Abstract
Phylogenetic distances of coexisting species differ greatly within plant communities, but their consequences for decomposers and decomposition remain unknown. We hypothesized that large phylogenetic distance of leaf litter mixtures increases differences of their litter traits, which may, in turn, result in increased resource complementarity or decreased resource concentration for decomposers and hence increased or decreased chemical transformation and reduction of litter. We conducted a litter mixture experiment including 12 common temperate tree species (evolutionarily separated by up to 106 Myr), and sampled after seven months, at which average mass loss was more than 50%. We found no effect of increased phylogenetic distance on litter mass loss or on abundance and diversity of invertebrate decomposers. However, phylogenetic distance decreased microbial biomass and increased carbon/nitrogen (C/N) ratios of litter mixtures. Consistently, four litter traits showed (marginally) significant phylogenetic signal and in three of these traits increasing trait difference decreased microbial biomass and increased C/N. We suggest that phylogenetic proximity of litter favours microbial decomposers and chemical transformation of litter owing to a resource concentration effect. This leads to a new hypothesis: closely related plant species occurring in the same niche should promote and profit from increased nutrient availability.
Keywords: complementarity versus resource concentration hypotheses, decomposer, litter degradation, niche, phylogenetic biodiversity ecosystem functioning, phylogenetic signal of functional traits
1. Introduction
Plant communities differ greatly in the phylogenetic distances between coexisting species. Different communities can be dominated by different evolutionary lineages [1] and harbour many or few of such lineages and thus may be composed of both closely and distantly related species [2,3]. Phylogenetically closely related species might on average share traits including functional traits that affect ecosystem processes [4,5]. Nevertheless, little is known about the consequences of variation in phylogenetic distance between coexisting species for ecosystem functioning. The few studies that exist focus on the consequences for the productivity of the vegetation itself and the stability of ecosystem functioning [4,6]. So far, no study has focused on consequences of phylogenetic distance of plants for ecosystem processes that result from the interaction between the vegetation and other trophic levels, such as the decomposer community that carries out decomposition of litter, i.e. its mass-reduction and chemical transformation (e.g. from high to low carbon/nitrogen; C/N).
Large phylogenetic distance between litter species might increase rates of mass loss and chemical transformation through complementarity effects [7–10], which we term the complementarity hypothesis (table 1). The importance of complementarity in decomposition studies has been previously documented in terms of the chemical and physical diversity of the litter [11–14]. The complementarity hypothesis states that microbes and detritivores might derive different resources from different types of litter, i.e. complementary resource use, when this litter is composed of chemically divergent leaf species [10]. Complementary resources might become available to a given organism via leaching or fungal activity, and might maximize decomposer net energy intake and allow the imbalances in carbon : nitrogen : phosphorus (C : N : P) ratios of leaf litter and the decomposer body tissues to be overcome [15,16]. Such complementary resources may increase the physiological performance and abundance of generalist decomposers. Moreover, complementary resources may permit multiple specialist decomposers to establish on different litters and these specialists in turn may have complementary effects on decomposition (a similar mechanism can also be seen in a pollination study [17]). More abundant or complementary decomposers may then accelerate transformation and reduction of the litter [10] resulting in higher rates of decomposition [8,9,18–21]. Large phylogenetic distance between litter species may imply more divergent litter traits (‘afterlife’ traits) and the resulting complementarity effects may increase decomposer abundance and diversity and accelerate litter transformation and reduction [5,22–25].
Table 1.
Predictions of hypotheses tested in this study, and explained in the Introduction. (Predictions that could be confirmed or partly confirmed in this study are given in italics.)
| prediction: mixtures of phylogenetically distant lineages show… | |
|---|---|
| complementarity hypothesis | resource concentration hypothesis |
| …higher trait differences | |
| …increased microbial biomass and abundance/diversity of invertebrate decomposers | …reduced microbial biomass and abundance/diversity of invertebrate decomposers |
| …faster mass loss | …slower mass loss |
| …faster decrease in C/N ratio | …slower decrease in C/N ratio |
Conversely, large phylogenetic distance between litter species may also decrease decomposers, litter transformation and litter reduction by reducing the concentration of each litter lineage as a resource for decomposers. This is termed the resource concentration hypothesis (table 1) and has previously been applied to phytophages feeding on plant monocultures and polycultures [26] or on phylogenetically distant plant species [27]. The resource concentration hypothesis states that herbivores are more likely to find hosts growing in monospecific stands and to maintain populations there. This hypothesis requires that consumers are not entirely generalists feeding on any plant, but are either mono- or oligophages as is the case for many phytophages [28,29]. Even decomposers may not be entirely generalists: some microbes tend to have limited ranges of optimal resources and even detritivores are not entirely generalists [30]. The resource concentration hypothesis is also an important aspect of the substrate–matrix interaction hypothesis of litter decomposition [31]. Again, coexisting phylogenetically distant plant species may be particularly different in functional traits and litter traits. Increased trait difference might dilute the optimal litter resources for non-generalist decomposers. This dilution may decrease the suitability of litter for those non-generalist decomposers and therefore result in lower rates of decomposition. Large phylogenetic distance between litter species via traits diluting optimal litter resources may hence decrease decomposer abundance and diversity and accelerate litter transformation and reduction.
To test which of these alternative hypotheses would best explain the relationships between phylogenetic distance and litter mixture effects, we investigated how phylogenetic distances of coexisting litter species influence the difference of litter traits known to be important drivers of decomposition [24,32–34]. We also investigated how phylogenetic distances of litter affect decomposer organisms and the transformation and reduction of litter mixtures. For this purpose, we used a litter mixture experiment to examine the effect of phylogenetic distance on: (i) the traits of litter species (phylogenetic signal), (ii) the microbial biomass, and the abundance and diversity of invertebrates in litter mixtures, and (iii) the mass loss, and the change in litter C/N ratios of litter mixtures (where a slow decrease in litter C/N-ratio corresponds to a low improvement of N-availability). We explored whether effects of phylogenetic distance on traits explained effects on decomposers or decomposition. We used two recently dated and resolved phylogenies with different resolution of age estimates for our particular study ecosystem. We ensured that colonizing decomposer species come from a phylogenetically unbiased environment, i.e. from a phylogenetic outgroup litter bed.
