Abstract
The OPEP coarse-grained protein model has been applied to a wide range of applications since its first release 15 years ago. The model, which combines energetic and structural accuracy and chemical specificity, allows studying single protein properties, DNA/RNA complexes, amyloid fibril formation and protein suspensions in a crowded environment. Here we first review the current state of the model and the most exciting applications using advanced conformational sampling methods. We then present the current limitations and a perspective on the on-going developments.
INTRODUCTION
Proteins, DNA and RNA carry out a variety of biochemical and biological tasks. These systems are very challenging experimentally and numerically due to their number of degrees of freedom, and the wide range of relevant time scales from nanoseconds to days associated with fluctuations about the native states, diffusion, folding and formation of harmful aggregates.
Classical atomistic molecular dynamics (MD) with explicit solvent and ions can complement experiments.1,2 With the specially MD-designed Anton computer, performing MD 100-500 times faster than the standard computer, it has been possible to break the millisecond barrier and gain insights on the mechanisms, thermodynamics and kinetics of the folding of diverse proteins with 10-80 amino acids.3 Anton has also proven useful in the development of allosteric inhibitors that target previously unknown binding sites.4 The dynamic processes of life at the molecular level require, however, knowledge of the structure, dynamics and thermodynamics of biomolecules in a crowded environment. Similarly, we cannot wait for faster computers to design engineered proteins with specific properties or new molecules able to interfere with protein-protein or protein-DNA/RNA complexes associated with disease functions as a result of sporadic mutations or genetic risk factors. For instance, neurodegenerative diseases such as Alzheimer, Parkinson, Huntington challenge our society.5 The V600E mutation in the BRAF protein is known to be responsible for 50% of melanoma cases,6 and women with mutations in the BRCA1 and BRCA2 genes have markedly elevated risks of breast and ovarian cancer.7
For all these reasons, it is necessary to design multiscale approaches, coarse-grained models and advanced sampling methods to converge rapidly to equilibrium and explore dynamics in the time scale of microseconds and beyond for large systems.
The development of coarse graining (CG) and multiscale modeling is not new. It was an exciting day for computational chemistry and biology when the 2013 Nobel Prize in Chemistry was awarded to Martin Karplus, Michael Levitt, and Arieh Warshel for the “development of multiscale models for complex chemical systems.” Among their contributions, Levitt and Warshel pioneered CG protein simulation, with atoms grouped into larger units or beads and normal modes to move the system on the energy landscape.8 Despite extensive efforts, coarse graining still remains a challenge and poses the problem of how to derive potentials for the selected number of beads that maintain the all-atom physical behavior in test tubes and cellular environments. Recently, two reviews summarized the state of coarse-graining of biomolecular systems so as to couple information from different scales.9,10 Marrink and Tieleman also presented the strengths and limitations of their CG Martini model, used for lipid membrane characterization, lipid polymorphism, membrane protein - lipid interplay, self-assembly of soluble proteins, and membrane protein oligomerization.11
The OPEP (Optimized Potential for Efficient protein structure Prediction) coarse-grained model with an implicit solvent model for soluble proteins in aqueous solution has evolved since its first version 15 years ago.12-24 This model which retains structural accuracy and chemical specificity is free of any biases in contrast to Martini that imposes secondary structure constraints. 11, 25, 26 While CG models have been developed by Sansom27,28 and Schulten29 for membrane proteins, Klein,30 Scheraga (UNRES),31-33 Baker (Rosetta),34,35 Voth,36 Dokhloyan,37 Hall (PRIME),38 Lavery (PaLaCe),39 Zacharias (ATTRACT),40,41 Feig (PRIMO),42 Shea,43 Papoian (AWSEM)44 and other scientists45-48 for soluble proteins, to the best of our knowledge, none of these models except Martini has been applied to a wide range of problems. Here, we present the design principles of the model and the state-of-the-art sampling techniques used. Furthermore this review provides an in-depth understanding on four timely topics in the chemical sciences.
The first topic is the self-assembly of amyloid proteins associated with neurodegenerative diseases. Alzheimer’s disease affects 24 million people and drug after drug has failed to slow its progression.49 The second topic is computer-assisted de novo design and structure prediction of peptides up to 52 amino acids, as they represent a source of novel antibiotics and therapeutics.50-52 To this end, we need a fast and accurate method able to play with he amino acid sequence. The third topic is RNA structure prediction which is still in its infancy compared to protein structure prediction.53 The small non-coding microRNA which regulate gene expression at a post-transcriptional level,54 or more generally all non-coding RNAs are increasingly attracting the attention of cancer investigators.55,56 The last topic concerns the effects of hydrodynamics and crowding which are mostly ignored in computer simulations and create a gap between the simulated and the real physics in the cell. We report an OPEP simulation of a system of unprecedented size and fully inclusive of hydrodynamic interactions, namely 18,000 flexible proteins and 70 million particles,57 a breakthrough compared to the largest simulation of 1000 rigid proteins ignoring hydrodynamics.58
Finally, we present the current OPEP limitations and we sketch some on-going developments or applications. These include for instance understanding the physics behind the difference in the stability of thermophilic and mesophilic proteins, determining the effect of external conditions such as shear flows on the dynamics, kinetics and thermodynamics of non-amyloid and amyloid proteins, and interacting directly on the CG model system using virtual reality to probe the mechanical properties of molecular structures.
Overall, the review gives a state-of-the-art account of the various subjects treated and a well-balanced assessment of the current literature, comparing with previous computational results and experimental data when available.
Design principles of the OPEP model
Granularity
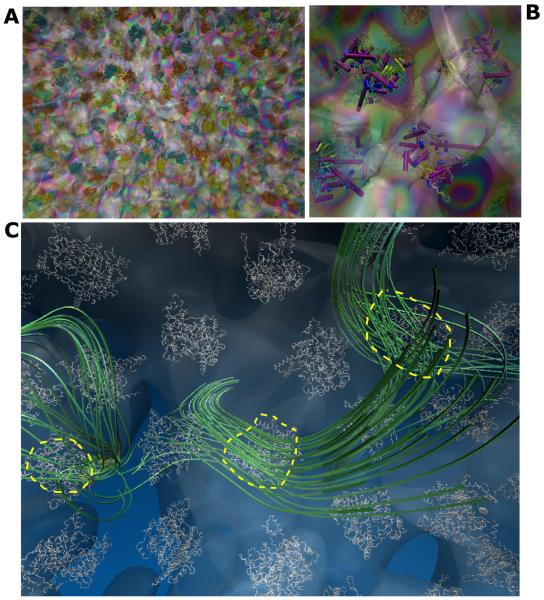
Various levels of granularity for amino acids have been developed ranging from two to six beads, and beyond. The OPEP CG model represents each amino acid by six centers of force: the side-chain is represented by a unique bead located at the center of mass of nonhydrogen atoms in the all-atom side chains of 2250 protein structures with sequence identity < 30%, while atomic resolution is used for the backbone that includes N, HN, Cα, C and O atoms. Proline is an exception represented by all its heavy atoms (Fig. 1A).20 The disulfide (S-S) bonds can be treated as two non-bonded beads or described at an atomic level using local terms. This OPEP CG strategy was chosen to represent a good compromise between energetic and structural accuracy and chemical specificity, even if this limits the use of large time steps compared to other CG models. Note that the Rosetta fragment assembly Monte Carlo program uses the same level of description in the first step of its hierarchical procedure.34,35
Fig.1.
OPEP CG model and enhanced sampling methods. (A) We use the peptide Ala-Lys-Phe-Pro-Val in its zwitterion form to show the details of the backbone and the side-chains. (B) The Activation-Relaxation Technique connecting local minima by first-order saddle points. (C) Example of a metadynamics simulation in a one-dimensional landscape with multiple metastable minima separated by energy barriers. Top panel: System trajectory in CV space as a function of simulation time. Bottom panel: Progressive filling (colored lines) of the underlying potential (black line) by bias. In both the panels color code is used to measure the simulation time. The system starts in the basin A1 and it is pushed by the bias to overcome the free-energy barriers and to visit basin B1 (t~100) and basin C1 (t~1500). In the second half of the simulation, the system can easily sample the whole landscape and the bias can be used to estimate the underlying free-energy surface.
Our level of granularity varies from the Martini model with coarse-grained solvent in which the main chain atoms of each residue are represented by a unique bead and, on average, four heavy side-chain atoms are represented by a single interaction center, with the exception of ring-like molecules.25,26 Our representation varies also from eight other CG models in implicit solvent: (i) Klein’s model where three to four heavy atoms are represented by a single CG bead. Most side-chains use one CG bead, except lysine and arginine with a hydrophobic and a hydrophilic site and the tyrosine, phenylalanine, and tryptophan residues represented by two, two, and three beads respectively,30 (ii) UNRES two-bead model with one unified bead for side-chain and the peptide center, p, located in the centers of Cα-Cα bonds,31-33 (iii) PRIMO using the Cα, Cβ and a combined CO particle for the backbone and one to several heavy side-chain atoms into CG sites,42 (iv) PRIME with the N, Cα, and carbonyl C backbone atoms and up to three side-chain beads,22 (v) Voth’s model with a Cα for the backbone and as many as four beads for the side-chains,36 (vi) ATTRACT with the N and O for the backbone and one or two beads for the side chains,40 though its most recent version is very similar to OPEP,41 (vii) AWSEM three-bead model with Cα, Cβ and O,44 and (viii) PaLaCe with one to three beads for the main non-bonded interactions, combined with atomistic peptide groups and some side-chain atoms.39 Scientists are also developing CG models for soluble proteins in simplified explicit solvent,59 or atomistic soluble proteins with CG water models.60,61
Optimization Procedure and Analytical expression
There are multiple approaches to derive the bonded and non-bonded potentials.9-11 The first approach, followed by PaLaCe and PRIMO, uses Boltzmann inversion of conformational probability distributions derived from a static or dynamic protein structure data set. The second one is to derive a CG potential from forces generated by atomistic simulations, referred to as force matching.36 The third thermodynamics-based approach consists of fitting and predicting free energies such as water/oil partitioning coefficients of the amino acid side-chain analogues (Martini),11 or density, surface tension (Klein)30 for the non-bonded interactions and by using the distributions of bonds, bending angles and dihedral angles from the Protein Data Bank (PDB,) to optimize the bonded interactions. Another approach is the factor expansion method where the pairwise potentials of mean force (PMFs) between side-chains are obtained from atomistic simulations, and the torsional, double-torsional, backbone-electrostatic and correlation terms are fitted on quantum-mechanical ab initio calculations.31-33 Other methods for force field derivation and optimization include minimization of relative entropy38 and simulations to test whether hexapeptides form non-amyloid or amyloid fibrils (PRIME)22 or proteins fold into their native states.62
The latest OPEP version uses a structure/thermodynamic/PMF approach, since the parameters and analytical forms are trained on bonded and non-bonded distances and angle distributions of native and non-native protein structures, are fitted to reproduce the experimental lowest free-energy conformations and the melting temperatures (TM) of a small set of peptides, and are derived from all-atom PMF simulations for the interactions between charged side-chains.23,24
The OPEP energy function is defined as a sum of local, non-bonded, and hydrogen bonding (H-bond) terms.15,21,24 All analytical expressions are given in Supplementary Material. The local interactions include bond length, angle bending, and improper and proper dihedral angles. The improper torsions maintain the desired chirality of amino acids, and control the out-of plane motion of the C=O and N-H bonds about the peptide bond. All these terms were modeled on the analytical form of the AMBER63 force field with an additional term for the Φ,Ψ dihedral angles to render realistic Ramachandran plots.21,24 H-bonds of backbone atoms are accounted for by two- and four-body potentials, rather than Coulomb interactions. The two-body term for each H-bond is the product of a 10-12 term dependent on the O-H distance by the square of the cosine of the N-H.. O angle.15,21 The four-body term takes the form of the product of two Gaussian functions each monitoring the existence of one H-bond on the basis of distance criteria, and represents a cooperative energy if tight conditions on sequence-separation, Δ, between four residues are verified. If (i, j) and (k, l) are the residues involved in the two H-bonds, Δ(ijkl) =1 if (k, l) = (i+1, j+1) (α-helix), or Δ(ijkl) =1 if (k, l) = (i+2 ,j−2) or (i+2, j+2) (β-sheets), otherwise Δ is set to 0. These conditions stabilize secondary structures, independently of the Φ, Ψ angles, but also any segment satisfying the conditions on ijkl.15,21,24
It is essentially the van der Waals potential that has evolved from OPEPv1 to OPEPv5 by distinguishing its form as a function of the center of forces.15,24 Each OPEP version does not have its own advantages, rather a new version is developed to solve unexpected failures of the previous version. In all versions, we use the 6-12 potential between the backbone atoms and between the backbone and side-chain atoms. In OPEPv3, the van der Waals energy between two side-chains was 6-12 if the interaction is hydrophobic or resulted from oppositely charged amino acids; otherwise an r−6 term was used.20 In OPEPv4, following our work on RNA, the r−6 term was replaced by r−8 for purely repulsive interactions; otherwise the 6-12 term was replaced by an analytical formulation to limit the energy values of the side-chains at longer distances. We also distinguished 11 side-chain – side-chain interactions depending on their sequence-separation to stabilize α-helices.23 From OPEPv4 to OPEPv5, we only changed the ion pair interactions from all-atom PMF potentials, characterized by one minimum for the pairs Lys-Asp and Lys-Glu and two minima for Arg-Asp and Arg-Glu.24
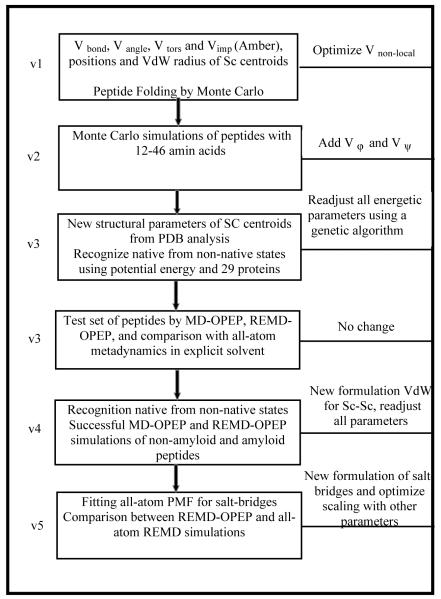
An overview of the optimization procedure is shown in the flowchart in Figure 2. The OPEPv1 and v2 parameters were adjusted by maximizing the energy gap between the native and misfolded states of six proteins, enabling the folding of 40 peptides of 12-46 amino acids consistent with NMR data in most cases.12-18 OPEPv3, which used a training and validating set of 13 and 16 proteins to optimize the parameters,20 was tested on a total of 11 proteins of 12-56 amino acids by MD,21 REMD21-22 or metadynamics64 starting from random or NMR structures. OPEPv4 passed two tests: 17 proteins of 37–152 amino acids remained within 3.1 Å root-mean-square deviation (RMSD) from their native states after 30-100 ns MD at 300 K, and REMD of five peptides with β-hairpin, α-helix or a WW domain, and REMD of the ccβ 51-residue peptide delivered structures consistent with experiment starting from random states.23
Fig.2.
Flowchart depicting the OPEP force field parametrization scheme.
Finally, the OPEPv5 parameters were tested on structurally diverse proteins differing in the number of charged residues by REMD.24 These include two 13-residue α-helix and 16-residue β-hairpin peptides and the ccβ-p2 peptide switching from a coiled-coil structure at low T to amyloid fibrils at higher T and concentration. We also verified that MD preserved the structures of proteins with 37-75 residues at 300 K. The final test involved an 85-residue protein with 19 charged amino acids. Running REMD of 24 replicas, each of 300 ns, the predicted TM is 360 K vs. 336 K experimentally. Overall, the OPEPv5 parameters, by refining the packing of the charged amino acids, impact the stability of secondary structure motifs and the population of intermediate states during temperature folding/unfolding; they also improve the aggregation propensity of peptides.24
In OPEPv5, the ε0 value in kcal.mol−1 or well depth at the minimum is 3.89 for the Ile-Ile contact and 4.05 for a Lys-Glu salt-bridge. The ε0 value, at the minimum, of an intramolecular H-bond is 3.3 and 2.7 kcal.mol−1 for (i, i+4) and (i, j ≥ i+5) interactions vs. 2.7 kcal.mol−1 for an intermolecular H-bond. The ε0 values of the 4-body H-bond terms are 1.4 and 3.6 kcal.mol−1 for α-helices and β-sheets, and any segments satisfying the conditions on ijkl. The two-body H-bond terms are cut off at an O-H distance of 0.3 nm and an angle N-H-O < 90° and the energy is modulated by a switching function of CHARMM-type65 from 0.25 to 0.3 nm. All other non-bonded interactions are cut off at 1.6 nm with a switching function starting at 1.3 nm.
Simulation Techniques
In what follows, we review the methods coupled to OPEP. These include the diffusion-controlled Monte Carlo (DCMC),12,16 the Activation-Relaxation Technique (ART-nouveau),66,67 molecular21 and Langevin dynamics,68 replica exchange (REMD)22 or Hamiltonian (H-REMD) MD,69 metadynamics,64,70 simulated tempering (ST)71, a greedy approach, 72,73 MUPHY57, and interactive MD simulations.
