Abstract
Preclinical studies show that epoxyeicosatrienoic acids (EETs) regulate cerebrovascular tone and protect against cerebral ischemia. We investigated the relationship between polymorphic genes involved in EET biosynthesis/metabolism, cytochrome P450 (CYP) eicosanoid levels, and outcomes in 363 patients with aneurysmal subarachnoid hemorrhage (aSAH). Epoxyeicosatrienoic acids and dihydroxyeicosatetraenoic acid (DHET) cerebrospinal fluid (CSF) levels, as well as acute outcomes defined by delayed cerebral ischemia (DCI) or clinical neurologic deterioration (CND), were assessed over 14 days. Long-term outcomes were defined by Modified Rankin Scale (MRS) at 3 and 12 months. CYP2C8*4 allele carriers had 44% and 36% lower mean EET and DHET CSF levels (P=0.003 and P=0.007) and were 2.2- and 2.5-fold more likely to develop DCI and CND (P=0.039 and P=0.041), respectively. EPHX2 55Arg, CYP2J2*7, CYP2C8*1B, and CYP2C8 g.36785A allele carriers had lower EET and DHET CSF levels. CYP2C8 g.25369T and CYP2C8 g.36755A allele carriers had higher EET levels. Patients with CYP2C8*2C and EPHX2 404del variants had worse long-term outcomes while those with EPHX2 287Gln, CYP2J2*7, and CYP2C9 g.816G variants had favorable outcomes. Epoxyeicosatrienoic acid levels were associated with Fisher grade and unfavorable 3-month outcomes. Dihydroxyeicosatetraenoic acids were not associated with outcomes. No associations passed Bonferroni multiple testing correction. These are the first clinical data demonstrating the association between the EET biosynthesis/metabolic pathway and the pathophysiology of aSAH.
Keywords: delayed cerebral ischemia, eicosanoid, epoxyeicosatrienoic acid, Modified Rankin Scale, stroke, subarachnoid hemorrhage
Introduction
In the United States, aneurysmal subarachnoid hemorrhage (aSAH) accounts for 5% of all strokes and 25% of stroke-related deaths.1 The outcome for patients with aSAH is often unfavorable with 40% to 50% 1-month mortality rates and ~50% of survivors unable to care for themselves.2 Prognostic factors of unfavorable outcomes after aSAH include the development of clinical neurologic deterioration (CND) and delayed cerebral ischemia (DCI), which typically occurs 3 to 14 days after the hemorrhage.3 Despite this time window for therapeutic intervention, strategies to improve outcomes have had limited success in part due to the lack of reliable and accurate methods to identify high-risk patients.
Previous studies report that eicosanoids derived from the cytochrome P450 (CYP) pathway of arachidonic acid metabolism regulate cerebrovascular tone and structure.4 Arachidonic acid is stored in the phospholipid membranes and released in response to various cellular stimuli including cerebral insults.5 In the brain, free arachidonic acid is oxidized to form epoxyeicosatrienoic acids (EETs) by CYP enzymes found in multiple brain regions.6 Epoxyeicosatrienoic acids consist of four regioselective isoforms (14,15-, 11,12-, 8,9-, and 5,6-EET) formed primarily by CYP2J2 and CYP2C8/9 enzymes.6 Epoxyeicosatrienoic acids dilate cerebral arteries, promote angiogenesis, and inhibit inflammation, apoptosis, and platelet aggregation.4 Epoxyeicosatrienoic acids can be further metabolized to inactive dihydroxyeicosatetraenoic acids (DHETs) by soluble epoxide hydrolase (sEH) present throughout the brain.7 Collectively, these studies suggest that increasing EET levels in the brain may improve the regulation of cerebral blood flow (CBF) and vascular homeostasis.
Moreover, preclinical evidence shows that EETs are important vascular regulators after cerebral injury resulting from ischemic or hemorrhagic stroke. Studies have shown that EET synthesis inhibition reduces baseline CBF in rats.8 Inhibition or gene deletion of sEH reduces infarct size and increases CBF in animal models of temporary focal ischemia.9, 10 In vitro studies show that EETs protect astrocytes against ischemic cell death.11 Also, variants in the EPHX2 gene, which codes for sEH, have been shown to affect sEH activity and neuronal survival after ischemic injury.12 These studies provide evidence that EETs alter CBF and cerebral injury in preclinical models and warrant further investigation in humans.
Given the preclinical evidence that EETs are involved in the pathophysiology of stroke, we investigated the relationship between polymorphic variants in the genes responsible for the formation or metabolism of these products, the resultant EET and DHET levels in cerebrospinal fluid (CSF), acute outcomes, and long-term outcomes in patients with aSAH. Furthermore, we compared genotype frequencies in our aSAH population with those reported in the general population using the Hapmap database to identify potential genetic markers for aSAH.
