Abstract
Mammalian target of rapamycin (mTOR) is a key protein kinase that regulates cell growth, metabolism, and autophagy to maintain cellular homeostasis. Its activity is inhibited by adverse conditions, including nutrient limitation, hypoxia, and DNA damage. In this study, we demonstrate that Che-1, a RNA polymerase II-binding protein activated by the DNA damage response, inhibits mTOR activity in response to stress conditions. We found that, under stress, Che-1 induces the expression of two important mTOR inhibitors, Redd1 and Deptor, and that this activity is required for sustaining stress-induced autophagy. Strikingly, Che-1 expression correlates with the progression of multiple myeloma and is required for cell growth and survival, a malignancy characterized by high autophagy response.
Keywords: mTOR, autophagy, multiple myeloma, Che-1
Introduction
Mammalian target of rapamycin (mTOR) is an evolutionarily conserved serine/threonine kinase that regulates cellular homeostasis by coordinating anabolic and catabolic processes in the presence of nutrients, energy, and oxygen as well as growth factor signaling (Sarbassov et al, 2005). mTOR forms two distinct signaling complexes called mTORC1 and mTORC2. The mTORC1 complex is responsible for controlling cell growth and protein synthesis, whereas mTORC2 regulates cell survival, metabolism, and cytoskeletal organization (Zoncu et al, 2011). The positive and negative control of mTORC1 activity is exerted through the tuberous sclerosis complex (TSC1/TSC2 complex), a GTPase-activating protein that negatively regulates the Rheb G-protein, which in turn positively regulates mTOR (Inoki et al, 2003; Tee et al, 2003). The 4EBP1 protein and the S6 kinase are two major substrates of mTOR (Ma & Blenis, 2009). The 4EBP1 protein binds and inactivates the translation initiation factor 4E, which is required for the translation of CAP-mRNAs; phosphorylation of 4EBP1 by mTOR inactivates 4EBP1 and allows efficient translation (Ma & Blenis, 2009). Phosphorylation and activation of S6 kinase modulate the functions of translational initiation factors during protein synthesis (Ma & Blenis, 2009). mTORC2 modulates cell survival and anabolism in response to growth factors by phosphorylating many AGC kinase, including Akt1, SGK1, and PKCα (Zoncu et al, 2011).
In all eukaryotes, mTOR is a master regulator that integrates the signals from nutrients and energy sensors with cell growth and proliferation. Nevertheless, mTOR not only controls the rate of protein synthesis, but also regulates transcriptional changes in response to a variety of conditions (Fingar & Blenis, 2004). In recent years, several stimuli have been shown to modify the activity of the mTOR cascade. In addition, decreased mTOR signaling activity has been associated with many types of stress, therefore suggesting that this pathway could play an important role in cells adapting to various stressful conditions (Reiling & Sabatini, 2006). Along with this line, when several types of cellular stress occur, the p53 and ATM pathways negatively regulate mTOR activity (Feng et al, 2007; Budanov & Karin, 2008; Cam et al, 2010). Redd1 and Deptor are two important inhibitors of mTOR activity (Brugarolas et al, 2004; Corradetti et al, 2005; Peterson et al, 2009). Redd1 acts in a TSC2-dependent manner to regulate mTORC1 activity (Brugarolas et al, 2004), whereas Deptor directly binds mTOR kinase and inhibits mTORC1 and mTORC2 activities (Peterson et al, 2009). The stability of Redd1 and Deptor is controlled by the SCFβ-TrCP E3 ubiquitin ligase (Katiyar et al, 2009; Duan et al, 2011; Gao et al, 2011; Zhao et al, 2011). Interestingly, although Deptor and Redd1 depletions strongly activate both mTORC1 and mTORC2 signalings, elevated expression of these genes unexpectedly relieves mTORC1-mediated inhibition of PI3K and activates Akt1 (Peterson et al, 2009; Jin et al, 2013). Furthermore, it has been described that mTOR generates a regulatory loop by controlling Deptor protein and mRNA expression (Peterson et al, 2009; Duan et al, 2011), thus highlighting the critical function of these proteins in mTOR pathway.
Che-1 (also named AATF and Traube) is a protein involved in the regulation of gene transcription by its interaction with RNA polymerase II (Passananti et al, 2007), and its expression is requested for proliferation in early embryogenesis (Thomas et al, 2000). Che-1 also exhibits a strong anti-apoptotic activity (Page et al, 1999; Guo & Xie, 2004; Hopker et al, 2012), and it was found down-regulated during apoptosis through its interaction with MDM2 (De Nicola et al, 2007) and NRAGE (Di Certo et al, 2007). Several findings support the hypothesis that Che-1 plays also an important role in protecting cells from different kinds of stress. In fact, in response to DNA damage, Che-1 is phosphorylated by ATM/Chk2 kinases and recruited on the p53 promoter, consequently activating transcription of this gene and contributing to increase p53 protein levels in response to genotoxic stress (Bruno et al, 2006). It has recently been shown that Che-1 sustains mutant p53 expression in human cancer cells, and its depletion activates DNA damage checkpoint and induces p73 transcription and apoptosis in these cells (Bruno et al, 2010). Moreover, Che-1 results as an antiapoptotic component of endoplasmic reticulum (ER) stress (Ishigaki et al, 2010). Thus, we asked whether Che-1 is also activated by other types of stress and whether Che-1 could be involved in regulating the mTOR pathway.
In this study, we demonstrate that Che-1 is phosphorylated in response to DNA damage, hypoxia, and glucose deprivation and then, once phosphorylated, regulates both mTORC1 and mTORC2 activities. Indeed, Che-1-depleted cells failed to inhibit mTOR activity and cell growth upon several stress conditions. In addition, we determined that Che-1 controls mTOR through the induction of Redd1 and Deptor, two important repressors of mTOR activity (Shoshani et al 2002; Brugarolas et al, 2004; Corradetti et al, 2005; Peterson et al, 2009). Furthermore, we found that Che-1, upon activation, regulates cell survival and autophagy. Finally, we provide evidence that Che-1 expression correlates with multiple myeloma progression, sustaining cell growth and survival. Hence, our study revealed a new link between cellular stress and mTOR signaling and suggests Che-1 to be a promising and attractive drug target for cancer therapy.
Results
Che-1 inhibits mTOR activity
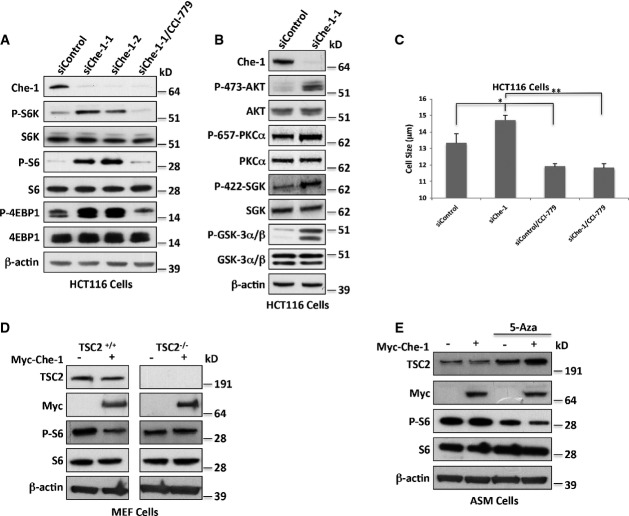
Che-1 was found to play an important role in DNA damage response and cell cycle checkpoint control (Bruno et al, 2006). To determine whether this protein also affects mTOR activity, we evaluated the effect of transient Che-1 overexpression on the phosphorylation of the key mTORC1 targets S6K, 4EBP1, and S6 ribosomal protein, the downstream target of S6K, in HCT116 cells. Ectopic expression of myc-tagged Che-1 dramatically inhibited endogenous T389 S6K and T37/T46 4EBP1 phosphorylation (Supplementary Fig S1A). Similarly, Che-1 also inhibited the phosphorylation of S235/S236 S6 (Supplementary Fig S1A).
To further support the physiological relevance of Che-1 in the regulation of mTOR, we measured the activity of mTORC1 pathway in cells with depleted Che-1 expression. HCT116 cells expressing either of two siRNAs targeting Che-1 showed an increase in S6K, S6, and 4EBP1 phosphorylation, and CCI-779 treatment, a specific mTORC1 inhibitor (Vignot et al, 2005), completely reversed the effect of Che-1 RNAi (Fig1A). The Che-1-mediated inhibition of mTOR activity was also observed in HeLa cells and mouse embryonic fibroblasts (MEF) (Supplementary Fig S1B), indicating that the effect is not specific to HCT116 cells. Notably, Che-1 depletion strongly activated mTORC2 activity as demonstrated by S473 Akt1, S422 GSK, S657 PKCα, and S21/9 GSK-3α/β phosphorylation (Fig1B). Consistent with these results, cells with reduced Che-1 expression exhibited a larger size than control cells, and CCI-779 treatment reversed this phenotype (Fig1C and Supplementary Fig S1C).
Figure 1. Che-1 inhibits mTOR activity.
- A, B Western blot (WB) analysis of total cell extracts (TCEs) from HCT116 cells transiently transfected with siRNA GFP (siControl) or two different siRNA Che-1 (siChe-1-1 and siChe-1-2) and treated where indicated with the mTOR inhibitor CCI-779 (100 nM).
- C Cell size analysis of HCT116 cells transiently transfected as in (A) and treated where indicated with CCI-779. Seventy-two hours after transfection, cell size was measured with a Coulter counter. The data represent the mean ± SD from three independent experiments performed in duplicate. *P < 0.03, **P < 0.001.
- D WB analysis with the indicated Abs of TCEs from TSC2+/+ and TSC2−/− MEF cells transiently transfected with Myc-Che-1 or pCS2-MT control vector.
- E WB with the indicated Abs of TCEs from ASM cells transiently transfected as in (D) and treated where indicated with 1 μM 5-Azacytidine (5-Aza).
Source data are available online for this figure.
Among the upstream regulators of mTORC1 activity, TSC1/TSC2 complex is considered to be the most important regulator, as it converges multiple upstream inputs to regulate mTORC1 activity (Sengupta et al, 2010). To examine the contribution of the TSC1/TSC2 complex to the regulation of mTOR function by Che-1, the effects of Che-1 transient overexpression on TSC2−/− MEFs were analyzed. As shown in Fig1D, Che-1 inhibited S6 phosphorylation in TSC2+/+ cells, whereas it failed to do so in TSC2-deficient cells. To further confirm these results in human cells, we overexpressed Che-1 in an angiomyolipoma cell line (ASM cells) carrying the methylation of the TSC2 gene promoter (TSC2−/Meth) (Lesma et al, 2009). In these cells, Che-1 overexpression did not affect S6 phosphorylation (Fig1E). Conversely, when the DNA methylase inhibitor 5-azacytidine reactivated TSC2 expression, Che-1 overexpression strongly inhibited mTOR activity (Fig1E). Altogether, these results indicate that Che-1 is an inhibitor of mTORC1 and mTORC2 activities.
