Abstract
There is compelling evidence that particulate matter (PM) increases lung cancer risk by triggering systemic inflammation, and leukocyte DNA hypomethylation. However, previous investigations focused on repeated element sequences from LINE-1 and Alu families. Tandem repeats, which display a greater propensity to mutate, and are often hypomethylated in cancer patients, have never been investigated in individuals exposed to PM. We measured methylation of three tandem repeats (SATα, NBL2, D4Z4) by polymerase chain reaction–pyrosequencing on blood samples from truck drivers and office workers (60 per group) in Beijing, China. We used lightweight monitors to measure personal PM2.5 (PM with aerodynamic diameter ≤2.5 µm) and elemental carbon (EC, a tracer of PM from vehicular traffic). Ambient PM10 data were obtained from air quality measuring stations. Overall, an interquartile increase in personal PM2.5 and ambient PM10 levels was associated with a significant covariate-adjusted decrease in SATα methylation (−1.35% 5-methyl cytosine [5mC], P = 0.01; and −1.33%5mC; P = 0.01, respectively). Effects from personal PM2.5 and ambient PM10 on SATα methylation were stronger in truck drivers (−2.34%5mC, P = 0.02; −1.44%5mC, P = 0.06) than office workers (−0.95%5mC, P = 0.26; −1.25%5mC, P = 0.12, respectively). Ambient PM10 was negatively correlated with NBL2 methylation in truck drivers (−1.38%5mC, P = 0.03) but not in office workers (1.04%5mC, P = 0.13). Our result suggests that PM exposure is associated with hypomethylation of selected tandem repeats. Measuring tandem-repeat hypomethylation in easy-to-obtain blood specimens might identify individuals with biological effects and potential cancer risk from PM exposure.
Keywords: Air pollution, epigenetics, epidemiology
INTRODUCTION
Particulate Matter (PM), a common airborne pollutant in urban environments, contributes to as many as 3.2 million premature deaths worldwide each year [Lim et al. 2013]. Several epidemiologic investigations have demonstrated the association between PM and lung cancer [Turner et al. 2011; Krewski et al. 2005]. Particles with an aerodynamic diameter <2.5 micrometers (PM2.5), are especially hazardous to cardiorespiratory health because they can penetrate into the airways and deposit in the respiratory bronchioles and alveoli [Kampa and Castanas 2008]. Each 10-µg/m3 elevation in chronic and acute PM2.5 has been associated with a ~8% increased risk of lung cancer mortality and a 2.8% increase in PM-related mortality, respectively [Pope et al. 2002; Kloog et al. 2013].
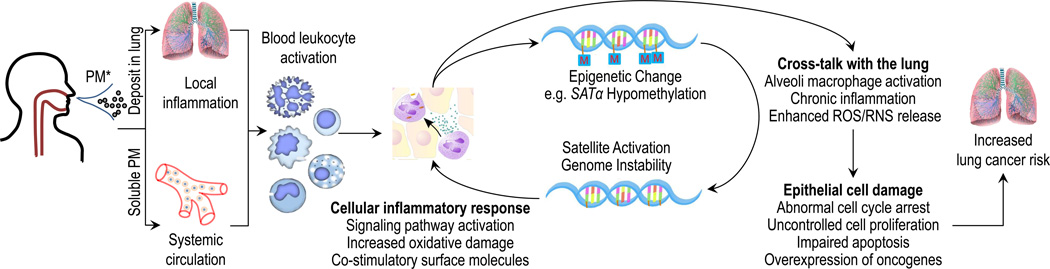
Several mechanistic pathways have been proposed to account for the link between PM and lung cancer, including altered gene expression, systemic inflammatory responses, and oxidative stress (Fig. 1) [Soberanes et al. 2012; Ghio et al. 2012; Delfino et al. 2010; Sava and Carlsten 2012]. PM triggers inflammation and induces the release of reactive oxygen species (ROS), both locally and systemically [Vattanasit et al. 2013; Torres-Ramos et al. 2011; Dagher et al. 2005; Zhao et al. 2013; Dagher et al. 2005], which – in turn – can contribute to epigenetic alterations and genomic instability [Saint-Georges et al. 2009]. DNA methylation is a reversible mechanism of epigenetic regulation that plays a key role in a wide variety of fundamental biological processes by affecting gene expression at the transcriptional level. Both in vitro and in vivo studies suggest that aberrant DNA methylation may lead to abnormal cell cycle arrest, uncontrolled cell proliferation and apoptosis, all of which are known risk factors for carcinogenesis, aggressive tumor neovascularization, angiogenesis, and metastasis [Lou et al. 2013; Valavanidis et al. 2013; Ghio et al. 2012]. In addition, dysregulation of DNA methylation has been linked with overexpression of pro-inflammatory cytokines, especially in leukocytes [Venza et al. 2012; Mishra et al. 2012]. The cross-talk between blood leukocytes and inflammatory cells in the lung enhances chronic inflammation and ROS/reactive nitrogen species (RNS) generation in the lung, which leads to chronic tissue injury and abnormal tumor immunity.
Figure 1.
Proposed conceptual model linking PM exposure, leukocyte DNA hypomethylation and lung cancer. PM, particulate matter; SATα, satellite alpha; ROS, reactive oxygen species; RNS, reactive nitrogen species.
Consistent evidence from in vitro and in vivo studies has shown that exposure to air pollution, including PM, triggers DNA hypomethylation, which can underlie the activation of inflammatory genes and repetitive sequences, including tandem elements and interspersed repeats [De Prins et al. 2013; Tarantini et al. 2009]; Madrigano et al. 2011; Bellavia et al. 2013]. Approximately half of the human genome consists of repetitive elements [Lander et al. 2001], which require DNA methylation to maintain its silencing. The association between decreased repetitive element methylation and exposure to air pollution has been clearly demonstrated in both general population [Baccarelli et al. 2009; Madrigano et al. 2011] and occupationally exposed individuals [Bollati et al. 2007; Tarantini et al. 2009; Pavanello et al. 2010]. However, repetitive elements are controlled through multiple mechanisms, and each element has unique transcription patterns in response to cellular stressors [Meaney and Szyf 2005].
Repetitive elements may encode proteins involved in their replication and insertion into new locations within the genome. These elements influence genome transcriptional output and aberrant epigenetic alteration of the neighboring genes [Byun et al. 2012; Faulkner et al. 2009; Prada et al. 2012]. Dysregulation of repeated elements, e.g. hypomethylation, could be associated with genomic instability and overexpression of oncogenes, and therefore contributes to tumorigenesis [Lamprecht et al. 2010]. Zhu and colleagues have shown that individuals with lower methylation in blood leukocytes repetitive elements, such as the long interspersed nucleotide elements (LINE)-1 are at high risk of developing and dying from cancer [Zhu et al. 2011].
Previous studies have focused exclusively on a limited set of repeated-element sequences in LINE-1 and Alu interspersed repeats. Tandem repeats, a highly represented family of repetitive elements, are hypervariable in mammals. Dysregulation of tandem repeats methylation has been repetitively observed in many types of cancers [Yoshida et al. 2011; Weisenberger et al. 2005; Nishiyama et al. 2005a; Nishiyama et al. 2005b; Tsumagari et al. 2008; Choi et al. 2009]. Various studies have pointed out a distinctive methylation pattern among different tandem repeats dependent on the type of cancer [Choi et al. 2009]. Growing evidence indicates that each tandem repeat may have specific functions and distinct causes of dysregulation, but also that changes in DNA methylation of tandem repeat elements could behave differently than that of interspersed repeats [Nishiyama et al. 2005a; Nishiyama et al. 2005b; Choi et al. 2009]. These findings indicate the need for investigating methylation patterns in different types of tandem repeats.
Multiple types of tandem repeats with different functions have been described in the human genome. Satellite α (SATα) tandem repeats, composed of 170-bp DNA sequences, represent the main DNA component of every human centromere [Lee et al. 1997]. Hypomethylation of satellite repeats has been shown to be tightly related to chromatin decondensation, as well as chromosomal breakages and abnormalities [Choi et al. 2009; Fabris et al. 2011; Yoshida et al. 2011; Prada et al. 2012]. Aberrant overexpression of satellite repeats is observed in mouse and human epithelial cancer, including lung cancer [Ting et al. 2011]. Non-satellite tandem repeats such as NBL2 and D4Z4, also exhibit cancer-linked hypomethylation in certain types of cancer [Ehrlich 2006; Choi et al. 2009].
Although the significance of epigenetic changes in tandem repeats on lung cancer development has been repeatedly demonstrated, the initial determinants of hypomethylation are not yet well understood [Xiang et al. 2012]. Little research has investigated the interplay among tandem repeats methylation, inflammatory responses, and cellular oxidative stress induced by ambient PM insult. Therefore, investigating the effect of PM exposure on tandem repeats methylation is critical to provide new hypotheses for the increased PM-related lung cancer risk, especially in high risk populations residing in heavily polluted environments.
Beijing has been ranked in the World Development Indicators as one of the 15 cities with the highest levels of air pollution [World Bank 2012]. In Beijing, PM is primarily from traffic sources aggravated by high population density and the recent rapid increase in vehicular traffic [Yu et al. 2011]. Particles transported from industrial sources and windblown dust are also major sources of air pollution in Beijing [Yu et al. 2011]. In China, a 1.63% increase in lung cancer incidence was recorded each year between 1988 and 2005 [Chen et al. 2010]. In 2008, lung cancer incidence and mortality ranked first among all cancers, both in Beijing and nationwide [Chang et al. 2012]. In 2009, 4598 new lung cancer cases were reported among the 7,645,186 Beijing residents [Chen et al. 2013].
We previously reported the methylation levels of SATα, NBL2, and D4Z4 in relation to several airborne toxic metals in truck drivers and office workers in Beijing, China, who were exposed to high levels of PM [Hou et al. 2013]. In this study, we further investigated the association between short-term PM exposure (personal PM2.5 and ambient PM10) and methylation of these three tandem repeats in peripheral blood leukocytes. Peripheral leukocytes are the first responders to inflammatory mediators released in the lung after environmental insult. DNA methylation regulates leukocyte functions at the transcription level [Kato et al. 2012] and therefore affects the recruitment and migration of leukocytes from the bloodstream into the lung. Blood DNA methylation has been shown as a sensitive marker for acute and short-term changes in the epigenome [Baccarelli et al. 2009; De Prins et al. 2013; Kile et al. 2013; Bellavia et al. 2013]. Therefore, evaluating blood tandem repeats methylation in relation to PM exposure may unveil novel molecular targets for PM effects and also provide novel biomarkers reflecting biological effects from PM exposures [Brennan and Flanagan 2012; Wu et al. 2012; Marsit and Christensen 2013].