DCMC and ART
The basic idea of DCMC is to limit the search to conformations that are thermodynamically accessible from a given conformation in a reasonable time.12 In principle; one has to determine the nearest saddle point, the energy barrier and the contribution of entropy. Here, we assume that the motion results from the diffusion in (Φ,Ψ) space and the transition time scales as Θ2/D’η, where Θ is the angular deviation of the residue from one state to another, D’ is a diffusion parameter and η is related the ruggedness of the energy landscape. DCMC was used to fold 40 structurally diverse proteins.13-18,74-75
ART-nouveau goes one step beyond by generating non-biased pathways connecting adjacent local minima via exact first-order saddle points and was first developed for hard spheres.66 Coupled to OPEP, the procedure works as follows (Fig. 1B). First, the system is deformed from its current minimum in a random direction (all atoms for a peptide up to 15 amino acids, and a subset of atoms for larger proteins or oligomers) until the lowest eigenvalue of the Hessian becomes negative, and the system is pushed along this direction while the energy is minimized in the orthogonal directions. Once the saddle point is reached, the system is relaxed to the other side of the barrier and minimized, and finally the move is accepted depending on the Metropolis criterion.67,76-85
The advantage of ART, as any activated methods,86-88 is that it is not sensitive to the energy barriers allowing the system to move rapidly on the energy landscape. Even if ART lacks a proper thermodynamics basis, in contrast to a discrete sampling method,89 ART-OPEP simulations revealed frustration in the energy landscape of the 60-residue protein A with multiple funnels,85 consistent with global optimization of a 69-residue protein using basin-hopping and genetic algorithms with Gō energy models.90 ART-OPEP also predicted conformations of the Aβ21-30 peptide consistent with NMR,83 and located new minima and mechanisms for amyloid oligomers that were validated experimentally.
Molecular and Langevin Dynamics
Newton’s equations of motion are integrated using the velocity-Verlet method. Each main atom has its standard mass while the side-chain beads have a mass equal to the total mass of their atomic constituents.21 MD runs are performed with a time step of 1 fs or 2 fs using the SHAKE algorithm.91 The system is first minimized and then heated to the desired temperature. Production runs in the NVT ensemble are performed either with Berendsen thermostat92 and a coupling parameter τ = 0.5 ps or the Langevin thermostat68 with a collision frequency γ = 1 ps−1. Note we found little variation in the equilibrium structures and heat capacity curves of two model monomer and trimer peptides using the two thermostats.68 Simulations can be performed either in a sphere with reflecting boundary conditions or in a box with periodic boundary conditions. Presently, the non-bonded interactions are updated at each time step, but the code can be easily improved in terms of CPU efficacy by using a multiple time step framework.
REMD and H-REMD
REMD simulations are carried out with a number of replica running in parallel and a temperature range dependent on the system size.22,93 For instance, we found that 8 replicas for 50 ns, 20 replicas for 600 ns and 22 replicas for 1200 ns are sufficient for the dimers of Aβ16-2294 and Aβ16-35,95 and the trimer of Aβ16-35,96 respectively to reach equilibrium. An exponential temperature distribution is used and exchanges between two consecutive replicas are attempted every 5.0 to 7.5 ps, leading on average to an acceptance ratio of 30-40%.22 To enhance sampling, it is useful to combine REMD with a Hamiltonian exchange procedure,97 where we use at the highest temperature several replicas with reduced non-bonded energies.69
We assess convergence of the simulations near the physiological temperature by using different time intervals and metrics. These metrics are the distributions of the radius of gyration and end-to-end distances, the secondary structure along the amino acid sequence and the total number of clusters.95,98 Convergence is also verified by the curves of the heat capacity and conformational entropy using different time windows.95,99
ST
In simulated tempering, temperature is a dynamical variable taking discrete values Tn. Standard ST requires the determination of a priori unknown weight parameters to ensure a random walk in T space, the Helmholtz free energies at Tn.100,101 Recently, we developed an ST algorithm with on-the-fly weight determination. The weights are self-updated via a trapezoid rule during the run,71 eliminating the need of trial simulations,102 or complicated update schemes.103,104 The advantage of our ST method over REMD was demonstrated using OPEP on Ala20 and the Aβ16-22 trimer.101 The same efficiency is observed in explicit solvent for Ala10, the 20-residue Trp-cage and the 37-residue WW-domain starting from random states and deviating by less than 0.2 nm RMSD from the NMR structure after less than 700 ns (in preparation).
Greedy Algorithm
This method differs from MC and genetic algorithms by growing a chain one fragment after another.105-107 Our procedure for structure prediction performs a rigid assembly of fragments of 4-residue length by superimposing the first three α-carbons of the new fragment onto the last three of the previously built structure. Our early version used forward (from N- to C-terminal) and backward (from C- to N-) operators to grow the chain.108,109 Our new version uses a zip operator to start the building process at any randomly chosen position, alternatively adding one residue at each side of the growing structure.110 At each position, the algorithm keeps 3000 states, the 1000 energetically best OPEP states and 2000 randomly selected ones in the pool of the remaining generated conformations.110
Metadynamics
Metadynamics is an advanced technique for enhancing sampling in MD simulations,70 with widespread applications in material science and chemical reactivity,111 protein-drug recognition,112 protein aggregation,113 and allosteric pathways.114 Enhanced sampling is achieved by introducing an external, history-dependent bias potential affecting few selected degrees of freedom, usually referred to as collective variables (CVs). The bias is adaptively constructed as a sum of Gaussians deposited along the system trajectory in CV space to discourage the system from revisiting regions that have already been explored (Fig. 1C). If the CVs capture all the slow, relevant degrees of freedom of the system, metadynamics provides a correct estimate of the system free-energy surface. An appropriate application of metadynamics requires the identification of a limited yet effective set of CVs. This may represent an intimidating task when dealing with extremely complex molecular processes. This limitation can be circumvented by combining metadynamics with replica exchange methods.115 This scheme, where several metadynamics runs are performed using the same CV set at different temperatures and are swapped following a Metropolis criterion, has been applied to assess the quality of the OPEPv3 potential with respect to the all-atom OPLS and AMBER99SB force fields in explicit solvent.64 Using two β-hairpin and α-helix peptides and an intrinsically disordered peptide, and by comparing the free energy surfaces (FES), the free energy differences between the folded and unfolded states and between the folded state and the transition state, we found remarkable agreement between the OPEP FES at 345-360 K and those using all-atom calculations in explicit solvent at 300 K.64 This information was used to improve the model during the refinement of OPEPv4 and OPEPv5.
MUPHY
This name refers to a way to embed molecules of generic complexity in a hydrodynamic solvent. The interaction of proteins with the surrounding solvent implies accounting for the hydrodynamic interactions exerted between particles of the macromolecules. As one particle moves in space, it creates a velocity field in the environment that acts on other particles, therefore generating effective, solvent-mediated interactions. One way to include hydrodynamics is the Brownian Dynamics technique developed by McCammon.116 However, because hydrodynamic interactions are in principle long-range, the basic technique has a computational cost proportional to cube of the number of particles, so that it is computationally extremely difficult even to handle a small set of proteins in suspension.
The MUPHY software has been developed to study generic biofluidics systems117 and, in order to specialize to protein suspensions, has implemented OPEP.57 MUPHY handles the dynamical evolution of generic fluids and particles treated via the dual mechanism, a mesh-based Lattice Boltzmann method for fluids, and specialized versions of MD for particles.117 Indeed, a powerful alternative to McCammon’s method is provided by explicitly solving for the evolution of the solvent, as encoded by the Navier-Stokes equations, by using the Lattice Boltzmann (LB) numerical method.118 In this approach, the coupling between fluid and particles takes place via specifically designed kernels based on kinetic modeling, significantly distinct from methods based on macroscopic hydrodynamics. Such a methodology is genuinely multiscale as it entails different levels of physical description (such as field-based for the fluid and particle-based for the proteins) within a single unifying framework.119
In LB, the solvent is described via the “populations” representing the probabilities of finding solvent molecules at a given position and time t and moving along a discrete direction p. The populations, represented over a mesh, evolve as: , where is the discrete Maxwell-Boltzmann equilibrium, associated to the weight Wp and discrete speed , with n = Σp fp and being the fluid density and velocity, respectively. The term Δfp is proportional to the drag force exerted by the fluid on the particle and vice versa, being a bidirectional of fluid and particles according to the action-reaction principle. Thus the drag force, together with a stochastic force, is included in the particles’ evolution besides the mechanical forces stemming from the OPEP force field.
The fluid-proteins concurrent evolution can be specialized to hybrid situations, such that the hydrodynamic interactions are decomposed into intra- and intermolecular components, the first ones evaluated analytically and the second ones handled via an under-resolved version of the LB fluid. Such a decomposition proves further advantageous in terms of CPU efficiency as it ideally balances the cost of computing intra- and intermolecular hydrodynamic interactions.120 Finally, MUPHY is fully parallel and distributes the computational load on multiple cores or multiple Graphical Processing Units (GPUs).
Interactive Molecular Dynamics (IMD)
Using our previously developed MDDriver software library,121 we rendered the CG simulation engine interactive by implementing the IMD network protocol and interactive steering modules. Using a TCP (transmission control protocol) network socket, any IMD-protocol-aware frontend is able to connect to the running OPEP/Hire-RNA simulation engine and inject additional user-forces to drive the experiment. For initial validation, we used both our custom UnityMol122 and the more widely distributed VMD123 frontends. Simulation and frontend may be run on the same machine or remotely to optimize performance and fluidity.
OPEP applications
Though OPEP was used to explore large-scale motions, such as the pathways from the holo to the apo states of two EF-hand proteins,124 or the conformations of 8-20 amino acid loops,125 we focus here on the following timely topics: self-assembly of amyloid proteins, structure prediction of linear and disulfide bonded cyclic peptides, thermodynamic properties of RNA, and protein dynamics in a crowded environment with hydrodynamics. Table 1 gives a summary of the different systems simulated with OPEP indicating the methodology adopted and the total time lengths.
Table 1.
Summary of the different systems studied with the CG force field.
| Applications | Methodology used | Total Time | References |
|---|---|---|---|
| Αβ1-40, Αβ1-42, | HT-REMD | 70.0 μs | 167 |
| WT(D23N) monomers and dimers | 97.5 μs | 168 | |
| Trimers of Αβ17-42 and interactions with drugs | REMD | 16.4 μs | 96 |
| Aggregation of 3- to 20-mers of amyloid fragments | MD ART REMD |
30.0 μs no 120.0 μs |
149, 150, 152 80,150 23, 68, 99, 149-153 |
| Size of the primary nucleus for fibril formation | MD REMD |
33.8 μs 90.0 μs |
158 99, 153 |
| Peptide Structure Prediction and Conformations of protein fragments | PEP-FOLD | no | 177, 178, 180, 182-189 |
| Protein-peptide interactions | PEP-FOLD | no | 190-194 |
| Design of immunogenic and antiviral peptides | PEP-FOLD | no | 195-201 |
| Impact of macromolecules and hydrodynamics | MUPHY/OPEP | no | this work |
| RNA and DNA folding | MD, REMD, ST | 85.0 μs | 231, 233, this work |
| Thermophilic and mesophilic proteins | REMD | 72.0 μs | this work |
| Impact of shear flow | MUPHY/OPEP | 30 ns | this work |
| Virtual reality | Interactive MD | 0.2 ns | this work |
Understanding the self-assembly of amyloid proteins
Alzheimer’s disease (AD) is marked by atrophy of cerebral cortex and loss of cortical and subcortical neurons. Autopsy 75 reveals accumulation of amyloid plaques and numerous neurofibrillary tangles made of filaments of the phosphorylated tau proteins. The major constituents of plaques are made of the amyloid-β (Aβ) peptides of 40 and then 42 amino acids formed from the amyloid precursor protein via the actions of the β- and γ-secretases.126 But many truncated variants, such as Aβ1-30 and Aβ1-26, and Aβ with proteolytic removal of D1 and A2 and subsequent cyclization of E3 to a pyroglutamate, have been detected by mass spectrometry in human AD brains.127,128 The human Aβ1-42 sequence, designated Aβ42, is DAEFRHDSGYEVHHQKLVFFAEDVGSNKGAIIGLMVGGVVIA with a charged N-terminus (A1-K16) and two hydrophobic patches L17-A21 (central hydrophobic core, CHC) and A30-A42 (C-terminus) separated by a hydrophilic patch E22-G29. Despite many clinical trials, drug after drug has failed to slow the progression of AD for three main reasons.49
While the experimental sigmoidal kinetics of amyloid formation with a lag-phase can be accounted for by means of primary classical nucleation theory (CNT) and/or secondary (fragmentation or lateral) nucleation processes,129,130 we lack information on the topology, structure and size of the primary nucleus (N*).
Secondly, though the low molecular weight (LMW) Aβ40/42 aggregates are the most critical players in the pathology, we have little information on their structure, rate and extent of formation. Due to their high aggregation propensity, the LMW oligomers are not amenable to solution nuclear magnetic resonance (NMR) and X-ray crystallography. As a result, only low-resolution structural data from circular dichroism (CD),131 Fourier transform infrared spectroscopy (FTIR),132 ion-mobility mass spectrometry (IM-MS),133 solid-state NMR,134 pulsed hydrogen-deuterium exchange coupled to MS,135 transmission electron (TEM) and atomic force microscopies (AFM) are available.136 The final Aβ40/42 products are insoluble and only solid-state NMR models are available. While fibrils of synthetic Aβ40/42 peptides display perfect U-shaped forms with β-strands spanning the CHC and the C-terminus, and the N-terminus disordered, fibrils of AD-brain derived Aβ40 peptides show deformed U-shaped states and, remarkably, the structure varies from one patient to another.137 A common feature of all fibrils is the inter-digitation of the side-chains, the so-called steric zipper.138
Thirdly, though the general consensus is that drugs are given too late,49 we lack the structures of Aβ40/42 peptides with known inhibitors of aggregation and toxicity, paving the way for the design of specific drugs with the highest affinities for Aβ40/42 oligomers. Overall, OPEP simulations have played a significant role to complement experiment on five aspects.
-
(1)
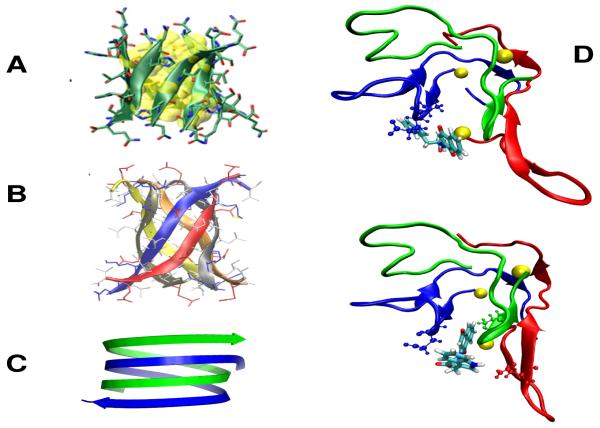
Independently of the force field and the sampling method, self-assembly starts by a hydrophobic collapse and the formation of molten oligomers, which is modulated by the degree of hydrophobicity of the peptide. Then, the H-bonds drive the system to highly flexible and transient β-rich oligomers.139-141 These β-rich oligomers have various topologies, and we were the first using OPEP simulations to (i) observe assemblies with various sheet-to-sheet pairing angles79,81,94 that were confirmed by structures of macrocyclic β-sheet mimics,142 and other force field calculations,43,140,141 (ii) evidence β-barrels (Fig. 3A)79,143 that were validated by the X-ray structure of a toxic hexamer of a 11-residue amyloid peptide (Fig. 3B),144 (iii) predict formation of antiparallel double stranded poly-L-glutamine nanotubes with 22 residues per turn (Fig. 3C),145 reminiscent of the water-filled model proposed by Perutz,146 and (iv) identify reptation moves of the β-strands in the late steps of aggregation78 that were validated by FTIR147 and atomistic simulations.140,148
-
(2)
The aggregates of 7- to 20-mers of GNNQQNY, NNQQ, Aβ16-22, KFFE and NHVTLSQ using OPEP are mostly amorphous and consist of a heterogeneous ensemble of β-rich states.99,149-153 In all systems, the transition at the melting temperature involves a change in the distributions of oligomer and β-sheet sizes, but mixed parallel/antiparallel (P/AP) β-strands dominate.99 This β-strand mismatch, observed with various force fields,38,139,154 provides strong evidence that one limiting-factor for fibril formation is the transition from mixed P/AP to fully P or AP strands. This was confirmed by bias-exchange metadynamics of 18Val8 and 18Aβ35-40 peptides in explicit solvent, where the crossing of the highest free energy involves the transition from mixed P/AP to P β-strands that can be accompanied by the formation of the steric zipper.113,155
-
(3)
A fundamental question pertains to the size of the primary nucleus, N*. Recently, two atomistic simulations in explicit solvent and one OPEP simulation have provided insights into N*. In the first study, an effective nucleus size on the order of 14 was proposed for 18 Val8 peptides by metadynamics.113 In the second atomistic study, the aggregation of 16Aβ37-42 peptides was investigated by REMD,156 and the population of 4-5 fully P β-strands, consistent with the fibril structure, was 1-2% at 300 K. Whether N* is around 15 for Aβ37-42 as for the Val8 system cannot be determined, due to finite-size effects and the fact that fibril formation is under kinetic and not thermodynamic control as evidenced experimentally157 and by Langevin dynamics of a mesoscopic model.48 Using unbiased MD-OPEP, we investigated the onset of aggregation in a 20-mer of GNNQQNY.158 Running 16.9 μs at 280 K and 300 K, we showed that aggregation follows the CNT and N* is 4-5 at 280 K and 5-6 at 300 K. The kinetics of growth cannot be fully described by the CNT, however, because there are important rearrangements after the nucleus is formed, as the aggregates attempt to optimize their organization.158
OPEP simulations do not systematically show fibril formation. One reason found for the peptide spanning the residues 144–153 of the prion protein is that oligomerization is not thermodynamically favorable, in agreement with a turbidimetric experiment.23 Another reason is the presence of a proline which can either destabilize the β-strand conformation of the monomer and totally prevent aggregation, or reduce the packing of β-sheets rendering fibril formation a slow process,80,150 consistent with experiments.159 Overall, many factors, in addition to pH and T, modulate amyloid formation (N* size, fibril topology and lag-phase) ranging from the energy landscape of the monomer,43,48,160 the entropy of the loops161 or the intrinsic disorder of the whole peptide,162,163 to the supersaturation of the protein solution.164,165
-
(4)
The solution NMR structure of the Aβ42 monomer reveals weak β-strand propensities at the CHC and the residues I31–V36 and V39–I41, and turns at D7–E11 and F20–S26.166 NMR relaxation data reveal that Aβ42 is more rigid at the C-terminus than Aβ40. IM-MS reports a collision cross-section of 1256 Å2 for Aβ42 dimers.133 Using different preparation methods, CD leads to a β-strand between 12% and 25% and an α-helix between 3% to 9% at 295 K, pH 7 and day 0, i.e. for an heterogeneous ensemble of oligomers.131,133 Remarkably, the Aβ40-D23N peptide forms fibrils with in-register antiparallel and parallel β-sheets under quiescent and strong agitations, respectively.157
To get insights into Aβ flexibility, we determined the free energy landscapes of the monomers167 and dimers168 of Aβ40, Aβ42, and with D23N using H-REMD-OPEP. We found that if the three monomeric alloforms are mostly disordered, in agreement with experimental data166 and confirmed by all-atom simulations,169-171 they display distinct morphologies. Aβ42 and Aβ40-D23N have higher β-strand propensities at residues 30–42 than Aβ40. D23N changes the Aβ40 structures; the residues 1–16 becoming more independent of the rest of the protein,167 which may explain in part why the kinetics and the final products vary between Aβ40 and Aβ40-D23N under quiescent agitation. Our results on the dimers showed that Aβ42 has a higher propensity than Aβ40 to form β-strands at the CHC and residues 30-42, explaining the higher Aβ42 aggregation kinetics.168 In none of the systems we observed any parallel β-sheet structure between the two CHC’s. D23N impacts the free energy landscape by increasing the population of states with higher β-strand propensities at the C-terminal and antiparallel β-sheet between the two C-termini, and this motif could be important in the nucleation of Aβ40-D23N toward parallel β-sheets. Our results also revealed many configurations stabilized by N-terminal interactions168 that were observed by single-molecule atomic force spectroscopy172 and all-atom REMD simulations.173
-
(5)
Based on the microcrystal structure of Aβ16-21 fibrils with the dye Orange G, Eisenberg designed compounds that reduce toxicity by preventing fragmentation of the Aβ42 fibrils without binding to the oligomers.174 Despite many experimental attempts, scientists have not succeeded to provide the structures of Aβ40/42 monomers or Aβ40/42 oligomers with inhibitors. Using a shorter fragment, Segal solved the NMR structure of NQTrp bound to the Aβ12-28 monomer, revealing three dominant binding sites between NQTrp and the Aβ18-21 region.175 As a first step toward understanding the interaction of Aβ oligomers with NQTrp we focused on the Aβ17-42 peptide also found in AD plaques and used a multiscale procedure.96 Our extensive OPEP-REMD simulation of the Aβ17-42 trimer, followed by all-atom docking of five molecules on the most populated Aβ structures, showed that NQTrp is a more favorable inhibitor than EGCG, 2002-H20 and resveratrol. In agreement with the NMR structure of NQTrp/Aβ12-28,175 NQTrp binds to Aβ through the side chains of F19 and F20 and the main chain atoms of F19-E22. Our simulations reveal, however, many transient binding sites (Fig. 3D),96 consistent with all-atom REMD of the Aβ1-42 dimer with 2NQTrp,176 indicating that the design of more efficient drugs targeting the Aβ42 dimer is not an easy task.