Che-1 regulates mTOR activity in response to cellular stress
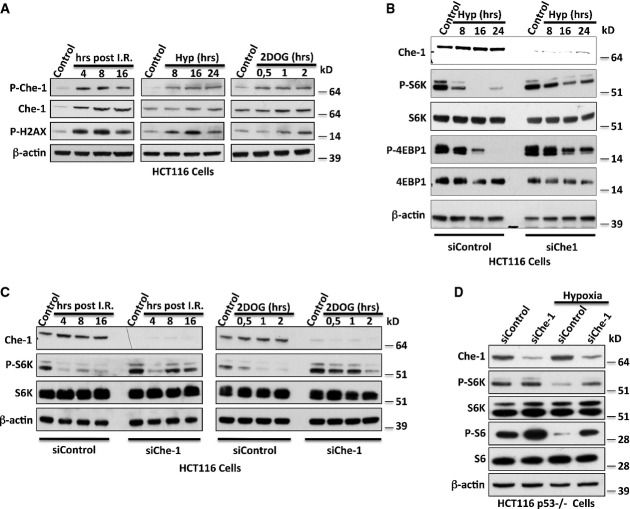
Based on these findings, we hypothesized an involvement of Che-1 in the regulation of mTOR signaling during the response to stress conditions. To test this, we evaluated the activation of Che-1 in response to different inducers of cellular stress (ionizing radiations, hypoxia, or glucose deprivation, the latter induced by using a synthetic glucose analog, i.e., 2-deoxy-glucose 2DOG) by employing a specific antibody directed against the phosphorylated S474 of Che-1 (Bruno et al, 2006). As shown in Fig2A, to a different extent, all these treatments activated Che-1. This effect was essentially the result of Che-1 phosphorylation, because Che-1 transcript was not significantly affected by these treatments, whereas treatments with KU55933, a specific ATM inhibitor, strongly reduced Che-1 protein levels (Supplementary Fig S2A and B). In agreement with these results, histone H2AX phosphorylation was also observed, confirming the induction of checkpoint kinases (Fig2A). Next, we tested whether Che-1 was required for stress-induced down-regulation of mTORC1 activity. For this purpose, HCT116 cells transfected with siRNA Che-1 or siRNA GFP as negative control were subjected to different stress conditions. In contrast to control cells, Che-1-depleted cells showed a reduced down-regulation of S6K and 4EBP1 phosphorylation in response to hypoxia (Fig2B), and similar results were obtained when cells were treated with ionizing radiations (I.R.) or 2-deoxy-glucose (2DOG) (Fig2C). These findings were further confirmed when HeLa and MEF cells were treated under the same conditions (Supplementary Fig S2C and D). Consistently, overexpression of a non-phosphorylable Che-1S4A mutant (Bruno et al, 2006) showed an impaired ability to inhibit S6K and S6 phosphorylation when compared to Che-1 wild-type (Supplementary Fig S2E) and to rescue the effects of endogenous Che-1 depletion on the mTORC1 pathway (Supplementary Fig S2F).
Figure 2. Che-1 regulates mTOR activity in response to cellular stress.
- A WB analysis with the indicated Abs of TCEs from HCT116 treated with I.R. (20 Gy) (left), hypoxia (1% O2) (center), or 2DOG (25 mM) (right).
- B, C WB analysis with the indicated Abs of TCEs from HCT116 cells with siRNA GFP (siControl) or siRNA Che-1 (siChe-1) and treated with hypoxia (B), I.R. (C, left), or 2DOG (C, right).
- D WB analysis of TCEs from HCT116 p53−/− cells treated or not with hypoxia (1% O2) for 16 h.
Source data are available online for this figure.
Several studies have demonstrated that the oncosuppressor p53 inhibits mTORC1 under stressful conditions via multiple mechanisms (Sengupta et al, 2010). Since Che-1 is required to sustain p53 expression in response to DNA damage (Bruno et al, 2006, 2010), we tested whether endogenous p53 was required for mTORC1 regulation by Che-1 using HCT116 p53−/− cells. As shown in Fig2D, HCT116 p53−/− control cells showed a down-regulation of S6K and S6 phosphorylations in response to hypoxia, which was much weaker when Che-1 was depleted, indicating that Che-1 controls mTOR functions by a p53-independent mechanism.
Che-1 induces Redd1 and Deptor expression
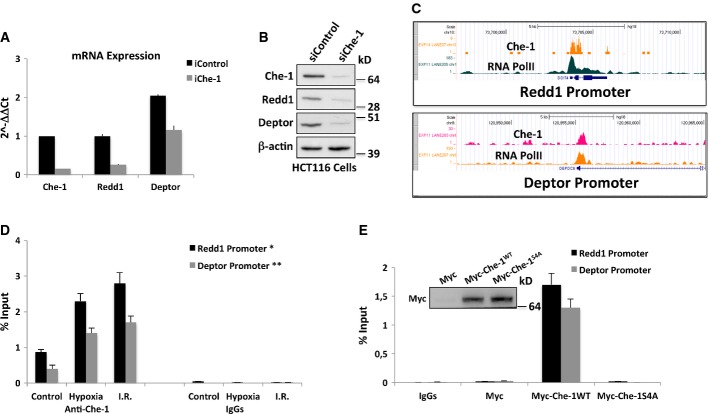
Since Che-1 is a RNA Pol II-binding protein, we hypothesized that it could modulate mTOR signaling in response to various stress conditions by regulating specific gene transcription. Thus, we performed a high-density Affymetrix microarray analysis using HCT116 transiently transfected with control siRNA or Che-1 siRNA (Supplementary Fig S3A). Interestingly, among the down-regulated genes in Che-1-depleted cells, we identified Redd1, Redd2, and Deptor, important genes involved in mTOR regulation (Supplementary Fig S3B) (Brugarolas et al, 2004; Corradetti et al, 2005; Peterson et al, 2009). These data were confirmed by our previous differential expression analysis performed in SKBR3 and MDA-MB468 human mammary carcinoma cells (Bruno et al, 2010; accession number GSE20622) and by quantitative real-time PCR (qRT–PCR) (Fig3A). Furthermore, Western blot analysis showed a significant reduction of Redd1 and Deptor protein levels in Che-1-depleted cells (Fig3B). Consistent with these results, the overexpression of Che-1 strongly induced mRNA and protein levels of both Redd1 and Deptor, whereas the mutant Myc-Che-1S4A did not exhibit any activity on these genes (Supplementary Fig S3C and D). ChIP-seq analysis performed with anti-Che-1 and anti-phospho-S5 RNA polymerase II demonstrated the co-localization of Che-1 and RNA Pol II onto Redd1 and Deptor promoters (Fig3C). Interestingly, quantitative ChIP assays confirmed that Che-1 is physically associated with Redd1 and Deptor promoters in normal proliferating conditions but also showed that its levels increase in response to energy and genotoxic stresses (Fig3D).
Figure 3. Che-1 induces Redd1 and Deptor expressions.
- Quantitative RT–PCR (qRT–PCR) for the indicated genes was performed after transient transfection of HCT116 cells with siRNA GFP (siControl) or siRNA Che-1 (siChe-1). Values were normalized to RPL19 expression. Error bars represent the standard error of three different experiments. P < 0.03
- WB analysis with the indicated Abs of TCEs from HCT116 cells transiently transfected as in (A).
- Genome browser snapshot of Che-1 and phospho-S5 RNA polymerase II ChIP-seq depicting the Redd1 (upper panel) and Deptor (lower panel) promoter regions.
- HCT116 cells treated with hypoxia (1% O2 for 4 h) or I.R. (20 Gy) were subjected to quantitative ChIP analysis (ChIP-qPCR) using anti-Che-1 antibody or control rabbit IgGs. Data are expressed as percent of input. Error bars represent the standard error of three different experiments. *P ≤ 0.001, **P = 0.01.
- HCT116 cells were transiently transfected with pCS2-MT control vector, Myc-Che-1 wild-type, or Myc-Che-1S4A expression vectors and treated with hypoxia (1% O2 for 16 h). Then, cells were subjected to ChIP-qPCR using anti-Myc Ab or control IgGs. Error bars represent the standard error of three different experiments.
To assess whether Che-1 activated the transcription of these genes, and whether this was dependent on its phosphorylation state, we compared the effects of wild-type Che-1 and Che-1S4A mutant on Redd1 and Deptor expression. As shown in Fig3E, under hypoxic conditions, wild-type Che-1 protein but not the mutant Che-1S4A was detectable on Redd1 and Deptor promoters, thus indicating that the phosphorylation of Che-1 is required for its presence on the promoters of these two genes.
Che-1 inhibits mTOR activity through Redd1 and Deptor inductions
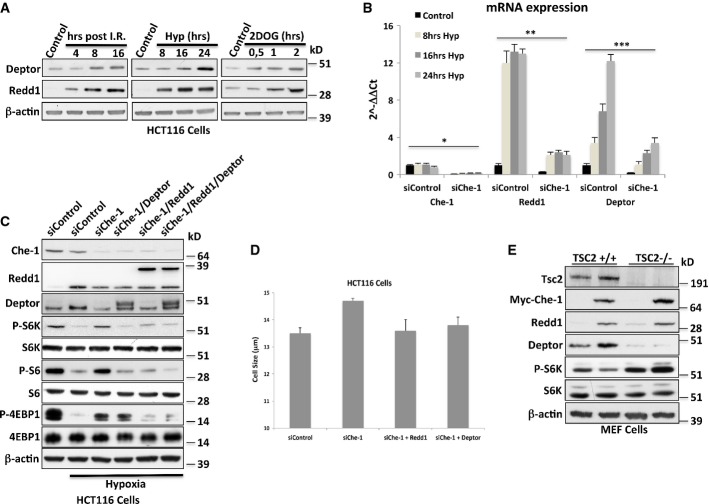
The data presented above indicate that Che-1 regulates the expression of Redd1 and Deptor; we therefore hypothesized that Che-1 might promote mTOR repression through the activation of these two genes. Western blot analysis from HCT116 cells treated with different kinds of stress confirmed Redd1 literature data and revealed that Deptor is also regulated in a similar manner (Fig4A). In accordance with our hypothesis, both Redd1 and Deptor mRNA levels increased in response to hypoxia, but this induction was not observed in Che-1-depleted cells (Fig4B). Consistent with these findings, Che-1, Redd1, or Deptor depletion showed similar effects on cell size and mTORC1 activity in response to hypoxia (Supplementary Fig S4A and B). Cytofluorimetric analysis demonstrated that these effects were not due to changes in cell cycle (Supplementary Fig S4C). Strikingly, restoration of Redd1 and Deptor protein levels counteracted mTOR activation induced by Che-1 ablation (Fig4C and 4D).
Figure 4. Che-1 inhibits mTOR activity through Redd1 and Deptor inductions.
- WB analysis with the indicated Abs of TCEs from HCT116 treated with I.R. (20 Gy) (left), hypoxia (1% O2) (center), or 25 mM 2DOG (right).
- qRT–PCR analysis of the indicated genes from HCT116 cells transiently transfected with siRNA GFP (siControl) or siRNA Che-1 (siChe-1) and exposed to hypoxia (1% O2) for the indicated times. Error bars represent the standard error of three different experiments. *P ≤ 0.01, **P ≤ 0.001, ***P ≤ 0.006.
- WB analysis with the indicated Abs of TCEs from HCT116 transiently transfected with siRNA GFP (siControl), siRNA Che-1 (siChe-1) and where indicated Flag-Deptor and Myc-Redd1 expression vectors. Cells were exposed to hypoxia (1%O2) for 16 h where indicated.