To examine the relationship between individual and environmental PM exposure and tandem-repeat methylation, we studied two groups of workers exposed to high levels of PM (truck drivers (n = 60) and office workers (n = 60)) in Beijing. To enhance the power to identify the effects of PM on tandem-repeat methylation, we studied each participant at two different time points, 1–2 weeks apart. We assessed their exposure to air pollution by using personal measures of PM2.5 and elemental carbon (EC, a surrogate for PM from vehicular traffic), as well as ambient levels of PM10. We hypothesized that individuals exposed to higher levels of PM will have differences in DNA methylation of SATα, NBL2, or D4Z4.
MATERIALS AND METHODS
Study Participants
The Beijing Truck Driver Air Pollution Study included 60 truck drivers and 60 office workers in Beijing, China, who participated between June 15 and July 27, 2008, as previously described [Baccarelli et al. 2011; Hou et al. 2013]. All participants lived in the Beijing metropolitan area and had been working at their current job for at least 2 years [Baccarelli et al. 2011]. Written informed Consent was obtained from all subjects. We examined all participants on two separate days 1–2 weeks apart. Therefore, the study was based on 120 observations for the 60 truck drivers and 120 observations for the 60 office workers, for a total of 240 observations. The truck driver and office worker groups were matched for smoking status and education. A self-administered questionnaire was used to collect detailed information on demographics, lifestyle, and other exposures. Information on time-varying factors, including tea and alcohol consumption and smoking habits, was obtained for both past lifestyles and their use on the examination days.
Personal Exposure Measurements
As previously reported [Baccarelli et al. 2011; Hou et al. 2013], we measured personal PM2.5 on both examination days by providing the study participants with a lightweight gravimetric sampler that they wore during the 8 hours of work. The sampler was carried in a belt pack with the inlet clipped near the breathing zone. Each sampler setup included an Apex pump (Casella Inc., Bedford, UK), a Triplex Sharp-Cut Cyclone separator (BGI Inc., Waltham, MA), and a 37-mm Teflon filter placed on top of a drain disc and inside a metal filter holder. We kept the filters in atmosphere-controlled conditions before and after sampling and were weighed with a microbalance (Mettler-Toledo Inc., Columbus, OH). A time-weighted average of PM2.5 concentration was recorded by dividing the change in filter weight before and after sampling by the volume of air sampled. We conducted technical validation tests of personal PM2.5 measures and found high reproducibility (r = 0.944) of our measurements. The blackness of the filters used to measure PM2.5 was assessed using an EEL Model M43D Smokestain Reflectometer (Diffusion Systems Ltd., London, UK), by applying standard black-smoke index calculations of the absorption coefficients based on reflectance (ISO 983). We assumed a factor of 1.0 for converting the absorption coefficient to EC mass [Kinney et al. 2000; Janssen et al. 2001], which was then divided by the sampled air volume to calculate average EC exposure concentration (ISO 983). EC is a combustion by-product contained in PM that has been used as a surrogate measure for PM from gasoline and especially diesel-powered motor vehicles [Kinney et al. 2000].
Ambient PM10 Data
During the study period, ambient PM10 (PM with an aerodynamic diameter ≤10 µm) data were obtained from the Beijing Municipal Environmental Bureau (http://www.bjepb.gov.cn/air2008/Air.aspx). To estimate the average PM10 level, we used daily averages of PM10 computed from data obtained from 27 monitoring stations throughout Beijing. We used these ambient PM10 data to assess exposure not only on the day blood was collected, but also on previous days in order to examine the distribution of the effect of air pollutants over time, i.e., the lag between the timing of exposure and the response (in our case tandem-repeat methylation). We also obtained daily outdoor temperature and humidity data for Beijing from the National Oceanic and Atmospheric Administration’s online database (http://www.noaa.gov/).
Sample Preparations and DNA Extraction
We used standardized protocols for blood collection and storage. Peripheral blood was collected from each participant on both examination days, and buffy coat was separated within 2 hours and stored locally at −80°C. DNA was extracted from buffy coat by using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI) according to the manufacturer’s instructions. Purified DNA was resuspended in the kit’s hydration solution, quantified, and stored at −20°C until analysis. Blood DNA samples were randomized across plates to limit potential bias from plate effects, and laboratory personnel were blinded to both exposure groups and exposure study.
DNA Methylation Analysis
As previously reported [Hou et al. 2013], 500 ng of DNA was treated with bisulfite using the EZ-96 DNA Methylation-Gold Kit (Zymo Research, Orange, CA) according to the manufacturer’s protocol to determine methylation. Final elution was performed with 30 µl of M-Elution Buffer. DNA methylation was quantified using bisulfite polymerase chain reaction (PCR) and pyrosequencing. The primers and conditions were described previously [Choi et al. 2009]. Briefly, a 30-µl PCR was carried out in 15 µl of GoTaq Hot Start Green Master Mix (Promega, Madison, WI), with 10 pmol of forward primer, 10 pmol of reverse primer, 1 µl of bisulfite-treated genomic DNA, and water. The amplicon size is in the range of 150–250 bp. Pyrosequencing was performed using the PyroMark Q96 MD Pyrosequencing System (QIAGEN, Germantown, MD). The percentage of methylated and unmethylated cytosines was quantified for three CpG sites (cytosine and guanine separated by one phosphate) from SATα, four CpG sites from NBL2, and four CpG sites from D4Z4. The degree of methylation is expressed as the percentage of methylated cytosines divided by the sum of methylated and unmethylated cytosines (%5mC) measured in each sample.
Measurement of Plasma Inflammatory Mediators
We measured plasma inflammatory mediators in all study subjects, using a pre-set Luminex Bio-Plex 27-plex panel (Bio-Rad, Hercules, CA). The multiplex bead-based Luminex Bio-Plex 27-plex panel permits the simultaneous quantification of 27 cytokines from previously stored plasma, includes IL7, IL8, IL12, GM-CSF, IL4, IL5, IL9, IL10, IL13, GM-CSF, IL1β, IL1ra, IL15, IL17, MCP1, MIP1α, MIP1β, PDGF-BB, VEGF, FGF, RANTES, IP-10, eotaxin, GCSF. 27 sets of fluorescently dyed beads were conjugated with target-specific reactants, and triggered biochemical reactions with target biomolecules, which were detected by flow cytometer. The fluorescent output was processed by a high-speed digital signal processor, and cytokine concentrations were automatically calculated by Bio-Plex Manager™ software using a standard curve. Samples were run in triplicates.
Statistical Analysis
The demographic characteristics of truck drivers and office workers were summarized using standard descriptive statistics (Table I). Because our data include two measures for each study participant, we used a mixed-effect regression model to account for the lack of independence between the two measures [McLean et al. 1991]. Our previous work [Hou et al. 2013] on exposure to airborne toxic metals and tandem repeat methylation used the average methylation levels across different positions for each tandem repeat. To avoid the assumptions of homogeneity-of-regression slopes across different CpG sites within a tandem repeat and achieve the best model fit for the entirety of the data presented, here we considered methylation measures on different positions in each tandem repeat as repeated measures. Therefore, in the present investigation, we used a multilevel model instead of a single-level model to incorporate a nested dependent structure. We first evaluated differences between groups (truck drivers vs. office workers) by using the following multilevel mixed-effect regression model:
| [1] |
where γijk is the methylation level of ith CpG sites at each tandem repeat investigated for the jth subject on the kth examination day (i = 1, 2, 3 for SATα, i = 1, 2, 3, 4 for NBL2 and D4Z4; j = 1,…,n; and k = 1, 2); β0 is the overall intercept; β1 is the regression coefficient for the group; β2…βp are the regression coefficients for the covariates, which were all fitted as fixed effects and included variables either not matched or not completely matched by design between the two groups, i.e., age, body mass index (BMI), number of cigarettes per examination day, and work hours per day; ν0k is the random effect at the examination day level; µ0jk is the random effect at the subject level, and e0ijk is the random effect at the CpG site level. We did not adjust for the matching variables sex and smoking, because they were nearly perfectly balanced between groups by matching. In contrast, Hou et al. adjusted for matching factors and other potential confounding variables such as the usage of central heating. The different approaches led to minor discrepancy in the results from between-group methylation comparisons [Hou et al. 2013].
Table I.
Characteristics of the Study Participants
| Characteristic | Office Workers (n = 60) |
Truck Drivers (n = 60) |
P valuea |
|---|---|---|---|
| Sex, n (%) | |||
| Male | 40 (66.7) | 40 (66.7) | |
| Female | 20 (33.3) | 20 (33.3) | 1.00 |
| Age [years], mean ± SD | 30.3 ± 8.0 | 33.5 ± 5.7 | <0.001 |
| Smoking status n (%) | |||
| Never smoked | 35 (58.4) | 34 (56.7) | |
| Ex-smoker | 2 (3.3) | 2 (3.3) | |
| Current smoker | 23 (38.3) | 24 (40.0) | 1.00 |
| Cigarettes smoked during the examination time, mean ± SDb | 0.5 ± 1.7 | 2.3 ± 4.2 | <0.001b |
| BMI [kg/m2], mean ± SD | 22.8 ± 3.4 | 24.3 ± 3.2 | 0.01 |
| Work hours per day, mean ± SDb | 7.9 ± 1.3 | 9.9 ± 1.7 | <0.001 |
P values were calculated using Student’s t-tests and Fisher’s exact tests for continuous and categorical variables, respectively, except for the variable indicated in footnote b below.
Measures from 1 study day. Mixed-effect regression was used in the statistical analyses to account for the lack of independence of the repeated measures and computed means, standard deviations, and P values.
SD, standard deviation; BMI, body mass index.