2. Material and methods
(a). Tree species
Litter of 12 common temperate tree species was sampled by hand before touching the ground from three open sites in the vicinity of Rennes, France. The region has an oceanic climate with a mean annual precipitation of 644 mm and a mean air temperature of 11.4°C. Different tree species were not in direct contact (more than 50 m distance) and hence did not directly interact with each other. The tree species were selected within four families: Fagaceae (Fagus sylvatica, Quercus robur, Quercus petraea), Salicaceae (Populus tremula, Salix cinerea, Salix caprea), Betulaceae (Betula pendula, Alnus incana, Alnus glutinosa) and Rosaceae (Malus sylvestris, Prunus serotina, Prunus avium). Within each family, two species are from the same genus, while the third species comes from another genus. These species were selected as they are highly represented in the study region. Litter was air-dried for two weeks after sampling and then stored in the laboratory at room temperature.
(b). Plant trait measurements
Initial litters of 10–50 pooled dead leaves per species were first immersed in tap water overnight, then wiped gently with paper tissue and measured for their water-saturated weight. Dry matter content was expressed as the ratio between oven-dry weight (g) and water-saturated weight (g). Leaf tensile strength was measured as the force needed to break the leaf, following Makkonen et al. [35]. The litter samples were also ground after oven-drying for 24 h at 80°C in order to measure the following litter traits: pH and concentration of lignin, total phenols, tannins, carbon (C) and nitrogen (N). The litter pH was measured following Cornelissen et al. [33]. Lignin concentration was determined as described in Poorter & Villar [36], briefly introduced by Freschet et al. [37]. Tannins and total phenols were measured using the modified Folin–Ciocalteau method [38]. The C- and N-concentrations of the litter were determined by dry combustion on a NA-1500 elemental analyser (Carlo Erba, Rodana, Italy). Two additional litter traits, phosphorus (P) concentration and specific leaf area (SLA) were taken from a database from Central-English populations [39].
(c). Experimental design
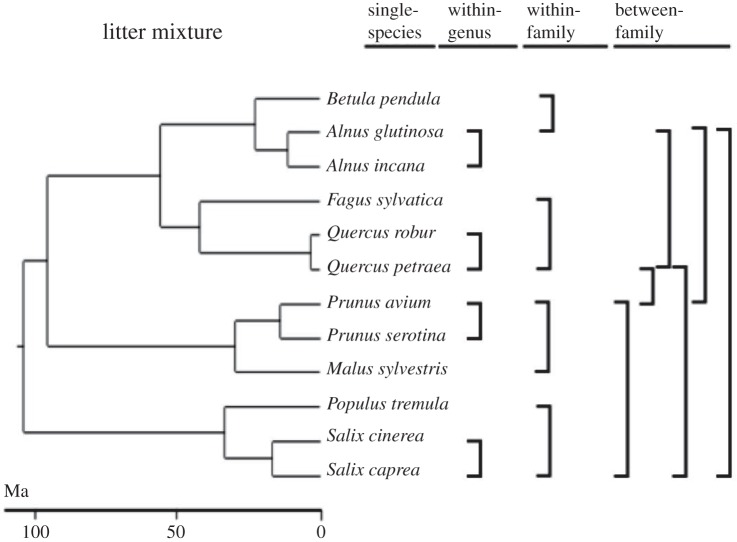
Each of the four families had one main representative species (Q. petraea, S. caprea, A. glutinosa or P. avium), which was the most abundant species in the study region. This main representative species was then combined in two species combinations with another representative species of the same genus (Q. robur, S. cinerea, A. incana or P. serotina), with another representative species of another genus from the same family (F. sylvatica, P. tremula, B. pendula or M. sylvestris), and with each of the other main representatives of the other three families. In addition, single species treatments were applied to each of the 12 species (figure 1). Numbers of replicates per level of phylogenetic distance were 12 single-species, four within-genus, four within-family and six between-family. We did deliberately involve only a core set of species across all levels of phylogenetic distance. Involving all species at all levels would result in numerous pairs of species that replicate almost the same branches. For instance, it would result in one pair of Q. petraea versus S. caprea and another pair of Q. robur versus S. capraea. Each pair would be treated as an independent data point but in reality they differ only in the short distance between Q. petrea and Q. robur.
Figure 1.
Experimental design. Phylogenetic classification was used to establish a balanced design with litter mixtures created as follows: single-species, within-genus, within-family and between-family. Note that a set of core species is present at all levels of phylogenetic distance each of which is mixed with successively more distantly related species. This design minimizes resampling of phylogenetic branches in different mixtures as it would occur if all combinations of all species were included. The analysis then accounts for the identity of species included in a litter mixture by calculating overyielding, e.g. mass loss of the species mixtures relative to mass losses of each of the species in isolation. Note also that each replicate of a given phylogenetic distance is a distinct species combination, avoiding pseudoreplication within combinations. Age estimates were based on the genus age-optimized tree and an alternative tree was analysed in addition (Material and methods).
Phylogenetic age distances were quantified between litter species, which extended up to 105.5 Myr. We used two recent angiosperm phylogenies with different age estimations: (i) a dated tree of the Dutch flora with age estimates resolved at the level of genera, i.e. the genus age-optimized tree [40], and (ii) a dated tree for the European flora with a large sample of species, i.e. the sample size-optimized tree [41]. We showed the results based on the genus age-optimized tree, and results based on the sample size-optimized tree can be seen in the electronic supplementary material, Appendices 1 and 2. All phylogenetic age distances were ln-transformed before analyses to improve the distribution of residuals.
Experimental plots were at the University of Rennes 1 campus (48°07′ N, 1°38′ W) and were shaded by surrounding trees during part of the day. To establish litter mixtures, we used litterbags of 25 × 20 cm size and the mesh size 5 × 5 mm, to ensure accessibility of litter to all soil invertebrates at the study site, while preventing the litter from dropping through the mesh due to physical transport. Each litter bag was filled with a consistent volume of litter (corresponding to 2–3 cm thickness) to ensure similar habitat size and proximity of experimental litter to the surrounding litter matrix, and approximately similar air-dry mass of 12 g, oven-drying being prohibitive for the litter exposed. Inevitably, constancy in volume, and weights being based on air-dry mass led to some variation in the oven-dry equivalents in the initial mass of litters, ranging from 6 to 12 g. In later analyses, initial mass was included when the respective dependent variable (marginally) significantly correlated with initial mass (simple regression model: for mass loss, p = 0.073; versus p > 0.248 for other variables). Note that initial mass did not correlate with variation in either of the phylogenetic age distances (simple regression model: p = 0.195 and 0.374), indicating phylogenetic distance was not confounded by the initial mass.