- Cell size analysis of HCT116 cells transiently transfected as in (C). Seventy-two hours after transfection, cell size was measured with a Coulter counter. The data represent the mean ± SD from three independent experiments performed in duplicate. P ≤ 0.02.
- WB with the indicated Abs of TCEs from TSC2+/+ and TSC2−/− MEF cells transiently transfected with Myc-Che-1 or pCS2-MT control vector.
Source data are available online for this figure.
Since Deptor associates with the mTOR complex (Peterson et al, 2009), it is difficult to reconcile the involvement of TSC2 in Che-1 effects on mTOR pathway. However, it has been demonstrated that mTOR generates an auto-amplification loop by regulating Deptor degradation (Duan et al, 2011; Gao et al, 2011). Consistent with these results, Peterson et al showed that TSC2−/− MEF cells exhibit very low levels of Deptor (Peterson et al, 2009). On the basis of these observations, we overexpressed Che-1 in TSC2+/+ and TSC2−/− MEF cells. As shown in Fig4E, Che-1 induced Deptor expression in TSC2+/+ cells, but not in TSC2−/− cells, thus confirming that TSC2 expression is required for mTORC1 inhibition by Che-1. Altogether, these results indicate that in response to cellular stress, Che-1 inhibits mTOR signaling by increasing Redd1 and Deptor expression.
Che-1 regulates autophagy through Deptor and Redd1
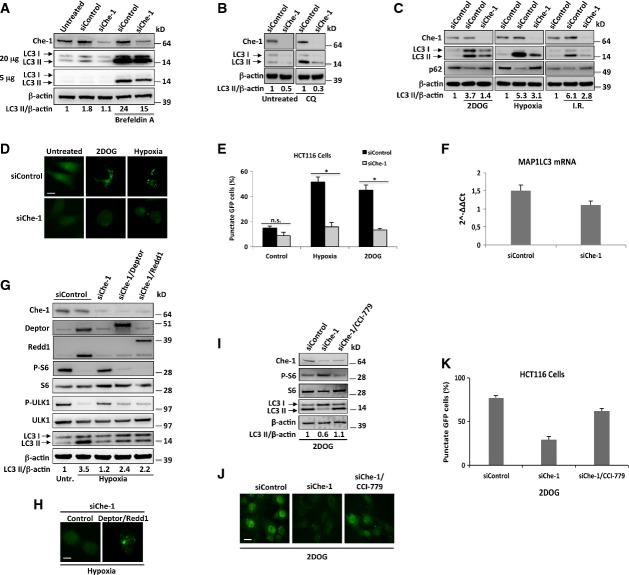
mTOR signaling pathway is a critical negative regulator of autophagy, and several kinds of stress such as glucose deprivation induce autophagy via inhibition of mTOR (Jung et al, 2010; Neufeld, 2010). Given that recent studies have reported that Deptor and Redd1 induced autophagy through suppression of mTOR activity in response to certain kinds of stress (Gao et al, 2011; Molitoris et al, 2011; Zhao et al, 2012), we evaluated whether Che-1 was involved in autophagy induction. For this purpose, Che-1-depleted HCT116 cells were treated with the autophagy inducer brefeldin A and the autophagy induction was evaluated by accumulation of LC3-II, a well-established hallmark of autophagy. As shown in Fig5A, Che-1 depletion reduced LC3-II accumulation and these effects were observed to be even greater in response to brefeldin A. To assess whether these results were due to a reduced activation of autophagy and not rather from an increased degradation of LC3-II, we treated HCT116 cells with chloroquine (CQ), an inhibitor of autophagic lysosomal protein degradation. Even in this case, Che-1-depleted cells exhibited a significant decrease of LC3-II accumulation (Fig5B), thus indicating that Che-1 is involved in autophagy induction. Next, we evaluated whether stress conditions required Che-1 expression to induce autophagy. As shown in Fig5C, glucose deprivation, hypoxia, and I.R. caused LC3-II accumulation in HCT116 cells concomitant with a decrease in p62/SQSTM1 (p62) abundance, as expected for cells experiencing an increase of autophagic flux (Klionsky et al, 2012). Notably, in response to each treatment, Che-1 depletion strongly reduced autophagy induction supported by the increased p62 levels concomitant with reduced LC3-II levels (Fig5C). These results were confirmed when HCT116 cells were transiently transfected with a GFP-LC3 expression vector, control siRNA, or Che-1 siRNA and treated with hypoxia or 2DOG. Cellular stresses strongly increased the percentage of cells with a punctate GFP-LC3 pattern in control siRNA cells, whereas Che-1 depletion significantly reduced this pattern (Fig5D and E). In these experiments, Che-1 depletion also produced a reduction of LC3-I, although lower than that observed for LC3II, suggesting that Che-1 could also control LC3 (MAP1LC3) gene expression. Data from Affymetrix analysis revealed a slight reduction of MAP1LC3 mRNA expression, and this was confirmed by qRT–PCR (Fig5F). To evaluate whether the reduction of autophagy after Che-1 inhibition involved Deptor and Redd1 down-regulations, we depleted Che-1 in HCT116 cells in the presence or in the absence of Deptor or Redd1 overexpression and treated cells with hypoxia. As shown in Fig5G, Che-1 depletion increased the phosphorylation of both S6 and ULK1, a target of mTOR involved in autophagic activation (Kim et al, 2011), and reduced autophagy induction. On the contrary, overexpression of Deptor or Redd1 restored mTOR inhibition and the induction of autophagy in response to hypoxia (Fig5G and H). Consistent with these results, CCI-779 treatment completely overcame the effects of Che-1 depletion in cells treated with 2DOG (Fig5I–K).
Figure 5. Che-1 regulates autophagy through Deptor and Redd1.
- A–C WB analysis with the indicated Abs of TCEs from HCT116 transiently transfected with siRNA GFP (siControl) or siRNA Che-1 (siChe-1) and treated where indicated with brefeldin A (5 μg/ml) (A), chloroquine (100 μM) (CQ) (B) or 2DOG (25 mM for 8 h) (left), hypoxia (1% O2 for 16 h) (center), or I.R. (20 Gy) (right) (C).
- D HCT116 cells transiently transfected with Stealth siRNA negative controls (siControl) or siRNA Che-1 (siChe-1) and GFP-LC3 expression vector were treated where indicated with 2DOG (25 mM for 8 h) or hypoxia (1% O2 for 16 h). Cells were fixed with 4% formaldehyde and analyzed by fluorescence microscopy. Scale bar, 10 μm.
- E HCT116 cells were transiently transfected and treated as in (D). Twenty-four hours later, cells were fixed and GFP-LC3 distribution was assessed by fluorescence microscopy and percentages represent punctate-LC3 expressing cells. The data represent the mean ± SD from three independent experiments performed in triplicate (n = 150 for each condition). n.s., not significant, *P ≤ 0.001.
- F Quantitative RT–PCR (qRT–PCR) for MAP1LC3 mRNA expression was performed after transient transfection of HCT116 cells with siRNA GFP or siRNA Che-1 (siChe-1). Values were normalized to RPL19 expression. Error bars represent the standard error of three different experiments. P = 0.02.
- G WB analysis with the indicated Abs of TCEs from HCT116 cells transiently transfected with Stealth siRNA negative controls (siControl), siRNA Che-1 (siChe-1), and, where indicated, with Flag-Deptor and Myc-Redd1 expression vectors, exposed to normoxia or hypoxia (1% O2 for 16 h).
- H Fluorescence microscopy analysis of HCT116 cells transiently transfected with siRNA Che-1 (siChe-1), GFP-LC3, and, where indicated, with Flag-Deptor and Myc-Redd1 expression vectors, treated with hypoxia. Scale bar, 10 μm.
- I WB analysis with the indicated Abs of TCEs from HCT116 transiently transfected with siRNA GFP (siControl) or siRNA Che-1 (siChe-1), treated with 2DOG (25 mM for 8 h), and, where indicated, with 100 nM CCI-779.
- J HCT116 cells were transiently transfected with GFP-LC3, and Stealth siRNA negative controls (siControl) or siRNA Che-1 (siChe-1), and treated as in (I). Cells were fixed with 4% formaldehyde and analyzed by fluorescence microscopy. Scale bar, 10 μm.
- K HCT116 cells were transiently transfected and treated as in (J). Twenty-four hours later cells were fixed, and GFP-LC3 distribution was assessed by fluorescence microscopy and percentages represent punctate-LC3 expressing cells. The data represent the mean ± SD from three independent experiments performed in triplicate (n = 140 for each condition). P ≤ 0.0002.
Source data are available online for this figure.
Altogether, these results indicate that Che-1 regulates autophagy in response to cellular stress, through the induction of Deptor and Redd1 expressions and mTOR inhibition.
Che-1 expression correlates with multiple myeloma progression
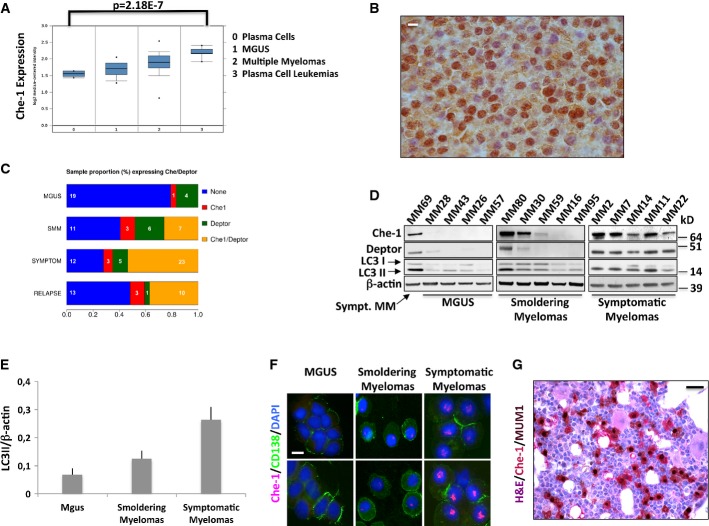
The data described above indicate that Che-1 is required to inhibit mTOR activity and cell growth in response to several stresses by regulating Redd1 and Deptor expressions. Surprisingly, Sabatini's group described that Deptor is overexpressed in a subset of human multiple myelomas (MM). Indeed, in this type of hematological malignancy, overexpression of Deptor is necessary for maintaining Akt1 activation and cell survival, most likely by inhibiting a negative feedback loop induced by mTORC1, which is also inhibited by Deptor (Peterson et al, 2009). These findings prompted us to investigate whether Che-1 overexpression might have the same effect on Akt1 activity. To this aim, HCT116 cells were transfected with Myc-Che-1 expression vector. As shown in Supplementary Fig S5A, overexpression of Che-1 led to an increase of mTORC2 signaling as monitored by the phosphorylation of its targets. Interestingly, in agreement with the previous results, the activation of Akt1 was not observed in cells transfected with Che-1S4A mutant (Supplementary Fig S5B). Next, we evaluated Che-1 expression in human MM cells. The analysis of Oncomine database (Agnelli et al, 2009) and other public datasets (GSE2658 and GSE5900 combined, Zhan et al, 2006) indicated that Che-1, Redd1, and Deptor mRNA expressions correlate with MM progression (Fig6A and Supplementary Fig S5C). In agreement with these findings, immunohistochemical analysis revealed high levels of Che-1 expression in MM cells (Fig6B). To further confirm these data, we analyzed Che-1 and Deptor expressions in 120 human primary MM samples. Western blot analysis of CD138-sorted plasma cell fractions derived from monoclonal gammopathies of undetermined clinical significance (MGUS) and MM patients revealed almost undetectable Che-1 and Deptor protein expression levels in the plasma cells from 23 MGUS samples, but their increasing and widespread expression in 9/27 (33%) of smoldering myeloma and 27/43 (62%) of symptomatic myeloma samples (Fig6C and D). Consistent with these results, the analysis of 559 MM from the above dataset (Zhan et al, 2006) revealed a linear correlation between Che-1 and Deptor mRNA expressions (Supplementary Fig S5D). A similar correlation was not found between Che-1 and Redd1 mRNA expressions (Supplementary Fig S5D); however, Western blot analysis of primary MM showed an increase of Redd1 expression during disease progression and its correlation with Che-1 levels (Supplementary Fig S5E). Notably, autophagy levels in these samples strongly correlated with Che-1 and Deptor expressions (Fig6E and Supplementary Fig S5F). Immunofluorescence analysis of 18 samples of CD138+ MM cells confirmed Western blot data, showing a progressive shift toward increased Che-1 expression in the majority of symptomatic myelomas in comparison with MGUS and smoldering myeloma samples (Fig6F). To further confirm the correlation between Che-1 and MM, we analyzed Che-1 expression in bone marrow samples from Vk*MYC mice. Indeed, these animals develop MM with age with the biological and clinical features highly characteristic of the human disease (Chesi et al, 2008). Immunohistochemical assay of these samples revealed high levels of Che-1 expression in MM cells, compared to the residual bone marrow (Fig6G). Altogether, these observations strongly suggest that increased levels of Che-1 may contribute to the development of MM.