Fig.3.
Amyloids. (A) The β-barrel of the β2m(82-87) peptide as predicted by OPEP.79,143 (B) The hexamer of the KV11 peptide consisting of six antiparallel β strands forming a barrel as determined by X-ray crystallography.144 (C) The predicted OPEP antiparallel double stranded poly-L-glutamine nanotube.145 (D) Two binding modes of the NQTrp drug to the Aβ17-42 trimer as predicted by our multiscale simulation. The yellow balls indicate the 17th amino acids and the drug is shown with all-atoms.96 Top view also shows all atoms of A21 and E22; bottom view shows all atoms of E22 (blue) and V39 (green).
Fast and Accurate 3D Peptide Structure Prediction
Peptides have regained considerable interest as they represent alternative ways to design therapeutics, vaccines or molecular probes. However, fast and accurate peptide structure determination remains a long-standing goal in structural biology and peptide engineering.34 Pep-Fold is an innovative approach aimed at de novo structure prediction of linear and disulfide bonded cyclic peptides with 9-52 amino acids (aa).177-178 Pep-Fold relies on a Hidden Markov Model derived structural alphabet (SA) of 27 letters to describe proteins as series of overlapping fragments of four aa.179 The SA letters can be assimilated to a generalized secondary structure, extending the number of states from 4 (α-helix, coil, turn or bend, and β-strand) to 27, but not all transitions are possible between two consecutive letters. The Pep-fold procedure consists in three steps. First, Pep-fold predicts a limited set of SA letters at each position from the sequence, and then performs a progressive assembly of the prototype fragments associated with each selected SA letter using our greedy algorithm108-110 driven by OPEP. As Pep-Fold uses a rigid assembly, we found necessary to smooth the OPEP side chain - side chain potential.177 The third step refines the CG models by Monte-Carlo before generating all-atom models and performing a clustering of all models returned by the simulations.
Pep-Fold1 efficiency was shown on 24 linear peptides of 9-25-aa in aqueous solution and neutral pH by predicting lowest-energy states with a mean 2.5 Å RMSD from the NMR rigid cores (RC, excluding the flexible parts).110 Pep-fold2, which revisited the prediction of the SA letters from the sequence and considers several filters to generate a variety of SA trajectories, was tested on peptide lengths up to 36-aa.178 The server allows the biologists or chemists to define S-S bonds or any residue-residue contact. Using 34 peptides with one to three S-S bonds, the best Pep-Fold2 models had a RMSD of 2.7 Å from the full NMR structures. Using 37 linear peptides, Pep-Fold2 located lowest-energy states with a 3 Å RMSD from the NMR RCs. We also showed the gain in the identification of the native state by filtering the Pep-Fold2 models using the backbone proton chemical shifts easily available from 2D NMR.180
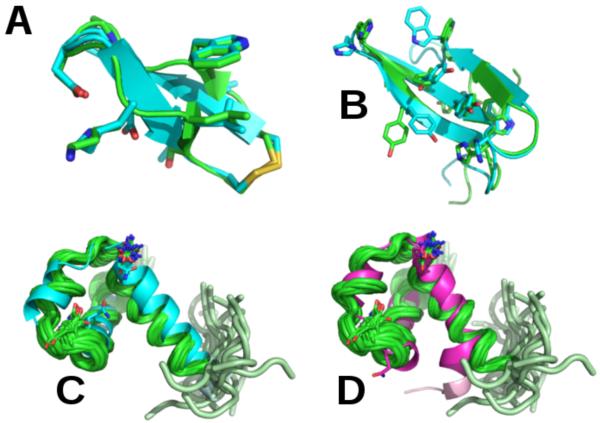
Finally, Pep-Fold2 was compared to the state-of-the-art Rosetta program on 56 peptides with 25-52-aa.180 Rosetta starts sampling with a CG model and fragment assembly MC, and then through successive steps, selects models for all-atom refinement.18-19 By using a total of 200 Rosetta and Pep-fold runs for each peptide, and a new Binet-Cauchy (BC) score,181 the mean BC score of the best models (lowest RMSD with respect to NMR) generated by Rosetta and Pep-fold are 0.83 and 0.87 (in preparation). While Rosetta generates high quality models (BC score > 0.9) for 34 targets vs. 29 for Pep-fold, suggesting that Pep-fold could benefit from an all-atom sampling refinement, Pep-fold generates near-native or native states for 53 peptides vs. 49 for Rosetta (BC score > 0.6). Fig. 4 shows the predicted structures of four peptides.
Fig.4.
Structure predictions superposed on the experimental structures. (A): Best Pep-fold model of the peptide code PDB 1n0a (11-aa, BC score = 0.94) with one S-S bond; (B): Best Pep-fold model of the peptide 1e0m (37-aa, BC score = 0.88). In (A) and (B), we show the all-atom representation of some side chains; (C) and (D): Best Pep-fold and Rosetta models of the peptide 2j8p (49-aa, BC scores = 0.93 and 0.88) superposed on the 20 NMR structures and showing the flexibility of one extremity. Green: experimental conformations.
Pep-Fold is freely available as a web server110,178 and has proven to be very useful by many scientists for different applications that can be broadly classified into six categories. The first application is predicting the conformations of protein fragments.182 Structural characterization of the C-terminal 27-aa tail of HIV gp41 remained relatively limited and contradictory. Pep-Fold tail models showed conserved α-helix structures despite significant sequence variations among diverse clades, and this is supported by CD.183 3D models of the N-terminal 20-aa of human cytochrome c and several cytochrome c2 variants from R. capsulatus were also generated by Pep-Fold and helped understand why the insertion of an alanine residue between Phe11 and Cys15 and substitution of residues Glu8 and Glu10 are critical for heme attachment by the mitochondrial protein holocytochrome c synthase.184 Pep-Fold was also used to generate protein N- and C-terminal conformations.185 Certain immune-driven mutations in HIV-1, such as those arising in p24Gag, decrease viral replicative capacity. In HIV-1 subtype B, the p24Gag M250I mutation is a rare variant, while in subtype C, it is a relatively common minor polymorphic variant (10 to 15%). The structural implications of M250I were predicted by Pep-Fold to be greater in subtype B versus C, providing a potential explanation for its lower frequency and enhanced replicative defects in subtype B.186 In addition, Pep-fold was used to model protein loops187 or protein linkers.188,189 A study on the linkers in a new class of modular alpha-amylases showed that the Pep-fold conformations are diverse, but match the data obtained from small-angle X-ray scattering.189
The second application is related to protein–peptide interaction in general, fundamentally important for signal transduction, transcription regulation and protein degradation.190-194 Wu used Pep-Fold to generate the structures of 13 peptides of 20-aa as initial structures for short MD and showed a very good correlation between the experimental and the calculated MM-PB/SA binding free energies for the peptides interacting with the vascular endothelial growth factor A.191 Kumar used the Pep-fold conformations of several 20-aa peptides to explore their binding mechanisms to calmodulin,192 Chopra used Pep-Fold for the design of a peptide able to bind to Bacillus anthraxis toxin-antitoxin module,193 while Stegman used Pep-fold prior to docking onto peptidyl-prolyl cis/trans isomerase PPIL1, a component of the human spliceosome.194
The third domain of application is the design of immunogenic peptides. Peptides play many roles in immunology, yet none are more important than their role as immunogenic epitopes driving the adaptive immune response against infectious disease.195 Peptide epitopes are mediated primarily by their interaction with major histocompatibility complexes (T-cell epitopes) and antibodies (B-cell epitopes). In this context, Wingren reported the first detailed analysis of antibody–peptide interaction characteristics, by combining large-scale experimental peptide binding data with the structural analysis of eight human recombinant antibodies and numerous peptides using Pep-fold, targeting tryptic mammalian and eukaryote proteomes.196
Another application concerns antiviral peptides (AVP) and vaccines. Pep-Fold contributed to the design of peptides inhibiting in vitro the Influenza A virus197 or other viruses198,199 and is now defined in AVPdb, a server allowing the design of AVP.200 Pep-fold was also used to design a DNA vaccine against human papillomavirus causing cervical cancer.201
Although OPEP has been optimized for aqueous solution, Pep-fold has been used on peptides in an apolar milieu,202-204 and in particular on antimicrobial peptides (AMP) regarded as one of the most promising alternatives to antibiotics affected by resistance mechanisms. Using in silico predictions including Pep-Fold and in vitro assays led to the discovery of potential AMPs with high activity and low toxicity from the entire human genome.204
Finally, Pep-fold has been found useful in understanding the solvent-dependent CD spectrum of a 24-aa peptide corresponding to the tubulin-binding site of the neurofilament light subunit205 and the effect of gold nanoparticle conjugation on peptide structure and dynamics.206 Pep-fold has been used in various design situations: new molecules for induction of bone formation,207 peptides binding lipids,208 peptides coating carbon nanotubes,209 and a peptide-based Hsp90 inhibitor leading to a novel anticancer agent210 that will enter preclinical trials conducted on patients with breast cancer, prostate cancer and skin cancer. To date, there is only one case of conflict between Pep-Fold and in vitro results. While Gautam designed 15-aa peptides with coil-turn CD, Pep-Fold predicts β-hairpins, but the experimental conditions (pH and ionic strength) are not reported.211
A framework for RNA and DNA coarse-grained models
In many vital cellular processes, especially in regulatory functions related to transcription and translation, proteins interact with nucleic acids. We recently developed a nucleic acid CG model, called Hire-RNA/DNA, by following the physical principles used for OPEP, in order to better understand the thermodynamics and dynamics of RNA/DNA.
The most widely used all-atom force field for nucleic acids is undoubtedly AMBER with ff99 achieving a good agreement with experiment for DNA double helices.63 The parameters are, however, constantly adjusted to better represent non-canonical structures in loops and bulges,212 and a new parameterization obtained by reproducing known thermodynamic and kinetic measurements of RNA monomers and dimers was just reported allowing de novo folding of three hyperstable RNA tetraloops to 1–3 Å RMSD from their experimental structures.213 Folding a single stranded RNA free of any biases remains, however, a computer challenge for nucleotide (nt) lengths > 20.61
Different strategies are applied to go beyond all-atom simulations and they can be organized into three categories:214 homology modeling, hybrid and ab initio methods. Homology modeling works well, if one can find a good template in the NDB, but this is typically not the case for single stranded RNAs.215,216 Hybrid methods based on knowledge-based energy functions vary from fragment reconstruction (MC-Fold and MC-Sym),217 fragment assembly (FARNA)218 to multiscale approaches relying on 2D structure predictions, CG 3D models based on the fragments selected from the NDB followed by all-atom minimization,219 or junction topology prediction and graph modeling followed by all-atom refinement.220 However, the best 2D structure prediction algorithms reach only 60% accuracy.221 Another bottleneck is the low population of non-canonical Watson-Crick (WC) base pairs in the NDB, and the prediction of pseudo-knots and junctions. This problem is also faced by ab initio CG force fields, built from atomistic simulations222,223 and electronic structure calculations223 or by using experimental data to assign parameters,224-226 e.g., iFold,225 or derive statistical potentials,227-229 e.g., NAST227. Most methods were recently evaluated in RNA-Puzzles and predicted a dimer of 46-nt with a RMSD from 0.34 to 0.69 nm, a 100-nt square of double-stranded RNA with a RMSD from 0.23 to 0.36 nm, and a 86-nt riboswitch domain with a RMSD from 0.72 to 2.3 nm.35
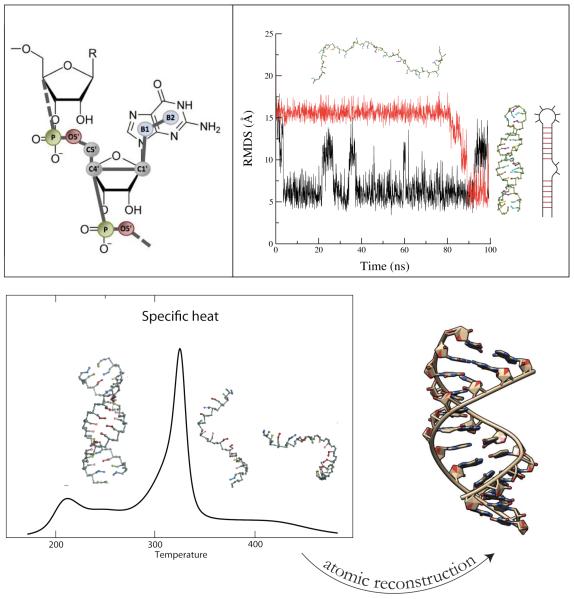
Following OPEP, our CG model has an energy function derived from physical intuition and parameters based on known structures. Among all CG models, our CG representation has the highest resolution, with an explicit representation of the heavy atoms of the sugar-phosphate backbone (P, O5’, C5’, C4’ and C1’), one bead for pyrimidine bases (C and U) and two beads for the purine bases (G and A), see Fig. 5. For comparison, the NAST,227 iFold225 and Xia228 models have 1, 3 and 5 beads, respectively. Note OH is not treated explicitly and therefore the distinction between RNA and DNA in our models is done solely on different equilibrium angles and torsions, with some angles and torsions allowed for one molecule but inaccessible to the other, and different base pairs, with DNA making only a subset of all RNA base pairs.230
Fig.5.
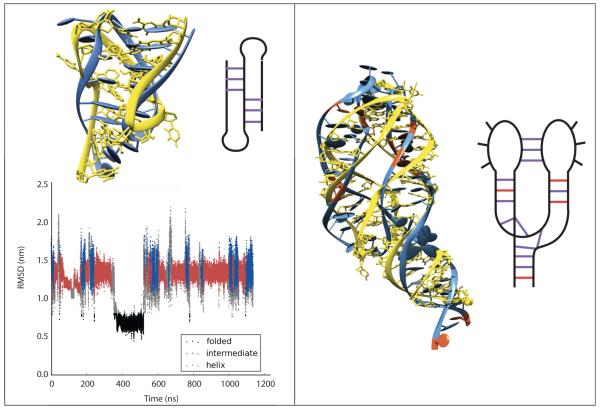
Hire-RNA model. Top, Left: Representation of a Guanine nucleotide with 7 beads. Top, Right: MD of the 36-nt 1N8X hairpin with Hire-RNAv1 recovering the native structure from a fully extended state. Secondary structure of the hairpin is shown on the right. Bottom, Left: Heat capacity plot of a 36-nt RNA duplex with HiRE-RNAv2. Bottom, Right: With our reconstruction algorithm, the predicted all-atom structure behaves similarly to the experimental structure using all-atom MD in explicit solvent.