Mixed-effect regression models were also used to evaluate the associations of personal PM2.5, personal EC, and ambient PM10 with tandem-repeat methylation adjusted for age, sex, BMI, smoking status, number of cigarettes per day, work hours per day, outdoor humidity, and temperature. In order to optimize power, we conducted primary analyses on the association between exposure measures and methylation levels of tandem repeats by fitting these models in all participants combined. We then evaluated associations in office workers or truck drivers separately. The mixed-effect model was:
| [2] |
where γijk is the methylation level of ith CpG sites at each tandem repeat investigated for the jth subject on the kth examination day (i = 1, 2, 3 for SATα, i = 1, 2, 3, 4 for NBL2 and D4Z4; j = 1,…,n; and k = 1, 2); β0 is the overall intercept; β1 is the regression coefficient for the exposure variable (PM2.5, EC, or PM10); β2…βp are the regression coefficients for the covariates, which included age, sex, BMI, smoking status, number of cigarettes smoked/examination day, work hours/day, outdoor humidity, and temperature; ν0k is the random effect at the examination day level; µ0jk is the random effect at the subject level, and e0ijk is the random effect at the CpG site level. Because personal exposure levels are expected to be higher in smokers, we also fit a second set of models in which smoking status and number of cigarettes smoked were excluded from the list of dependent variables. These models generated regression coefficients for air pollution exposures very similar to the full models with no differences in statistical significance.
To explore the timing of the relationship between the exposure and tandem-repeat methylation, we further analyzed ambient PM10 measures, which—because they were available daily throughout the study period—could be used not only to assess exposure on the day of blood collection, but also on previous days (i.e., lags). To this end, we used ambient PM10 data up to 7 days before the blood collection in unconstrained distributed lag models. All tests were two-sided, and P < 0.05 was considered significant.
RESULTS
Characteristics of the Study Participants and Exposure to Air Particulate Matter
The characteristics of the 60 office workers and 60 truck drivers were described previously [Baccarelli et al. 2011; Hou et al. 2013]. Briefly, truck drivers were moderately older than office workers, had higher BMI, smoked more cigarettes during the 8 hours of the study, and worked more hours per day. Mean personal PM2.5 was 126.8 µg/m3 for truck drivers and 94.6 µg/m3 for office workers (P < 0.001) (Table II). Average personal EC was 17.2 µg/m3 for truck drivers and 13.0 µg/m3 for office workers (P < 0.001). As expected, the levels of ambient PM10, outdoor temperature, and relative humidity on the day of examination did not differ between truck drivers and office workers (Table II). Personal PM2.5 exposure was higher in smokers compared to nonsmokers among office workers and truck drivers, as well as in all participants combined (Supplementary Table S1). Personal PM2.5 exposure was also significantly correlated with the number of cigarettes smoked during the 8 hours of monitoring (r = 0.37; P < 0.001). Personal EC exposure was higher in smokers in all participants combined and among truck drivers but not among office workers (Supplementary Table S1). EC was also significantly correlated with the number of cigarettes smoked during the 8 hours of monitoring (r = 0.29; P = 0.005).
Table II.
Level of Personal Exposure to PM2.5 and Elemental Carbon (EC) During Work Hours and Level of Ambient PM10, Outdoor Temperature, and Humidity on the Examination Days
| Time window | Office Workers (60 participants, 2 measures per participant)a |
Truck Drivers (60 participants, 2 measures per participant)a |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
# Obs |
Mean | SD | 10th pct |
25th pct |
Median | 75th pct |
90th pct |
# Obs |
Mean | SD | 10th pct |
25th pct |
Median | 75th pct |
90th pct |
P value | |
| Personal PM2.5b (µg/m3) on the examination days, from personal monitors | |||||||||||||||||
| 8 hoursa | 120 | 94.6 | 64.9 | 22.4 | 48.5 | 86.2 | 126.6 | 183.4 | 119 | 126.8 | 68.8 | 46.3 | 73.9 | 116.8 | 160.5 | 213.9 | <0.001 |
| Personal ECb (µg/m3) on the examination days, from personal monitors | |||||||||||||||||
| 8 hoursa | 118 | 13.0 | 4.0 | 7.3 | 10.1 | 13.2 | 15.7 | 18.4 | 120 | 17.2 | 6.6 | 9.2 | 12.9 | 16.7 | 20.8 | 25.5 | <0.001 |
| Ambient PM (µg/m3) from ambient monitors on the day of the study | |||||||||||||||||
| 24 hoursc | 120 | 116.7 | 50.2 | 60.0 | 82.0 | 114.0 | 141.0 | 181.0 | 120 | 123.5 | 50.1 | 72.0 | 88.0 | 116.0 | 150.0 | 188.0 | 0.29 |
| Outdoor relative humidity (%) on the day of the study | |||||||||||||||||
| 24 hoursc | 120 | 20.6 | 2.1 | 17.5 | 20.0 | 21.0 | 21.5 | 23.0 | 120 | 20.6 | 2.1 | 17.0 | 20.0 | 21.0 | 22.0 | 23.0 | 0.93 |
| Outdoor temperature (°C) on the day of the study | |||||||||||||||||
| 24 hoursc | 120 | 25.4 | 2.5 | 22.0 | 23.0 | 26.0 | 28.0 | 29.0 | 120 | 25.3 | 2.5 | 22.0 | 23.0 | 26.0 | 28.0 | 28.0 | 0.96 |
Each of the truck drivers (n = 60) and office workers (n = 60) was studied on two different examination days for a total of 120 observations in each of the two groups. Mixed-effect regression was used in the statistical analyses to account for the lack of independence of the repeated measures and to compute P values.
Measured during the work hours of examination days using lightweight personal monitors.
24-hour average of ambient PM10, relative humidity, or temperature on the examination days.
Obs, number of observations; SD, standard deviation; pct, percentile
DNA Methylation of Tandem-Repeat Sequences in Truck Drivers and Office Workers
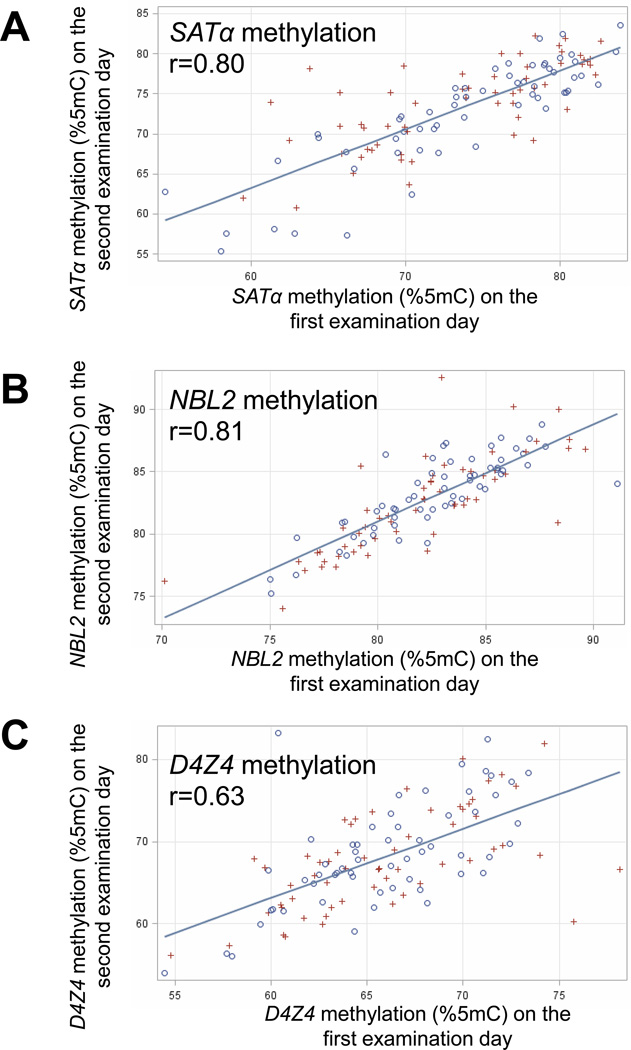
Tandem-repeat methylation levels exhibited varying degrees of within-participant correlation between the two examination days, which were separated by 1–2 weeks (Fig. 2). In all participants combined, Pearson’s correlation coefficients (r) were 0.80 for SATα methylation, 0.81 for NBL2 methylation, and 0.63 for D4Z4 methylation. There were no significant differences in methylation levels of SATα, NBL2, and D4Z4 between truck drivers and office workers in unadjusted analyses, as well as in analyses adjusted for age, BMI, number of cigarettes, and work hours/day (Table III). Further analyses of DNA methylation levels at individual CpG sites within each of the sequences did not show any statistically significant differences between truck drivers and office workers (Supplementary Table S2).
Figure 2.
Scatterplots of the within-participant correlations in blood tandem-repeat methylation between the two examination days. Circles and crosses represent truck drivers and office workers, respectively.
Table III.
Average DNA Methylation of Tandem-Repeat Sequences in Office Workers and Truck Drivers
| DNA Methylation (%5mC) |
|||||
|---|---|---|---|---|---|
| Marker | Office Workers (60 participants, 2 measures per participant)a |
Truck Drivers (60 participants, 2 measures per participant)a |
P value | ||
| Mean | (SD) | Mean | (SD) | ||
| Unadjusted | |||||
| SATα | 73.9 | (15.1) | 73.1 | (15.1) | 0.17 |
| NBL2 | 82.2 | (5.7) | 82.6 | (5.7) | 0.34 |
| D4Z4 | 66.7 | (6.5) | 67.3 | (6.5) | 0.52 |
| Adjusted for age, BMI, number of cigarettes, and work hours/dayb | |||||
| SATα | 79.6 | (15.3) | 79.7 | (15.3) | 0.93 |
| NBL2 | 86.1 | (6.0) | 87.0 | (6.1) | 0.11 |
| D4Z4 | 69.6 | (7.6) | 70.8 | (7.8) | 0.26 |
Each of the truck drivers (n = 60) and office workers (n = 60) was studied on 2 different examination days for a total of 120 observations in each of the two groups. Mixed-effect regression was used in the statistical analyses to account for the lack of independence of the repeated measures and to compute means, standard deviations (SDs), and P values.
Adjusted means were computed by holding covariates fixed at their average values. Office workers and truck drivers were matched by sex and smoking status (never, former, current). Only unmatched variables were adjusted for in the regression models.
SD, standard deviation; BMI, body mass index.