Figure 6. Che-1 expression correlates with multiple myeloma progression.
- Data from Agnelli et al (2009) provided by the Oncomine database and reanalyzed to show expression levels of Che-1 in normal bone marrow, MGUS, MM, and plasma cell leukemia (n = 158). The associated P-value is shown above. Box and whisker plot shows the upper and lower quartiles (25–75%) with a line at the median, whiskers extend from the 10th to the 90th percentile, and dots correspond to the minimal and maximal values.
- Immunohistochemical analysis for Che-1 expression was performed on bone marrow section from a symptomatic myeloma. Scale bar, 20 μm.
- Percentage of positive Che-1 and Deptor protein expression in plasma cells isolated from the bone marrow of 120 MM patient samples using CD138 magnetic beads.
- Representative WB analysis with the indicated Abs of TCEs from CD138-purified MM primary tumors.
- LC3II/β-actin expression ratio of 120 MM patient samples. The data represent the mean ± SD from the ratio of every single sample.
- Immunofluorescence analysis of Che-1 expression in plasma cells purified from primary myelomas. Nuclei were visualized by staining with Hoechst dye. Scale bar, 20 μm.
- Immunohistochemical analysis for Che-1 expression was performed on bone marrow section from a 113-week-old Vk*MYC mouse. Che-1 is stained in red and MUM1 in brown. Scale bar, 50 μm.
Source data are available online for this figure.
Che-1 sustains survival in multiple myeloma cells
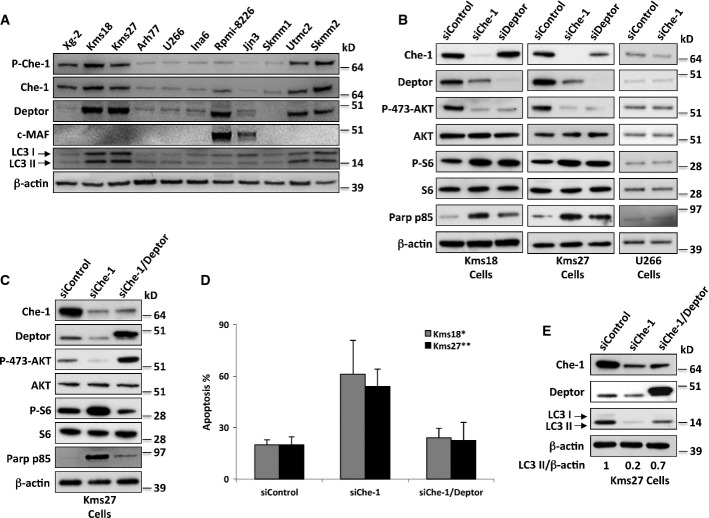
On the basis of the correlation between Che-1 levels and MM progression as well as its involvement in the control of mTOR pathway and autophagy induction, we evaluated its relevance in MM growth and survival. To this aim, we analyzed Che-1 levels in different MM cell lines and we found that lines with high levels of Che-1 (i.e., Kms18, Kms27, Utmc2, and Skmm2) showed higher levels of Deptor expression and autophagy (Fig7A and Supplementary Fig S6A).
Figure 7. Che-1 sustains survival in multiple myeloma.
- WB analysis with the indicated Abs of TCEs from the indicated human MM cell lines.
- WB analysis with the indicated Abs of TCEs from Kms18, Kms27, and U266 cells transiently transfected where indicated with siRNA GFP (siControl), siRNA Che-1 (siChe-1), or siRNA Deptor (siDeptor).
- Kms27 cells were transiently transfected with siRNA GFP (siControl), siRNA Che-1 (siChe-1), and, where indicated, Flag-Deptor expression vector. WB analysis of TCEs from these cells was performed with the indicated Abs.
- Kms27 and Kms18 cells were transfected as in (C). Cell death was assayed by trypan blue staining 20 h after the transfection, and percentages represent trypan blue-incorporating cells. Data are presented as the mean ± SD from three independent experiments performed in duplicate. *P < 0.001, **P < 0.002.
- Kms27 cells were transiently transfected as in (C). WB analysis of TCEs from these cells was performed with the indicated Abs.
Source data are available online for this figure.
Deptor was found up-regulated in a subset of newly diagnosed myelomas with cMaf/MafB translocations (Peterson et al, 2009). To evaluate a possible correlation between Che-1 and c-MAF translocations in this disease, we analyzed the same public datasets described above (Zhan et al, 2006). From this analysis, we did not observe a significant Che-1 up-regulation in this particular subset (Supplementary Fig S6B). However, when we expanded the analysis of multiple myeloma cell lines, we found that several cell lines lacking c-MAF overexpression (Hurt et al, 2004) exhibited high levels of Che-1 and Deptor (Fig7A). These results were confirmed when we evaluated c-MAF expression in symptomatic myeloma patients expressing Che-1 and Deptor (Supplementary Fig S6C), indicating that Deptor expression is activated in multiple myeloma by different mechanisms. These findings were consistent with a cytogenetic analysis, which showed that only 20% of MM patients with high levels of Che-1 and Deptor displayed c-MAF translocations (M. Fanciulli, unpublished observation). Next, we investigated whether Che-1 controls autophagy and survival in MM cells. Inactivation of Che-1 or Deptor in Kms18 and Kms27 cells produced similar effects, increasing mTORC1 activity and inactivating mTORC2 complex (Fig7B), but Akt1 activity was not repressed when Kms27 Che-1-depleted cells were treated with CCI-779 (Supplementary Fig S6D). Accordingly, Che-1 depletion in these cells increased cell size (Supplementary Fig S6E) and induced apoptosis (Fig7B and D, and Supplementary Fig 6F). Notably, overexpression of Deptor in Che-1-depleted Kms27 cells reversed mTORC1 activation, mTORC2 inhibition, cell growth, and apoptosis (Fig7C–E, Supplementary Fig S6E and F). In agreement with these results, both Che-1 overexpression and its depletion did not affect mTOR activity and apoptosis in U266 cells, which exhibited low levels of Deptor (Fig7B, Supplementary Fig S6G and H). In these cells, ChIP experiments demonstrated the absence of Che-1 on the Deptor promoter (Supplementary Fig S6I). Next, we evaluated whether Che-1 could be involved in the autophagy observed in MM cells. To this aim, MM cells were transfected with control siRNA or Che-1 siRNA. As shown in Fig7E and Supplementary Fig S6J, Che-1 depletion strongly reduced autophagy levels in these cells. Consistently, the rescue of Deptor expression in Che-1-depleted cells restored high levels of LC3-II (Fig7E and Supplementary Fig S6K). Finally, to confirm that Che-1 regulates autophagy by inhibiting mTORC1 activity, we depleted Che-1 expression in Kms27 cells and treated them with different concentrations of CCI-779. As shown in Supplementary Fig S6L and D, treatments with low doses of this mTORC1 inhibitor were able to protect MM cells from death induced by Che-1 knockdown, whereas high doses of CCI-779 exhibited a strong toxicity.
Among the most effective class of compounds used in the treatment of multiple myeloma, there are proteasome inhibitors that are capable of inducing cell death through different mechanisms (Hideshima et al, 2011), including prominently the induction of ER stress, leading to the activation of apoptosis (Obeng et al, 2006). Thus, we investigated whether Che-1 silencing might enhance the effect of these compounds on MM cells. Che-1 depletion in Kms27 cells strongly sensitized these cells to bortezomib, a proteasome inhibitor largely utilized in the treatment of MM (Supplementary Fig S6M), and similar results were obtained from Kms18 cells (T. Bruno & M. Fanciulli, unpublished observation). Altogether, these data strongly indicate a critical role of Che-1 in MM survival.