The particles in Hire-RNAv1231 interact via standard local terms for covalent bonds, bond angles and dihedral rotations, an electrostatic repulsion between phosphate groups, a modified Lennard-Jones potential for long range van der Waals interactions as used in OPEPv4-v5 and base pair terms. Base pairing is the most crucial interaction and we treat it with more detail than all other top-down ab initio models. A two-body term, as in OPEP, depends on the relative distance and angles formed by two base beads interacting through their WC sides, see Westhof’s classification.232 All the bases can form, and not just A-U and G-C, with different strengths. A three-body repulsive term prevents different bases to simultaneously interact, even though transient multiple pairs can form, and a four-body term, as in OPEP, helps stabilize pairs of consecutive bases.
With Hire-RNAv1, we folded two RNA of 26- and 40-nt into hairpins from fully extended states by MD (Fig. 5). Running REMD, we showed that the NMR configuration is the most populated structure at low T.231 In a second study, we slightly modified the form of the two-body and four-body terms for base pairs and with Hire-RNAv2, we examined the assembly of DNA and RNA duplexes by REMD.233 For the two RNA and the DNA consisting of 36- and 24-nt, we calculated the heat capacity curves and found one transition from an assembled state (RMSD of 0.18-0.26 nm with respect to the crystal structure) to disassembled states (Fig. 5). In principle, RNA could fold on itself and form a hairpin, but this is not a favorable free energy state for our nt sequences. The melting temperatures we find for the three systems deviate at most by 17 K from the values obtained by the HyTher algorithm, a reference in the field.234 Overall, the same energetic parameters perform well for single- and double-stranded systems of 40-nt, and based on our algorithm generating an all-atom model from a CG state (Fig. 5), we showed the equivalence of MD results using AMBER ff99 with explicit ions and water starting from our REMD-predicted and the experimental structures for both RNA and DNA.233
Simulations of protein suspensions with hydrodynamics
In the last few years macromolecular crowding has been the subject of several investigations since it has crucial implications on cell functioning.235,236 There is increasing evidence that macromolecular crowding exerts large effects on the protein mobility, association and stability.237,238 It is generally thought that crowding serves as a means of confining proteins in space, where enzymatic activity is undertaken. Also, data suggest that at high concentrations proteins non-specifically enhance association rates, with in vivo and in vitro rates and equilibria differing by orders of magnitude. Macromolecular crowding also affects hydration structure and dynamics,239 and protein conformational change.240 Evolution has fined-tune microtubule-based motor proteins to deliver cargoes rapidly and reliably throughout the cytoplasm by having molecular properties that prevent them from forming traffic jams.241
Hydrodynamic and excluded volume interactions are likely the two main factors that account for the large reduction of protein diffusivity in crowding conditions.242 While hydrodynamics interactions do not alter the equilibrium distribution of states of a system, they potentially affect the local dynamics as well as the escape from metastable states characterizing the spatially and energetically heterogeneous crowded system. For example, hydrodynamics has a primary role on the transport properties, as for the translational and rotational diffusivities, and in general the importance on the dynamics of suspensions is well known. Along with simulations, diffusional data can be accessed experimentally via quasi-elastic neutron scattering, single molecule tracking, fluorescence correlation spectroscopy and fluorescence-recovery-after-photobleaching,243 so the multiscale approach can be directly compared with in vitro and in vivo data.
One large-scale MUPHY/OPEP application is offered here to illustrate the potentiality of the coupling of the CG force field with hydrodynamics interactions. A large system composed of 17576 Rat1 yeast proteins in solution is simulated at 300 K for 30 ns. Rat1 is a 666-aa protein that functions primarily in the nucleus and plays an important role in transcription.244 Altogether we consider a system of 70 million particles, each Rat1 having 4013 particles. To account for the solvent, a hybrid LB/Brownian Dynamics scheme with a time step of 1 fs was used on the Titan supercomputer, exploiting 17576 GPUs in parallel.120 The highest volume fraction considered (40%) emulates the crowding conditions found in the cytoplasm, typically with 20-30% of the cytoplasmic volume occupied by proteins, nucleic acids and other macromolecules. As a result, the distance between proteins is comparable to the size of the proteins.235
During the evolution of the Rat1 suspension, proteins move and tumble together. Fig. 6 shows the typical protein configuration in the suspension, in particular by highlighting the hydrodynamic “bubble” that each protein carries along, representing the isosurface of constant velocity surrounding proteins. Each bubble is further distorted and connected with those generated by neighboring proteins. Visual inspection of the flow streamlines reveals that, as proteins move, they generate a substantial accompanying drain on the solvent. Even at physiological concentrations, the streamlines travel mostly undisturbed over several protein sizes, that is, distant proteins effectively experience solvent mediated mutual interactions.
Fig.6.
MUPHY/OPEP suspension. (A) Snapshot highlighting the Rat1 proteins and the solvent velocity field shown in transparency. (B) A zoom showing the Rat1 secondary structures and the local solvent velocity field. (C) Flow streamlines generated by three selected proteins in the suspension in a single timeframe.
Crowding is generally thought to induce sub-diffusive and slow dynamics on the short timescale and diffusive dynamics at longer times.245 In principle, the coherent long-ranged organization of the solvent flow field can act on the suspension as a lubricant, in order to facilitating the protein motion. On the other hand, the hydrodynamic field can interfere with the protein motion, since viscous dissipation can drain momentum away from the suspension.
In the following, we illustrate simulation data for the Rat1 suspension for the translational diffusion coefficients that pertain to the short-time dynamics (10 ns). The diffusion coefficient is shown in Fig. 7 and is evaluated via the integration of the protein center of mass velocity autocorrelation function. For the sake of comparison, experimental data on the translational coefficient obtained by quasi-elastic neutron scattering for the bovine serum albumin protein are also shown.
Fig.7.
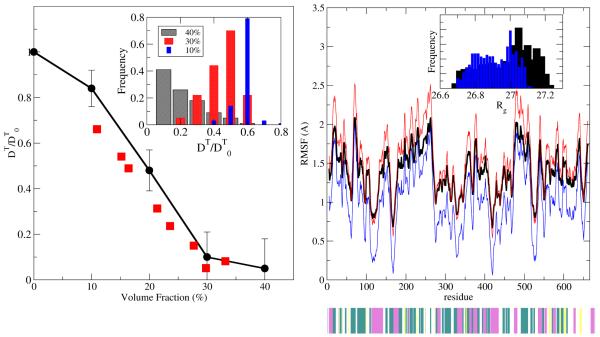
MUPHY/OPEP results. Left: Translational diffusion coefficients at various volume fractions (black circles) are compared to the experimental data of bovine serum albumin246 (red squares). The diffusion coefficient is normalized by the value at virtually zero volume fraction, and the solid line is a guide to the eye. Inset: Histogram of diffusivity stemming from the ensemble of proteins. Right: RMS fluctuations from the crystallographic structure at 40% volume fraction (black curve). Structures with maximal (red) and minimal (blue) departures from the average value are shown. Secondary structure is indicated as a lower bar with colour green (turn), yellow (β-strand), magenta (α-helix) and white (coil). Inset: Histogram of Rg values.
On the considered timescale, diffusion shows anomalous behavior, in the sense that the effective mean square displacement does not scale linearly with time but is rather subdiffusive (data not shown). As Fig. 7 shows, the translational diffusion coefficients provide similar, although systematically larger values than the experimental ones, probably related to the larger temporal scale accessed by the simulation as compared to the one pertinent to the scattering spectra (3.5ns < τexpt < 5 ns).246 This temporal window exceeds the hydrodynamic one, which arises from the propagation of vorticity over the protein linear size (~100 ps) and slightly slows down the protein self-diffusion. The drop of the translational diffusion coefficient for volume fractions comprised between 10% and 30% signals the onset of caging effects on account of steric interactions. At larger volume fractions the diffusivity drops to one order of magnitude smaller than that at the high-dilution value, with proteins possessing some residual mobility. Analysis of the trajectories of the macromolecules, in particular by focusing on the intermolecular contacts, highlights that during their erratic encounters proteins display structural heterogeneity, with several non-specific and specific interactions. Heterogeneity reflects the presence of small clusters made of two or three proteins, together with the presence of isolated (singlet) proteins. At the same time, the simulations show a certain dispersion of the diffusion coefficient increasing with the level of volume fraction.
On the structural side, crowding conditions are usually considered to stabilize protein structures, due to the concomitant presence of specific and non-specific intermolecular interactions. At the same time, entropic effects due to the suppression of available space can destabilize the macromolecular scaffolding. Simulations of the Trp cage protein in cavity-like environments highlighted the thermodynamic shifts induced by polar (destabilizing) vs. non-polar (stabilizing) interactions between the protein and the confining surface.237b It was also reported that, when confined in a reverse micelle, atomistic fluctuations are reduced.237c Fig. 7 reports the Rat1 RMS fluctuations from their initial states. As crowding induces a larger number of intermolecular contacts, a mild destabilization of the proteins takes place, in agreement with recent experimental247 and computational studies237d on other proteins. However, the Rat1 system shows that the enhanced fluctuations do not induce substantial departures from the initial structures and are in the conventional range of values. Analysis of the whole ensemble of proteins (Rg inset, Fig. 7) shows that a small heterogeneity of structures is detected at the highest volume fraction considered, indicating that the presence of small clusters in the suspension does not trigger partial unfolding of the molecules.
This simulation of unprecedented size made possible by a multiscale methodology, bringing together the OPEP CG and a consistent treatment of the hydrodynamic interactions, is a first step towards simulating the real physics in the cell. The coupling of proteins and solvent reveals the interplay between specific and non-specific intermolecular interactions, and the role of hydrodynamic forces on the structural and diffusional properties of proteins in crowded environments (in preparation).
OPEP Limitations
The OPEP force field has several limitations, as is the case for any other CG or all-atom force field. Some are easy to alleviate and are the subject of on-going projects. Other issues are more delicate. It is important to be aware of the strengths, weaknesses, and limitations so as to use OPEP for the right questions.
Buffer Conditions and pH
We can block the N-terminus by an acetyl group (CH3-CO) and the C-terminus by an NH2 group, or block one end while the other is in its zwitterion form. Alternatively, the proteins can be in their zwitterion forms. The charged residues are parameterized for neutral pH. This means the N-terminus is NH3+, the C-terminus is CO2−, the Arg and Lys residues are treated as positively charged (NH3+), the Glu and Asp residues are treated as negatively charged (CO2−), and the His residues are neutral. So, OPEP can be safely used only in the pH range of 6-7. The pH effect can be illustrated on the Aβ12-24 peptide forming amyloid fibrils very rapidly at pH ≤ 5 and very slowly at pH 8.4 using the same in vitro conditions.248 Another aspect to be known is that the non-bonded parameters have been parameterized in “normal” aqueous solution. So we expect deviations with experiments at high ionic strengths or buffers made of H2KPO4 and adjusted with H2SO4, or with DMSO.
Non-natural Amino Acids and Small Molecules
OPEP has been extensively tested for the 20 standard natural L-amino acids, but D-amino acids can be used as well. The three-proteinogenic amino acids occurring in all kingdoms of life, selenocysteine, pyrrolysine and N-formylmethione, cannot be treated. S-S bonds can be treated at a bead level using a 6-12 potential when folding peptides with Pep-Fold, or described at an atomic level using standard local terms and Amber parameters for the bond angles and dihedral angles. While the N-methylated amino acids have been parameterized using quantum mechanics calculations,129 many non-canonical amino acids cannot be used. These include peptoids,249 β-amino acids for designing antibiotics where the amino group is bonded to the β carbon rather than the α carbon,250 γ-amino acids where the amino group at the third carbon atom is after the carboxyl carbon atom, such as γ-aminobutyric acid the most important neurotransmitter in the central nervous system,251 side chains with cyclo-hexyl groups to design inhibitors of Aβ40/42 aggregation,252 and Aβ40/42 with a pyroglutamate.127 For all these systems, it is now straightforward within the framework of the OPEPv5 code to derive effective potentials and forces from all-atom simulations.24 Generating OPEP parameters for small drugs is out of reach since the explicit representation of H-bond donors or acceptors is an essential requirement of the rule of “five”,253 but we can imagine a multi-resolution method on the fly with an all-atom representation of the protein and the drug in the regions of interest.
Effective Time Scale and Long-time Dynamics
In OPEP, the solvent contributions are treated through effective non-bonded interactions and a single bead replaces most side chains. So why does OPEP use 2 fs for integrating the equations of motion? For comparison, Deserno uses 100 fs with a resolution model of four beads,46 Klein 25 fs, Martini simulations 20-40 fs,11 Shea 10 fs with a three-bead model,43 PaLaCe and UNRES 5 and 4.9 fs,39,33 PRIMO 4 fs,42 and Voth uses 2 fs.36 The reason is that OPEP explicitly represents the N-H bond and its vibrational mode254,255 at 3600 cm−1 which limits the time step for conserving the total energy in the NVE ensemble. The second reason is that augmenting the time step to 3-4 fs by changing the mass of the hydrogen atom would introduce dynamics perturbations compared to all-atom simulations.
Using an all-atom force field in implicit solvent, Rao showed that folding of three peptides is accelerated by two orders of magnitude.256 The relationship between the OPEP-MD simulation time and the experimental time varies with the system. Poly-L-alanine and poly-L-proline have the same number of degrees of freedom in OPEP and in an all-atom model, while poly-L-valine has not. The implicit solvent and CG side chains do not affect the motions uniformly and even if dynamics were investigated by Langevin simulations, we would miss important dynamical contributions as a result of the momentum transfer that would occur through the solvent. Overall, our experience suggests a 5- to 10-fold speed-up compared to all-atom MD in explicit solvent, and the OPEP-generated dynamics cannot totally reflect the dynamics in explicit solvent. The OPEP-MD time is therefore smaller than the CG-DMD time37,38 and the Martini-MD11 time, preventing the self-assembly of large oligomers of Aβ42257 or diphenylalanine258 peptides using reasonable computer time.
Short-time Dynamics
Using multiple MD trajectories of 30-100 ns at 300 K, the RMS deviations of all proteins are 0.15 nm higher than in all-atom MD simulations in explicit solvent,23-24 though all-atom force fields do not describe similarly the folded259 or unfolded260 proteins. While the secondary structures are well preserved and display RMS fluctuations consistent with NMR, the loops display higher mobility. This results from their intrinsic flexibility and the simplified side chains, but more importantly from the absence of interactions between the loop residues and the solvent. As is the case for all-atom and CG force fields, OPEP has limitations in describing the vibrational modes.255,261,262
Thermodynamic Properties
The heat capacity and the melting temperature play a major role in relating microscopic and macroscopic properties of proteins. Their accurate predictions by simulations remain a significant challenge due to the complex and dynamic nature of protein structures, their solvent environment, and conformation averaging. Constructing the heat capacity curves, CV, as a function of T from REMD, ST or metadynamics simulations is an easy task using PTWHAM263 or MBAR.264 OPEPv5 was optimized to fit the experimental TM of a β-hairpin (297 K).24 Using the same parameters, the monomer of the ccβ-p2 monomer has a calculated TM of 275 K and a α-helix content of 70% fully consistent with the Agadir program. Experimentally, the peptide displays a α-helical CD signal at 277 K, suggesting a TM within 290-300 K. Finally, we predicted a TM of 360 K vs. 336 K experimentally for the 85-residue HPr protein. Although a larger test set of proteins is needed, there is a systematic deviation of ± 25 K between the OPEPv5 calculated and experimental melting temperatures.
Four points are worth noting: (i) few CG models report TM values. While earlier UNRES simulations found TM of 1000 K,31 the last UNRES version reports 297 and 317 K for the trpzip1 and trpzip2 peptides vs. 323 and 345 K experimentally.33 No other systems are, however, available for validation. Note that by using OPEPv4 parameters, we found a TM of 360 K for trpzip2.23 In contrast, Voth’s model finds TM values 120 K lower than experiments for trpzip2 and Trp-cage, 36 (ii) OPEPv322 and other calculations265 showed that an overestimation of TM can result from the absence of a desolvation energy barrier; (iii) even all-atom force fields in explicit solvent overestimate TM by 30–40 K,3 and (iv) due to this TM shift, we recommend to start with a minimal T of 260 K for ST- or REMD-OPEP simulations.
A second aspect to be aware of is that the OPEP heat capacities above TM, are smaller than the experimental values. This is not surprising since three major terms account for the absolute heat capacity of a protein: one first term depending on the covalent structure and the contributions from all internal vibrational modes; a second term arising from non-covalent interactions of the 2D and 3D structures; and a third term from hydration. For a typical globular protein in solution the heat capacity at 25°C is given by the covalent structure term (85%) and the hydration term (15%). In contrast, the change in heat capacity upon unfolding results from the increase in the hydration term (95%) and then the loss of non-covalent interactions (5%).266 Simplified side chains and the implicit solvent in OPEP make it difficult therefore to estimate the hydration contribution accurately above TM. Although free energy differences may fit experimental data, a breakdown of free energies into enthalpies and entropies can be reliable for the backbone, but is not accurate for the side chains. We can however envision running a number of all-atom simulations in explicit solvent from a selected list of poses in the folded and unfolded states.