Association of Personal PM2.5, Personal EC, and Ambient PM10 with Methylation Levels of Tandem Repeats
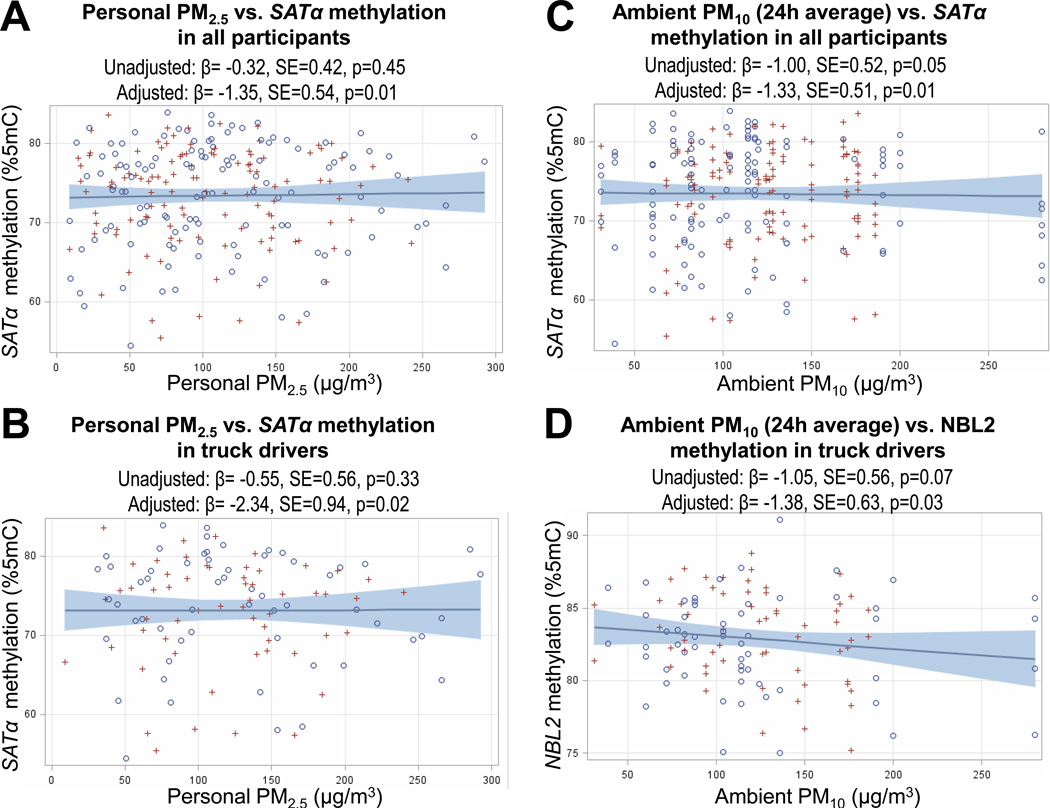
In covariate-adjusted analyses of all participants combined, personal PM2.5 exposure exhibited a significant negative association with SATα methylation. An interquartile increase in personal PM2.5 exposure was associated with a 1.35%5mC decrease in SATα methylation (P = 0.01) in a model adjusted for age, sex, BMI, smoking status, number of cigarettes smoked during the 8 hours of the study, work hours/day, and outdoor humidity and temperature on the examination days. The negative correlation between personal PM2.5 exposure and SATα methylation was found in both groups, although it was more pronounced and statistically significant (P = 0.02) only in truck drivers (Table IV).
Table IV.
Effects of Airborne Particulate Matter on DNA Methylation of Tandem-Repeat Sequencesa
| Marker | Exposure | All Participants (120 participants, 2 measures per participant)b |
Office Workers (60 participants, 2 measures per participant)b |
Truck Drivers (60 participants, 2 measures per participant)b |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| β | (SE) | P value | β | (SE) | P value | β | (SE) | P value | ||
| SATα | Personal PM2.5c | −1.35 | (0.54) | 0.01 | −0.95 | (0.83) | 0.26 | −2.34 | (0.94) | 0.02 |
| Personal ECc | −0.37 | (0.49) | 0.45 | −0.84 | (0.90) | 0.36 | −0.54 | (0.68) | 0.43 | |
| Ambient PM10d | −1.33 | (0.51) | 0.01 | −1.26 | (0.79) | 0.12 | −1.44 | (0.75) | 0.06 | |
| NBL2 | Personal PM2.5c | −0.05 | (0.46) | 0.92 | 0.23 | (0.71) | 0.75 | −0.88 | (0.84) | 0.30 |
| Personal ECc | 0.03 | (0.40) | 0.94 | −0.44 | (0.77) | 0.57 | 0.09 | (0.58) | 0.88 | |
| Ambient PM10d | −0.09 | (0.44) | 0.84 | 1.04 | (0.66) | 0.13 | −1.38 | (0.63) | 0.03 | |
| D4Z4 | Personal PM2.5c | 0.74 | (0.91) | 0.42 | 0.36 | (1.43) | 0.80 | 1.02 | (1.73) | 0.56 |
| Personal ECc | 0.15 | (0.80) | 0.85 | 0.36 | (1.54) | 0.81 | −0.83 | (1.17) | 0.49 | |
| Ambient PM10d | −0.13 | (0.87) | 0.88 | 0.88 | (1.36) | 0.52 | −1.34 | (1.33) | 0.32 | |
Estimates (βs) and standard errors (SEs) adjusted for age, sex, BMI, smoking status, number of cigarettes smoked during the 8 hours of the study, work hours/day, and outdoor humidity and temperature on the examination days. Estimates are scaled to an interquartile range increase in exposure.
Each of the truck drivers (n = 60) and office workers (n = 60) was studied on two different examination days for a total of 120 observations in each group. Mixed-effect regression was used in the statistical analyses to account for the lack of independence of the repeated measures and to compute β coefficients, standard errors (SEs), and P values.
Measured during the work hours of examination days using lightweight personal monitors.
24-hour average measure of ambient PM10 on the examination days
Ambient PM10 levels also showed a significant negative association with SATα methylation; an interquartile increase in ambient PM10 levels was associated with a covariate-adjusted 1.33%5mC decrease in SATα methylation in all participants combined (P = 0.01). The negative correlation between ambient PM10 exposure and SATα methylation was found in both groups, although it was moderately more pronounced and borderline statistically significant (P = 0.06) only in truck drivers (Table IV). Personal EC exposure did not show any significant associations with SATα methylation (Table IV).
In all participants combined, NBL2 and D4Z4 showed no associations with the levels of personal PM2.5, personal EC, or ambient PM10 (Table IV). However, ambient PM10 levels showed a significant negative association with NBL2 methylation in truck drivers but not in office workers: an interquartile increase in ambient PM10 levels was associated with a covariate-adjusted 1.38%5mC decrease in NBL2 methylation among truck drivers (P = 0.03).
To complement the results of the covariate-adjusted analyses, we present scatterplots of the raw data and unadjusted correlations corresponding to the significant findings described above (Fig. 3). Consistent with the covariate-adjusted analyses, all the unadjusted correlations were negative, although the degree of significance varied: in all participants combined, an interquartile increase in personal PM2.5 was associated with a nonsignificant unadjusted 0.32%5mC decrease in SATα methylation (P = 0.45, Fig. 3A); among truck drivers, an interquartile increase in personal PM2.5 was associated with a nonsignificant unadjusted 0.55%5mC decrease in SATα methylation (P = 0.33, Fig. 3B); in all participants combined, an interquartile increase in ambient PM10 was associated with an unadjusted 1.00%5mC decrease in SATα methylation (P = 0.05; Fig. 2, Fig. 3C); and in truck drivers, an interquartile increase in ambient PM10 levels was associated with an unadjusted 1.05%5mC decrease in NBL2 methylation (P = 0.07; Fig. 3D).
Figure 3.
Scatterplots of the crude correlations between particulate matter exposures and tandem-repeat methylation for the four significant findings from adjusted multivariable analyses. Circles and crosses represent observations from the first and second examination days, respectively. Unadjusted and adjusted regression estimates obtained from mixed-effect regression models are shown. Mixed-effect regression was used to account for the lack of independence of the repeated measures and to compute β coefficients, standard errors (SEs), and P values. Adjusted estimates were computed from models fitting the following independent variables: age, sex, BMI, smoking status, number of cigarettes smoked during the 8 hours of the study, work hours/day, and outdoor humidity and temperature on the examination days. Estimates are scaled to an interquartile range (IQR) increase in exposure.
To examine the timing of the relationship between PM exposure and tandem-repeat methylation, we used unconstrained distributed lag models to further analyze ambient PM10. PM10 measures were available daily throughout the study period and could be used not only to assess exposure on the day of blood collection, but also on the previous days (i.e., lags). As reported in Table V, results using the distributed lag model showed that the effects of PM10 observed on examination days were distributed across several lags 3–6 days before the day of blood collection. For instance, the negative correlation between ambient PM10 and SATα methylation was observed between 3-day and 6-day lags, with the most negative estimate corresponding to the 6-day lag (Table V).
Table V.