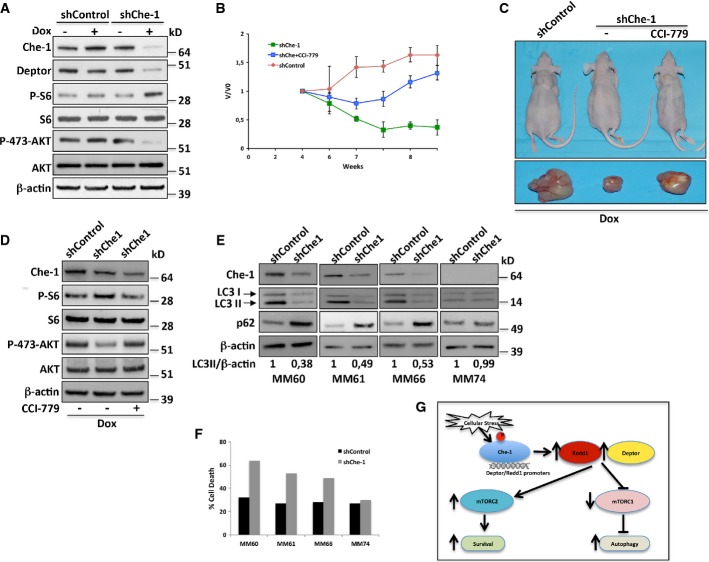
Che-1 depletion suppresses multiple myeloma growth in vivo
The results described above indicate that Che-1 is required to sustain autophagy and to maintain survival in MM cells. To confirm in vivo these results, we generated a conditional RNAi model by infecting Kms27 and U266 with either a doxycycline (Dox)-inducible lentiviral vector carrying a specific hairpin (sh) RNA against Che-1 (Kms27 ind-shChe-1) or with a vector carrying a control hairpin (Kms27 ind-shControl) (Bruno et al, 2010). When these cells were treated with Dox, Kms27 ind-shChe-1, but not Kms27 ind-shControl cells, exhibited a strong decline in Che-1 and Deptor levels and confirmed Che-1 involvement in mTOR activity (Fig8A). On the contrary, Che-1 depletion in U266 cells did not produce any effect (Supplementary Fig S7A). Next, either ind-shChe-1- or ind-shControl-engineered cells were implanted subcutaneously in nude immunodeficient mice. Once injected cells had generated tumor nodules (0.8 cm3), animals were treated with Dox (2.0 g/l) for 5 weeks. As shown in Fig8B and C, ablation of Che-1 in Kms27 cells had an impact on tumor growth in comparison to control mice, producing a considerable reduction in tumor size. Notably, this effect was strongly counteracted by treating mice with CCI-779 (Fig8B and C), thus confirming in vivo that Che-1 sustains MM cell growth by inhibiting mTORC1 activity. Consistent with these findings, a Western blot analysis of these tumors revealed an increase of p-S6 levels with a concomitant decrease of p-Akt1 levels in Che-1-depleted cells, but these effects were partially reversed in CCI-779-treated mice (Fig8D). On the contrary, Che-1 depletion in U266 cells did not exhibit similar results (Supplementary Fig S7B and C). To substantiate these results, we isolated CD138+ neoplastic cells from patients with symptomatic myeloma and co-cultured the neoplastic plasma cells with autologous stromal cells for 24 h before infecting them with shChe-1 or shControl lentiviral vectors (Supplementary Fig S7D). In addition to Che-1 depletion, shChe-1 transduction in Che-1-expressing cells produced a reduction in autophagy induction (Fig8E) with a concomitant increased rate in cell death (Fig8E). These effects were not observed in a MM sample in which Che-1 was not expressed (Fig8E and F). Taken together, these findings strongly indicate that Che-1 maintains the viability of myeloma cells and suggest it as a candidate therapeutic target of this disease.
Figure 8. Che-1 depletion suppresses multiple myeloma growth in vivo.
- A Kms27 cells were infected with LV-THsh/Che-1 (shChe-1) or LV-THsh/Control (shControl) and LV-tTR-KRAB lentiviruses and TCEs from cells induced (+Dox) or not (−Dox) were analyzed by WB with the indicated Abs.
- B–D (B) Engineered Kms27 ind-shChe-1 and Kms27 ind-shControl cell lines were implanted subcutaneously into nu/nu mice, and animals were treated as reported (see text). Animals were weekly monitored as reported in Materials and Methods. Relative tumor growth was estimated by the formula V/V0, where V represents tumor volume at different times post-treatment, whereas V0 represents tumor volume before treatment. The graphic is representative of two independent experiments with similar results. (C) Representative mice (top) or excised tumors (bottom) from induced (+Dox) Kms27 ind-shChe-1 and Kms27 ind-shControl implanted cells treated or not with CCI-779. (D) Western blot analysis of excised tumors performed by using the indicated Abs.
- E, F CD138+ neoplastic cells from patients with symptomatic myeloma were cocultured with stromal cells from the same patient for 24 h before infecting them with shChe-1 or shControl lentiviral vectors. After 96 h, TCEs from MM primary cells transfected with siControl or siChe-1 were analyzed by WB with the indicated Abs (E), and the relative number of viable myeloma cells was quantified by FACS (F).
- G Model to explain Che-1 involvement in the control of mTOR activity. In response to cellular stress, Che-1 is activated and recruited onto Redd1 and Deptor promoters inducing their expression. This results in a simultaneous decrease of mTORC1 activity and increase of mTORC2 activity, allowing induction of autophagy and survival.
Source data are available online for this figure.
Discussion
The mTOR signaling pathway plays a central role in cell growth and survival control (Laplante & Sabatini, 2012). Therefore, it is not surprising that several stress conditions regulate mTOR activity, thereby allowing cells to survive under non-optimal conditions (Sengupta et al, 2010). In particular, it has been demonstrated that the tumor suppressor p53 upon several kinds of stress can repress mTOR through multiple mechanisms (Ellisen et al, 2002; Feng et al, 2007; Budanov & Karin, 2008).
In the present study, we provide evidence that Che-1 negatively controls mTOR activity in a p53-independent way, consequently requiring TSC2. We show that Che-1 is activated by several stress conditions and inhibits mTOR pathway by inducing Redd1 and Deptor expression, two important inhibitors of mTOR. Furthermore, inhibition of Che-1 strongly decreases autophagy leading to apoptosis. We also document overexpression of Che-1 during MM progression and a positive correlation between Che-1 and Deptor expressions in several human symptomatic myelomas, thus providing evidence that Che-1 sustains cell growth and survival in MM cells.
Our results demonstrate that not only DNA damage but also other stress conditions lead to Che-1 phosphorylation and activation (Fig2A and Supplementary Fig S2B). Interestingly, we found histone H2AX phosphorylation in response to hypoxia and 2DOG treatments (Fig2A), thus suggesting that this kind of stresses can activate DNA damage response. Several studies have demonstrated that hypoxia can activate ATM and Chk2 kinase activities (Gibson et al, 2005; Cam et al, 2010), whereas the effects of glucose deprivation on DNA integrity have not yet been clarified. Nevertheless, we can speculate that metabolic alterations generated by 2DOG treatment might produce oxidative DNA damage and activate checkpoint kinases.
We show that Che-1 regulates Redd1 and Deptor gene expression (FigA and B, Supplementary Fig S3A, C and D). We found that Che-1 is recruited onto their promoters together with RNA polymerase II (Fig3C) and that this recruitment is mediated by phosphorylation, which increases in response to stress (Fig3D and E). Strikingly, overexpression of Redd1 and Deptor in Che-1-depleted cells completely restored mTOR inhibition (Fig4C and D). Interestingly, a recent analysis of the mTOR-dependent phosphoproteome has identified the murine ortholog of Che-1 as a potential mTOR effector (Yu et al, 2011). These findings were consistent with a direct interaction between Che-1 and mTOR (T. Bruno & M. Fanciulli, unpublished observation), allowing to speculate that Che-1 like Deptor might be a mTOR target and participate in the mTOR auto-amplification loop recently described (Duan et al, 2011). Che-1 inhibits mTORC1 in a TSC2-dependent manner (Fig1C and D). While this finding is in agreement with Redd1 induction by Che-1, it is not easily reconcilable with Deptor activity. Nevertheless, Peterson et al showed that TSC2−/− MEF cells exhibit very low levels of Deptor (Peterson et al, 2009). Further supporting these findings, we showed that Che-1 induces Deptor expression in TSC2+/+ cells, but not in TSC2−/− cells, which presented almost undetectable levels of Deptor even before Che-1 overexpression (Fig4E).
Our findings demonstrate that Che-1 inhibition decreases autophagy activation in response to cellular stress (Fig5A and B), and this phenomenon is lost when Redd1 and Deptor expression are restored (Fig5G and H) or when cells are treated with a mTORC1 inhibitor (Fig5I–K). This indicates that Che-1 plays a role in the autophagy induction, partially through regulating the activity of mTOR signaling, a central regulator of this pathway (Jung et al, 2010; Neufeld, 2010). Autophagy is a tightly regulated pathway involving the massive degradation of cellular organelles or cytosolic components (Kroemer et al, 2010). This pathway constitutes a major mechanism that allows cells to survive in response to several stressors, and many molecular events support a mutual exclusion between autophagy and apoptosis (Kroemer et al, 2010). In agreement with this concept, our results enable us to propose a model in which Che-1 is an important regulator of the balance between autophagy and apoptosis in response to cellular stress, promoting growth arrest and survival and inhibiting cell death (Fig8F). At the same time, Che-1 may contribute to prevent energy crisis under stress conditions, through maintaining low mTORC1 activity and high Akt1 activity, thus, reducing energy consumption while promoting energy production.
We further report overexpression of Che-1 during MM progression (Agnelli et al, 2009). This finding is rather intriguing, as Che-1 has been described to play a relevant role in protecting cells in ER stress/unfolded protein response (UPR) (Ishigaki et al, 2010), which are able to render MM cells resistant to proapoptotic stresses (Shapiro-Shelef & Calame, 2004). Therefore, the identification of a new important mechanism that favors MM-specific survival may be critical in gaining further insights into the disease. Our results show that Che-1 and Deptor are poorly expressed in most monoclonal gammopathies and smoldering myelomas, whereas in about 62% of symptomatic diseases, we found high levels of these proteins. Therefore, it is tempting to speculate that high levels of Che-1 and Deptor are required for the survival of MM cells, and additionally, they may be considered useful markers to monitor disease evolution.
Deptor is up-regulated in a subset of newly diagnosed myelomas with cMAF/MAFB overexpression (Peterson et al, 2009). Che-1 expression was not found increased in this particular subset (Supplementary Fig S6A). Nevertheless, our results indicate that both Che-1 and Deptor are up-regulated in several MM cell lines and patients samples without cMAF overexpression (Fig7A and Supplementary Fig S6B), thus suggesting that several mechanisms may contribute to the high levels of Deptor observed in MM.
We found a high level of autophagy in symptomatic myelomas (Fig6D). This agrees with the evidence showing that ER stress triggers autophagy to sustain cell viability, especially in cancer cells (Yorimitsu et al, 2006; Suh et al, 2012). In addition, recent studies have demonstrated that autophagy is specifically required for plasma cell homeostasis by limiting ER expansion (Pengo et al, 2013) and that inhibition of autophagy cell death through caspase 10 is essential for myeloma cell viability (Lamy et al, 2013). Consistent with these observations, high levels of Che-1 in MM may be required for modulating autophagy response, preventing it from inducing cell death (Ishigaki et al, 2010). Therefore, it is possible to hypothesize a scenario in which increasing levels of ER stress/UPR during MM progression induce Che-1 and Deptor expression to inhibit mTORC1 activity, thereby activating autophagy and promoting cell survival by Akt1 activation (Fig8G). Moreover, it has been recently demonstrated that inactivation of Xbp1, an important gene involved in UPR, contributes to therapeutic resistance in MM (Leung-Hagesteijn et al, 2013). Since Che-1 was found less expressed in relapsed samples than in untreated samples (Fig6C), it is also possible to speculate that MM cells, in which Che-1 is poorly expressed or inactivated, can acquire an advantage in response to chemotherapeutic treatments.
In conclusion, these findings highlight the important role that Che-1 plays in response to many stress conditions, regulating processes such as cell growth, autophagy, and apoptosis, further strengthening the notion that Che-1 could be considered a valid target for novel therapeutic approaches.