On-Going Projects and Developments
Physics behind thermophilic and mesophilic proteins
The capability of OPEP to simulate protein folding/unfolding and temperature melting makes the model a powerful tool to study the elementary stabilizing forces in biomolecules. In this regard, proteins from thermophilic organisms are ideal study-cases. These proteins are stable and functional up to 100°C.267,268 While the general mechanisms that sustain such an extreme behavior remain to be determined, some molecular peculiarities have been singled out. A comparative structural analysis indicates that short loops are important motifs for stability269 and de novo protein design based on the ROSETTA force field successfully predicted enhanced stability of proteins with minimal loops.270 At the level of chemical composition, thermophiles have a systematic higher population of charged amino acids and salt-bridges, thus optimized electrostatics are thought to be a key ingredient for enhanced stability. Optimizing these interactions at the protein surface, based on the simplified Kirkwood-Tanford electrostatic model, allowed the design of proteins with increased stability.271 However playing with electrostatics is not always an effective route to enhance stability, because mutations designed to introduce ion pairs can compromise stability due to the large desolvation penalty associated with buried ionic groups.272-274
The OPEPv4 model was used to explore the thermal stability of two homologues, the G-domains of EF-Tu and 1α proteins. These 200-aa domains were simulated by REMD using 24 replicas spanning 260-580 K, each for 300 ns. The specific heats of unfolding, reported in Fig. 8, show two main peaks. Though convergence is not reached, remarkably, the curve of the hyper-thermophilic protein is systematically shifted to higher temperatures (see horizontal arrows) mirroring its enhanced thermal stability. The calculated shift between the two homologues is 35 K, comparing favorably with the experimental difference of 40 K. Extended simulations and tests with OPEPv5 that includes improved potentials for salt bridges are in progress.
Fig.8.
Specific heat of unfolding Cv for the mesophilic (green) and hyperthermophilic (red) domains of the EF-Tu and 1α proteins, respectively calculated from OPEP-REMD simulations. The structural homology of the two proteins is highlighted in the bottom right panel. The presence of two main peaks in the Cv profile is caused by the unfolding events of different secondary structure motifs. In a single two-state model, the Cv is expected to show a single peak at the melting temperature TM at which the population of the folded (pf) and unfolded (pu) states are equal. The melting temperature indicates the zero of the stability curve, see the upper inset graph. Several mechanisms can cause the increase of the TM of thermophiles, i.e. the upshift, the right shift or the broadening of the curve.268,275-277.
This preliminary result shows that OPEP with REMD can shed light on the intriguing problem of thermal stability. First, it is possible to obtain at a reasonable computational cost the melting temperature of homologues either by monitoring the peak of the heat capacity or by reconstructing the stability curve, ΔGf/u =−kT·ln (pu/pf), where pf(u) is the probability to occupy the folded and unfolded states. This latter strategy could be crucial to understand the thermodynamic mechanism sustaining protein activity at high temperatures. 268,275-277
The extensive sampling of conformations in both the folded and unfolded states then would provide key information on the protein flexibility at ambient condition and the presence of motifs in the unfolded state. Moreover, the decomposition of the free energy gap into enthalpy and entropy, here clearly limited by the nature of the CG to the behavior of the backbone, could provide extra information on the stability mechanism.278-280
Effect of shear flow on protein folding and amyloid formation
It has been reported that hydrodynamic interactions accelerate collapse during polymer coil-to-globule transition281 or protein folding,282-283 and affect the kinetics of lipid membrane self-assembly.284 Thanks to the MUPHY/OPEP coupling it is now possible to explore the behavior of proteins under shear flows. Assessing the effect of shear flow on the stability of proteins is of interest for biotechnological applications because proteins might be degraded due to filtering or injection processes.
Thus far, experimental studies have reached contradictory conclusions about the minimal shear-rate needed to perturb globular proteins.285 Computational studies have also tackled this problem using simplified (generally Gō-like) models and showed that under strong uniform or elongation flow, proteins do unfold.283 However, the minimal shear-rate generating unfolding or the necessary time for cumulating shear stress remains an open issue.285 We are applying OPEP to shed light on theses issues. In Fig. 9 we present preliminary results of the MD-OPEPv5 dynamics of a β-hairpin peptide in a strong laminar shear flow, . We see that after a few nanoseconds the peptide suddenly unfolds as marked by the RMSD increase, and explores several configurations that extend along the velocity gradient.
Fig.9.
Time evolution of the RMSD and snapshots of a β-hairpin peptide (PDB code 1PGB, fragment 41-56) simulated in laminar shear flow. The velocity gradient is generated along the Z direction and corresponds in our simulation to a shear rate of . We show the detailed structures of the peptide prior to unfolding (I) and at various unfolding stages (II-IV), with the velocity field represented in background and a colour scale given in the top of the figure.
A systematic study of shear-induced unfolding is appealing also for probing mechanical stability as compared to atomic force spectroscopy experiments; in the former case the drag force is sensed in different locations due to the thermal motion of the protein while in the latter the external pulling applies only along the end-to-end distance axis. Moreover, as for the glycoprotein Ibα receptor, conformational change induced by shear flow can be essential for function, i.e. the binding to the cofactor.286
The effect of shear was also appreciated in the context of amyloid fibril formation.287-290 For instance, it has been observed that in an uniform laminar flow generated in a Couette cell, Aβ1-40 sample forms fibrils within 15 hours at 37°C while in the absence of shear, the process requires at least 1 month.288 This acceleration corresponds to a decrease of the activation barrier of 4.3 kT or the loss of one hydrogen bond per monomer in solution.288 A possible mechanism for the effect of shear is that it may lead to the alignment of aggregates, which in turn facilitates their assembly into fibrils. It was further probed that changing the nature of the shear flow, i.e. a heterogeneous field generated by a magnetic stirrer bar, enhances the formation of protofibrils and the growth of fibrils288 and affects the rate of fragmentation.290 We are investigating the shear-induced effects on amyloid peptides using OPEP.
Buffer- and pH-dependent OPEP force field
We are currently using all-atom MD simulations and Boltzmann inversion to generate salt-bridge potentials between Lys, Arg and Asp and Glu as a function of pH. Similarly, we are deriving OPEP potentials for polypeptides at various high ionic strengths and in buffers made of H2KPO4, H2SO4 and DMSO so as to mimic as closely as possible the in vitro conditions used to form amyloids. These potentials will be useful for Pep-fold predictions.
Hire-RNA version 3
To predict complex RNA topologies, two features are critical. The first is electrostatics, RNAs being highly charged and ions playing important roles in the structures and thermodynamics. Many models consider ion screening at long distances between phosphate groups via the Debye-Hückel treatment of electrostatic interactions. Other models go more into details by considering either one layer of explicit ions surrounding RNA, referred to as ion condensation,291 some implicit “structural ions”,228,292 or explicit ions and solvent.293 In Hire-RNAv3, we added a Debye-Hückel term and the screening parameter is being calibrated against experimental melting temperatures of duplexes as a function of ionic concentrations. We are also exploring the presence of explicit ions.
The second issue is the treatment of non-canonical W-C interactions never taken into account by ab initio models. Our new force field allows two bases to interact on all sides, giving rise to about 30 recognition motifs, each one with its specific geometry and strength. Overall, Hire-RNAv3 consists of local terms, excluded volume, ionic screening electrostatics, a proper stacking interaction depending on base position and orientation, and terms accounting for both canonical and non-canonical interactions on the three base sides and also for the co-planarity of the interacting bases (in preparation).
With Hire-RNAv3, we are now able to predict complex RNA structures. As a first benchmark, we studied three systems starting from fully extended states: a 22-nt pseudo-knot, a 49-nt telomerase triple-helix pseudo-knot, and a 79-nt riboswitch with a kissing loop. For the 22-nt RNA, we recovered the native state by running 1.2 μs ST simulation with 15 discrete temperatures from 300 to 450 K (Fig. 10). With a REMD simulation of 64 replicas, each for 0.5 μs, we recovered the native topology of the 49-nt RNA with a small shift of the base pairs, however. For the 79-nt riboswitch, we implemented the possibility of including some restraints. Information about base pairing is easily obtained by preliminary NMR data and is not sufficient to assign the full 3D structure. Imposing 4 restraints in the three helices, we were able to fold this molecule to its NMR topology by a MD trajectory of 0.6 μs at 300 K and recovered the kissing hairpin configuration, although the guanine ligand was not considered (Fig. 10).
Fig.10.
Hire-RNAv3 results. Left, Folding of the 22-nt pseudo-knot (2G1W) using ST simulation with the predicted state (blue) superposed on the NMR structure (yellow). The RMSD with respect to the NMR structure over time is shown. Right, MD-predicted structure of the 79-nt guanine riboswitch 1Y26 (blue) superposed on the experimental structure (yellow) using our four restraints (in red). In both panels the secondary structure of the system is shown on the right.
Virtual reality and interactive simulations
The use of haptic manipulations of molecular models has been well described.294-295 The technical requirements are modest; and it is nowadays easy to setup interactive simulations.296 Even quantum chemistry applications are within reach.297 For the manipulation of complex biological assemblies, coarser methods are preferable and have been exploited notably for fitting models into experimentally determined envelopes.298 Generally speaking such approaches build on the idea to render accurate molecular models more real and tangible to the scientists.299
We previously pointed out that CG models play a particular role in virtual interactive experiments.121,300 CG descriptions represent an excellent compromise between simulation speed and biological fidelity. Furthermore our experience suggests that CG-level simulations are generally more robust with respect to user interactions than computations carried out at an all-atom level.
OPEP and Hire-RNA are of particular interest in this context and provide original features that we could not address previously, due to their relatively high resolution of the backbone representation and the presence of directional bonded terms. Both OPEP and Hire-RNA simulation engines were extended for interactive manipulation as described in the methods section. Here, we will mainly brush over the potential benefits of such an approach and restrict ourselves to present a very first, simple toy application, as the validation of this recent MDDriver/IMD implementation is still ongoing.
Generally speaking, the interactive approach opens up perspectives to guide simulations via user input, for example using a haptic device, within a dedicated graphical environment. Hence, the user feels an immediate force feedback by a straightforward combination of classical molecular modeling and virtual reality. An instant benefit is to gather an intuitive understanding of the causal relationship between the theoretical model and its chemically and biologically relevant properties.
These hands-on investigations echo recent experimental ventures into the mechanical properties of molecular structures, and can be associated with the term mechanochemistry. Experiments, such as AFM pulling and allosteric spring probes, can be reproduced on the fly. Multiple forces may be applied simultaneously to reproduce complex deformations and assemble or disassemble several molecules. Such an interactive exploration can provide insight into the key interactions that govern the mechanical properties of molecular structures and is a unique tool to probe mechanochemistry at a molecular level.
We previously carried out such investigations at a CG level using elastic network models as in the studies of the SNARE complex,121 RecA nucleofilament301 and dystrophin fibril.302 Such spring-based CG models do however preclude any significant changes in the underlying molecular structure that may occur upon tension, a limitation that we are able to lift using OPEP.
In order to illustrate these enhanced possibilities, we interactively manipulated a Hire-RNA model of an RNA hairpin (Fig. 11). By pulling on one or both ends of the structure, it is fairly easy to control the successive detachment of the base pairs. When the added external forces are released, the structure may either progressively return to the initial hairpin state or feature a base shift. This reversible process takes place on the order of a few hundred picoseconds, depending on how much the ends were torn apart. This numerical experiment provides insight on how the hairpin behaves under such stress, similarly to what can be probed experimentally with optical tweezers.303
Fig.11.
Interactive force unfolding of an RNA hairpin modelled by Hire-RNA. Visualization and interaction were performed within VMD. Starting from an initial folded conformation (top left), the user tears apart both ends by applying forces (blue arrows) on the C4’ beads of the end bases. Two scenarios were observed after releasing the forces, either the structure refolds and returns to a full hairpin state (1) or a base shift occurs (2). A cumulated view of simulation snapshots is shown on the very right, coloured from white (start of the simulation) to black (final snapshot). The end base C4’-C4’ distance curve over time is shown for both experiments.
One apparent caveat is the relatively short time step (0.1 to 2 fs) and/or low temperature (100 K) that have to be used to reduce vibrations in order to allow for accurate manipulation. This inconvenience could be lifted by adding the ability to interactively change these parameters during the virtual experiment: using low time-step/temperature values when selecting and applying the forces, then going back to standard values to observe the resulting effects. This kind of simple manipulation can be useful to quickly probe features of the force field or to generate non-trivial starting structures. When carried out more rigorously, the approach may be used to interpret experimental results.
Future extensions of the interactive approach similar to those previously reported298,302 will enable the use of OPEP and Hire-RNA models to integrate low-resolution experimental data, from small-angle X-ray scattering (SAXS) or Cryo-EM, where both the force field and the user intuition will guide the refinement.
Conclusions
We have presented some of the good applications of OPEP and what OPEP-based simulations can tell us about the structures, dynamics, kinetics and thermodynamics of single proteins, amyloid fibril formation, proteins in a crowded environment with hydrodynamics and RNA/DNA complexes. Whether OPEP can reproduce the effects of a single mutation on protein energy landscapes remains to be determined.163,304 Compared to the nine CG models described in this report, the OPEP CG strategy with inclusion of the amide hydrogen allows generating more accurate melting temperatures than Voth’s model consistent with experimental values and all-atom simulations with growing accuracy of the force fields. OPEP is also free of any bioinformatics-based information (AWSEW), restraints on the backbone (ATTRACT, Klein’s model and MARTINI) and has been extensively used on amyloid and non-amyloid systems in contrast to the PRIME, PRIMO, UNRES and Palace systems. The main OPEP disadvantage is that it can only use 2 fs time step. OPEP is coupled to many unbiased advanced conformational sampling methods and interactive virtual reality approaches. We have briefly sketched the main on-going applications and developments. Others, that have just started, include flexible protein/protein and protein/RNA-DNA docking with the necessity to couple the protein and nucleic acid force fields, and the use of the basin hopping method to locate the global energy minimum and calculate disconnectivity graphs to visualize the energy landscape.305 All these studies will help improve the OPEP parameters and gain a better understanding of how living systems function, and how these functions can be perturbed by internal or external factors.
Supplementary Material
Acknowledgements
Fabio Sterpone and Maria Kalimeri thank funding from the European Research Council under the European Community’s Seventh Framework Program (FP7/2007-2013) Grant Agreement no.258748. Part of this work used HPC resources from GENCI [CINES and TGCC] (Grant 2012 c2012086818 and 2013 x201376818). Simone Melchionna thanks M. Bernaschi, M. Bisson, M. Fatica, C. Pierleoni and U. Marconi for discussions, and the Oak Ridge Leadership Computing Facility and CINECA supercomputing center (ISCRA grants KINPROT and FLEXPROT). Pierre Tuffery thanks support of ANR IA “BipBip” and IBiSA for funding the RPBS platform. Alessandro Barducci thanks Massimiliano Bonomi for discussion and the Swiss National Science Foundation for financial support under the Ambizione grant PZ00P2_136856. Marc Baaden thanks the French Agency for Research Grant “ExaViz” (ANR-11-MONU-003), and CNRS (Grant PEPS BMI 2012). Normand Mousseau and Philippe Derreumaux thank the Alzheimer Society of Canada and its 2005 postdoc program. Finally, Philippe Derreumaux thanks support of University of Aix-Marseille II (1999-2003), University of Paris Diderot from 2003, ANR SIMI7 GRAL 12-BS07-0017, ANR LABEX Grant “DYNAMO” (ANR-11-LABX-0011), 6th European PRCD (Immunoprion, FP6-Food023144), Institut Universitaire de France, French/Singapore Merlion PhD program (Grant 5.08.10), Pierre de Gilles de Gennes Foundation and its international PhD grant, Fudan University in China, CNRS - Académie Polonaise des Sciences (Grant 168836), and Institut de Chimie du CNRS over all these 15 years.
Footnotes
The full terms of the OPEP energy function and the accessibility of the parameters are available at the electronic supplementary information (ESI).