Results of Distributed-Lag Models Examining the Timing of the Effects of PM10 Before the Examination Days on DNA Methylation of Tandem-Repeat Sequencesa
| Marker | Exposure | All Participants (120 participants, 240 examination days) |
Office Workers (60 participants, 120 examination days) |
Truck Drivers (60 participants, 120 examination days) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| β | (SE) | P-value | β | (SE) | P-value | β | (SE) | P-value | ||
| SATα | Ambient PM10 (1-day lag) | 0.35 | (0.41) | 0.39 | 1.31 | (0.58) | 0.026 | −0.64 | (0.59) | 0.27 |
| Ambient PM10 (2-day lag) | 0.36 | (0.43) | 0.41 | −0.92 | (0.64) | 0.16 | 1.20 | (0.59) | 0.045 | |
| Ambient PM10 (3-day lag) | −0.34 | (0.35) | 0.33 | 0.16 | (0.48) | 0.74 | −1.46 | (0.51) | 0.005 | |
| Ambient PM10 (4-day lag) | −0.42 | (0.42) | 0.33 | −1.02 | (0.57) | 0.07 | 0.21 | (0.63) | 0.74 | |
| Ambient PM10 (5-day lag) | −0.23 | (0.44) | 0.61 | 0.38 | (0.68) | 0.57 | −0.88 | (0.61) | 0.16 | |
| Ambient PM10 (6-day lag) | −0.50 | (0.48) | 0.30 | −1.57 | (0.67) | 0.021 | 0.14 | (0.69) | 0.84 | |
| Ambient PM10 (7-day lag) | 0.12 | (0.43) | 0.79 | 0.18 | (0.60) | 0.77 | −0.84 | (0.64) | 0.19 | |
| NBL2 | Ambient PM10 (1-day lag) | −0.09 | (0.15) | 0.58 | 0.10 | (0.26) | 0.71 | −0.24 | (0.20) | 0.23 |
| Ambient PM10 (2-day lag) | 0.20 | (0.16) | 0.22 | 0.10 | (0.29) | 0.72 | 0.20 | (0.19) | 0.30 | |
| Ambient PM10 (3-day lag) | −0.06 | (0.13) | 0.65 | 0.03 | (0.21) | 0.90 | −0.24 | (0.17) | 0.15 | |
| Ambient PM10 (4-day lag) | 0.33 | (0.16) | 0.041 | 0.48 | (0.25) | 0.06 | 0.07 | (0.21) | 0.73 | |
| Ambient PM10 (5-day lag) | −0.09 | (0.17) | 0.58 | −0.06 | (0.31) | 0.85 | −0.29 | (0.21) | 0.16 | |
| Ambient PM10 (6-day lag) | 0.03 | (0.18) | 0.85 | −0.23 | (0.30) | 0.46 | 0.31 | (0.23) | 0.18 | |
| Ambient PM10 (7-day lag) | 0.09 | (0.16) | 0.60 | 0.09 | (0.27) | 0.74 | −0.12 | (0.22) | 0.60 | |
| D4Z4 | Ambient PM10 (1-day lag) | −0.16 | (0.27) | 0.56 | −0.19 | (0.39) | 0.62 | −0.11 | (0.40) | 0.78 |
| Ambient PM10 (2-day lag) | 0.40 | (0.29) | 0.17 | 0.64 | (0.43) | 0.14 | 0.25 | (0.41) | 0.55 | |
| Ambient PM10 (3-day lag) | 0.07 | (0.23) | 0.77 | 0.00 | (0.32) | 0.99 | 0.24 | (0.35) | 0.49 | |
| Ambient PM10 (4-day lag) | −0.45 | (0.28) | 0.11 | −0.64 | (0.38) | 0.09 | −0.37 | (0.44) | 0.40 | |
| Ambient PM10 (5-day lag) | 0.24 | (0.30) | 0.42 | −0.10 | (0.45) | 0.83 | 0.58 | (0.43) | 0.18 | |
| Ambient PM10 (6-day lag) | −0.28 | (0.32) | 0.39 | −0.23 | (0.45) | 0.61 | −0.59 | (0.49) | 0.23 | |
| Ambient PM10 (7-day lag) | −0.14 | (0.28) | 0.63 | 0.04 | (0.40) | 0.92 | −0.05 | (0.44) | 0.92 | |
Estimates adjusted for age, sex, BMI, smoking status, number of cigarettes smoked during the eight hours of the study, work hours/day, and outdoor humidity and temperature on the examination days. For each marker, seven lag-day PM10 variables were evaluated at the same time in one model.
a Estimates (βs) and standard errors (SEs) adjusted for age, sex, BMI, smoking status, number of cigarettes smoked during the 8 hours of the study, work hours/day, and outdoor humidity and temperature on the examination days. Estimates are scaled to an interquartile range (IQR) increase in exposure.
Each of the truck drivers (n = 60) and office workers (n = 60) was studied on 2 different examination days for a total of 120 observations in each of the two groups. Mixed-effect regression was used in the statistical analyses to account for the lack of independence of the repeated measures and to compute β coefficients, standard errors (SEs), and P values.
Correlation of Tandem-Repeat Methylation with Inflammatory Markers
We analyzed DNA methylation of tandem repeats in relation to plasma levels of 27 cytokines measured using a premade platform (Bio-Plex Cytokine 27-plex Pro Human Cytokine; Bio-Rad, Hercules, CA). As shown in Supplementary Table S3, 9 of the 27 cytokines were negatively correlated with SATα methylation at a false discovery rate (FDR) lower than 0.05. Neither NBL2 nor D4Z4 methylation showed positive correlations with cytokine levels at the same FDR threshold (Table VI).
Table VI.
Correlation of Tandem-Repeat DNA Methylation with Plasma Levels of 27 Cytokines
|
SATα |
NBL2 |
D4Z4 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cytokine | β | SE |
P value |
FDR | β | SE |
P value |
FDR | β | SE |
P value |
FDR |
| EOTAXIN | −0.345 | 0.365 | 0.348 | 0.47 | −0.202 | 0.673 | 0.765 | 0.892 | 0.477 | 0.296 | 0.11 | 0.269 |
| FGF_BASIC | −0.272 | 0.138 | 0.051 | 0.107 | 0.267 | 0.285 | 0.351 | 0.892 | 0.123 | 0.152 | 0.418 | 0.538 |
| GM_CSF | −0.185 | 0.107 | 0.086 | 0.154 | −0.017 | 0.216 | 0.938 | 0.938 | 0.069 | 0.109 | 0.525 | 0.621 |
| G_CSF | −0.122 | 0.094 | 0.196 | 0.294 | 0.379 | 0.19 | 0.049 | 0.892 | 0.138 | 0.094 | 0.143 | 0.297 |
| IFN_G | −1.452 | 0.595 | 0.016 | 0.049 | 1.304 | 1.231 | 0.292 | 0.892 | 1.423 | 0.607 | 0.021 | 0.157 |
| IL_10 | −0.018 | 0.006 | 0.007 | 0.032 | −0.006 | 0.014 | 0.669 | 0.892 | 0.002 | 0.007 | 0.783 | 0.844 |
| IL_12_P70 | 0.047 | 0.124 | 0.705 | 0.762 | −0.068 | 0.211 | 0.748 | 0.892 | 0.08 | 0.08 | 0.32 | 0.5 |
| IL_13 | −0.115 | 0.082 | 0.164 | 0.261 | −0.031 | 0.174 | 0.859 | 0.892 | 0.047 | 0.086 | 0.588 | 0.662 |
| IL_15 | −0.025 | 0.013 | 0.052 | 0.107 | −0.013 | 0.026 | 0.632 | 0.892 | 0.009 | 0.014 | 0.529 | 0.621 |
| IL_17 | −0.248 | 0.09 | 0.007 | 0.032 | −0.059 | 0.19 | 0.757 | 0.892 | 0.087 | 0.103 | 0.401 | 0.538 |
| IL_1B | −0.033 | 0.012 | 0.008 | 0.032 | 0.04 | 0.026 | 0.123 | 0.892 | 0.027 | 0.013 | 0.037 | 0.166 |
| IL_1RA | −1.856 | 0.691 | 0.008 | 0.032 | 0.683 | 1.454 | 0.639 | 0.892 | 0.981 | 0.71 | 0.169 | 0.305 |
| IL_2 | −0.033 | 0.071 | 0.64 | 0.72 | 0.028 | 0.137 | 0.84 | 0.892 | 0.07 | 0.072 | 0.333 | 0.5 |
| IL_4 | −0.036 | 0.012 | 0.002 | 0.032 | 0.027 | 0.025 | 0.277 | 0.892 | 0.025 | 0.012 | 0.051 | 0.167 |
| IL_5 | −0.061 | 0.024 | 0.01 | 0.035 | 0.01 | 0.049 | 0.837 | 0.892 | 0.036 | 0.024 | 0.138 | 0.297 |
| IL_6 | −0.06 | 0.039 | 0.124 | 0.21 | 0.018 | 0.077 | 0.813 | 0.892 | 0.032 | 0.037 | 0.395 | 0.538 |
| IL_7 | −0.154 | 0.054 | 0.005 | 0.032 | 0.193 | 0.113 | 0.091 | 0.892 | 0.153 | 0.057 | 0.008 | 0.11 |
| IL_8 | −0.139 | 0.047 | 0.004 | 0.032 | 0.058 | 0.097 | 0.552 | 0.892 | 0.106 | 0.046 | 0.023 | 0.157 |
| IL_9 | −0.115 | 0.505 | 0.82 | 0.852 | −0.282 | 0.927 | 0.762 | 0.892 | −0.055 | 0.377 | 0.884 | 0.884 |
| IP_11 | −1.247 | 2.596 | 0.632 | 0.72 | 2.265 | 5.097 | 0.658 | 0.892 | 2.772 | 2.693 | 0.305 | 0.5 |
| MCP_1_MCAF | −0.342 | 0.148 | 0.023 | 0.061 | 0.178 | 0.285 | 0.534 | 0.892 | 0.253 | 0.127 | 0.048 | 0.167 |
| MIP_1A | 0.019 | 0.04 | 0.638 | 0.72 | 0.019 | 0.075 | 0.803 | 0.892 | 0.073 | 0.033 | 0.029 | 0.157 |
| MIP_1B | −0.592 | 0.279 | 0.036 | 0.088 | 0.313 | 0.526 | 0.554 | 0.892 | 0.651 | 0.238 | 0.007 | 0.11 |
| PDGF_BB | −8.466 | 4.446 | 0.059 | 0.115 | 7.201 | 9.139 | 0.432 | 0.892 | 6.8 | 4.79 | 0.158 | 0.305 |
| RANTES | −19.952 | 17.723 | 0.263 | 0.373 | 13.553 | 35.181 | 0.701 | 0.892 | 30.901 | 16.827 | 0.069 | 0.186 |
| TNF_A | −0.068 | 0.423 | 0.873 | 0.873 | −0.481 | 0.703 | 0.495 | 0.892 | −0.069 | 0.288 | 0.813 | 0.844 |
| VEGF | −0.071 | 0.1 | 0.478 | 0.615 | −0.111 | 0.187 | 0.553 | 0.892 | 0.155 | 0.08 | 0.056 | 0.167 |
SE, standard error; FDR, false discovery rate.
DISCUSSION
In the present investigation of truck drivers and office workers in Beijing, China, who were exposed to high levels of PM, we found significant associations between both personal PM2.5 and ambient PM10 levels and lower levels of SATα methylation in DNA from blood. The associations were more pronounced in truck drivers than in office workers. There were no associations between EC exposure and tandem repeat methylation.
Air pollution has been consistently associated with adverse health outcomes, including lung cancer, in several epidemiologic studies [Krewski et al. 2005; Vineis et al. 2006; Merlo et al. 2010; Raaschou-Nielsen et al. 2010]. Although the mechanisms underlying air pollution-related lung carcinogenesis remain to be fully clarified, many carcinogens and co-carcinogens found in tobacco smoke, such as polycyclic aromatic hydrocarbons, transition metals, benzene, and aldehydes, are also present in PM [Feng et al. 2006].