Materials and Methods
Cell lines, constructs, and transfections
HCT116 wild-type and p53−/−, 293T packaging cells, HeLa, MEF, MEF TSC2+/+ p53−/−, and MEF TSC2−/− p53−/− cells (kindly provided by Dr. D. Kwitakowski, Harvard Medical School) were cultured in DMEM supplemented with 10% FBS. Angiomiolipoma cells TSC2−/meth ASM (9C) were cultered in DMEM/F12 supplemented with 15% FBS, EGF 10 ng/ml, and hydrocortisone 0.1 nM. U266, Kms18, Kms27, Rpmi-8226, Arh77, Xg-2, Jjn3, Ina6, Skmm1, Utcm2, and Skmm2 multiple myeloma cell lines were cultered in Optimem supplemented with 15% FBS. All cell lines were periodically tested for mycoplasma contamination by RT–PCR analysis. For hypoxia exposure, culture dishes were placed in a hypoxia chamber (Fisher Scientific) allowing the formation of a hypoxic environment of 5% CO2, 95% N2. Unless stated otherwise, these hypoxic levels (1% of oxygen concentration, 16 h) were used in all experiments. Transfections were carried out by Lipofectamine 2000 (Life Technologies) following the manufacturer's instructions. The following plasmids were used in transfection experiments: pRK5 human Flag-Deptor (Addgene, ID 21334), Myc-tagged Che-1, and its mutant Myc-Che-1S4A were described elsewhere (Bruno et al, 2006). Myc-tagged Redd1 was generated by cloning Redd1 cDNA into the pCS2-MT vector. GFP-LC3 expression vector was kindly provided from Dr. Hovard (Martens et al, 2005). The mTOR inhibitor CCI-779 was purchased from Chemocare, whereas 2DOG, 5-azacytidine, doxycycline, chloroquine (diphosphate salt), brefeldin A, and KU55933 were purchased from Sigma.
Cell extracts and Western blot analysis
Cell extracts purifications and Western blotting analyses were performed as described (Bruno et al, 2006) by using the following antibodies:
Rabbit polyclonal antibodies: anti-Che-1 (Fanciulli et al, 2000) WB 1:1,000, IF 1:500; PARP-1 p85 fragment (Parp p85) (Promega, G7341) WB 1:1,000; Deptor (Upstate/Millipore, 09-463) WB 1:500, IF 1:100; Redd1 (Proteintech group, 10638-1-AP) WB 1:1,200; Ulk1 (Santa Cruz, sc-33182, clone H-240) WB 1:500; SGK phospho-S422 (Santa Cruz, sc-16745) WB 1:250; PKCα phospho-S657 (Santa Cruz, sc-12356) WB 1:250; c-MAF (Santa Cruz, sc-7866, clone M-153) WB 1:500; S6K phospho-T389 (Cell Signaling, Sampler Kit #9862, clone 108D2) WB 1:1,000; 4E-BP1 phospho-T37/T46 (Cell Signaling, Sampler Kit #9862) WB 1:1,000; S6 p-S235/236 (Cell Signaling, #4858, clone D57.2.2E) WB 1:1,000; Akt1 phospho-S473 (Cell Signaling, #4063, clone D9E) WB 1:1,000; Akt1 phospho-T308 (Cell Signaling, #13038, clone D25E6) WB 1:1,000; GSK α/β phospho-S21/9 (Cell Signaling, #4858, clone D57.2.2E) WB 1:1,000; Ulk1 phospho-S757 (Cell Signaling, #6888) WB 1:1,000; TSC2 (Cell Signaling, #3612) WB 1:1,000; 4E-BP1 (Cell Signaling, #9644, clone 53H11) WB 1:1,000; Akt1 (Cell Signaling, #2938, clone C73H10) WB 1:1,000; S6K (Cell Signaling, #2708, clone 49D7) WB 1:1,000; ATM (Cell Signaling, #2873, clone D2E2) WB 1:1,000; and LC3B (Cell Signaling, #3868S, clone D11) WB 1:1,000, IF 1:200. Mouse monoclonal antibodies: anti-Myc (Life Technologies, #13-2500, clone 9e10) WB 1:1,000; β-actin (Sigma, A5441, clone AC-15) WB 1:10,000; Flag M2 (Sigma, F1804) WB 1:1,000; SGK (Santa Cruz, sc-377360, clone G-4) WB 1:250; PKCα (Santa Cruz, sc-8393, clone H-7) WB 1:250; p62/SQSTM1 (Santa Cruz, sc-28359, clone D-3) WB 1:200; ATM phospho-S1981 (Cell Signaling, #4526, clone 10H11.E12) WB 1:1,000; histone H2A.X phospho-S139 (Merck-Millipore, 05-636, clone JBW301) WB 1:1,000; S6 (Cell Signaling, #2317, clone 54D2) WB 1:1,000; and CD138, (Dako, M7228, clone MI15) IF 1:25. Goat polyclonal MUM1 (Santa Cruz, sc-139011, clone N-18) IHC 1:200. Affinity-purified polyclonal anti-P474-Che-1 was produced by Phosphosolutions. Secondary antibodies used were goat anti-mouse and goat anti-rabbit, conjugated to horseradish peroxidase (Bio-Rad). Immunostained bands were detected by the chemiluminescent method (Pierce).
Immunofluorescence
Immunofluorescence analyses of anti-Che1, anti-Deptor, or anti-LC3 on CD138+ positive cells were performed using cytospin preparations of cells mounted on glass slides using a Thermo Shandon cytospin 2 (Thermo Fisher Scientific), whereas transfected HCT116 cells were subjected to immunofluorescence assays 48 h following transfection. Briefly, cells were fixed in 4% formaldehyde for 15 min and then permeabilized with 0.1% Triton X-100 in phosphate-buffered saline (PBS) for 5 min. Primary antibodies were used for immunostaining, followed by Alexa-Fluor-594-conjugated and Alexa-Fluor-488-conjugated anti-rabbit IgG (Life Technologies). Nuclei were visualized by staining with 1 μg/ml Hoechst dye 33258 (Sigma). As a control, the primary antibodies were omitted (data not shown). Immunofluorescence analysis was performed using the microscope Axioskop 2 plus, and fluorescence signals were analyzed by recording images using a CCD camera (Zeiss, Oberkochen, Germany).
Immunohistochemistry
For double immunohistochemical staining, the following procedure was adopted. Briefly, formalin-fixed, paraffin-embedded 4-μm-thick sections underwent staining with IRF4/MUM1 monoclonal antibody (Santa Cruz, clone sc-56713, dil 1:1500, Ag retrieval with Tris-EDTA, pH 9, at 98°C for 10 min); slides were washed in TBS and developed in diaminobenzidine. Subsequently, polyclonal anti-Che1 was applied (dil 1:400, Ag retrieval with citrate, pH 6, at maximum MW for 5 min). Sections were counterstained with hematoxylin. Proper negative controls were used.
siRNA
The 22-nt siRNA duplexes corresponding to nucleotides 1062–1083 (siChe-1-1), 1473–1492 (siChe-1-2) and 2093–2111 (siChe-1-3) of human Che-1 sequence, and to nucleotides 122–143 of the negative control green fluorescent protein (GFP), were synthesized in vitro by using the Silencer siRNA construction kit (Ambion), following the manufacturer's instruction. Stealth Deptor, Redd1, and siRNA Negative Controls double-stranded RNA oligonucleotides were purchased from Life Technologies. RNA interference experiments were performed by transfecting RNA oligos using Lipofectamine 2000 (Life Technologies).
Chromatin immunoprecipitation assays (ChIP)
Chromatin immunoprecipitation assays in HCT116, Kms27, and U266 cells were performed as previously described (Bruno et al, 2002) by using anti-Che-1 and anti-Myc antibodies. Immunoprecipitations with no specific immunoglobulins (Santa Cruz) were performed as negative controls. For quantitative ChIP analysis (ChIP-qPCR), 1 μl of purified DNA was used for amplification on an Applied Biosystems 7500 Fast Real Time PCR system (with Applied Biosystem SYBR GREEN). The following human promoter-specific primers were employed in PCR amplifications:
Deptor forward 5′-ATGGCTGAGGTCTTGGTCAC-3′
Deptor reverse 5′- GTACACGGGAAGACGGACAG-3′
Redd1 forward 5′-CAGGAGAGAACGTTGCTTACG-3′
Redd1 reverse 5′-AGCCGCTGTAAGACAAGAGG-3′
ChIP-sequencing
Chromatin immunoprecipitations were performed as previously described (Bruno et al, 2002) by using rabbit polyclonal antibodies anti-Che-1 and anti-RNA polymerase II S-5P (Abcam, ab5131) and Dynabeads Protein G (Life Technologies). The quantity of the immunoprecipitated material was determined by Qubit 2.0 fluorometer (Life Technologies). 10 ng of the immunoprecipitated chromatin was used to prepare the libraries for sequencing following the manufacturer's instructions including DNA end repairing, adaptor ligation, and amplification (Illumina). Fragments of about 100–180 bp (without linkers) were isolated from agarose gel and used for sequencing using the Illumina GA IIx. (36 bp, 21–26 million quality-filtered and uniquely aligned reads per sample).
Data analysis
The Fastq files generated by the Illumina pipeline were mapped to the human genome using Bowtie allowing up to 2 mismatches. Only uniquely mapped reads were kept. The peaks of the ChIP-seq were identified using MACS version 1.4 (Zhang et al, 2008). An input control library served as a negative control. The false discovery rate allowed was < 1%. The setting for peak calling was tsize = 36, bw = 250, mfold = 10,30, and P-value at least 1e-5. The reference genome used throughout was the human genome assembly hg18. All gene and transcript data, such as transcription start site positions, came from UCSC (http://genome.ucsc.edu).
Microarray analysis
Total RNA was extracted from HCT116 cells by TRIzol (Invitrogen), purified from genomic DNA contamination through a DNase I (Qiagen) digestion step and further enriched by Qiagen RNeasy columns for gene expression profiling (Qiagen). Quantity and integrity of the extracted RNA were assessed by NanoDrop Spectrophotometer (NanoDrop Technologies) and by Agilent 2100 Bioanalyzer (Agilent Technologies), respectively. Expression profiles were determined by using the Human Gene 1.0 ST arrays (Affymetrix) according to the manufacturer's instructions. Briefly, the Affymetrix GeneChip® Whole Transcript Sense Target Labelling Assay was used to amplify and reverse-transcribe total RNA, and to biotinylate sense-strand DNA targets. Arrays were hybridized with the labeled target hybridization cocktail by rotation in the Affymetrix Gene Chip hybridization oven at 45°C for 16 h, washed in the Affymetrix GeneChip Fluidics station FS 450, and scanned by Affymetrix Gene Chip scanner 3000 7G system. Scanned image files (CEL) were processed, normalized (RMA-Sketch Quantile), and log2-transformed by Expression Console Software (Affymetrix website). Normalized intensity files were statistically analyzed to calculate t-statistics and P-values, and adjusted by the false discovery rate (FDR< 5%) approach.