References
- 1.McCammon JA, Gelin BR, Karplus M. Nature. 1977;267:585. doi: 10.1038/267585a0. [DOI] [PubMed] [Google Scholar]
- 2.Duan Y, Kollman P. Science. 1998;282:740. doi: 10.1126/science.282.5389.740. [DOI] [PubMed] [Google Scholar]
- 3.Lindorff-Larsen K, Piana S, Dror RO, Shaw DE. Science. 2011;334:517. doi: 10.1126/science.1208351. [DOI] [PubMed] [Google Scholar]; Piana S, Lindorff-Larsen K, Shaw DE. Proc Natl Acad Sci U S A. 2013;110:5915. doi: 10.1073/pnas.1218321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dror RO, Green HF, Valant C, Borhani DW, Valcourt JR, Pan AC, Arlow DH, Canals M, Lane JR, Rahmani R, Baell JB, Sexton PM, Christopoulos A, Shaw DE. Nature. 2013;503:295. doi: 10.1038/nature12595. [DOI] [PubMed] [Google Scholar]
- 5.Stefani M, Dobson CM. J Mol Med (Berl) 2003;81:678. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]
- 6.Lito P, Rosen N, Solit DB. Nat Med. 2013;19:1401. doi: 10.1038/nm.3392. [DOI] [PubMed] [Google Scholar]
- 7.Maxwell KN, Domchek SM. Nat Rev Clin Oncol. 2012;9:520. doi: 10.1038/nrclinonc.2012.123. [DOI] [PubMed] [Google Scholar]
- 8.Levitt M, Warshel A. Nature. 1975;253:694. doi: 10.1038/253694a0. [DOI] [PubMed] [Google Scholar]
- 9.Saunders MG, Voth GA. Annu Rev Biophys. 2013;42:73. doi: 10.1146/annurev-biophys-083012-130348. [DOI] [PubMed] [Google Scholar]
- 10.Noid WG. J Chem Phys. 2013;139:090901. doi: 10.1063/1.4818908. [DOI] [PubMed] [Google Scholar]
- 11.Marrink SJ, Tieleman DP. Chem. Soc. Rev. 2013;42:6801. doi: 10.1039/c3cs60093a. [DOI] [PubMed] [Google Scholar]
- 12.Derreumaux P. J. Chem. Phys. 1997;107:1941. [Google Scholar]
- 13.Derreumaux P. J. Chem. Phys. 1997;106:5260. [Google Scholar]
- 14.Derreumaux P. J. Chem. Phys. 1998;109:1567. [Google Scholar]
- 15.Derreumaux P. J. Chem. Phys. 1999;111:2310. [Google Scholar]
- 16.Derreumaux P. Phys. Rev. Lett. 2000;85:206. doi: 10.1103/PhysRevLett.85.206. [DOI] [PubMed] [Google Scholar]
- 17.Forcellino F, Derreumaux P. Proteins. 2001;45:159. doi: 10.1002/prot.1135. [DOI] [PubMed] [Google Scholar]
- 18.Derreumaux P. J. Chem. Phys. 2002;117:3499. [Google Scholar]
- 19.Wei G, Derreumaux P, Mousseau N. J. Chem. Phys. 2003;119:6403. [Google Scholar]
- 20.Maupetit J, Tuffery P, Derreumaux P. Proteins. 2007;69:394. doi: 10.1002/prot.21505. [DOI] [PubMed] [Google Scholar]
- 21.Derreumaux P, Mousseau N. J. Chem. Phys. 2007;126:025101. doi: 10.1063/1.2408414. [DOI] [PubMed] [Google Scholar]
- 22.Chebaro Y, Dong X, Laghaei R, Derreumaux P, Mousseau N. J. Phys. Chem. B. 2009;113:267. doi: 10.1021/jp805309e. [DOI] [PubMed] [Google Scholar]
- 23.Chebaro Y, Pasquali S, Derreumaux P. J. Phys. Chem. B. 2012;116:8741. doi: 10.1021/jp301665f. [DOI] [PubMed] [Google Scholar]
- 24.Sterpone F, Nguyen PH, Kalimeri M, Derreumaux P. J. Chem. Theory Comput. 2013;9:4574. doi: 10.1021/ct4003493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monticelli L, Kandasamy SK, Periole X, Larson RG, Tieleman DP, Marrink SJ. J. Chem. Theor. Comput. 2008;4:819. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- 26.de Jong DH, Singh G, Bennett WFD, Arnarez C, Wassenaar TA, Schafer LV, Periole X, Tieleman DP, Marrink SJ. J. Chem. Theor. Comput. 2013;9:687. doi: 10.1021/ct300646g. [DOI] [PubMed] [Google Scholar]
- 27.Bond PJ, Sansom MSP. J. Am. Chem. Soc. 2006;128:2697. doi: 10.1021/ja0569104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parton DL, Tek A, Baaden M, Sansom MS. PLoS Comput Biol. 2013;9:e1003034. doi: 10.1371/journal.pcbi.1003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shih AY, Arkhipov A, Freddolino PL, Schulten K. J Phys Chem B. 2006;110:3674. doi: 10.1021/jp0550816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Devane R, Shinoda W, Moore PB, Klein ML. J. Chem. Theory Comput. 2009;5:2115. doi: 10.1021/ct800441u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nanias M, Czaplewski C, Scheraga HA. J Chem Theor Comput. 2006;2:513. doi: 10.1021/ct050253o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liwo A, He Y, Scheraga HA. Phys Chem Chem Phys. 2011;13:16890. doi: 10.1039/c1cp20752k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makowski M, Liwo A, Scheraga HA. J Phys Chem B. 2011;115:6130. doi: 10.1021/jp111259e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simons K, Kooperberg C, Huang E, Baker D. J Mol Biol. 1997;268:209. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 35.Leaver-Fay A, O’Meara MJ, Tyka M, Jacak R, Song Y, Kellogg EH, Thompson J, Davis IW, Pache RA, Lyskov S, Gray JJ, Kortemme T, Richardson JS, Havranek JJ, Snoeyink J, Baker D, Kuhlman B. Methods Enzymol. 2013;523:109. doi: 10.1016/B978-0-12-394292-0.00006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hills RD, Jr, Lu L, Voth GA. PLoS Comput Biol. 2010;6:e1000827. doi: 10.1371/journal.pcbi.1000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng S, Ding F, Urbanc B, Buldyrev SV, Cruz L, Stanley HE, Dokholyan NV. Phys Rev E Stat Nonlin Soft Matter Phys. 2004;4:041908. doi: 10.1103/PhysRevE.69.041908. [DOI] [PubMed] [Google Scholar]
- 38.Cheon M, Chang I, Hall CK. Proteins. 2010;78:2950. doi: 10.1002/prot.22817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pasi M, Lavery R, Ceres N. J Chem Theory Comput. 2013;9:785. doi: 10.1021/ct3007925. [DOI] [PubMed] [Google Scholar]
- 40.Zacharias M. Protein Sci. 2003;12:1271. doi: 10.1110/ps.0239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zacharias M. Proteins. 2013;81:81. doi: 10.1002/prot.24164. [DOI] [PubMed] [Google Scholar]
- 42.Kar P, Gopal SM, Cheng YM, Predeus A, Feig M. J Chem Theory Comput. 2013;9:3769. doi: 10.1021/ct400230y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bellesia G, Shea JE. J. Chem. Phys. 2009;130:145103. doi: 10.1063/1.3108461. [DOI] [PubMed] [Google Scholar]
- 44.Davtyan A, Schafer NP, Zheng W, Clementi C, Wolynes PG, Papoian GA. J Phys Chem B. 2012;116:8494. doi: 10.1021/jp212541y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Basdevant N, Borgis D, Ha-Duong T. J Phys Chem B. 2007;111:9390. doi: 10.1021/jp0727190. [DOI] [PubMed] [Google Scholar]; Basdevant N, Borgis D, Ha-Duong T. J Chem Theory Comput. 2013;9:803. doi: 10.1021/ct300943w. [DOI] [PubMed] [Google Scholar]
- 46.Bereau T, Deserno M. J. Chem. Phys. 2009;130:235106. doi: 10.1063/1.3152842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Májek P, Elber R. Proteins. 2009;76:822. doi: 10.1002/prot.22388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pellarin R, Schuetz P, Guarnera E, Caflisch A. J. Am. Chem. Soc. 2010;132:14960?. doi: 10.1021/ja106044u. [DOI] [PubMed] [Google Scholar]
- 49.Aisen PS, Vellas B, Hampel H. Nat Rev Drug Discov. 2013;4:324. doi: 10.1038/nrd3842-c1. [DOI] [PubMed] [Google Scholar]
- 50.Vlieghe P, Lisowski V, Martinez J, Khrestchatisky M. Drug Discov Today. 2010;15:40. doi: 10.1016/j.drudis.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 51.Lohan S, Bisht GS. Mini Rev Med Chem. 2013;13:1073. [PubMed] [Google Scholar]
- 52.Jamieson AG, Boutard N, Sabatino D, Lubell WD. Chem Biol Drug Des. 2013;81:148. doi: 10.1111/cbdd.12042. [DOI] [PubMed] [Google Scholar]
- 53.Cruz JA, Blanchet MF, Boniecki M, Bujnicki JM, Chen SJ, Cao S, Das R, Ding F, Dokholyan NV, Flores SC, Huang L, Lavender CA, Lisi V, Major F, Mikolajczak K, Patel DJ, Philips A, Puton T, Santalucia J, Sijenyi F, Hermann T, Rother K, Rother M, Serganov A, Skorupski M, Soltysinski T, Sripakdeevong P, Tuszynska I, Weeks KM, Waldsich C, Wildauer M, Leontis NB, Westhof E. RNA. 2012;4:610. doi: 10.1261/rna.031054.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Allmer J. Methods Mol Biol. 2014;1107:157. doi: 10.1007/978-1-62703-748-8_9. [DOI] [PubMed] [Google Scholar]
- 55.Crea F, Clermont PL, Parolia A, Wang Y, Helgason CD. Cancer Metastasis Rev. 2013 doi: 10.1007/s10555-013-9455-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv XW, Li J. Cancer Lett. 2014;344:20. doi: 10.1016/j.canlet.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 57.Bernaschi M, Bisson M, Fatica M, Melchionna S, Succi S. Comm. Comput. Phys. 2012;184:329. [Google Scholar]
- 58.McGuffee SR, Elcock AH. PLoS Comput Biol. 2010;6:e1000694. doi: 10.1371/journal.pcbi.1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharma S, Kumar SK, Buldyrev SV, Debenedetti PG, Rossky PJ, Stanley HE. Sci Rep. 2013;3:1841. doi: 10.1038/srep01841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gonzalez HC, Darré L, Pantan SJ. J Phys Chem B. 2013;117:14438. doi: 10.1021/jp4079579. [DOI] [PubMed] [Google Scholar]
- 61.Han W, Schulten K. J Chem Theory Comput. 2012;8:4413. doi: 10.1021/ct300696c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kapoor A, Travesset A. Proteins. 2014;82:505. doi: 10.1002/prot.24416. [DOI] [PubMed] [Google Scholar]
- 63.Cheatham TE, III, Case DA. Biopolymers. 2013;99:969. doi: 10.1002/bip.22331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barducci A, Bonomi M, Derreumaux P. J. Chem. Theory Comput. 2011;7:1928. doi: 10.1021/ct100646f. [DOI] [PubMed] [Google Scholar]
- 65.Brooks BR, Brooks CL, III, Mackerell AD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. J. Comp. Chem. 2009;30:1545. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Malek R, Mousseau N. Phys Rev. E. 2000;62:7723. doi: 10.1103/physreve.62.7723. [DOI] [PubMed] [Google Scholar]
- 67.Wei G, Mousseau N, Derreumaux P. J. Chem. Phys. 2002;117:113749. [Google Scholar]
- 68.Spill YG, Pasquali S, Derreumaux P. J. Chem. Theory Comput. 2011;7:1502. doi: 10.1021/ct100619p. [DOI] [PubMed] [Google Scholar]
- 69.Laghaei R, Mousseau N, Wei G. J. Phys. Chem. B. 2010;114:7071. doi: 10.1021/jp100205w. [DOI] [PubMed] [Google Scholar]
- 70.Laio A, Parrinello M. Proc. Natl. Acad. Sci. U.S.A. 2002;99:12562. doi: 10.1073/pnas.202427399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nguyen PH, Okamoto Y, Derreumaux P. J. Chem. Phys. 2013;138:061102. doi: 10.1063/1.4792046. [DOI] [PubMed] [Google Scholar]
- 72.Tuffery P, Derreumaux P. Proteins. 2005;61:732. doi: 10.1002/prot.20698. [DOI] [PubMed] [Google Scholar]
- 73.Tuffery P, Guyon F, Derreumaux P. J. Comput. Chem. 2005;26:506. doi: 10.1002/jcc.20181. [DOI] [PubMed] [Google Scholar]
- 74.Derreumaux P. Biophys J. 2001;81:1657. doi: 10.1016/S0006-3495(01)75819-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Derreumaux P. J. Chem. Phys. 2003;119:4940. [Google Scholar]
- 76.Wei G, Mousseau N, Derreumaux P. Proteins. 2004;3:464. doi: 10.1002/prot.20127. [DOI] [PubMed] [Google Scholar]
- 77.Santini S, Wei G, Mousseau N, Derreumaux P. Structure. 2004;12:1245. doi: 10.1016/j.str.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 78.Santini S, Mousseau N, Derreumaux P. J. Am. Chem. Soc. 2004;126:11509. doi: 10.1021/ja047286i. [DOI] [PubMed] [Google Scholar]
- 79.Wei G, Mousseau N, Derreumaux P. Biophys. J. 2004;87:3648. doi: 10.1529/biophysj.104.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Melquiond A, Boucher G, Mousseau N, Derreumaux P. J. Chem. Phys. 2005;122:174904. doi: 10.1063/1.1886725. [DOI] [PubMed] [Google Scholar]
- 81.Mousseau N, Derreumaux P. Acc Chem Res. 2005;38:885. doi: 10.1021/ar050045a. [DOI] [PubMed] [Google Scholar]
- 82.Melquiond A, Mousseau N, Derreumaux P. Proteins. 2006;65:180. doi: 10.1002/prot.21100. [DOI] [PubMed] [Google Scholar]
- 83.Chen W, Mousseau N, Derreumaux P. J. Chem. Phys. 2006;125:084911. doi: 10.1063/1.2337628. [DOI] [PubMed] [Google Scholar]
- 84.Mousseau N, Derreumaux P. Front. Biosci. 2008;13:4495. doi: 10.2741/3019. [DOI] [PubMed] [Google Scholar]
- 85.St-Pierre JF, Mousseau N, Derreumaux P. J. Chem. Phys. 2008;128:045101. doi: 10.1063/1.2812562. [DOI] [PubMed] [Google Scholar]
- 86.Doye JPK, Wales DJ. Z. Phys. D. 1997;40:194. [Google Scholar]
- 87.Olsen RA, Kroes GJ, Henkelman G, Arnaldsson A, Jónsson H. J. Chem. Phys. 2004;121:9776. doi: 10.1063/1.1809574. [DOI] [PubMed] [Google Scholar]
- 88.Yun MR, Mousseau N, Derreumaux P. J Chem Phys. 2007;126:105101. doi: 10.1063/1.2710270. [DOI] [PubMed] [Google Scholar]
- 89.Evans DA, Wales DJ. J. Chem. Phys. 2003;119:9947. [Google Scholar]
- 90.Oakley MT, Wales DJ, Johnston RL. J Phys Chem B. 2011;115:11525–9. doi: 10.1021/jp207246m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Andersen HC. J. Comput. Phys. 1983;52:24. [Google Scholar]
- 92.Berendsen HJC, Postma JPM, van Gusnteren WF, DiNola A, Haak JR. J. Chem. Phys. 1984;81:3668. [Google Scholar]
- 93.Sugita Y, Okamoto Y. Chem. Phys. Lett. 1999;314:141. [Google Scholar]
- 94.Wei GH, Mousseau N, Derreumaux P. Prion. 2007;1:3. doi: 10.4161/pri.1.1.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chebaro Y, Mousseau N, Derreumaux P. J. Phys. Chem. B. 2009;113:7668. doi: 10.1021/jp900425e. [DOI] [PubMed] [Google Scholar]
- 96.Chebaro Y, Jiang P, Zhang T, Mu Y, Nguyen PH, Mousseau N, Derreumaux P. J. Phys. Chem. B. 2012;116:8412. doi: 10.1021/jp2118778. [DOI] [PubMed] [Google Scholar]
- 97.Fukunishi H, Watanabe O, Takada S. J. Chem. Phys. 2002;116:9058. [Google Scholar]
- 98.Mo Y, Lu Y, Wei G, Derreumaux P. J Chem Phys. 2009;130:125101. doi: 10.1063/1.3097982. [DOI] [PubMed] [Google Scholar]
- 99.Lu Y, Wei G, Derreumaux P. J. Chem. Phys. 2012;137:025101. doi: 10.1063/1.4732761. [DOI] [PubMed] [Google Scholar]
- 100.Hansmann UH, Okamoto Y. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 1996;5:5863. doi: 10.1103/physreve.54.5863. [DOI] [PubMed] [Google Scholar]
- 101.Rosta E, Hummer G. J Chem Phys. 2010;132:034102. doi: 10.1063/1.3290767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Park S, Pande V. Phys Rev E. 2007;E76:016703. doi: 10.1103/PhysRevE.76.016703. [DOI] [PubMed] [Google Scholar]
- 103.Zhang C, Ma J. J. Chem. Phys. 2008;129:134112. doi: 10.1063/1.2988339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu Y, Strümpfer J, Freddolino PL, Gruebele M, Schulten K. J Phys Chem Lett. 2012;3:1117. doi: 10.1021/jz300017c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Park BH, Levitt M. J Mol Biol. 1995;249:493. doi: 10.1006/jmbi.1995.0311. [DOI] [PubMed] [Google Scholar]
- 106.Kolodny R, Koehl P, Guibas L, Levitt M. J Mol Biol. 2002;323:297. doi: 10.1016/s0022-2836(02)00942-7. [DOI] [PubMed] [Google Scholar]
- 107.Vendruscolo M, Kussell E, Domany E. Fold Des. 1997;2:295. doi: 10.1016/S1359-0278(97)00041-2. [DOI] [PubMed] [Google Scholar]
- 108.Tuffery P, Derreumaux P. Proteins. 2005;61:732. doi: 10.1002/prot.20698. [DOI] [PubMed] [Google Scholar]
- 109.Tuffery P, Guyon F, Derreumaux P. J. Comput. Chem. 2005;26:506. doi: 10.1002/jcc.20181. [DOI] [PubMed] [Google Scholar]
- 110.Maupetit J, Derreumaux P, Tufféry P. Nucleic Acids Res. 2009;37:W498. doi: 10.1093/nar/gkp323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Barducci A, Bonomi M, Parrinello M. Wiley Interdiscip Rev Comput Mol Sci. 2011;1:826. [Google Scholar]