The litter bags were incubated (10 cm apart) below the surface of a ‘common garden’ litter bed [23] of 1.3 by 2.0 m. The litter bed was composed of a 20 cm deep layer of a 50 : 50 mixture of two phylogenetic outgroup species (Tilia platyphyllos and Platanus × hybrid). We used these two species as they are abundant in the study region and phylogenetically equally distant to each of the litter species in the litterbags, i.e. a phylogenetic outgroup. This ‘outgroup’ litter bed ensures that the pool of decomposers is not biased towards any of the lineages represented in the litter bags. Litter bags were incubated starting on 30 November 2009 and harvested after seven months (comparable sampling approach as in [42]). This harvest time was determined by the mass of the remaining litters in the litterbags: if decomposition had lasted longer than seven months, the mass of remaining litters for some species would have been too small to examine the microbial biomass per litter species and to detect the effect of phylogenetic distance [9]. Note that the winter is mild in our study site, and frost is rare and temperatures and moisture are hence favourable for substantial decomposition. In fact, there was on average more than 50% mass loss across all the species combinations after seven months and 8 out of 26 species combinations had more than 80% mass loss. For comparison, in a temperate area in Central England with broadly corresponding climate mass loss was on average faster during an even shorter period including ‘winter’ [43].
(d). Harvest, microbial biomass and soil invertebrates
After seven months, litterbags were retrieved and transported in individual plastic bags to the laboratory. Small subsamples of component litter species in litter mixtures were taken and attached mineral soil was brushed off. Microbial biomass of each component species was analysed using the substrate-induced respiration (SIR) method [44]. The microbial respiratory response was measured in an electrolytic O2 microcompensation apparatus at hourly intervals for 24 h at 22°C [45]. Microbial biomass was measured after the addition of glucose to saturate the catabolic activity of microorganisms. The maximum initial respiratory response (MIRR: ml O2 g−1 dw h−1) was calculated as the mean of the lowest three readings within the first 10 h and microbial biomass was calculated as Cmic = 38 × MIRR (μg Cmic g−1 dw; [46]). The microbial biomass of a litter mixture was calculated as the mean microbial biomass of two component litter species. Invertebrates were extracted by heat and stored in saturated salt solution (NaCl) at 10°C [47]. Animals were counted and identified by light microscopy. For each group (as defined in the electronic supplementary material, Appendix 3), we calculated total abundance and the Simpson index D, and transformed-logD [48].
(e). Weighing and chemical analysis of remaining litters
After extracting the soil fauna, all the harvested litters were oven-dried at 65°C for one week and weighed. Litter mass loss (ΔM) was calculated as ΔM(%) = ((m0 – m1)/m0) × 100, where m0 is the (estimated) oven-dry weight of initial litter and m1 refers to the oven-dry weight of remaining litter after seven months. However, we did not oven-dry the litter prior to exposure to avoid destroying leaf characters essential for decomposition. Instead we oven-dried five samples per litter species and used the air-dry/oven-dry mass ratio to estimate the oven-dry masses of the litter we had exposed. Finally, the C- and N-concentrations of the remaining litter mixtures were determined as above in the section of plant trait measurements. The change in C/N ratios was then calculated as the pre-exposure C/N ratios divided by the post-exposure C/N ratios.
(f). Statistical analysis
We characterized each litter mixture by its overyielding in mass loss, i.e. the proportional differences between the mass loss observed in the mixture (yp) and the mean of the mass losses in the corresponding two single-species treatments (ym) expressed as log ratio: ln(yp/ym) [49,50]. In addition, overyielding was calculated for change in C/N ratios, microbial biomass and for abundances and diversities of invertebrates. Then, we related overyieldings to the phylogenetic distances of the mixed species using general regression models with best subset search (adjusted R² criterion optimizing R² while accounting for numbers of variables, Statistica v. 7.0). As covariables, we included the representation of Betulaceae and Salicaceae in the litter mixture as these families most strongly affected overyieldings. We also included the overyielding of mass loss, as different stages of the decomposition process may correspond to different stages in decomposer colonization, a correspondence that would be maintained after standardizing decomposition and colonization in the calculation of overyielding. When the overyielding of mass loss was the dependent variable, we included initial mass as a covariable to control for any possible relationship between initial mass and the overyielding of mass loss. Note that the overyielding has to be zero for a phylogenetic distance of zero (there cannot be an effect of species mixing in the absence of a second species) and intercepts were hence set to zero. We illustrated the slope of the effect of phylogenetic distance identified in the multiple regression analysis using partial residual plots. In addition, we calculated tolerances of independent variables [51]. Tolerances were relatively high (more than 0.42), indicating that multicolinearity among independent variables was not a problem and effects of these variables could be discerned [51].
The overyielding approach controls for species identity as values observed for species mixtures are compared to those expected for the very same species based on monocultures. The shortcoming of overyielding, however, is that it does not permit inclusion of the level of shortest phylogenetic distances, i.e. the within-species treatment. We hence conducted an additional analysis in which we related phylogenetic distance (including 0) to observed mass loss, C/N-change and microbial biomass (without correcting observed for expected values). In these analyses, we again explored covariables first and found that the proportion of all four families correlated with different decomposition variables. Therefore, we included proportions of all families (one being excluded as its effect is redundant with the combination of all other families). We also included mass loss as a covariable for reasons explained above. Also, we again calculated the tolerance values and the smallest tolerance value was 0.21, indicating again that multicolinearity among independent variables was only weak.