RNA isolation and quantitative RT–PCR analysis
Cells were harvested 36 h after transfection and total RNA isolated using TRIzol reagent (Life Technologies) in accordance with the manufacturer's instructions, and the first-strand cDNA was synthesized with random primers and SuperScript II reverse transcriptase (Life Technologies). The cDNA was used for quantitative real-time PCR (qRT–PCR) experiments carried out in an 7500 Fast Real Time PCR System (Applied Biosystem) using SYBR GREEN PCR Master Mix (Applied Biosystem). ΔΔCt values were normalized with those obtained from the amplification of the endogenous RPL19 gene. The following human-specific primers were employed in PCR amplifications:
Che1 forward 5′-CCGGAATTCGGATAAGACCAAACTGGCT -3′
Che1 reverse 5′-CCGCTCGAGGAGTTCTCGAAGGAGCTG-3′
Deptor forward 5′-AGCTTTGCCACCGGCTTAT-3′
Deptor reverse 5′-GGCAGAAGGGACTGTCATGAG-3′
Redd1 forward 5′-CACCCCAAAAGTTCAGTCGT-3′
Redd1 reverse 5′-TGTTTAGCTCCGCCAACTCT-3′
RPL19 forward 5′- CGGAAGGGCAGGCACAT-3′
RPL19 reverse 5′-GGCGCAAAATCCTCATTCTC-3′
MAP1LC3B forward 5′-AGCAGCATCCAACCAAAATC-3′
MAP1LC3B reverse 5′-CTGTGTCCGTTCACCAACAG-3′
Myc-Che-1 forward 5′-ATGGAGAGCTTGGGCGACCTCACC-3′
Myc-Che-1 reverse 5′-TCATTCCATGCTTTTCTA3′
Design and cloning of shRNA
shChe-1 sequence (nucleotides 824–842):
5′-gatccccAAAGTTTCTGAGGAAGTGGttcaagagaCCACTTCCTCAGAAACTTTtttttggaaa-3′ and 5′-agcttttccaaaaaAAAGTTTCTGAGGAAGTGGtctcttgaaCCACTTCCTCAGAAACTTTggg-3′;
shControl sequence:
5′-cgcgtCTATAACGGCGCTCGATATttcaagagaATATCGAGCGCCGTTATAGtttttggaaat-3′, and 5′-cgatttccaaaaaCTATAACGGCGCTCGATATtctcttgaaATATCGAGCGCCGTTATAGa-3′. Oligos were annealed and cloned in pLV-THM vector (Wiznerowicz & Trono, 2003), MluI/ClaI (Boehringer Mannheim) digested, generating the new lentiviral vector pLV-TH shChe-1.
Viral vectors
Lentiviral vectors pLV-THM and pLV-tTR-KRAB (Wiznerowicz & Trono, 2003) were produced as previously described (Bruno et al, 2010). Lentiviral stocks were titered following standard protocols (Wiznerowicz & Trono, 2003), and routinely, a viral titer of 106 transducing units per ml (TU/ml) was achieved.
In vivo experiments
Kms27 and U266 cells were transduced as previously described (Bruno et al, 2010).
Groups of female CD-1 nude (nu/nu) mice (6–8 weeks old and 22–24 g in body weight) (Charles River Laboratories, Lecco, Italy) were maintained in sterile environment. Engineered cells (1.0 × 107 cells/mouse) were injected with matrigel (BD Biosciences) in intra-scapular area. Doxycycline was delivered to the mice through drinking water [tap water + 3,0% sucrose (Sigma)] in dark stained bottles and renewed every 4 days. To rescue Che1-depletion effects in vivo, a randomly allocated subgroup of ind-sh/Che1 (8 mice/group) tumor-bearing mice were treated with CCI-779, whereas control ind-sh/Che1 and ind-sh/Control tumor-bearing mice were delivered with vehicle solution. CCI-779 was diluted in 5% Tween-80 and 5% polyethylene glycol-400 (Sigma) to the final concentration of 5 g/l. The CCI-779 drug solution (20 μl) was administrated intraperitoneally two times a week (total of 10 injections). Xenograft tumor growth delay was monitored twice a week by calliper measurements, and tumors volumes [TV (cm3)] were estimated by the formula: TV = a × (b2)/2, where a and b are tumor length and width, respectively, and results reported as relative tumor growth (V/V0). At the end of treatments, animals were sacrificed following standard protocols, tumors excised, and sections frozen in nitrogen liquid. The group number was determined by assuming an alpha = 0.05 and power (1-beta) of 80%, with a standard deviation of 3%, that allowed to reveal growth difference by using the formula of Shah (2011). Pre-established criteria were as follows: Inclusion, animals with established tumor, with volume range from 0.15 to 0.30 cm3; exclusion, tumor-free animals and tumor-bearing animals, with volume out of reported range. The animal facility staff performed random group allocation, treatments, and tumor measurements in blinding condition.
All procedures involving animals and their care were conducted in accordance with Institutional guidelines and Italian Health Ministry approvals.
Patients' material
Monoclonal gammopathies of undetermined clinical significance and MM patient samples were collected as part of routine clinical examination. Written informed consent to participate in this study was provided by all subjects in accordance with the Declaration of Helsinki and the Mayo Clinic Institutional Review Board. The study was approved by the Regina Elena Cancer Institute Ethical committee. MGUS and MM patients were determined according to the international working group classification based on the level/concentration of serum M-protein and percentage of bone marrow plasma cells. The distinction between symptomatic and asymptomatic myeloma relies on the presence or absence of myeloma-related organ or tissue impairment (The International Myeloma Working Group, 2003). MGUS patients had < 10% BM clonal PCs and < 3 g/dl of serum M-protein, while patients diagnosed with MM had 10% or greater clonal BM-PCs and/or > 3 g/dl of serum M-protein. Bone marrow aspirates were enriched for plasma cells by magnetic cell separation using a human CD138 positive selection kit (Miltenyi Biotec, Germany) and Macs Separator (Macs Miltenyi Biotec, Germany).
Statistical analysis
Statistical analysis was performed by using the Student's two-tailed t-test to compare in vitro experiments. All statistical tests were carried out using GraphPad Prism version 5.0 for Windows, GraphPad Software, San Diego California, USA (www.graphpad.com). Probability value of < 0.05 was considered statistically significant.
Accession number
The microarray data from this publication have been submitted to the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) and assigned the identifier accession number GSE45009 (submitter M. Fanciulli).
Acknowledgments
We acknowledge Mrs. Tania Merlino for the English language revision of the text. We thank Dr. Daniela Kovacs and Dr. Barbara Benassi for their precious assistance. This work was supported by the Italian Association for Cancer Research (A.I.R.C.) (M.F. 11356, and G.B. 14455). Agata Desantis is the recipient of the ‘Giaime Fiumanò’ A.I.R.C. fellowship.
Author contributions
All the authors discussed the experiments and contributed to the text of the manuscript. AD, TB, SI, and CS performed most of the experiments and analyzed data. VC performed autophagy experiments. FDN and FG performed microarrays and ChIP-seq analyses. MP carried out immunohistochemical analysis. GBo performed the lentiviral delivery experiments. EL carried out experiments in angiomyolipoma cells. FLR, PDDM, and TC did bioinformatics and statistical analysis. MC and LB performed experiments in Vk*-Myc mice. FP, MRR, VF, and MTP provided patient material. GT analyzed patient data and participated in manuscript writing. AF, CP, and GBl participated in data analysis and manuscript writing. MF conceived the project, secured funding, supervised the experiments, and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6A–D
Supplementary Figure S6E–I
Supplementary Figure S6J–M
Supplementary Figure S7
Supplementary Legends
Review Process File
Source data for Figure 1
Source data for Figure 2
Source data for Figure 4
Source data for Figure 5
Source data for Figure 6
Source data for Figure 7
Source data for Figure 8
References
- Agnelli L, Mosca L, Fabris S, Lionetti M, Andronache A, Kwee I, Todoerti K, Verdelli D, Battaglia C, Bertoni F, Deliliers GL, Neri A. A SNP microarray and FISH-based procedure to detect allelic imbalances in multiple myeloma: an integrated genomics approach reveals a wide gene dosage effect. Genes Chromosom Cancer. 2009;48:603–614. doi: 10.1002/gcc.20668. [DOI] [PubMed] [Google Scholar]
- Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG., Jr Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18:2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno T, De Angelis R, De Nicola F, Barbato C, Di Padova M, Corbi N, Libri V, Benassi B, Mattei E, Chersi A, Soddu S, Floridi A, Passananti C, Fanciulli M. Che-1 affects cell growth by interfering with the recruitment of HDAC1 by Rb. Cancer Cell. 2002;2:387–399. doi: 10.1016/s1535-6108(02)00182-4. [DOI] [PubMed] [Google Scholar]
- Bruno T, De Nicola F, Iezzi S, Lecis D, D'Angelo C, Di Padova M, Corbi N, Dimiziani L, Tannini L, Jekimovs C, Scarsella M, Porrello A, Chersi A, Crescenzi M, Leonetti C, Khanna KK, Soddu S, Floridi A, Passananti C, Delia D, et al. Che-1 phosphorylation by ATM and Chk2 kinases activates p53 transcription and the G2/M checkpoint. Cancer Cell. 2006;10:479–486. doi: 10.1016/j.ccr.2006.10.012. [DOI] [PubMed] [Google Scholar]
- Bruno T, Desantis A, Bossi G, Di Agostino S, Sorino C, De Nicola F, Iezzi S, Franchitto A, Benassi B, Galanti S, La Rosa F, Floridi A, Bellacosa A, Passananti C, Blandino G, Fanciulli M. Che-1 promotes tumor cell survival by sustaining mutant p53 transcription and inhibiting DNA damage response activation. Cancer Cell. 2010;18:122–134. doi: 10.1016/j.ccr.2010.05.027. [DOI] [PubMed] [Google Scholar]
- Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam H, Easton JB, High A, Houghton PJ. mTORC1 signaling under hypoxic conditions is controlled by ATM-dependent phosphorylation of HIF-1α. Mol Cell. 2010;40:509–520. doi: 10.1016/j.molcel.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesi M, Robbiani DF, Sebag M, Chng WJ, Affer M, Tiedemann R, Valdez R, Palmer SE, Haas SS, Stewart AK, Fonseca R, Kremer R, Cattoretti G, Bergsagel PL. AID-dependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of post-germinal malignancies. Cancer Cell. 2008;13:167–180. doi: 10.1016/j.ccr.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradetti MN, Inoki K, Guan KL. The stress-inducted proteins RTP801 and RTP801L are negative regulators of the mammalian target of rapamycin pathway. J Biol Chem. 2005;280:9769–9772. doi: 10.1074/jbc.C400557200. [DOI] [PubMed] [Google Scholar]
- De Nicola F, Bruno T, Iezzi S, Di Padova M, Floridi F, Passananti C, Del Sal G, Fanciulli M. The prolyl isomerase Pin1 affects Che-1 stability in response to apoptotic DNA damage. J Biol Chem. 2007;282:19685–19691. doi: 10.1074/jbc.M610282200. [DOI] [PubMed] [Google Scholar]
- Di Certo MG, Corbi N, Bruno T, Iezzi S, De Nicola F, Desantis A, Ciotti MT, Mattei E, Floridi A, Fanciulli M, Passananti C. NRAGE associates with the anti-apoptotic factor Che-1 and regulates its degradation to induce cell death. J Cell Sci. 2007;120:1852–1858. doi: 10.1242/jcs.03454. [DOI] [PubMed] [Google Scholar]
