PINP1 is a predicted DEAH-box RNA helicase that acts as a general regulator of distinct classes of small RNAs in plants. Importantly, a Phytophthora RNA-silencing suppressor, PSR1, directly targets PINP1 to interfere with the accumulation of small RNAs. The function of PINP1 in RNA silencing was not previously identified in plants. Using PSR1 as a molecular probe, we are able to define the essential role of this conserved protein family in RNA silencing and immunity. This study also highlights the identification of a novel class of effector targets and sheds mechanistic insight into the pathogenesis of the notorious Phytophthora diseases.
Materials and Methods
Plant Growth Conditions.
Arabidopsis was grown at 23 °C with a 10/14 light/dark regime. N. benthamiana was grown at 22 °C with a 16/8 light/dark regime. Arabidopsis seedlings for D-body observation were grown on Murashige and Skoog agar containing 3% (wt/vol) sucrose.
Phytophthora Growth Conditions.
Phytophthora strains used in this study are listed in Table S2. P. capsici isolate LT263 was grown on 10% (vol/vol) V8 medium at 25 °C in the dark. P. infestans isolate 1306 was grown on rye sucrose agar plates.
Protein Pull-Down Assays.
For in vitro pull-down, GST–PSR1 and MBP–PINP1–HIS were expressed in E. coli strain BL21 (DE3). Coprecipitation of PINP1 with PSR1 was examined by Western blotting before (input) and after affinity purification (pull-down) using glutathione agarose beads (Pierce). Anti-GST and -HIS antibodies were purchased from Santa Cruz Biotechnology. For in planta pull-down, 3× FLAG–PSR1 and PINP1–YFP were coexpressed in N. benthamiana by Agro-infiltration. Total proteins were extracted using an IP buffer [10% (vol/vol) glycerol, 25 mM Tris, pH 7.5, 1 mM EDTA, 150 mM NaCl, 10 mM DTT, 2% (wt/vol) PVPP, 1× protease inhibitor mixture (Roche), 1 mM PMSF, and 0.15% Nonidet P-40], and then incubated with anti-FLAG affinity gel (Sigma-Aldrich) at 4 °C. Coprecipitation of PINP1 with PSR1 was detected by using an anti-GFP antibody (Clontech).
Gene Silencing in Arabidopsis and N. benthamiana.
amiRNAs were designed to silence PINP1 in Arabidopsis by using the WMD online tool (wmd3.weigelworld.org). The amiRNA was cloned into the vector pRS300 (36), and the complete silencing cassette was then cloned into pEG100 (37) for Arabidopsis transformation. The PINP1 homologous genes, NbPINP1a and NbPINP1b, were silenced by the TRV system as described (38) by using antisense fragments, NbPINP1i-1 and NbPINP1i-2.
Visualization of DCL1- and HYL1-Containing Nuclear Bodies.
Subcellular localization of DCL1 and HYL1 were determined by following the procedure described in refs. 22 and 26. pUBQ10–PSR1 and pUBQ10–amiRPINP1 were introduced into Arabidopsis eco. Col-0 expressing p35S–DCL1–YFP or p35S–HYL1–YFP (22). Transgenic seedlings were grown on MS medium, and the number of DCL1- and HYL1-containing speckles in root cells was evaluated by using a Leica SP5 Laser Confocal Microscope.
qRT-PCR.
Primers used to amplify pri-miRNAs, pre-miRNAs, and small RNA-target genes are listed in Table S3.
Phytophthora Infection Assays.
NbPINP1a/b-silenced leaves were detached 3 wk after the expression of the VIGS constructs and inoculated with 30 μL of zoospores suspension (4 × 104 zoospores per mL) of P. infestans isolate 1306 as described in ref. 15. Disease symptoms and lesion sizes were examined at 5 dpi. Adult leaves of Arabidopsis were detached and inoculated with P. capsici isolate LT263 by using 10 μL of zoospores suspension (1 × 105 zoospores per mL) as described in refs. 23 and 24. Disease severity was evaluated at 3 dpi using a disease index based on hyphae extension (24).