Our study investigated a group of healthy individuals exposed to high levels of PM, well above the levels that have been documented to cause lung carcinogenicity [Pope et al. 2004]. We found that SATα methylation measured in blood leukocytes was negatively associated with personal PM2.5 and ambient PM10 levels. Personal exposure to EC, used as a tracer of PM from vehicular traffic, was not significantly associated with tandem-repeat methylation. Taken together, these findings indicate that SATα methylation is sensitive to overall particle levels, as traced by personal PM2.5 and ambient PM10, rather than specifically to PM from vehicular traffic. Consistent with this conclusion, we found negative correlations of personal PM2.5 and ambient PM10 with SATα methylation among truck drivers and office workers, although they were moderately stronger in truck drivers. Our previous study found a positive association between NBL2 methylation and concentrations of Si and Ca in truck drivers, and a positive association between SATα methylation and concentrations of S in office workers. The opposite directions of associations in two sets of pollutants suggest that the patterns of epigenetic regulation of tandem repeat are not only element-specific, but also exposure-specific. We did not find any significant association between D4Z4 methylation and PM exposures. A subgroup association of ambient PM10 was observed with NBL2 methylation. Consistent with our previous finding, SATα and NBL2 methylation demonstrate a better sensitivity than D4Z4 methylation to the overall particle and elemental components. Whether D4Z4 and NBL2 methylation in DNA from blood specimens or other tissues is sensitive to PM exposure should be further evaluated in future investigations using larger samples of exposed individuals.
Extensive evidence indicates that decreased methylation of repetitive elements in cancer tissues is causally involved in carcinogenesis [Ehrlich 2009; Wolff et al. 2010]. Initial findings also indicate that repetitive element methylation measured in blood leukocyte DNA might provide a biomarker for cancer risk, thus supporting epigenetic analyses of blood samples. For instance, in a recent study of 722 elderly participants from the Normative Aging Study cohort, Zhu et al. showed that repetitive element hypomethylation in blood leukocyte DNA was related to higher incidence of and mortality from cancer, including lung cancer [Zhu et al. 2011]. A recent study by Friso et al. showed that low levels of DNA methylation in blood, which is largely repetitive element methylation, was associated with cancer risk [Friso et al. 2013]. In addition, the sensitivity of peripheral blood DNA methylation profile after PM exposure suggests that blood DNA could serves as an appropriate biomarker to monitor effects of PM on the epigenome [Baccarelli et al. 2009; De Prins et al. 2013; Kile et al. 2013].
Tandem repeats are a distinct family of repetitive elements that, although extensively present in the human genome and involved in cancer etiology, have not yet been studied in human investigations of environmental carcinogens. Relative to most regions of the genome, tandem repeats display a greater propensity to mutate and variable methylation may influence their mutagenicity rates. Our results suggest that different tandem repeats have differential sensitivity to DNA methylation. In particular, we found the strongest associations between PM exposure and lower methylation of SATα. This finding is consistent with the biological behavior of different types of tandem repeats in carcinogenesis. Mutation rates in tandem repeats have been shown to be highly dependent on the structure and length of each specific, repetitive element [Brinkmann et al. 1998]. Similarly, the patterns of epigenetic regulation of each type of tandem repeat are specific to each element [Meaney and Szyf 2005]. In fact, although the three tandem repeats evaluated in our study have all been shown to contribute to chromosomal instability if dysregulated, they are controlled through different mechanisms and have different transcription patterns in response to cellular stressors [Meaney and Szyf 2005]. In particular, the SATα family of tandem-repeated sequences, which are found in all normal human centromeres, has been shown to be methylated via the action of the DNA methyltransferase 3B (DNMT3B), which plays a critical role in maintaining chromosomal stability [Ehrlich, Sanchez et al. 2008]. A constitutive centromeric protein, centromere protein C (CENP-C), has been shown to interact specifically with DNMT3B and to recruit it to methylate SATα repeats [Gopalakrishnan, Sullivan et al. 2009]. A recent study based on next-generation digital gene expression analysis in cancer cells, including lung cancer, showed that satellite hypomethylation is associated with massive expression of noncoding RNAs (ncRNAs) that may have active roles in dysregulated cell growth and cancer progression [Ting, Lipson et al. 2011]. It remains to be determined whether the relation between PM exposure and SATα DNA hypomethylation is mediated by effects on the DNMT3B/CENP-C interaction and by increased ncRNA expression.
Our experimental data showed that the decreased methylation of SATα, but not of NBL2 and D4Z4, was correlated with higher levels of several cytokines in plasma. This finding demonstrates that SATα hypomethylation is associated with both increased immunoregulatory and pro-inflammatory cytokines levels. However, our results are observational and therefore not sufficient to explain their roles as inflammatory mediators after PM exposure. Future mechanistic studies are needed to unravel the mechanism of oxidative stress and inflammatory responses in the hypomethylation of the SATα sequences and subsequent lung carcinogenesis.
DNA methylation is a reversible mechanism of epigenetic regulation and can reflect the dynamic gene-environmental interaction. Reduced blood DNA methylation has been repetitively reported after short-term PM exposure[Dioni, Hoxha et al. 2011; Bind, Baccarelli et al. 2012; Bellavia, Urch et al. 2013; Kile, Fang et al. 2013], suggesting that this condition may affect the epigenome. In our study, we evaluated DNA methylation of tandem repeats only in relation to personal PM2.5 levels of exposure measured on the same day of the examinations. In addition, because ambient PM10 data were available throughout the study period, we used distributed lag models to explore the timing of the relationship between PM10 exposure and tandem-repeat methylation.
In Table V, the reduction in SATα methylation associated with the 3-day lag was highly significant in truck drivers. For office workers, the negative estimates corresponded to both the 4-day and 6-day lags. We observed increased SATα methylation with marginal significance in the 1-day and 2-day lags for the office worker group and the truck driver group respectively. Whereas our lag model results do not provide conclusive evidence as to the temporal relationships linking PM exposure to SATα methylation, the fluctuation in methylation level across the exposure period suggest a relatively high degree of variability in DNA methylation. However, these results appear to indicate that the effect of PM exposures on tandem-repeat methylation is not solely due to the PM levels on the day of blood collection but rather, PM exposure may be distributed over the preceding several days. The most significant reduction in SATα methylation corresponded to 3-day and 6-day lag in the truck drivers and office workers, respectively. This result suggests that the difference in DNA methylation response between two groups might reflect the magnitude of environmental exposure.
As mentioned above, DNA hypomethylation is usually considered a reversible epigenetic change. However, the consequences of hypomethylation, such as uncontrolled inflammation responses, ROS production, abnormal cell cycle arrest, enhanced blood-to-lung signaling, and genomic instability may lead to irreversible alterations and are associated with both the duration and intensity of exposure.
Given the slow clearance rate of PM from the respiratory system, combined with the fact that the effects of chronic PM pollution exist even at very low exposure, it is plausible that PM induced inflammation, ROS damage, and tissue damage are cumulative [Brunekreef and Holgate 2002; Daniels, Dominici et al. 2000]. These events and changes are important risk factors for lung carcinogenesis. In addition, inflammation and ROS stress exacerbates DNA hypomethylation. As those stimuli continues, the methylation and stress/damage in the lung act in a positive feedback manner. As such, capturing this reversible epigenetic change associated with short-term PM exposure may represent a target for early intervention, which is especially important in lung cancer research due to the lack of early stage biomarker.
As previously noted [Baccarelli, Barretta et al. 2011], although we matched subjects for smoking status and education and used multivariable models to control for potential confounders, we cannot exclude residual confounding from unmeasured variables. One limitation of the study is that we did not measure the differential blood counts. Also, the averaging of ambient PM10 from many stationary monitors might not accurately estimate personal outdoor exposure to particles. Simulation studies have shown that the error introduced by using data from stationary monitors is highly unlikely to bias away from the null hypothesis, but rather is expected to underestimate the air pollution [Zeger, Thomas et al. 2000]. In addition, serial measures of ambient PM concentrations have been shown to be representative of variations in personal exposures [Wilson and Brauer 2006], particularly in the presence of high ambient PM levels [Avery, Mills et al. 2010].
In summary, our results indicate that tandem-repeat hypomethylation in SATα can be detected in an easily obtainable DNA source, such as blood leukocytes. Future studies are warranted to determine whether PM-induced SATα blood hypomethylation is associated with risk of lung cancer.
Supplementary Material
ACKNOWLEDGEMENTS
The study was supported by the National Institute of Environmental Health Sciences (NIEHS) [grants R21ES020010, and R21ES020984]. This project used resources and infrastructure from the Harvard School of Public Health (HSPH)-NIEHS Center for Environmental Health [P30ES000002] and United States Environmental Protection Agency (USEPA) grant RD 83479801. The contents of the manuscript are solely the responsibility of the grantees and do not necessarily represent the official views of the funding agencies. Further, the funding agencies do not endorse the purchase of any commercial products or services mentioned in the publication.
Footnotes
STATEMENT OF AUTHOR CONTRIBUTIONS
C.D., J.P.M., Z.F., and B.M. planned and supervised exposure assessment, sample collection, and participant recruitment. A.A.B., L.H., J.S., and S.W. designed the Beijing Truck Air Pollution Study. L.G. performed the methylation experiments and wrote the manuscript. H.M.B. and V.M. contributed to the set-up and optimization of the methylation assays, as well as quality control procedures. J.Z. performed the manuscript revision. S.C. and Y.Z. performed the data analysis. J.B. handled data management and bioinformatics. All authors provided edits and comments to the manuscript, and all authors read and approved the final manuscript.