For all traits, we calculated phylogenetic signal, i.e. an increase in trait distance with phylogenetic distance, implying that mixtures of phylogenetically distant litters are more dissimilar than mixtures of phylogenetically close litters. We used Blomberg's K as a measure of phylogenetic signal [52,53]. This measure is widely used and permits comparison to other functional trait studies [54,55]. We used the approach of Blomberg et al. [52] to test for statistical significance, based on observed variance of (unsigned) phylogenetically independent contrasts as compared to a null expectation. Lower than expected variance reflects close relatives being more similar than expected by chance. For these phylogenetic signal tests, we used the ‘phylosignal’ function in the package ‘picante’ (R software v. 2.13.0). Four traits yielded an at least marginally significant phylogenetic signal and were hence candidates for explaining effects of phylogenetic distance identified by the above analyses. For these traits, we calculated (unsigned) differences between species in each litter mixture. We then related these trait differences of litter mixtures to the corresponding overyieldings of microbial biomass and C/N ratio changes, i.e. the dependent variables for which we had identified a negative effect of phylogenetic distance in the above analyses. As covariables, we included the overyielding of mass loss for reasons explained above, and the mean trait values as trait differences are bound to be small if trait means are small. We also included the ‘difference × mean’ interaction term as a covariable as we found that the effect of difference of a trait may change with the mean value of that trait. Note that we did not apply a best subset search here as best subset search sometimes excluded one or two component variables of the interaction term while maintaining the interaction term, which would be meaningless. In all analyses, we verified residual distribution graphically using predicted/residual plots and normal probability plots. At most one extreme outlier was excluded.
3. Results
Our species showed strong phylogenetic signal in phosphorus and lignin concentrations (table 2; for phosphorus: K = 0.616, p = 0.019; for lignin: K = 0.916, p = 0.001), and marginally significant phylogenetic signal in SLA and non-tannin phenols (table 2; for SLA: K = 0.482, p = 0.062; for lignin: K = 0.489, p = 0.063). No strong phylogenetic signal was found in the other leaf or litter traits (table 2). In other words, larger phylogenetic distance of litter mixtures should result in more dissimilar trait values in the concentrations of phosphorus, lignin, non-tannins, as well as SLA.
Table 2.
Phylogenetic signals of species traits, and effect of traits with signal on C/N ratio change and microbial biomass, which in turn relate to phylogenetic distance (figure 1). (The left part of the table gives Blomberg's K (which increases with the strength of phylogenetic signal), and associated p-values (which compare observed variances of phylogenetically independent contrasts of a trait to a null expectation). Significant results are shown in bold, and marginally significant results are shown in italic. The right part of the table focuses on the traits that show at least a marginally significant phylogenetic signal and explores how differences of these traits relate to C/N changes and microbial biomass, accounting for trait means, their interaction with trait difference and mass loss (see Material and methods).)
| relationship of trait difference to |
||||||
|---|---|---|---|---|---|---|
| C/N ratio change |
microbial biomass |
|||||
| phylogenetic signal traits | K | p-values | t | p | t | p |
| SLA | 0.482 | 0.062 | −3.059 | 0.018 | 1.527 | 0.171 |
| toughness | 0.164 | 0.779 | ||||
| P concentration | 0.616 | 0.019 | 2.709 | 0.030 | −1.646 | 0.144 |
| pH | 0.315 | 0.317 | ||||
| Dry matter content | 0.128 | 0.861 | ||||
| lignin concentration | 0.916 | 0.001 | −1.386 | 0.208 | −0.498 | 0.634 |
| N concentration | 0.246 | 0.506 | ||||
| C concentration | 0.174 | 0.683 | ||||
| C/N | 0.260 | 0.417 | ||||
| total phenolics | 0.206 | 0.635 | ||||
| non-tannin phenols | 0.489 | 0.063 | −0.435 | 0.677 | −2.310 | 0.054 |
| tannins | 0.149 | 0.838 | ||||
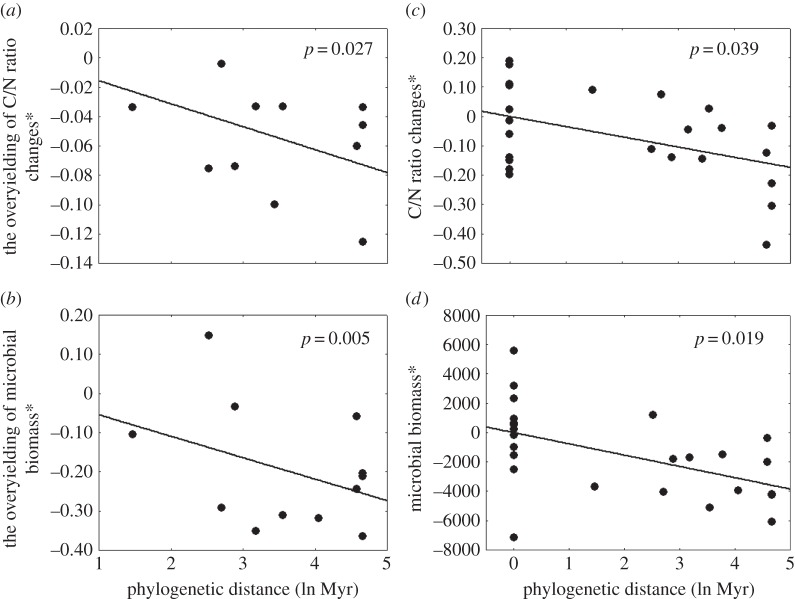
The overyielding of microbial biomass was negatively related to phylogenetic distance (figure 2, N = 13, t = −3.641, p = 0.005), and so was the overyielding in C/N ratio changes (figure 2, N = 12, t = −2.799, p = 0.027). By contrast, overyieldings of mass loss, or the abundances or the diversities of invertebrates were not affected by phylogenetic distance of litter mixtures (best subset search excluded phylogenetic distance from the regression model). Analyses based on raw values instead of overyieldings (see above) confirmed these results: phylogenetic distance had negative effects on microbial biomass and on C/N ratio changes (figure 2; for microbial biomass: N = 26, t = −2.573, p = 0.019; for C/N ratio changes: N = 26, t = −2.223, p = 0.039).
Figure 2.
Effects of phylogenetic distance of litter mixtures on C/N-ratio changes and microbial biomass. Analyses (a,b) are based on overyielding, i.e. standardized observations for mixtures of litter species by the performance of each species in isolation. Analyses (c,d) are based on raw data, i.e. unstandardized but permitting inclusion of zero-distances into the analysis. Asterisks (*) denote dependent variables are presented as partial residuals from a multiple regression analysis, i.e. accounting for covariables as outlined in Material and methods (partial residuals are dimensionless; see the electronic supplementary material, Appendices 4 and 5 for original data). Phylogenetic distances were based on the genus age-optimized tree. See the electronic supplementary material, Appendices 1 and 2 for analyses based on the sample size-optimized tree. Note that we do not present mass loss here as phylogenetic distance was excluded from the model for the overyielding analyses.