- 112.Limongelli V, Marinelli L, Cosconati S, La Motta C, Sartini S, Mugnaini L, Da Settimo F, Novellino E, Parrinello M. Proc Natl Acad Sci USA. 2012;109:1467. doi: 10.1073/pnas.1112181108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Baftizadeh F, Biarnés X, Pietrucci F, Affinito F, Laio A. J. Am. Chem. Soc. 2012;134:3886. doi: 10.1021/ja210826a. [DOI] [PubMed] [Google Scholar]
- 114.Palazzesi F, Barducci A, Tollinger M, Parrinello M. Proc Natl Acad Sci USA. 2013;110:14237. doi: 10.1073/pnas.1313548110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bussi G, Gervasio FL, Laio A, Parrinello M. J Am Chem Soc. 2006;128:13435. doi: 10.1021/ja062463w. [DOI] [PubMed] [Google Scholar]
- 116.Ermak EL, McCammon JA. J. Chem. Phys. 1978;69:1352. [Google Scholar]
- 117.Bernaschi M, Melchionna S, Succi S, Fyta M, Kaxiras E, Sircar JK. Comput. Phys. Comm. 2009;180:1495. [Google Scholar]
- 118.Benzi R, Succi S, Vergassola M. Phys. Rep. 1992;222:145. [Google Scholar]
- 119.Fyta M, Melchionna S, Kaxiras E, Succi S. Comput. Sci. & Eng. 2008;10:10. [Google Scholar]
- 120.Bernaschi M, Bisson M, Fatica M, Melchionna S. Proc. ACM/IEEE Intl. Conf. for High Performance Computing, SC13; 2013. [Google Scholar]
- 121.Delalande O, Férey N, Grasseau G, Baaden M. J. Comput. Chem. 2009;30:2375. doi: 10.1002/jcc.21235. [DOI] [PubMed] [Google Scholar]
- 122.Lv Z, Tek A, Da Silva F, Empereur-mot C, Chavent M, Baaden M. PLoS ONE. 2013;8:e57990. doi: 10.1371/journal.pone.0057990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Humphrey W, Dalke A, Schulten K. J. Mol. Graph. 1996;14:33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 124.Dupuis L, Mousseau N. J Chem Phys. 2012;136:035101. doi: 10.1063/1.3671986. [DOI] [PubMed] [Google Scholar]
- 125.St-Pierre JF, Mousseau N. Proteins. 2012;80:1883. doi: 10.1002/prot.24085. [DOI] [PubMed] [Google Scholar]
- 126.Selkoe DJ. Nature. 2003;426:900. doi: 10.1038/nature02264. [DOI] [PubMed] [Google Scholar]
- 127.Masters CL, Selkoe DJ. Cold Spring Harb. Perspect. Med. 2012;2:a006262. doi: 10.1101/cshperspect.a006262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Moore BD, Chakrabarty P, Levites Y, Kukar TL, Baine AM, Moroni T, Ladd TB, Das P, Dickson DW, Golde TE. Alzheimers Res Ther. 2012;4:18. doi: 10.1186/alzrt121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Knowles TP, Waudby CA, Devlin GL, Cohen SI, Aguzzi A, Vendruscolo M, Terentjev EM, Welland ME, Dobson CM. Science. 2009;326:1533. doi: 10.1126/science.1178250. [DOI] [PubMed] [Google Scholar]
- 130.Cohen SI, Linse S, Luheshi LM, Hellstrand E, White DA, Rajah L, Otzen DE, Vendruscolo M, Dobson CM, Knowles TP. Proc Natl Acad Sci U S A. 2013;110:975. doi: 10.1073/pnas.1218402110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kirkitadze MD, Condron MM, Teplow DB. J. Mol. Biol. 2001;312:1103. doi: 10.1006/jmbi.2001.4970. [DOI] [PubMed] [Google Scholar]
- 132.Sarroukh R, Goormaghtigh E, Ruysschaert JM, Raussens V. Biochim Biophys Acta. 2013;1828:2328. doi: 10.1016/j.bbamem.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 133.Bernstein SL, Dupuis NF, Lazo ND, Wyttenbach T, Condron MM, Bitan G, Teplow DB, Shea JE, Ruotolo BT, Robinson CV, Bowers MT. Nat Chem. 2009;1:326. doi: 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tay WM, Huang D, Rosenberry TL, Paravastu AK. J Mol Biol. 2013;425:2494. doi: 10.1016/j.jmb.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang Y, Rempel DL, Zhang J, Sharma AK, Mirica LM, Gross ML. Proc Natl Acad Sci U S A. 2013;110:14604. doi: 10.1073/pnas.1309175110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Garcia S, Cuscó C, Brissos RF, Torrents E, Caubet A, Gamez P. J Inorg Biochem. 2012;116:26. doi: 10.1016/j.jinorgbio.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 137.Lu J-X, Qiang W, Yau WM, Schwieters CD, Meredith SC, Tycko R. Cell. 2013;154:1257. doi: 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJ, McFarlane HT, Madsen A, Riekel C, Eisenberg D. Nature. 2007;447:453. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 139.Matthes D, Gapsys V, de Groot BL. J. Mol. Biol. 2012;421:390. doi: 10.1016/j.jmb.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 140.Li DW, Mohanty S, Irback A, Huo S. PLoS Comput. Biol. 2008;4:e1000238. doi: 10.1371/journal.pcbi.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.De Simone A, Derreumaux P. J. Chem. Phys. 2010;132:165103. doi: 10.1063/1.3385470. [DOI] [PubMed] [Google Scholar]
- 142.Liu C, Sawaya MR, Cheng PN, Zheng J, Nowick JS, Eisenberg D. J. Am. Chem. Soc. 2011;133:6736. doi: 10.1021/ja200222n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Song W, Wei G, Mousseau N, Derreumaux P. J. Phys. Chem. B. 2008;112:4410. doi: 10.1021/jp710592v. [DOI] [PubMed] [Google Scholar]
- 144.Laganowsky A, Liu C, Sawaya MR, Whitelegge JP, Park J, Zhao M, Pensalfini A, Soriaga AB, Landau M, Teng PK, Cascio D, Glabe C, Eisenberg D. Science. 2012;335:1228. doi: 10.1126/science.1213151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Laghaei R, Mousseau N. J. Chem. Phys. 2010;132:165102. doi: 10.1063/1.3383244. [DOI] [PubMed] [Google Scholar]
- 146.Perutz MF, Finch JT, Berrilan J, Lesk A. Proc Natl Acad Sci U S A. 2002;99:5591. doi: 10.1073/pnas.042681399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Petty SA, Decatur SM. Proc Natl Acad Sci U S A. 2005;102:14272. doi: 10.1073/pnas.0502804102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kittner M, Knecht V. J. Phys. Chem. B. 2010;114:15288. doi: 10.1021/jp1065264. [DOI] [PubMed] [Google Scholar]; Whittleston CS, Wales DJ. J Am Chem Soc. 2007;129:16005. doi: 10.1021/ja075346p. [DOI] [PubMed] [Google Scholar]
- 149.Chebaro Y, Derreumaux P. Proteins. 2009;75:442. doi: 10.1002/prot.22254. [DOI] [PubMed] [Google Scholar]
- 150.Wei G, Song W, Derreumaux P, Mousseau N. Front Biosci. 2008;13:5681. doi: 10.2741/3109. [DOI] [PubMed] [Google Scholar]
- 151.Song W, Wei G, Mousseau N, Derreumaux P. J. Phys. Chem. B. 2008;112:4410. doi: 10.1021/jp710592v. [DOI] [PubMed] [Google Scholar]
- 152.Lu Y, Derreumaux P, Guo Z, Mousseau N, Wei G. Proteins. 2009;75:954. doi: 10.1002/prot.22305. [DOI] [PubMed] [Google Scholar]
- 153.Nasica-Labouze J, Meli M, Derreumaux P, Colombo G, Mousseau N. PLoS Comput. Biol. 2011;7:e1002051. doi: 10.1371/journal.pcbi.1002051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Barz B, Wales DJ, Strodel B. J Phys Chem B. 2014;118:1003. doi: 10.1021/jp412648u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Baftizadeh F, Pietrucci F, Biarnés X, Laio A. Phys Rev Lett. 2013;110:168103. doi: 10.1103/PhysRevLett.110.168103. [DOI] [PubMed] [Google Scholar]
- 156.Nguyen PH, Derreumaux P. J. Phys. Chem. B. 2013;117:5831. doi: 10.1021/jp401563n. [DOI] [PubMed] [Google Scholar]
- 157.Tycko R, Wickner RB. Acc. Chem. Res. 2013;46:1487. doi: 10.1021/ar300282r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Nasica-Labouze J, Mousseau N. PLos Comp. Biol. 2012;8:e1002782. doi: 10.1371/journal.pcbi.1002782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Middleton CT, Marek P, Cao P, Chiu C, Singh S, Woys AM, de Pablo JJ, Raleigh DP, Zanni MT. Nature Chem. 2012;4:355. doi: 10.1038/nchem.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Straub JE, Thirumalai D. Annu Rev Phys Chem. 2011;62:437. doi: 10.1146/annurev-physchem-032210-103526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.De Simone A, Kitchen C, Kwan AH, Sunde M, Dobson CM, Frenkel D. Proc Natl Acad Sci U S A. 2012;109:6951. doi: 10.1073/pnas.1118048109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Qiao Q, Bowman GR, Huang X. J. Am. Chem. Soc. 2013;135:16092. doi: 10.1021/ja403147m. [DOI] [PubMed] [Google Scholar]
- 163.Nguyen PH, Tarus B, Derreumaux P. J Phys Chem B. 2014;118:501. doi: 10.1021/jp4115404. [DOI] [PubMed] [Google Scholar]
- 164.Nguyen PH, Derreumaux P. Acc. Chem. Res. 2014;47:603. doi: 10.1021/ar4002075. [DOI] [PubMed] [Google Scholar]
- 165.Ciryam P, Tartaglia GG, Morimoto RI, Dobson CM, Vendruscolo M. Cell Rep. 2013;5:781. doi: 10.1016/j.celrep.2013.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Laurents DV, Gorman PM, Guo M, Rico M, Chakrabartty A, Bruix M. J. Biol. Chem. 2005;280:3675. doi: 10.1074/jbc.M409507200. [DOI] [PubMed] [Google Scholar]
- 167.Côté S, Derreumaux P, Mousseau N. J. Chem. Theory Comput. 2011;7:2584. doi: 10.1021/ct1006967. [DOI] [PubMed] [Google Scholar]
- 168.Cote S, Laghaei R, Derreumaux P, Mousseau N. N. J. Phys. Chem. B. 2012;116:4043. doi: 10.1021/jp2126366. [DOI] [PubMed] [Google Scholar]
- 169.Lin YS, Pande VS. Biophys J. 2012;103:L47. doi: 10.1016/j.bpj.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Rosenman DJ, Connors CR, Chen W, Wang C, Garcia AE. J. Mol. Biol. 2013;425:3338. doi: 10.1016/j.jmb.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ball KA, Phillips AH, Wemmer DE, Head-Gordon T. Biophys. J. 2013;104:2714. doi: 10.1016/j.bpj.2013.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lv Z, Roychaudhuri R, Condron MM, Teplow DB, Lyubchenko YL. Sci. Rep. 2013;3:2880. doi: 10.1038/srep02880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhang T, Zhang J, Derreumaux P, Mu Y. J. Phys. Chem. B. 2013;117:3993. doi: 10.1021/jp312573y. [DOI] [PubMed] [Google Scholar]
- 174.Jiang L, Liu C, Leibly D, Landau M, Zhao M, Hughes MP, Eisenberg DS. Elife. 2013;2:e00857. doi: 10.7554/eLife.00857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Scherzer-Attali R, Pellarin R, Convertino M, Frydman-Marom A, Egoz-Matia N, Peled S, Levy-Sakin M, Shalev DE, Caflisch A, Gazit E, Segal D. PLoS One. 2010;5:e11101. doi: 10.1371/journal.pone.0011101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zhang T, Xu W, Mu Y, Derreumaux P. ACS Chem. Neurosci. 2014;5:148. doi: 10.1021/cn400197x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Maupetit J, Derreumaux P, Tufféry P. J. Comput. Chem. 2010;31:726. doi: 10.1002/jcc.21365. [DOI] [PubMed] [Google Scholar]
- 178.Thévenet P, Shen Y, Maupetit J, Guyon F, Derreumaux P, Tufféry P. Nucleic Acids Res. 2012;40:W288. doi: 10.1093/nar/gks419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Camproux AC, Gautier R, Tuffery P. J. Mol. Biol. 2004;339:591. doi: 10.1016/j.jmb.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 180.Thévenet P, Shen Y, Maupetit J, Guyon F F, Padilla A, Derreumaux P, Tuffery P. J. Peptide Sci. 2012;18:60. doi: 10.1093/nar/gks419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Guyon F, Tuffery P. Bioinformatics. 2014;30:784. doi: 10.1093/bioinformatics/btt618. [DOI] [PubMed] [Google Scholar]
- 182.Shen Y, Picord G, Guyon F, Tuffery P. PLoS One. 2013;8:e80493. doi: 10.1371/journal.pone.0080493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Steckbeck JD, Craigo JK, Barnes CO, Montelaro RC. J. Biol. Chem. 2011;286:27156. doi: 10.1074/jbc.M111.258855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.San Francisco B, Bretsnyder EC, Kranz RG. Proc Natl Acad Sci U S A. 2013;110:E788. doi: 10.1073/pnas.1213897109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Horjales S, Schmidt-Arras D, Limardo RR, Leclercq O, Obal G, Prina E, Turjanski AG, Späth GF, Buschiazzo A. Structure. 2012;20:1649. doi: 10.1016/j.str.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 186.Chopera DR, Cotton LA, Zawaira A, Mann JK, Ngandu NK, Ntale R, Carlson JM, Mlisana K, Woodman Z, de Assis Rosa D, Martin E, Miura T, Pereyra F, Walker BD, Gray CM, Martin DP, Ndung’u T, Brockman MA, Karim SA, Brumme ZL, Williamson C, C CAPRISA 002 Study Team J Virol. 2012;86:13423. doi: 10.1128/JVI.02171-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Mouscadet JF, Arora R, André J, Lambry JC, Delelis O, Malet I, Marcelin AG, Calvez V, Tchertanov L. J. Mol. Recognit. 2009;22:480. doi: 10.1002/jmr.970. [DOI] [PubMed] [Google Scholar]
- 188.Malik-Chaudhry HK, Saavedra A, Lia J. Biotechnology and Bioengineering. 2014 doi: 10.1002/bit.25183. DOI: 10.1002/bit.25183. [DOI] [PubMed] [Google Scholar]
- 189.Feller G, Dehareng D, Lage JL. FEBS J. 2011;278:2333. doi: 10.1111/j.1742-4658.2011.08154.x. [DOI] [PubMed] [Google Scholar]
- 190.Li M, Chang S, Yang L, Shi J, McFarland K, Yang X, Moller A, Wang C, Zou X, Chi C, Cui J. J. Biol. Chem. 2014;289:4735. doi: 10.1074/jbc.M113.535898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wu G, Han K, Lv F. J. Theor. Biol. 2013;317:293. doi: 10.1016/j.jtbi.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 192.Kumar M, Ahmad S, Ahmad E, Saifi MA, Khan RH. PLoS One. 2012;7:e36770. doi: 10.1371/journal.pone.0036770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Chopra N, Agarwal S, Verma S, Bhatnagar S, Bhatnagar R. J. Comput. Aided Mol. Des. 2011;25:275. doi: 10.1007/s10822-011-9419-z. [DOI] [PubMed] [Google Scholar]
- 194.Stegmann CM, Lührmann R. PLoS One. 2010;5:e10013. doi: 10.1371/journal.pone.0010013. M. C. and Wahl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Flower DR. Nature Chem. Biol. 2013;9:749. doi: 10.1038/nchembio.1383. [DOI] [PubMed] [Google Scholar]
- 196.Olsson N, Wallin S, James P, Borrebaeck CA, Wingren C. Protein Sci. 2012;21:1897. doi: 10.1002/pro.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.López-Martínez R, Ramírez-Salinas GL, Correa-Basurto J, Barrón BL. PLoS One. 2013;8:e76876. doi: 10.1371/journal.pone.0076876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Jesús T, Rogelio L, Abraham C, Uriel L, J-Daniel G, Alfonso MT, Lilia BB. Bioinformation. 2012;8:870. doi: 10.6026/97320630008870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Deng W, Guan C, Liu K, Zhang X, Feng X, Zhou B, Su X, Chen P. Virus Research. 2013;13:00369. doi: 10.1016/j.virusres.2013.10.022. [DOI] [PubMed] [Google Scholar]
- 200.Qureshi A, Thakur N, Tandon H, Kumar M. Nucleic Acids Res. 2014;42:D1147. doi: 10.1093/nar/gkt1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Gupta SK, Singh A, Srivastava M, Gupta SK, Akhoon BA. Vaccine. 2009;28:120. doi: 10.1016/j.vaccine.2009.09.095. [DOI] [PubMed] [Google Scholar]
- 202.Teixeira ML, Dalla Rosa A, Brandelli A. Microbiology. 2013;159:980. doi: 10.1099/mic.0.062828-0. [DOI] [PubMed] [Google Scholar]
- 203.Lin J, Alexander-Katz A. ACS Nano. 2013;7:10799. doi: 10.1021/nn4040553. [DOI] [PubMed] [Google Scholar]
- 204.Yan L, Yan Y, Liu H, Lv Q. BioSystems. 2013;113:1. doi: 10.1016/j.biosystems.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 205.Berges R, Balzeau J, Takahashi M, Prevost C, Eyer J. PLoS One. 2012;7:e49436. doi: 10.1371/journal.pone.0049436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lee KH, Ytreberg FM. Entropy. 2012;14:630. [Google Scholar]
- 207.Ramis JM, Rubert M, Vondrasek J, Gayà A, Lyngstadaas SP, Monjo M. Tissue Eng. Part A. 2012;18:1253. doi: 10.1089/ten.tea.2011.0404. [DOI] [PubMed] [Google Scholar]