REFERENCES
- Avery CL, Mills KT, Williams R, McGraw KA, Poole C, Smith RL, Whitsel EA. Estimating error in using ambient PM2.5 concentrations as proxies for personal exposures: A review. Epidemiology. 2010;21:215–223. doi: 10.1097/EDE.0b013e3181cb41f7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A, Barretta F, Dou C, Zhang X, McCracken JP, Diaz A, Bertazzi PA, Schwartz J, Wang S, Hou L. Effects of particulate air pollution on blood pressure in a highly exposed population in Beijing, China: A repeated-measure study. Environ Health. 2011;10:108. doi: 10.1186/1476-069X-10-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, Suh HH, Zanobetti A, Sparrow D, Vokonas PS, Schwartz J. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med. 2009;179:572–578. doi: 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellavia A, Urch B, Speck M, Brook RD, Scott JA, Albetti B, Behbod B, North M, Valeri L, Bertazzi PA, et al. DNA hypomethylation, ambient particulate matter, and increased blood pressure: Findings from controlled human exposure experiments. J Am Heart Assoc. 2013;2:e000212. doi: 10.1161/JAHA.113.000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bind MA, Baccarelli A, Zanobetti A, Tarantini L, Suh H, Vokonas P, Schwartz J. Air pollution and markers of coagulation, inflammation, and endothelial function: Associations and epigene-environment interactions in an elderly cohort. Epidemiology. 2012;23:332–340. doi: 10.1097/EDE.0b013e31824523f0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, Byun HM, Jiang J, Marinelli B, Pesatori AC, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67:876–880. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- Brennan K, Flanagan JM. Epigenetic epidemiology for cancer risk: Harnessing germline epigenetic variation. Methods Mol Biol. 2012;863:439–465. doi: 10.1007/978-1-61779-612-8_27. [DOI] [PubMed] [Google Scholar]
- Brinkmann B, Klintschar M, Neuhuber F, Huhne J, Rolf B. Mutation rate in human microsatellites: Influence of the structure and length of the tandem repeat. Am J Hum Genet. 1998;62:1408–1415. doi: 10.1086/301869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun HM, Heo K, Mitchell KJ, Yang AS. Mono-allelic retrotransposon insertion addresses epigenetic transcriptional repression in human genome. J Biomed Sci. 2012;19:13. doi: 10.1186/1423-0127-19-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Dai M, Ren JS, Chen YH, Guo LW. Estimates and prediction on incidence, mortality and prevalence of lung cancer in China in 2008. Zhonghua Liu Xing Bing Xue Za Zhi. 2012;33:391–394. [PubMed] [Google Scholar]
- Chen W, Zhang S, Zou X. Estimation and projection of lung cancer incidence and mortality in China. Zhongguo Fei Ai Za Zhi. 2010;13:488–493. doi: 10.3779/j.issn.1009-3419.2010.05.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Zheng R, Zhang S, Zou X, Zhao P, He J. Lung cancer incidence and mortality in China, 2009. Thoracic Cancer. 2013;4:102–108. doi: 10.1111/1759-7714.12025. [DOI] [PubMed] [Google Scholar]
- Choi SH, Worswick S, Byun HM, Shear T, Soussa JC, Wolff EM, Douer D, Garcia-Manero G, Liang G, Yang AS. Changes in DNA methylation of tandem DNA repeats are different from interspersed repeats in cancer. Int J Cancer. 2009;125:723–729. doi: 10.1002/ijc.24384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagher Z, Garcon G, Gosset P, Ledoux F, Surpateanu G, Courcot D, Aboukais A, Puskaric E, Shirali P. Pro-inflammatory effects of Dunkerque city air pollution particulate matter 2.5 in human epithelial lung cells (L132) in culture. J Appl Toxicol. 2005;25:166–175. doi: 10.1002/jat.1050. [DOI] [PubMed] [Google Scholar]
- De Prins S, Koppen G, Jacobs G, Dons E, Van de Mieroop E, Nelen V, Fierens F, Int Panis L, De Boever P, Cox B, et al. Influence of ambient air pollution on global DNA methylation in healthy adults: A seasonal follow-up. Environ Int. 2013;59:418–424. doi: 10.1016/j.envint.2013.07.007. [DOI] [PubMed] [Google Scholar]
- Delfino RJ, Staimer N, Tjoa T, Arhami M, Polidori A, Gillen DL, George SC, Shafer MM, Schauer JJ, Sioutas C. Associations of primary and secondary organic aerosols with airway and systemic inflammation in an elderly panel cohort. Epidemiology. 2010;21:892–902. doi: 10.1097/EDE.0b013e3181f20e6c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dioni L, Hoxha M, Nordio F, Bonzini M, Tarantini L, Albetti B, Savarese A, Schwartz J, Bertazzi PA, Apostoli P, et al. Effects of short-term exposure to inhalable particulate matter on telomere length, telomerase expression, and telomerase methylation in steel workers. Environ Health Perspect. 2011;119:622–627. doi: 10.1289/ehp.1002486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M. Cancer-linked DNA hypomethylation and its relationship to hypermethylation. Curr Top Microbiol Immunol. 2006;310:251–274. doi: 10.1007/3-540-31181-5_12. [DOI] [PubMed] [Google Scholar]
- Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009;1:239–259. doi: 10.2217/epi.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Sanchez C, Shao C, Nishiyama R, Kehrl J, Kuick R, Kubota T, Hanash SM. ICF, an immunodeficiency syndrome: DNA methyltransferase 3B involvement, chromosome anomalies, and gene dysregulation. Autoimmunity. 2008;41:253–271. doi: 10.1080/08916930802024202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabris S, Bollati V, Agnelli L, Morabito F, Motta V, Cutrona G, Matis S, Grazia Recchia A, Gigliotti V, Gentile M, et al. Biological and clinical relevance of quantitative global methylation of repetitive DNA sequences in chronic lymphocytic leukemia. Epigenetics. 2011;6:188–194. doi: 10.4161/epi.6.2.13528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- Feng Z, Hu W, Hu Y, Tang MS. Acrolein is a major cigarette-related lung cancer agent: Preferential binding at p53 mutational hotspots and inhibition of DNA repair. Proc Natl Acad Sci U S A. 2006;103:15404–15409. doi: 10.1073/pnas.0607031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso S, Udali S, Guarini P, Pellegrini C, Pattini P, Moruzzi S, Girelli D, Pizzolo F, Martinelli N, Corrocher R, et al. Global DNA hypomethylation in peripheral blood mononuclear cells as a biomarker of cancer risk. Cancer Epidemiol Biomarkers Prev. 2013;3:348–355. doi: 10.1158/1055-9965.EPI-12-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghio AJ, Carraway MS, Madden MC. Composition of air pollution particles and oxidative stress in cells, tissues, and living systems. J Toxicol Environ Health B Crit Rev. 2012;15:1–21. doi: 10.1080/10937404.2012.632359. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan S, Sullivan BA, Trazzi S, Della Valle G, Robertson KD. DNMT3B interacts with constitutive centromere protein CENP-C to modulate DNA methylation and the histone code at centromeric regions. Hum Mol Genet. 2009;18:3178–3193. doi: 10.1093/hmg/ddp256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Zhang X, Zheng Y, Wang S, Dou C, Guo L, Byun HM, Motta V, McCracken J, Díaz A, et al. Altered methylation in tandem repeat element and elemental component levels in inhalable air particles. Environ Mol Mutagen. 2013 doi: 10.1002/em.21829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen NAH, Van Vliet PHN, Aarts F, Harssema H, Brunekreef B. Assessment of exposure to traffic related air pollution of children attending schools near motorways. Atmospheric Environment. 2001;35:3875–3884. [Google Scholar]
- Kampa M, Castanas E. Human health effects of air pollution. Environ Pollut. 2008;151:362–367. doi: 10.1016/j.envpol.2007.06.012. [DOI] [PubMed] [Google Scholar]
- Kato T, Suzuki K, Okada S, Kamiyama H, Maeda T, Saito M, Koizumi K, Miyaki Y, Konishi F. Aberrant methylation of PSD disturbs Rac1-mediated immune responses governing neutrophil chemotaxis and apoptosis in ulcerative colitis-associated carcinogenesis. Int J Oncol. 2012;40:942–950. doi: 10.3892/ijo.2011.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile ML, Fang S, Baccarelli AA, Tarantini L, Cavallari J, Christiani DC. A panel study of occupational exposure to fine particulate matter and changes in DNA methylation over a single workday and years worked in boilermaker welders. Environ Health. 2013;12:47. doi: 10.1186/1476-069X-12-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney PL, Aggarwal M, Northridge ME, Janssen NA, Shepard P. Airborne concentrations of PM(2.5) and diesel exhaust particles on Harlem sidewalks: A community-based pilot study. Environ Health Perspect. 2000;108:213–218. doi: 10.1289/ehp.00108213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloog I, Ridgway B, Koutrakis P, Coull BA, Schwartz JD. Long- and short-term exposure to PM2.5 and mortality: Using novel exposure models. Epidemiology. 2013;24:555–561. doi: 10.1097/EDE.0b013e318294beaa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krewski D, Burnett R, Jerrett M, Pope CA, Rainham D, Calle E, Thurston G, Thun M. Mortality and long-term exposure to ambient air pollution: ongoing analyses based on the American Cancer Society cohort. J Toxicol Environ Health A. 2005;68:1093–1109. doi: 10.1080/15287390590935941. [DOI] [PubMed] [Google Scholar]
- Lamprecht B, Walter K, Kreher S, Kumar R, Hummel M, Lenze D, Kochert K, Bouhlel MA, Richter J, Soler E, et al. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat Med. 2010;16:571–579. doi: 10.1038/nm.2129. 571p following 579. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lee C, Wevrick R, Fisher RB, Ferguson-Smith MA, Lin CC. Human centromeric DNAs. Hum Genet. 1997;100:291–304. doi: 10.1007/s004390050508. [DOI] [PubMed] [Google Scholar]
- Lim SS, Carnahan E, Danaei G, Vos T, Lopez AD, Murray CJ, Ezzatti M. Annual deaths attributable to physical inactivity: Whither the missing 2 million? Lancet. 2013;381:993. doi: 10.1016/S0140-6736(13)60706-0. - Authors’ reply. [DOI] [PubMed] [Google Scholar]
- Lou J, Wang Y, Yao C, Jin L, Wang X, Xiao Y, Wu N, Song P, Song Y, Tan Y, et al. Role of DNA Methylation in Cell Cycle Arrest Induced by Cr (VI) in Two Cell Lines. PLoS One. 2013;8:e71031. doi: 10.1371/journal.pone.0071031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrigano J, Baccarelli A, Mittleman MA, Wright RO, Sparrow D, Vokonas PS, Tarantini L, Schwartz J. Prolonged exposure to particulate pollution, genes associated with glutathione pathways, and DNA methylation in a cohort of older men. Environ Health Perspect. 2011;119:977–982. doi: 10.1289/ehp.1002773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsit C, Christensen B. Blood-derived DNA methylation markers of cancer risk. Adv Exp Med Biol. 2013;754:233–252. doi: 10.1007/978-1-4419-9967-2_12. [DOI] [PubMed] [Google Scholar]