The observed negative effect of phylogenetic distance on the microbial biomass can be partly explained by the trait difference in non-tannin concentrations, which tended to have a negative effect on the overyielding of microbial biomass (table 2, N = 12, t = −2.310, p = 0.054). The negative effect of phylogenetic distance on C/N ratio changes can be partly explained by the trait difference of SLA, which also had a negative effect on the C/N ratio changes (table 2, N = 12, t = −3.059, p = 0.018). However, the trait difference of phosphorus had a positive effect on C/N ratio changes (table 2, N = 12, t = 2.709, p = 0.030), and no effect of trait difference in lignin concentration was found on either microbial biomass or C/N ratio changes.
4. Discussion
We found phylogenetic signal in litter concentrations of phosphorus, lignin and non-tannins, and SLA and hence difference of these traits in litter mixtures will increase with the phylogenetic distance of the mixed species. Phylogenetic distances of litter mixtures, in turn, did not affect litter mass loss or abundances or diversities of invertebrates. However, phylogenetic distances of litter mixtures did affect the microbial biomass and litter transformation: microbial biomass was low and the C/N ratio remained high in treatments of larger phylogenetic distance. These effects of phylogenetic distance on C/N-ratio change and microbial biomass can partly be attributed to differences in traits showing phylogenetic signal. Our results were robust to different analytical approaches of quantifying the effect of phylogenetic distance. To disentangle effects of mixing per se from effects of species identity, we had used an overyielding approach (and we confirmed our results on microbial biomass by an approach suggested by Loreau & Hector [7], electronic supplementary material, Appendix 6). Overall, at least across the 12 tree species concerned, phylogenetic distances of litter mixtures had either no effects or negative effects on decomposers or decomposition, contrary to what has been shown for plant productivity [50]. This difference might be due to the absence of competition and a pressure on niche complementarity among our plant litters and likely among the tree species producing them (species were samples from isolated or monospecific stands). The difference might also reflect the presence of strong trophic interactions in the relationship between plant phylogenetic distances and decomposition.
(a). Testing for the complementarity hypothesis
Our results do not confirm any of the predictions of the complementarity hypothesis except for the differences in four traits, which it shares with the resource concentration hypothesis. One possible explanation is that the complementarity effect of litter species on decomposition was proposed to be due to nutrient transfer by leaching or hyphal transport between complementary dead leaves [9,10,14]. Studies discussing litter complementarity usually considered traits that are leachable or transferable between litter species, such as N and P which are more ‘mobile’ traits. More dissimilar trait values of litter N and P concentrations within litter mixtures will provide opportunities for microbes and detritivores to optimize nutrient acquisition, become more abundant and hence increase decomposition rates [10,14]. Our results indeed showed that larger trait differences in P concentration, which is relative mobile and leachable, led to a faster decline in C/N ratios. However, structure-related traits not clearly linked to mobile chemistry, such as SLA and lignin concentration, showed no evidence of complementarity increasing decomposition. Low SLA usually indicates toughness and slows down litter decomposition rates [23,32,56] but the mixing of dead leaves differing in structure might not provide any complementarity advantage [57].
(b). Testing for the resource concentration hypothesis
Our results do support several predictions of the resource concentration hypothesis on the microbial biomass and the change in C/N ratios, but not on the total mass loss or abundances or diversities of invertebrates: increasing phylogenetic distance between litter species may lead to decreased suitability of litter species for microbes, reducing microbial biomasses. This decreased microbial biomass may have caused the weaker declines in C/N ratios. The resource concentration effect assumes that decomposers are not entirely generalists that feed on any plant but tend to show a preference for optimal resources. This assumption may be true for microbes, because fungi and bacteria are relatively immobile. This implies that each microbe is forced to be specialized on the respective species of the leaf-litter it is using, and might have been under selection pressure by this particular leaf-litter for many generations [58,59]. Dilution of optimal resources among very different, phylogenetically distant litters may then explain lower microbial biomass. For invertebrates, however, our results do not support the resource concentration hypothesis. This may be owing to their relatively high degree of generalism [60,61]. Additionally, it should be noted that the mobility of invertebrates may also decrease the power to detect any effect of phylogenetic distance as one harvest represents only one spotlight on a highly mobile community.
There are several mutually non-exclusive explanations for the observed absence of resource concentration effects on mass loss despite the presence of such effects on C/N ratio changes and microbial biomass: (i) the reduced biomass of microbes on phylogenetically distant litter mixtures might be compensated by an increased efficiency in reducing litter mass. This increased efficiency might possibly be due to phylogenetically distant litter being used by dissimilar microbes which then triggers a complementarity effect on decomposition at the level of microbes; (ii) C/N ratio changes depend on leaching of N instead of respiration of C and are thus independent of mass loss. A relatively strong decline of C/N would hence reflect a relatively weak leaching of N. This, however, would result in low N concentration in the soil below litter bags with strong decline in C/N ratios, which we did not find (rather the opposite: r = 0.37, p = 0.07, results not shown); (iii) litter mass might be increasingly replaced by microbial biomass, specifically by fungi, i.e. the material of higher N-content. In that case, the true litter mass would be overestimated in phylogenetically close litter mixtures and true litter mass might indeed have declined in phylogenetically close litter mixtures; and (iv) we only measured the total mass loss of litter mixtures. It is possible that one component species lost mass faster and another lost mass slower than in monoculture, therefore there would be no net effect on total mass loss. If the more rapidly decaying species is one of particularly high C/N ratios, then the overall C/N ratio of the litter mixture might still decline. However, species of high C/N ratio usually decompose more slowly than species of low C/N.
We stress that our study did not cover the full range of angiosperm clades but was restricted to species from the rosids clade. We found the phylogenetic signal for a part of the traits but not for all. It is possible that phylogenetic signal would be more frequent if one considered a much larger clade such as the entire spermatophytes. In that case, numerous monocots and gymnosperms would be included besides rosids and the ancient differentiation of leaf traits between these groups [62] would strongly influence the calculated phylogenetic signal. Overall, we suggest that in the future the ecosystem consequences of mixing distantly related lineages should be identified for clades other than rosids, including ones that are more or less integrative than rosids.