- Duan S, Skaar JR, Kuchay S, Toschi A, Kanarek N, Ben-Neriah Y, Pagano M. mTOR generates an auto-amplification loop by triggering the βTrCP- and CK1α-dependent degradation of DEPTOR. Mol Cell. 2011;44:317–324. doi: 10.1016/j.molcel.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellisen LW, Ramsayer KD, Johannessen CM, Yang A, Beppu H, Minda K, Oliner JD, McKeon F, Haber DA. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol Cell. 2002;10:995–1005. doi: 10.1016/s1097-2765(02)00706-2. [DOI] [PubMed] [Google Scholar]
- Fanciulli M, Bruno T, Di Padova M, De Angelis R, Iezzi S, Iacobini C, Floridi A, Passananti C. Identification of a novel partner of RNA polymerase II subunit 11, Che-1, which interacts with and affects the growth suppression function of Rb. FASEB J. 2000;7:904–912. doi: 10.1096/fasebj.14.7.904. [DOI] [PubMed] [Google Scholar]
- Feng Z, Hu W, de Stanchina E, Teresky AK, Jin S, Lowe S, Levine AJ. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 2007;67:3043–3053. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- Fingar DC, Blenis J. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene. 2004;23:3151–3171. doi: 10.1038/sj.onc.1207542. [DOI] [PubMed] [Google Scholar]
- Gao D, Inuzuka H, Tan MK, Fukushima H, Locasale JW, Liu P, Wan L, Zhai B, Chin YR, Shaik S, Lyssiotis CA, Gygi SP, Toker A, Cantley LC, Asara JM, Harper JW, Wei W. mTOR drives its own activation via SCF(βTrCP)-dependent degradation of the mTOR inhibitor DEPTOR. Mol Cell. 2011;44:290–303. doi: 10.1016/j.molcel.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson SL, Bindra RS, Glazer PM. Hypoxia-induced phosphorylation of Chk2 in an ataxia telangiectasia mutated-dependent manner. Cancer Res. 2005;65:10734–10741. doi: 10.1158/0008-5472.CAN-05-1160. [DOI] [PubMed] [Google Scholar]
- Guo Q, Xie J. AATF inhibits aberrant production of amyloid beta peptide 1-42 by interacting directly with Par-4. J Biol Chem. 2004;279:4596–4603. doi: 10.1074/jbc.M309811200. [DOI] [PubMed] [Google Scholar]
- Hideshima T, Richardson PG, Anderson KC. Mechanism of action of proteasome inhibitors and deacetylase inhibitors and the biological basis of synergy in multiple myeloma. Mol Cancer Ther. 2011;10:2034–2042. doi: 10.1158/1535-7163.MCT-11-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopker K, Hagmann H, Khurshid S, Chen S, Hasskamp P, Seeger-Nukpezah T, Schilberg K, Heukamp L, Lamkemeyer T, Sos ML, Thomas RK, Lowery D, Roels F, Fischer M, Liebau MC, Resch U, Kisner T, Röther F, Bartram MP, Müller RU, et al. AATF/Che-1 acts as a phosphorylation-dependent molecular modulator to repress p53-driven apoptosis. EMBO J. 2012;31:1–15. doi: 10.1038/emboj.2012.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt EM, Wiestner A, Rosenwald A, Shaffer AL, Campo T, Bergsagel PL, Kuehl WM, Staudt LM. Overexpression of c-maf is a frequent oncogenic event in multiple myeloma that promotes proliferation and pathological interactions with bone marrow stroma. Cancer Cell. 2004;5:191–199. doi: 10.1016/s1535-6108(04)00019-4. [DOI] [PubMed] [Google Scholar]
- Inoki K, Li Y, Xu T, Guan KL. Rebh GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki S, Fonseca SG, Oslowski CM, Jurczyk A, Shearstone JR, Zhu LJ, Permutt MA, Greiner DL, Bortell R, Urano F. AATF mediates an antiapoptotic effect of the unfolded protein response through transcriptional regulation of AKT1. Cell Death Differ. 2010;17:774–786. doi: 10.1038/cdd.2009.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H-O, Hong S-E, Kim J-H, Choi H-N, Kim K, An S, Choe T-B, Hwang C-S, Lee J-H, Kim J-I, Kim HA, Kim EK, Noh WC, Hong YJ, Hong SI, Lee JK, Park IC. Sustained overexpression of Redd1 leads to Akt activation involved in cell survival. Cancer Lett. 2013;336:319–324. doi: 10.1016/j.canlet.2013.03.021. [DOI] [PubMed] [Google Scholar]
- Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar S, Liu E, Knutzen CA, Lombardo CR, Sankar S, Toth JI, Petroski MD, Ronai Z, Chiang GG. REDD1, an inhibitor of mTOR signalling, is regulated by the CUL4A-DDB1 ubiquitin ligase. EMBO Rep. 2009;10:866–872. doi: 10.1038/embor.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan K-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, Ahn HJ, Ait-Mohamed O, Ait-Si-Ali S, Akematsu T, Akira S, Al-Younes HM, Al-Zeer MA, Albert ML, Albin RL, Alegre-Abarrategui J, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamy L, Ngo VN, Tolga Emre NC, Shaffer AL, III, Yang Y, Tian E, Nair V, Kruhlak MJ, Zingone A, Landregren O, Staudt LM. Control of autophagic death by caspase-10 in multiple myeloma. Cancer Cell. 2013;23:435–449. doi: 10.1016/j.ccr.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesma E, Sirchia SM, Ancona S, Carelli S, Bosari S, Ghelma F, Montanari E, Di Giulio AM, Gorio A. The methylation of the TSC2 promoter underlies the abnormal growth of TSC2 angiomyolipoma-derived smooth muscle cells. Am J Pathol. 2009;174:2150–2159. doi: 10.2353/ajpath.2009.080799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung-Hagesteijn C, Erdmann N, Cheung G, Keats JJ, Stewart AK, Reece DE, Chung KC, Tiedemann RE. Xbp1s-negative tumor B cells and pre-plasmablasts mediate therapeutic proteasome inhibitor resistance in multiple myeloma. Cancer Cell. 2013;24:289–304. doi: 10.1016/j.ccr.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- Martens S, Parvanova I, Zerrahn J, Griffiths G, Schell G, Reichmann G, Howard JC. Disruption of Toxoplasma gondii parasitophorous vacuoles by the mouse p47-resistance GTPases. PLoS Pathog. 2005;1:e24. doi: 10.1371/journal.ppat.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molitoris JK, McColl KS, Swerdlow S, Matsuyama M, Lam M, Finkel TH, Matsuyama S, Distelhorst CW. Glucocorticoid elevation of dexamethasone-induced gene 2 (Dig2/RTP801/REDD1) protein mediates autophagy in lymphocytes. J Biol Chem. 2011;286:30181–30189. doi: 10.1074/jbc.M111.245423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld TP. TOR-dependent control of autophagy: biting the hand that feeds. Curr Opin Cell Biol. 2010;22:157–168. doi: 10.1016/j.ceb.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeng EA, Carlson LM, Gutman DM, Harrington WJ, Jr, Lee KP, Bolse LH. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood. 2006;107:4907–4916. doi: 10.1182/blood-2005-08-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page G, Lodige I, Kogel D, Scheidtmann KH. AATF, a novel transcription factor that interacts with Dlk/ZIP kinase and interferes with apoptosis. FEBS Lett. 1999;462:187–191. doi: 10.1016/s0014-5793(99)01529-x. [DOI] [PubMed] [Google Scholar]
- Passananti C, Floridi A, Fanciulli M. Che-1/AATF, a multivalent adaptor connecting transcriptional regulation, checkpoint control, and apoptosis. Biochem Cell Biol. 2007;85:477–483. doi: 10.1139/O07-062. [DOI] [PubMed] [Google Scholar]
- Pengo N, Scolari M, Oliva L, Milan E, Mainoldi F, Raimondi A, Fagioli C, Merlini A, Mariani E, Pasqualetto E, Orfanelli U, Ponzoni M, Sitia R, Casola S, Cenci S. Plasma cells require autophagy for sustainable immunoglobulin production. Nat Immunol. 2013;14:298–305. doi: 10.1038/ni.2524. [DOI] [PubMed] [Google Scholar]
- Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiling JH, Sabatini DM. Stress and mTORture signaling. Oncogene. 2006;25:6373–6383. doi: 10.1038/sj.onc.1209889. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell. 2010;40:310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah H. How to calculate the sample size for animal studies. Technical Notes. Natl J Physiol Pharm Pharmacol. 2011;1:35–39. [Google Scholar]
- Shapiro-Shelef M, Calame K. Plasma cell differentiation and multiple myeloma. Curr Opin Immunol. 2004;16:226–234. doi: 10.1016/j.coi.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Shoshani T, Faerman A, Mett I, Zelin E, Tenne T, Gorodin S, Moshel Y, Elbaz S, Budanov A, Chajut A, Kalinski H, Kamer I, Rozen A, Mor O, Keshet E, Leshkowitz D, Einat P, Skaliter R, Feinstein E. Identification of a novel hypoxia-inducible factor 1-responsive gene, RTP801, involved in apoptosis. Mol Cell Biol. 2002;22:2283–2293. doi: 10.1128/MCB.22.7.2283-2293.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh DH, Kim M-K, Kim HS, Chung HH, Song YS. Unfolded protein response for anticancer therapy. Ann NY Acad Sci. 2012;1271:20–32. doi: 10.1111/j.1749-6632.2012.06739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase- activating protein complex toward Rheb. Curr Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- The International Myeloma Working Group. Criteria for the classification of monoclonal gammopathies, multiple myeloma and related disorders: a report of the International Myeloma Working Group. Br J Haematol. 2003;121:749–757. [PubMed] [Google Scholar]
- Thomas T, Voss AK, Petrou P, Gruss P. The murine gene, Traube, is essential for the growth of preimplantation embryos. Develop Biol. 2000;227:324–342. doi: 10.1006/dbio.2000.9915. [DOI] [PubMed] [Google Scholar]
- Vignot S, Faivre S, Aguirre D, Raymond E. mTOR-targeted therapy of cancer with rapamycin derivatives. Ann Oncol. 2005;16:525–537. doi: 10.1093/annonc/mdi113. [DOI] [PubMed] [Google Scholar]
- Wiznerowicz M, Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J Virol. 2003;77:8957–8961. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Nair U, Yang Z, Klionsky DJ. Endoplasmic reticulum stress triggers autophagy. J Biol Chem. 2006;281:30299–30304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Oh YoonS, Poulogiannis G, Yang Q, Max MaX, Villen J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, Blenis J. Quantitative phosphoproteomic analysis identifies the adaptor protein Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan F, Huang Y, Stewart JP, Hanamura I, Gupta S, Epstein J, Yaccoby S, Sawyer J, Burington B, Anaissie E, Hollmig K, Pineda-Roman M, Tricot G, van Rhee F, Walker R, Zangari M, Crowley J, Barlogie B, Shaughnessy JD., Jr The molecular classification of multiple myeloma. Blood. 2006;108:2020–2028. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model basis analysis of ChIP (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Xiong X, Jia L, Sun Y. Targeting Cullin-RING ligases by MLN4924 induces autophagy via modulating the HIF1-REDD1-TSC1-mTORC1-DEPTOR axis. Cell Death Dis. 2012;3:e386. doi: 10.1038/cddis.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Xiong X, Sun Y. DEPTOR, an mTOR inhibitor, is a physiological substrate of SCF(βTrCP) E3 ubiquitin ligase and regulates survival and autophagy. Mol Cell. 2011;44:304–316. doi: 10.1016/j.molcel.2011.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6A–D
Supplementary Figure S6E–I
Supplementary Figure S6J–M
Supplementary Figure S7
Supplementary Legends
Review Process File
Source data for Figure 1
Source data for Figure 2
Source data for Figure 4
Source data for Figure 5
Source data for Figure 6
Source data for Figure 7
Source data for Figure 8