- 208.Friedrich MG, Lam J, Truscott RJW. J. Biol. Chem. 2012;287:39012. doi: 10.1074/jbc.M112.391565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Chen X, Yu X, Liu Y, Zhang J. J. Mol. Graph. Model. 2013;46:83. doi: 10.1016/j.jmgm.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 210.Gupta UK, Mahanta S, Subhankar P. Medical Hypotheses. 2013;81:853. doi: 10.1016/j.mehy.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 211.Gautam S, Loh KC. Separation and purification technology. 2013;102:173. [Google Scholar]
- 212.Zgarbova M, Otyepka M, Sponer J, Mladek A, Banas P, Cheatham TE, Jurecka P. J Chem Theory Comput. 2011;7:2886. doi: 10.1021/ct200162x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Chen AA, Garcia AE. Proc Natl Acad Sci U S A. 2013;110:16820. doi: 10.1073/pnas.1309392110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Rother K, Rother M, Boniecki M, Puton T, Bujnicki JM. J Mol Model. 2011;17:2325. doi: 10.1007/s00894-010-0951-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Rother K, Rother M, Puton T, Bujnicki JM. Nucleic Acids Res. 2011;39:4007. doi: 10.1093/nar/gkq1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Coulbourn S, Flores X, Altman RB. RNA. 2010;16:1769. doi: 10.1261/rna.2112110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Parisien M, Major F. Nature. 2008;452:51. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 218.Das R, Baker D. Proc Natl Acad Sci U S A. 2011;104:14664. doi: 10.1073/pnas.0703836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Cao S, Chen SJ. J. Phys. Chem B. 2011;115:4216. doi: 10.1021/jp112059y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Laing C, Jung S, Kim N, Elmetwaly S, Zahran M, Schlick T. PLoS One. 2013;8:e71947. doi: 10.1371/journal.pone.0071947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Rivas E. RNA Biol. 2013;10:1185. doi: 10.4161/rna.24971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Morris-Andrews A, Rottler J, Plotkin SS. J. Chem. Phys. 2010;132:35105. doi: 10.1063/1.3269994. [DOI] [PubMed] [Google Scholar]
- 223.He Y, Maciejczyk M, Oldziej S, Scheraga HA, Liwo A. Phys Rev Lett. 2013;110:098101. doi: 10.1103/PhysRevLett.110.098101. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hsu CW, Fyta M, Lakatos G, Melchionna S, Kaxiras E. J. Chem. Phys. 2012;137:105102. doi: 10.1063/1.4748105. [DOI] [PubMed] [Google Scholar]
- 224.Ouldridge TE, Louis AA, Doye JPK. J. Chem. Phys. 2011;134:085101. doi: 10.1063/1.3552946. [DOI] [PubMed] [Google Scholar]
- 225.Ding F, Sharma S, Chalasani P, Demidov VV, Broude NE, Dokholyan NV. RNA. 2008;14:1164. doi: 10.1261/rna.894608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Hyeon C, Thirumalai D. Proc Natl Acad Sci U S A. 2005;102:6789. doi: 10.1073/pnas.0408314102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Jonikas MA, Radmer RJ, Laederach A, Das R, Pearlman S, Herschlag D, Altman RB. RNA. 2009;15:189. doi: 10.1261/rna.1270809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Xia Z, Gardner DP, Gutell RR, Ren P. J. Phys Chem B. 2010;114:13497. doi: 10.1021/jp104926t. [DOI] [PMC free article] [PubMed] [Google Scholar]; Xia Z, Bell DR, Shi Y, Ren P. J Phys Chem B. 2013;117:3135. doi: 10.1021/jp400751w. [DOI] [PubMed] [Google Scholar]
- 229.Bernauer J, Huang X, Sim AY, Levitt M. RNA. 2011;17:1066. doi: 10.1261/rna.2543711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Denning EJ, MacKerell AD., Jr. J. Am. Chem. Soc. 2012;134:2800. doi: 10.1021/ja211328g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Pasquali S, Derreumaux P. J. Phys. Chem B. 2010;114:11957. doi: 10.1021/jp102497y. [DOI] [PubMed] [Google Scholar]
- 232.Leontis NB, Stombaugh J, Westhof E. NAR. 2002;30:3497. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Cragnolini T, Derreumaux P, Pasquali S. J. Phys. Chem B. 2013;117:8047. doi: 10.1021/jp400786b. [DOI] [PubMed] [Google Scholar]
- 234.Santa Lucia J. Proc Natl Acad Sci USA. 1998;95:1460. [Google Scholar]
- 235.Ellis RJ. Trends Biochem. Sci. 2001;26:597. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]; Zimmerman SB, Minton AP. Ann. Rev. Biophys. & Biomol. Struct. 1993;22:27. doi: 10.1146/annurev.bb.22.060193.000331. [DOI] [PubMed] [Google Scholar]; Zhou H-X, Qin S. Biophys. Rev. 2013;5:207. doi: 10.1007/s12551-013-0101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Frembgen-Kesner T, Elcock AH. Biophys Rev. 2013;5:109. doi: 10.1007/s12551-013-0110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Harada R, Tochio N, Kigawa T, Sugita Y, Feig M. J Am Chem Soc. 2013;135:3696. doi: 10.1021/ja3126992. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Tian J, Garcia AE. J. Am. Chem. Soc. 2011;133:15157. doi: 10.1021/ja2054572. [DOI] [PubMed] [Google Scholar]; c) Tian J, Garcia AE. J. Chem. Phys. 2011;134:225101. doi: 10.1063/1.3592712. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) McGuffee SR, Elcock AH. J. Am. Chem. Soc. 2006;128:12098. doi: 10.1021/ja0614058. [DOI] [PubMed] [Google Scholar]
- 238.Kurniawan NA, Enemark S, Rajagopalan R. J Am Chem Soc. 2012;134:10200. doi: 10.1021/ja302943m. [DOI] [PubMed] [Google Scholar]
- 239.Harada R, Sugita Y, Feig M. J Am Chem Soc. 2012;134:4842. doi: 10.1021/ja211115q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Dong H, Qin S, Zhou HX. PLoS Comput Biol. 2010;6:e1000833. doi: 10.1371/journal.pcbi.1000833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Leduc C, Padberg-Gehle K, Varga V, Helbing D, Diez S, Howard J. Proc Natl Acad Sci U S A. 2012;109:6100. doi: 10.1073/pnas.1107281109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Ando T, Skolnick J. Proc. Natl. Acad. Sci. USA. 2010;107:18457. doi: 10.1073/pnas.1011354107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Mika JT, Poolman B. Curr. Opin. Biotech. 2011;22:117. doi: 10.1016/j.copbio.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 244.Qu L-H, Henras A, Lu Y-J, Zhou H, Zhou W-X, Zhu Y-Q, Zhao J, Henry Y, Caizergues-Ferrer M, Bachellerie J-P. Mol. & Cell. Biol. 1999;19:1144. doi: 10.1128/mcb.19.2.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Weiss M, Elsner M, Kartberg F, Nilsson T. Biophys. J. 2004;87:3518. doi: 10.1529/biophysj.104.044263. [DOI] [PMC free article] [PubMed] [Google Scholar]; Banks DS, Fradin C. Biophys. J. 2005;89:296. doi: 10.1529/biophysj.104.051078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Roosen-Runge F, Hennig M, Zhang F, Jacobs RMJ, Sztucki M, Schober H, Seydel T, Schreiber F. Proc. Natl. Acad. Sci USA. 2011;108:11815. doi: 10.1073/pnas.1107287108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Benton LA, Smith AE, Young GB, Pielak GJ. Biochem. 2012;51:9773. doi: 10.1021/bi300909q. [DOI] [PubMed] [Google Scholar]
- 248.Xu W, Zhang C, Derreumaux P, Gräslund A, Morozova-Roche L, Mu Y. PLoS One. 2011;6:e24329. doi: 10.1371/journal.pone.0024329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Chongsiriwatana NP, Patch JA, Czyzewski AM, Dohm MT, Ivankin A, et al. Proc Natl Acad Sci USA. 2008;105:2794. doi: 10.1073/pnas.0708254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Appella DH, Christianson LA, Karle IL, Powell DR, Gellman SH. J. Am. Chem. Soc. 1996;118:13071. [Google Scholar]
- 251.Tian J, Dang H, Chen Z, Guan A, Jin Y, Atkinson MA, Kaufman DL. Diabetes. 2013;62:3760. doi: 10.2337/db13-0931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Amijee H, Bate C, Williams A, Virdee J, Jeggo R, Spanswick D, Scopes DI, Treherne JM, Mazzitelli S, Chawner R, Eyers CE, Doig AJ. Biochemistry. 2012;51:8338. doi: 10.1021/bi300415v. [DOI] [PubMed] [Google Scholar]
- 253.Baell J, Congreve M, Leeson P, Abad-Zapatero C. Future Med Chem. 2013;5:745. doi: 10.4155/fmc.13.61. [DOI] [PubMed] [Google Scholar]
- 254.Derreumaux P, Vergoten G, Lagant P. J. Comput. Chem. 1990;11:560. [Google Scholar]
- 255.Derreumaux P, Vergoten G. J. Chem. Phys. 1995;102:8586. [Google Scholar]
- 256.Rao F, Settanni G, Guarnera E, Caflisch A. J. Chem. Phys. 2005;122:184901. doi: 10.1063/1.1893753. [DOI] [PubMed] [Google Scholar]
- 257.Urbanc B, Betnel M, Cruz L, Bitan G, Teplow DB. J Am Chem Soc. 2010;132:4266. doi: 10.1021/ja9096303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Guo C, Luo Y, Zhou R, Wei G. ACSNano. 2012;6:3907. doi: 10.1021/nn300015g. [DOI] [PubMed] [Google Scholar]
- 259.Lindorff-Larsen K, Maragakis P, Piana S, Eastwood MP, Dror RO, Shaw DE. PLoS One. 2012;7:e32131. doi: 10.1371/journal.pone.0032131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Nguyen PH, Li MS, Derreumaux P. Phys. Chem. Chem. Phys. 2011;13:9778. doi: 10.1039/c1cp20323a. [DOI] [PubMed] [Google Scholar]
- 261.Derreumaux P, Dauchez M, Vergoten G. J. Mol. Struct. 1993;295:203. [Google Scholar]
- 262.Derreumaux P, Wilson KJ, Vergoten G, Peticolas WL. J. Phys. Chem. 1989;93:1338. [Google Scholar]
- 263.Ferrenberg AM, Swendsen RH. Phys. Rev. Lett. 1988;61:2635. doi: 10.1103/PhysRevLett.61.2635. [DOI] [PubMed] [Google Scholar]
- 264.Shirts MR, Chodera JD. J. Chem. Phys. 2008;129:124105. doi: 10.1063/1.2978177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Liu Z, Chan HS. J Mol Biol. 2005;349:872. doi: 10.1016/j.jmb.2005.03.084. [DOI] [PubMed] [Google Scholar]
- 266.Cooper A, Cameron D, Jakus J, Pettigrew GW. Biochem Soc Trans. 2007;35:1547. doi: 10.1042/BST0351547. [DOI] [PubMed] [Google Scholar]
- 267.Vieille C, Zeikus GJ. Microbiol.Mol.Biol.Rev. 2001;65:1. doi: 10.1128/MMBR.65.1.1-43.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Sterpone F, Melchionna S. Chem.Soc.Rev. 2012;41:1665. doi: 10.1039/c1cs15199a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Balasco N, Esposito L, De Simone A, Vitigliano L. Protein Sci. 2013;22:1016. doi: 10.1002/pro.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Koga N, Tatsumi-Koga R, Liu G, Xiao R, Acton TB, Montelione GT, Baker D. Nature. 2012;491:222. doi: 10.1038/nature11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Gribenko AV, Patel MM, Liu J, McCallum SA, Wang C, Makhatadze GI. Proc. Natl. Acad. Sci. USA. 2009;106:2601. doi: 10.1073/pnas.0808220106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Xiao L, Honig B. J. Mol. Biol. 1999;289:1435. doi: 10.1006/jmbi.1999.2810. [DOI] [PubMed] [Google Scholar]
- 273.Kumar S, Ma B, Tsai CJ, Nussinov R. Proteins. 2000;38:368. doi: 10.1002/(sici)1097-0134(20000301)38:4<368::aid-prot3>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 274.Xiao S, Patsalo V, Shan B, Bi Y, Green D, Raleigh DP. Proc Natl Acad Sci USA. 2013;110:11337. doi: 10.1073/pnas.1222245110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Nojima H, Ikai A, Oshima T, Noda H. J. Mol .Biol. 1977;116:429. doi: 10.1016/0022-2836(77)90078-x. [DOI] [PubMed] [Google Scholar]
- 276.Razvi A, Scholtz JM. Protein Sci. 2006;15:1569. doi: 10.1110/ps.062130306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Feller G. J Phys : Condens Matter. 2010;22:323101. doi: 10.1088/0953-8984/22/32/323101. [DOI] [PubMed] [Google Scholar]
- 278.Liu CC, Licata VJ. Proteins. 2013 DOI:10.1002/prot.24458. [Google Scholar]
- 279.Kalimeri M, Rahaman O, Melchionna S, Sterpone F. J Phys Chem B. 2013;117:13775. doi: 10.1021/jp407078z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Dagan S, Hagai T, Gavrilov Y, Kapon R, Levy Y, Reich Z. Proc Natl Acad Sci USA. 2013;110:10628. doi: 10.1073/pnas.1302284110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Kamata K, Araki T, Tanaka H. Phys Rev Lett. 2009;102:108303. doi: 10.1103/PhysRevLett.102.108303. [DOI] [PubMed] [Google Scholar]
- 282.FrembgenKesner T, Elcock AH. J Chem Theory Comput. 2009;5:242. doi: 10.1021/ct800499p. [DOI] [PubMed] [Google Scholar]
- 283.Szymczak P, Cieplak M. J Phys: Condens Matter. 2011;23:033102. doi: 10.1088/0953-8984/23/3/033102. [DOI] [PubMed] [Google Scholar]
- 284.Ando T, Skolnick J. Biophys. J. 2013;104:96. doi: 10.1016/j.bpj.2012.11.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Jaspe J, Hagen SJ. Biophys J. 2006;91:3415. doi: 10.1529/biophysj.106.089367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Zou X, Liu Y, Chen Z, Cárdenas-Jirón GI, Schulten K. Biophys J. 2010;99:1182. doi: 10.1016/j.bpj.2010.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Hamilton-Brown P, Bekard I, Ducker WA, Dunstan D, Dave E. J Phys Chem B. 2008;112:16249. doi: 10.1021/jp805257n. [DOI] [PubMed] [Google Scholar]
- 288.Dunstan D, Hamilton-Brown P, Asimakis P, Ducker W, Bertolini J. Protein Eng Des Sel. 2009;22:741. doi: 10.1093/protein/gzp059. [DOI] [PubMed] [Google Scholar]
- 289.Lee C, Bird S, Shaw M, Jean L, Vaux DJ. J Biol Chem. 2012;287:38006. doi: 10.1074/jbc.M112.400580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Qiang W, Kelley K, Tycko R. J Am Chem Soc. 2013;135:6860. doi: 10.1021/ja311963f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Denesyuk NA, Thirumalai D. J Phys Chem B. 2013;117:4901. doi: 10.1021/jp401087x. [DOI] [PubMed] [Google Scholar]
- 292.Draper DE. RNA. 2004;10:335. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.DeMille RC, Cheatham TE, III, Molinero V. J. Phys. Chem B. 2011;115:132. doi: 10.1021/jp107028n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Ricci A, Anthopoulos A, Massarotti A, Grimstead I, Brancale A. Future Medicinal Chemistry. 2012;4:1219. doi: 10.4155/fmc.12.60. [DOI] [PubMed] [Google Scholar]
- 295.Wollacott AM, Merz KM., Jr. J. Mol. Graphics Model. 2007;25:801. doi: 10.1016/j.jmgm.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 296.Stone JE, Kohlmeyer A, Vandivort KL, et al. ADVANCES IN VISUAL COMPUTING, PT II Book Series: Lecture Notes in Computer Science. 2010;6454:382. [Google Scholar]
- 297.Haag MP, Reiher M. Int. J. Quantum Chem. 2013;113:8. [Google Scholar]
- 298.Birmanns S, Rusu M, Wriggers W. J. Struct. Biol. 2011;173:428. doi: 10.1016/j.jsb.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Gillet A, Sanner M, Stoffler D, Olson A. Structure. 2005;13:483. doi: 10.1016/j.str.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 300.Baaden M, Marrink SJ. Current Opin. Struct. Biol. 2013;23:878. doi: 10.1016/j.sbi.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 301.Saladin A, Amourda C, Poulain P, Férey N, Baaden M, Zacharias M, Delalande O, Prévost C. Nucleic Acids Res. 2010;38:6313. doi: 10.1093/nar/gkq459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Molza AE, Ferey N, Czjzek M, Le Rumeur E, Hubert JF, Tek A, Laurent B, Baaden M, Delalande O. Faraday Discuss. 2014 doi: 10.1039/c3fd00134b. DOI: 10.1039/C3FD00134B. [DOI] [PubMed] [Google Scholar]
- 303.Manosas M, Wen JD, Li PT, Smith SB, Bustamante C, Tinoco I, Jr, Ritort F. Biophys J. 2007;92:3010. doi: 10.1529/biophysj.106.094243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Viet MH, Nguyen PH, Ngo ST, Li MS, Derreumaux P. ACS Chem. Neurosci. 2013;4:1446. doi: 10.1021/cn400110d. [DOI] [PMC free article] [PubMed] [Google Scholar]; Huet A, Derreumaux P. Biophys. J. 2006;91:3829. doi: 10.1529/biophysj.106.090993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Doye J, Wales D. J. Phys. Chem. A. 1997;101:5111. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.