5. Conclusion
Our study is, to our knowledge, the first exploration of consequences of phylogenetic distance for decomposers and decomposition. We found no effect of phylogenetic distance on litter mass loss, but we found an effect of phylogenetic distance on microbial biomass and litter transformation. More specifically, coexistence of closely related species litters might facilitate microbes and thereby contribute to a faster decline in C/N ratios. This might be due to closely related species belonging to the same plant lineage providing more concentrated litter resources for non-generalist decomposers that are capable of using this lineage. The necessary condition for co-occurrence of closely related species in a patch of the same niche is phylogenetic signal of niches [63]. Plants persisting in a shared, likely ancestral, niche might hence profit from an increased biomass of soil microbes and availability of N. For the time being, this relationship between phylogenetic signal of niches, co-occurrence, ecosystem processes and performance of species remains entirely hypothetical. We suggest it be tested in future research. This research should also include feedback mechanisms not accounted for in this study, notably how co-occurrence among close relatives affects their traits [22] and thereby the decomposition of litter.
Supplementary Material
Acknowledgements
We are grateful to Richard van Logtestijn for guiding us with the trait measurements, and for J. Vamosi, S. Ibanez and anonymous referees for comments that considerably improved our manuscript.
Data accessibility
Data are available in the electronic supplementary material.
Funding statement
M.D. was partly funded by NSFC (grant no. 31261120580) and the Innovative R & D grant of Hangzhou Normal University (grant no. 201203). A.P. was supported by CNRS-ATIP grant. I.V.B. was supported by Purkine Fellowship. X.P. was supported by the two mobility grants from UEB and CNRS-CAS, as well as by KNAW-CEP grant no. 12CDP007 to J.H.C.C.
References
- 1.Grandcolas P. 1998. Phylogenetic analysis and the study of community structure. Oikos 82, 397–400. ( 10.2307/3546983) [DOI] [Google Scholar]
- 2.Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. 2002. Phylogenies and community ecology. Annu. Rev. Ecol. Syst. 33, 475–505. ( 10.1146/annurev.ecolsys.33.010802.150448) [DOI] [Google Scholar]
- 3.Cavender-Bares J, Ackerly DD, Baum DA, Bazzaz FA. 2004. Phylogenetic overdispersion in Floridian oak communities. Am. Nat. 163, 823–843. ( 10.1086/386375) [DOI] [PubMed] [Google Scholar]
- 4.Cadotte MW, Cavender-Bares J, Tilman D, Oakley TH. 2009. Using phylogenetic, functional and trait diversity to understand patterns of plant community productivity. PLoS ONE 4, e5695 ( 10.1371/journal.pone.0005695) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Srivastava DS, Cadotte MW, MacDonald AAM, Marushia RG, Mirotchnick N. 2012. Phylogenetic diversity and the functioning of ecosystems. Ecol. Lett. 15, 637–648. ( 10.1111/j.1461-0248.2012.01795.x) [DOI] [PubMed] [Google Scholar]
- 6.Cadotte MW, Dinnage R, Tilman D. 2012. Phylogenetic diversity promotes ecosystem stability. Ecology 93, S223–S233. ( 10.1890/11-0426.1) [DOI] [Google Scholar]
- 7.Loreau M, Hector A. 2001. Partitioning selection and complementarity in biodiversity experiments. Nature 412, 72–76. ( 10.1038/35083573) [DOI] [PubMed] [Google Scholar]
- 8.Heemsbergen DA, Berg MP, Loreau M, van Hal JR, Faber JH, Verhoef HA. 2004. Biodiversity effects on soil processes explained by interspecific functional dissimilarity. Science 306, 1019–1020. ( 10.1126/science.1101865) [DOI] [PubMed] [Google Scholar]
- 9.Hättenschwiler S, Tiunov AV, Scheu S. 2005. Biodiversity and litter decomposition in terrestrial ecosystems. Ann. Rev. Ecol. Evol. Syst. 36, 191–218. ( 10.1146/annurev.ecolsys.36.112904.151932) [DOI] [Google Scholar]
- 10.Gessner MO, Swan CM, Dang CK, McKie BG, Bardgett RD, Wall DH, Hattenschwiler S. 2010. Diversity meets decomposition. Trends Ecol. Evol. 25, 372–380. ( 10.1016/j.tree.2010.01.010) [DOI] [PubMed] [Google Scholar]
- 11.Wardle DA, Nilsson MC, Zackrisson O, Gallet C. 2003. Determinants of litter mixing effects in a Swedish boreal forest. Soil Biol. Biochem. 35, 827–835. ( 10.1016/S0038-0717(03)00118-4) [DOI] [Google Scholar]
- 12.Meier CL, Bowman WD. 2008. Links between plant litter chemistry, species diversity, and below-ground ecosystem function. Proc. Natl Acad. Sci. USA 105, 19 780–19 785. ( 10.1073/pnas.0805600105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoorens B, Coomes D, Aerts R. 2010. Neighbour identity hardly affects litter-mixture effects on decomposition rates of New Zealand forest species. Oecologia 162, 479–489. ( 10.1007/s00442-009-1454-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vos VA, Ruijven J, Berg M, Peeters EHM, Berendse F. 2013. Leaf litter quality drives litter mixing effects through complementary resource use among detritivores. Oecologia 173, 269–280. ( 10.1007/s00442-012-2588-1) [DOI] [PubMed] [Google Scholar]
- 15.Martinson H, Schneider K, Gilbert J, Hines J, Hambäck P, Fagan W. 2008. Detritivory: stoichiometry of a neglected trophic level. Ecol. Res. 23, 487–491. ( 10.1007/s11284-008-0471-7) [DOI] [Google Scholar]
- 16.Hladyz S, Gessner MO, Giller PS, Pozo J, Woodward GUY. 2009. Resource quality and stoichiometric constraints on stream ecosystem functioning. Freshwater Biol. 54, 957–970. ( 10.1111/j.1365-2427.2008.02138.x) [DOI] [Google Scholar]