Supplementary Material
Acknowledgments
We thank Dr. Xuemei Chen for Arabidopsis mutants dcl1-7 and dcl1-9, Dr. Xiaofeng Cao for Arabidopsis mutant not2a-1 2b-1 and transgenic lines expressing DCL1–YFP and HYL1–YFP, Dr. Doil Choi for TRV-based vectors, Dr. Howard Judelson for P. infestans strain 1306, and Dr. Kurt Lamour for P. capsici isolate LT263. This work was supported by US Department of Agriculture (USDA)–National Institute of Food and Agriculture Grant 2013-02974 and USDA Agriculture Experimental Station Grant CA-R-PPA-5075-H (to W.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1421475112/-/DCSupplemental.
References
- 1.Spoel SH, Dong X. How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol. 2012;12(2):89–100. doi: 10.1038/nri3141. [DOI] [PubMed] [Google Scholar]
- 2.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444(7117):323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 3.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell. 2006;124(4):803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Boller T, He SY. Innate immunity in plants: An arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science. 2009;324(5928):742–744. doi: 10.1126/science.1171647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dou D, Zhou J-M. Phytopathogen effectors subverting host immunity: Different foes, similar battleground. Cell Host Microbe. 2012;12(4):484–495. doi: 10.1016/j.chom.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Dangl JL, Horvath DM, Staskawicz BJ. Pivoting the plant immune system from dissection to deployment. Science. 2013;341(6147):746–751. doi: 10.1126/science.1236011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haas BJ, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461(7262):393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 8.Tyler BM, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313(5791):1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 9.Tyler BM. Phytophthora sojae: Root rot pathogen of soybean and model oomycete. Mol Plant Pathol. 2007;8(1):1–8. doi: 10.1111/j.1364-3703.2006.00373.x. [DOI] [PubMed] [Google Scholar]
- 10.Petre B, Kamoun S. How do filamentous pathogens deliver effector proteins into plant cells? PLoS Biol. 2014;12(2):e1001801. doi: 10.1371/journal.pbio.1001801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pais M, et al. From pathogen genomes to host plant processes: The power of plant parasitic oomycetes. Genome Biol. 2013;14(6):211. doi: 10.1186/gb-2013-14-6-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haldar K, Kamoun S, Hiller NL, Bhattacharje S, van Ooij C. Common infection strategies of pathogenic eukaryotes. Nat Rev Microbiol. 2006;4(12):922–931. doi: 10.1038/nrmicro1549. [DOI] [PubMed] [Google Scholar]
- 13.Bozkurt TO, Schornack S, Banfield MJ, Kamoun S. Oomycetes, effectors, and all that jazz. Curr Opin Plant Biol. 2012;15(4):483–492. doi: 10.1016/j.pbi.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 14.Giraldo MC, Valent B. Filamentous plant pathogen effectors in action. Nat Rev Microbiol. 2013;11(11):800–814. doi: 10.1038/nrmicro3119. [DOI] [PubMed] [Google Scholar]
- 15.Qiao Y, et al. Oomycete pathogens encode RNA silencing suppressors. Nat Genet. 2013;45(3):330–333. doi: 10.1038/ng.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bologna NG, Voinnet O. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu Rev Plant Biol. 2014;65:473–503. doi: 10.1146/annurev-arplant-050213-035728. [DOI] [PubMed] [Google Scholar]
- 17.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130(3):413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruiz-Ferrer V, Voinnet O. Roles of plant small RNAs in biotic stress responses. Annu Rev Plant Biol. 2009;60:485–510. doi: 10.1146/annurev.arplant.043008.092111. [DOI] [PubMed] [Google Scholar]
- 19.Wong J, et al. Roles of small RNAs in soybean defense against Phytophthora sojae infection. Plant J. 2014;79(6):928–940. doi: 10.1111/tpj.12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu-Scharf D, Jeong B, Zhang C, Cerutti H. Transgene and transposon silencing in Chlamydomonas reinhardtii by a DEAH-box RNA helicase. Science. 2000;290(5494):1159–1162. doi: 10.1126/science.290.5494.1159. [DOI] [PubMed] [Google Scholar]