- Meaney MJ, Szyf M. Environmental programming of stress responses through DNA methylation: life at the interface between a dynamic environment and a fixed genome. Dialogues Clin Neurosci. 2005;7:103–123. doi: 10.31887/DCNS.2005.7.2/mmeaney. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo DF, Stagi E, Fontana V, Consonni D, Gozza C, Garrone E, Bertazzi PA, Pesatori AC. A historical mortality study among bus drivers and bus maintenance workers exposed to urban air pollutants in the city of Genoa, Italy. Occup Environ Med. 2010;67:611–619. doi: 10.1136/oem.2009.050377. [DOI] [PubMed] [Google Scholar]
- Mishra A, Liu S, Sams GH, Curphey DP, Santhanam R, Rush LJ, Schaefer D, Falkenberg LG, Sullivan L, Jaroncyk L, et al. Aberrant overexpression of IL-15 initiates large granular lymphocyte leukemia through chromosomal instability and DNA hypermethylation. Cancer Cell. 2012;22:645–655. doi: 10.1016/j.ccr.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama R, Qi L, Lacey M, Ehrlich M. Both hypomethylation and hypermethylation in a 0.2-kb region of a DNA repeat in cancer. Mol Cancer Res. 2005a;3:617–626. doi: 10.1158/1541-7786.MCR-05-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama R, Qi L, Tsumagari K, Weissbecker K, Dubeau L, Champagne M, Sikka S, Nagai H, Ehrlich M. A DNA repeat, NBL2, is hypermethylated in some cancers but hypomethylated in others. Cancer Biol Ther. 2005b;4:440–448. doi: 10.4161/cbt.4.4.1622. [DOI] [PubMed] [Google Scholar]
- Pavanello S, Pesatori AC, Dioni L, Hoxha M, Bollati V, Siwinska E, Mielzynska D, Bolognesi C, Bertazzi PA, Baccarelli A. Shorter telomere length in peripheral blood lymphocytes of workers exposed to polycyclic aromatic hydrocarbons. Carcinogenesis. 2010;31:216–221. doi: 10.1093/carcin/bgp278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope CA, Burnett RT, 3rd, Thun MJ, Calle EE, Krewski D, Ito K, Thurston GD. Lung cancer, cardiopulmonary mortality, and long-term exposure to fine particulate air pollution. JAMA. 2002;287:1132–1141. doi: 10.1001/jama.287.9.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope CA, 3rd, Burnett RT, Thurston GD, Thun MJ, Calle EE, Krewski D, Godleski JJ. Cardiovascular mortality and long-term exposure to particulate air pollution: Epidemiological evidence of general pathophysiological pathways of disease. Circulation. 2004;109:71–77. doi: 10.1161/01.CIR.0000108927.80044.7F. [DOI] [PubMed] [Google Scholar]
- Prada D, Gonzalez R, Sanchez L, Castro C, Fabian E, Herrera LA. Satellite 2 demethylation induced by 5-azacytidine is associated with missegregation of chromosomes 1 and 16 in human somatic cells. Mutat Res. 2012;729:100–105. doi: 10.1016/j.mrfmmm.2011.10.007. [DOI] [PubMed] [Google Scholar]
- Raaschou-Nielsen O, Bak H, Sorensen M, Jensen SS, Ketzel M, Hvidberg M, Schnohr P, Tjonneland A, Overvad K, Loft S. Air pollution from traffic and risk for lung cancer in three Danish cohorts. Cancer Epidemiol Biomarkers Prev. 2010;19:1284–1291. doi: 10.1158/1055-9965.EPI-10-0036. [DOI] [PubMed] [Google Scholar]
- McLean R, Sanders WL, Stroup WW. A Unified Approach to Mixed Linear Models. The American Statistician. 1991;45:54–64. [Google Scholar]
- Saint-Georges F, Garcon G, Escande F, Abbas I, Verdin A, Gosset P, Mulliez P, Shirali P. Role of air pollution Particulate Matter (PM(2.5)) in the occurrence of loss of heterozygosity in multiple critical regions of 3p chromosome in human epithelial lung cells (L132) Toxicol Lett. 2009;187:172–179. doi: 10.1016/j.toxlet.2009.02.016. [DOI] [PubMed] [Google Scholar]
- Sava F, Carlsten C. Respiratory health effects of ambient air pollution: An update. Clin Chest Med. 2012;33:759–769. doi: 10.1016/j.ccm.2012.07.003. [DOI] [PubMed] [Google Scholar]
- Soberanes S, Gonzalez A, Urich D, Chiarella SE, Radigan KA, Osornio-Vargas A, Joseph J, Kalyanaraman B, Ridge KM, Chandel NS, et al. Particulate matter Air Pollution induces hypermethylation of the p16 promoter Via a mitochondrial ROS-JNK-DNMT1 pathway. Sci Rep. 2012;2:275. doi: 10.1038/srep00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarantini L, Bonzini M, Apostoli P, Pegoraro V, Bollati V, Marinelli B, Cantone L, Rizzo G, Hou L, Schwartz J, et al. Effects of particulate matter on genomic DNA methylation content and iNOS promoter methylation. Environ Health Perspect. 2009;117:217–222. doi: 10.1289/ehp.11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting DT, Lipson D, Paul S, Brannigan BW, Akhavanfard S, Coffman EJ, Contino G, Deshpande V, Iafrate AJ, Letovsky S, et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331:593–596. doi: 10.1126/science.1200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Ramos YD, Montoya-Estrada A, Guzman-Grenfell AM, Mancilla-Ramirez J, Cardenas-Gonzalez B, Blanco-Jimenez S, Sepulveda-Sanchez JD, Ramirez-Venegas Hicks JJ. Urban PM2.5 induces ROS generation and RBC damage in COPD patients. Front Biosci (Elite Ed) 2011;3:808–817. doi: 10.2741/e288. [DOI] [PubMed] [Google Scholar]
- Tsumagari K, Qi L, Jackson K, Shao C, Lacey M, Sowden J, Tawil R, Vedanarayanan V, Ehrlich M. Epigenetics of a tandem DNA repeat: Chromatin DNaseI sensitivity and opposite methylation changes in cancers. Nucleic Acids Res. 2008;36:2196–2207. doi: 10.1093/nar/gkn055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner MC, Krewski D, Pope CA, 3rd, Chen Y, Gapstur SM, Thun MJ. Long-term ambient fine particulate matter air pollution and lung cancer in a large cohort of never-smokers. Am J Respir Crit Care Med. 2011;184:1374–1381. doi: 10.1164/rccm.201106-1011OC. [DOI] [PubMed] [Google Scholar]
- Valavanidis A, Vlachogianni T, Fiotakis K, Loridas S. Pulmonary Oxidative Stress, Inflammation and Cancer: Respirable Particulate Matter, Fibrous Dusts and Ozone as Major Causes of Lung Carcinogenesis through Reactive Oxygen Species Mechanisms. Int J Environ Res Public Health. 2013;10:3886–3907. doi: 10.3390/ijerph10093886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattanasit U, Navasumrit P, Khadka MB, Kanitwithayanun J, Promvijit J, Autrup H, Ruchirawat M. Oxidative DNA damage and inflammatory responses in cultured human cells and in humans exposed to traffic-related particles. Int J Hyg Environ Health. 2013 doi: 10.1016/j.ijheh.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Venza I, Visalli M, Fortunato C, Ruggeri M, Ratone S, Caffo M, Caruso G, Alafaci C, Tomasello F, Teti D, et al. PGE2 induces interleukin-8 derepression in human astrocytoma through coordinated DNA demethylation and histone hyperacetylation. Epigenetics. 2012;7:1315–1330. doi: 10.4161/epi.22446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vineis P, Hoek G, Krzyzanowski M, Vigna-Taglianti F, Veglia F, Airoldi L, Autrup H, Dunning A, Garte S, Hainaut P, et al. Air pollution and risk of lung cancer in a prospective study in Europe. Int J Cancer. 2006;119:169–174. doi: 10.1002/ijc.21801. [DOI] [PubMed] [Google Scholar]
- Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, Ehrlich M, Laird PW. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–6836. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson WE, Brauer M. Estimation of ambient and non-ambient components of particulate matter exposure from a personal monitoring panel study. J Expo Sci Environ Epidemiol. 2006;16:264–274. doi: 10.1038/sj.jes.7500483. [DOI] [PubMed] [Google Scholar]
- Wolff EM, Byun HM, Han HF, Sharma S, Nichols PW, Siegmund KD, Yang AS, Jones PA, Liang G. Hypomethylation of a LINE-1 promoter activates an alternate transcript of the MET oncogene in bladders with cancer. PLoS Genet. 2010;6:e1000917. doi: 10.1371/journal.pgen.1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Bank. World Development Indicators. 2012 [Google Scholar]
- Wu HC, Delgado-Cruzata L, Flom JD, Perrin M, Liao Y, Ferris JS, Santella RM, Terry MB. Repetitive element DNA methylation levels in white blood cell DNA from sisters discordant for breast cancer from the New York site of the Breast Cancer Family Registry. Carcinogenesis. 2012;33:1946–1952. doi: 10.1093/carcin/bgs201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang C, Gao H, Meng L, Qin Z, Ma R, Liu Y, Jiang Y, Dang C, Jin L, He F, et al. Functional variable number of tandem repeats variation in the promoter of proto-oncogene PTTG1IP is associated with risk of estrogen receptor-positive breast cancer. Cancer Sci. 2012;103:1121–1128. doi: 10.1111/j.1349-7006.2012.02266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Yamashita S, Takamura-Enya T, Niwa T, Ando T, Enomoto S, Maekita T, Nakazawa K, Tatematsu M, Ichinose M, et al. Alu and Satalpha hypomethylation in Helicobacter pylori-infected gastric mucosae. Int J Cancer. 2011;128:33–39. doi: 10.1002/ijc.25534. [DOI] [PubMed] [Google Scholar]
- Yu Y, Schleicher N, Norra S, Fricker M, Dietze V, Kaminski U, Cen K, Stuben D. Dynamics and origin of PM2.5 during a three-year sampling period in Beijing, China. J Environ Monit. 2011;13:334–346. doi: 10.1039/c0em00467g. [DOI] [PubMed] [Google Scholar]
- Zeger SL, Thomas D, Dominici F, Samet JM, Schwartz J, Dockery D, Cohen A. Exposure measurement error in time-series studies of air pollution: concepts and consequences. Environ Health Perspect. 2000;108:419–426. doi: 10.1289/ehp.00108419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Gao Z, Tian Z, Xie Y, Xin F, Jiang R, Kan H, Song W. The biological effects of individual-level PM(2.5) exposure on systemic immunity and inflammatory response in traffic policemen. Occup Environ Med. 2013;70:426–431. doi: 10.1136/oemed-2012-100864. [DOI] [PubMed] [Google Scholar]
- Zhu ZZ, Sparrow D, Hou L, Tarantini L, Bollati V, Litonjua AA, Zanobetti A, Vokonas P, Wright RO, Baccarelli A, et al. Repetitive element hypomethylation in blood leukocyte DNA and cancer incidence, prevalence, and mortality in elderly individuals: The Normative Aging Study. Cancer Causes Control. 2011;22:437–447. doi: 10.1007/s10552-010-9715-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.