- 17.Fontaine C, Dajoz I, Meriguet J, Loreau M. 2005. Functional diversity of plant–pollinator interaction webs enhances the persistence of plant communities. PLoS Biol. 4, e0040001 ( 10.1371/journal.pbio.0040001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bardgett RD, Shine A. 1999. Linkages between plant litter diversity, soil microbial biomass and ecosystem function in temperate grasslands. Soil Biol. Biochem. 31, 317–321. ( 10.1016/S0038-0717(98)00121-7) [DOI] [Google Scholar]
- 19.Hooper DU, et al. 2000. Interactions between aboveground and belowground biodiversity in terrestrial ecosystems: patterns, mechanisms, and feedbacks. BioScience 50, 1049–1061. ( 10.1641/0006-3568(2000)050[1049:IBAABB]2.0.CO;2) [DOI] [Google Scholar]
- 20.Mikola J, Bardgett RD, Hedlund K. (ed.). 2002. Biodiversity, ecosystem functioning and soil decomposer food webs. Oxford, UK: Oxford University Press. [Google Scholar]
- 21.Schädler M, Brandl R. 2005. Do invertebrate decomposers affect the disappearance rate of litter mixtures? Soil Biol. Biochem. 37, 329–337. ( 10.1016/j.soilbio.2004.07.042) [DOI] [Google Scholar]
- 22.Prinzing A, Reiffers R, Braakhekke WG, Hennekens SM, Tackenberg O, Ozinga WA, Schaminée JHJ, Van Groenendael JM. 2008. Less lineages—more trait variation: phylogenetically clustered plant communities are functionally more diverse. Ecol. Lett. 11, 809–819. ( 10.1111/j.1461-0248.2008.01189.x) [DOI] [PubMed] [Google Scholar]
- 23.Cornwell WK, et al. 2008. Plant species traits are the predominant control on litter decomposition rates within biomes worldwide. Ecol. Lett. 11, 1065–1071. ( 10.1111/j.1461-0248.2008.01219.x) [DOI] [PubMed] [Google Scholar]
- 24.Pérez-Harguindeguy N, Díaz S, Cornelissen JHC, Vendramini F, Cabido M, Castellanos A. 2000. Chemistry and toughness predict leaf litter decomposition rates over a wide spectrum of functional types and taxa in central Argentina. Plant Soil 218, 21–30. ( 10.1023/A:1014981715532) [DOI] [Google Scholar]
- 25.Eiland F, Klamer M, Lind AM, Leth M, Bååth E. 2001. Influence of initial C/N ratio on chemical and microbial composition during long term composting of straw. Microbial Ecol. 41, 272–280. ( 10.1007/s002480000071) [DOI] [PubMed] [Google Scholar]
- 26.Root RB. 1973. Organization of a plant-arthropod association in simple and diverse habitats: the fauna of Collards (Brassica oleracea). Ecol. Monogr. 43, 95–124. ( 10.2307/1942161) [DOI] [Google Scholar]
- 27.Yguel B, Bailey R, Tosh ND, Vialatte A, Vasseur C, Vitrac X, Jean F, Prinzing A. 2011. Phytophagy on phylogenetically isolated trees: why hosts should escape their relatives. Ecol. Lett. 14, 1117–1124. ( 10.1111/j.1461-0248.2011.01680.x) [DOI] [PubMed] [Google Scholar]
- 28.Winkler IS, Mitter C. 2008. The phylogenetic dimension of insect-plant interactions: a review of recent evidence. In Specialization, speciation and radiation: the evolutionary biology of herbivorous insects (ed. Tilmon KJ.), pp. 240–263. Berkeley, CA: University of California Press. [Google Scholar]
- 29.Menken SB, Boomsma JJ, Van Nieukerken EJ. 2010. Large-scale evolutionary patterns of host plant associations in the Lepidoptera. Evolution 64, 1098–1119. ( 10.1111/j.1558-5646.2009.00889.x) [DOI] [PubMed] [Google Scholar]
- 30.Ott D, Rall BC, Brose U. 2012. Climate change effects on macrofaunal litter decomposition: the interplay of temperature, body masses and stoichiometry. Phil. Trans. R. Soc. B 367, 3025–3032. ( 10.1098/rstb.2012.0240) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freschet GT, Aerts R, Cornelissen JHC. 2012. Multiple mechanisms for trait effects on litter decomposition: moving beyond home-field advantage with a new hypothesis. J. Ecol. 100, 619–630. ( 10.1111/j.1365-2745.2011.01943.x) [DOI] [Google Scholar]
- 32.Cadisch G, Giller KE. 1997. Driven by nature: plant litter quality and decomposition. Wallingford, UK: CAB International. [Google Scholar]
- 33.Cornelissen JHC, Quested HM, Logtestijn RSP, Pérez-Harguindeguy N, Gwynn-Jones D, Díaz S, Callaghan TV, Press MC, Aerts R. 2006. Foliar pH as a new plant trait: can it explain variation in foliar chemistry and carbon cycling processes among subarctic plant species and types? Oecologia 147, 315–326. ( 10.1007/s00442-005-0269-z) [DOI] [PubMed] [Google Scholar]
- 34.Freschet GT, Aerts R, Cornelissen JHC. 2012. A plant economics spectrum of litter decomposability. Funct. Ecol. 26, 56–65. ( 10.1111/j.1365-2435.2011.01913.x) [DOI] [Google Scholar]
- 35.Makkonen M, Berg MP, van Logtestijn RSP, van Hal JR, Aerts R. 2012. Do physical plant litter traits explain non-additivity in litter mixtures? A test of the improved microenvironmental conditions theory. Oikos 122, 987–997. ( 10.1111/j.1600-0706.2012.20750.x) [DOI] [Google Scholar]
- 36.Poorter H, Villar R. 1997. The fate of acquired carbon in plants: chemical composition and construction costs. In Plant resource allocation (eds Bazzaz FA, Grace J.), pp. 39–72. San Diego, CA: Academic Press. [Google Scholar]
- 37.Freschet GT, Cornelissen JHC, Van Logtestijn RSP, Aerts R. 2010. Evidence of the ‘plant economics spectrum’ in a subarctic flora. J. Ecol. 98, 362–373. ( 10.1111/j.1365-2745.2009.01615.x) [DOI] [Google Scholar]
- 38.Makkar HPS. 2003. Quantification of tannins in tree and shrub foliage: a laboratory manual. Dordrecht, The Netherlands: Kluwer Academic. [Google Scholar]
- 39.Cornelissen JHC, Cerabolini B, Castro-Díez P, Villar-Salvador P, Montserrat-Martí G, Puyravaud JP, Maestro M, Werger MJA, Aerts R. 2003. Functional traits of woody plants: correspondence of species rankings between field adults and laboratory-grown seedlings? J. Veg. Sci. 14, 311–322. ( 10.1111/j.1654-1103.2003.tb02157.x) [DOI] [Google Scholar]