- 21.Linder P, Owttrim GW. Plant RNA helicases: Linking aberrant and silencing RNA. Trends Plant Sci. 2009;14(6):344–352. doi: 10.1016/j.tplants.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Wang L, et al. NOT2 proteins promote polymerase II-dependent transcription and interact with multiple MicroRNA biogenesis factors in Arabidopsis. Plant Cell. 2013;25(2):715–727. doi: 10.1105/tpc.112.105882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Bouwmeester K, van de Mortel JE, Shan W, Govers F. A novel Arabidopsis-oomycete pathosystem: Differential interactions with Phytophthora capsici reveal a role for camalexin, indole glucosinolates and salicylic acid in defence. Plant Cell Environ. 2013;36(6):1192–1203. doi: 10.1111/pce.12052. [DOI] [PubMed] [Google Scholar]
- 24.Xiong Q, et al. Phytophthora suppressor of RNA silencing 2 is a conserved RxLR effector that promotes infection in soybean and Arabidopsis thaliana. Mol Plant Microbe Interact. 2014;27(12):1379–1389. doi: 10.1094/MPMI-06-14-0190-R. [DOI] [PubMed] [Google Scholar]
- 25.Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci USA. 2004;101(34):12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang Y, Spector DL. Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr Biol. 2007;17(9):818–823. doi: 10.1016/j.cub.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song L, Han M-H, Lesicka J, Fedoroff N. Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc Natl Acad Sci USA. 2007;104(13):5437–5442. doi: 10.1073/pnas.0701061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pumplin N, Voinnet O. RNA silencing suppression by plant pathogens: Defence, counter-defence and counter-counter-defence. Nat Rev Microbiol. 2013;11(11):745–760. doi: 10.1038/nrmicro3120. [DOI] [PubMed] [Google Scholar]
- 29.Navarro L, Jay F, Nomura K, He SY, Voinnet O. Suppression of the microRNA pathway by bacterial effector proteins. Science. 2008;321(5891):964–967. doi: 10.1126/science.1159505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhai J, et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011;25(23):2540–2553. doi: 10.1101/gad.177527.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agorio A, Vera P. ARGONAUTE4 is required for resistance to Pseudomonas syringae in Arabidopsis. Plant Cell. 2007;19(11):3778–3790. doi: 10.1105/tpc.107.054494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhattacharjee S, et al. Virus resistance induced by NB-LRR proteins involves Argonaute4-dependent translational control. Plant J. 2009;58(6):940–951. doi: 10.1111/j.1365-313X.2009.03832.x. [DOI] [PubMed] [Google Scholar]
- 33.Yu A, et al. Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc Natl Acad Sci USA. 2013;110(6):2389–2394. doi: 10.1073/pnas.1211757110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moy RH, et al. Stem-loop recognition by DDX17 facilitates miRNA processing and antiviral defense. Cell. 2014;158(4):764–777. doi: 10.1016/j.cell.2014.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia D, et al. Ago hook and RNA helicase motifs underpin dual roles for SDE3 in antiviral defense and silencing of nonconserved intergenic regions. Mol Cell. 2012;48(1):109–120. doi: 10.1016/j.molcel.2012.07.028. [DOI] [PubMed] [Google Scholar]
- 36.Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18(5):1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Earley KW, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45(4):616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 38.Dong Y, Burch-Smith TM, Liu Y, Mamillapalli P, Dinesh-Kumar SP. A ligation-independent cloning tobacco rattle virus vector for high-throughput virus-induced gene silencing identifies roles for NbMADS4-1 and -2 in floral development. Plant Physiol. 2007;145(4):1161–1170. doi: 10.1104/pp.107.107391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.