- 40.Hermant M, Hennion F, Bartish IV, Yguel B, Prinzing A. 2012. Disparate relatives: life histories vary more in genera occupying intermediate environments. Persp. Plant Ecol. Evol. Syst. 14, 283–301. ( 10.1016/j.ppees.2012.02.001) [DOI] [Google Scholar]
- 41.Durka W, Michalski SG. 2012. Daphne: a dated phylogeny of a large European flora for phylogenetically informed ecological analyses. Ecology 93, 2297 ( 10.1890/12-0743.1) [DOI] [Google Scholar]
- 42.Hättenschwiler S, Jørgensen HB. 2010. Carbon quality rather than stoichiometry controls litter decomposition in a tropical rain forest. J. Ecol. 98, 754–763. ( 10.1111/j.1365-2745.2010.01671.x) [DOI] [Google Scholar]
- 43.Cornelissen JHC. 1996. An experimental comparison of leaf decomposition rates in a wide range of temperate plant species and types. J. Ecol. 84, 573–582. ( 10.2307/2261479) [DOI] [Google Scholar]
- 44.Anderson JPE, Domsch KH. 1978. A physiological method for the quantitative measurement of microbial biomass in soils. Soil Biol. Biochem. 10, 215–221. ( 10.1016/0038-0717(78)90099-8) [DOI] [Google Scholar]
- 45.Scheu S. 1992. Automated measurement of the respiratory response of soil microcompartments: active microbial biomass in earthworm faeces. Soil Biol. Biochem. 24, 1113–1118. ( 10.1016/0038-0717(92)90061-2) [DOI] [Google Scholar]
- 46.Beck T, Joergensen RG, Kandeler E, Makeschin F, Nuss E, Oberholzer HR, Scheu S. 1997. An inter-laboratory comparison of ten different ways of measuring soil microbial biomass C. Soil Biol. Biochem. 29, 1023–1032. ( 10.1016/S0038-0717(97)00030-8) [DOI] [Google Scholar]
- 47.Macfadyen A. 1961. Improved funnel-type extractors for soil arthropods. J. Anim. Ecol. 30, 171–184. ( 10.2307/2120) [DOI] [Google Scholar]
- 48.Rosenzweig ML. 1995. Species diversity in space and time. New York, NY: Cambridge University Press. [Google Scholar]
- 49.Cardinale BJ, Wright JP, Cadotte MW, Carroll IT, Hector A, Srivastava DS, Loreau M, Weis JJ. 2007. Impacts of plant diversity on biomass production increase through time because of species complementarity. Proc. Natl Acad. Sci. USA 104, 18 123–18 128. ( 10.1073/pnas.0709069104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cadotte MW, Cardinale BJ, Oakley TH. 2008. Evolutionary history and the effect of biodiversity on plant productivity. Proc. Natl Acad. Sci. USA 105, 17 012–17 017. ( 10.1073/pnas.0805962105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neter J, Wasserman W, Kutner MH. 1985. Multicollinearity, influential observations, and other topics in regression analysis—II. In Applied statistical linear models (ed. Richard D.), pp. 390–393, 2nd edn Homewood, IL: Irwin, Inc. [Google Scholar]
- 52.Blomberg SP, Garland T, Ives AR. 2003. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57, 717–745. ( 10.1111/j.0014-3820.2003.tb00285.x) [DOI] [PubMed] [Google Scholar]
- 53.Münkemüller T, Lavergne S, Bzeznik B, Dray S, Jombart T, Schiffers K, Thuiller W. 2012. How to measure and test phylogenetic signal. Methods Ecol. Evol. 3, 743–756. ( 10.1111/j.2041-210X.2012.00196.x) [DOI] [Google Scholar]
- 54.Kraft NJB, Ackerly DD. 2010. Functional trait and phylogenetic tests of community assembly across spatial scales in an Amazonian forest. Ecol. Monogr. 80, 401–422. ( 10.1890/09-1672.1) [DOI] [Google Scholar]
- 55.Ackerly D. 2009. Conservatism and diversification of plant functional traits: evolutionary rates versus phylogenetic signal. Proc. Natl Acad. Sci. USA 106(Suppl. 2), 19 699–19 706. ( 10.1073/pnas.0901635106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Coq S, Souquet JM, Meudec E, Cheynier V, Hattenschwiler S. 2010. Interspecific variation in leaf litter tannins drives decomposition in a tropical rain forest of French Guiana. Ecology 91, 2080–2091. ( 10.1890/09-1076.1) [DOI] [PubMed] [Google Scholar]
- 57.Gartner TB, Cardon ZG. 2004. Decomposition dynamics in mixed-species leaf litter. Oikos 104, 230–246. ( 10.1111/j.0030-1299.2004.12738.x) [DOI] [Google Scholar]
- 58.Hanson C, Allison S, Bradford M, Wallenstein M, Treseder K. 2008. Fungal taxa target different carbon sources in forest soil. Ecosystems 11, 1157–1167. ( 10.1007/s10021-008-9186-4) [DOI] [Google Scholar]
- 59.Quinn C, Wyant K, Wangeline A, Shulman J, Galeas M, Valdez J, Self J, Paschke M, Pilon-Smits E. 2011. Enhanced decomposition of selenium hyperaccumulator litter in a seleniferous habitat: evidence for specialist decomposers? Plant Soil 341, 51–61. ( 10.1007/s11104-010-0446-7) [DOI] [Google Scholar]
- 60.Ponge JF. 2000. Vertical distribution of Collembola (Hexapoda) and their food resources in organic horizons of beech forests. Biol. Fertil. Soils 32, 508–522. ( 10.1007/s003740000285) [DOI] [Google Scholar]
- 61.Murray PJ, Clegg CD, Crotty FV, de la Fuente Martinez N, Williams JK, Blackshaw RP. 2009. Dissipation of bacterially derived C and N through the micro- and macrofauna of a grassland soil. Soil Biol. Biochem. 41, 1146–1150. ( 10.1016/j.soilbio.2009.02.021) [DOI] [Google Scholar]
- 62.Wright IJ, et al. 2004. The worldwide leaf economics spectrum. Nature 428, 821–827. ( 10.1038/nature02403) [DOI] [PubMed] [Google Scholar]
- 63.Cavender-Bares J, Kozak KH, Fine PVA, Kembel SW. 2009. The merging of community ecology and phylogenetic biology. Ecol. Lett. 12, 693–715. ( 10.1111/j.1461-0248.2009.01314.x) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available in the electronic supplementary material.