Materials and methods
Design and Participants
Participants were prospectively recruited from patients admitted to the University of Pittsburgh Medical Center neurovascular intensive care unit. The protocol was approved by the Institutional Review Board at the University of Pittsburgh and informed consent was obtained from the patient or their proxy in accordance with the ethical standards of the University of Pittsburgh Institutional Review Board as well as with the Helsinki Declaration of 1975 (and as revised in 1983). The study included 363 adult patients (age 21 to 75) with aSAH diagnosed via head computed tomography (CT) and classified as Fisher grade >1. Aneurysmal subarachnoid hemorrhage was diagnosed via head CT and angiography was used to determine the presence of aneurysm and location. Patients were not enrolled if they had a history of debilitating neurologic disease or SAH from trauma, mycotic aneurysm, or arteriovenous malformation. The number of patients was estimated based on the expected variances and differences between CYP eicosanoid levels in outcome groups. Cerebrospinal fluid from 269 patients was available for analysis of CYP eicosanoid levels. We limited our genetic analyses to 304 Caucasians in an attempt to address population stratification.13 All patients received standard nursing and medical care in the neurovascular intensive care unit.13
Data/Sample Collection
Sociodemographics, including sex, age, race, and clinical data were collected from the medical record. Severity of injury measured by traditional Fisher grade (hemorrhage burden)14 and Hunt and Hess (HH) score (symptom burden)15 were documented by the attending neurosurgeon on admission. Fisher grade and HH scores have been strongly associated with acute and long-term outcomes, respectively.16 Plasma and CSF samples for genotyping and CSF analysis of CYP eicosanoid levels were collected approximately every 12 hours from consent through day 14 after hemorrhage. External ventricular drains were placed as standard of care in aSAH patients with hydrocephalus and poor or declining neurologic status. Cerebrospinal fluid samples were pulled from the collection bag of existing external ventricular drains (while a drainage device is in place), aliquoted and frozen at −80°C. DNA was extracted from a venous blood sample that and white cells were isolated using a salting out procedure.17 All DNA is stored in 1X TE buffer at 4°C. All analysis was performed by individuals blinded to patient outcomes.
Analysis of Cytochrome P450 Eicosanoid Levels
Chemical standards were purchased from Cayman Chemical Company (Ann Arbor, MI, USA). High purity organic solvents were purchased from VWR (West Chester, PA, USA) and all other chemicals were purchased from Sigma-Aldrich (St Louis, MO, USA). Cerebrospinal fluid samples were withdrawn from collection bags on external ventricular drains approximately every 12 hours during the inpatient stay (up to 14 days). Samples were processed using solid phase extraction (SPE). Sample volumes of 2.0 to 3.0 mL were loaded onto 3 cc Oasis HLB SPE cartridges (Waters, Milford, MA, USA) that were conditioned and equilibrated with 2 mL of MeOH and dH2O, respectively. Columns were washed and eluted with 3 mL of 5% MeOH and 100% MeOH, respectively. Samples were dried under nitrogen gas and were reconstituted in 50 μL of 80:20 MeOH:ddH2O. Quantitation of EETs and DHETs was performed using a previously described UPLC-MS/MS method with minor modifications.18 Concentrations of EETs and DHETs were determined from the standard curve of the ratio of their peak areas to internal standard peak areas of 14,15-EET-d11 and 14,15-DHET-d11, respectively, over a linear range of 0.007 to 8.88 ng/mL.
Genetic Analysis
Candidate genes in the EET metabolic pathway included CYP2C8, CYP2C9, CYP2J2, and EPHX2. Tagging single-nucleotide polymorphisms (tSNPs) were selected using the CEU population from Hapmap database that fully captured the variability in the gene and its flanking regions (Release 27; www.hapmap.org) and criteria included r2>0.8 and minor allele frequency ⩾20%. Functional SNPs (fSNPs) were defined as those previously reported to affect mRNA transcription, protein expression, or enzyme activity in vitro. Known fSNPs were selected regardless of allele frequency, but we excluded those that were not informative enough for analyses (variant genotype frequencies <1% in our aSAH population). Also, rs11572080 and rs71220599 were excluded from the analysis because these SNPs were in 100% linkage disequilibrium with rs10509681 and rs71553864, respectively. Our genetic analysis included 14 tSNPs (rs11572133, rs11572139, rs1934952, rs1934953, rs12772884, rs1934967, rs2253635, rs4086116, rs4918766, rs9332104, rs1155002, rs7515289, rs2071575, and rs7816586), eight fSNPs (rs10509681, rs1058930, rs1799853, rs1057910, rs890293, rs41507953, rs751141, and rs71553864) and one fSNP/tSNP (rs7909236) as shown in Supplementary Table 1. Genotyping was performed using Taqman allele discrimination assay with ABI Prism 7000 Sequence Detection System (Applied Bioscience, Carlsbad, CA, USA) for rs1057910, Affymetrix Human Genome-wide SNP Array 6.0 (Affymetrix, Santa Clara, CA, USA) for rs1934952, and iPLEX MassArray (Sequenom, San Diego, CA, USA) for all other SNPs. Consistency and integrity of genotyping data was checked by inclusion of duplicate CEPH controls on each plate for internal as well as plate-to-plate consistency, using genotype call rate criteria of >85%, comparing the observed and Hapmap Caucasian (CEU) frequencies, and performing checks for Hardy–Weinberg Equilibrium (HWE) consistency. Analyses involving genetic data included both genotype groups (codominant model) and the presence or absence of the variant allele (dominant model) groups. The SNPstats software was used to assign haplotypes for candidate genes and combinations of fSNPs on different candidate genes.19
Outcomes Assessment
Acute outcomes included the presence or absence of CND and DCI during the inpatient stay (up to 14 days).20 Clinical neurologic deterioration was determined by a decline in neurologic exam for >1 hour evidenced by a documented global or focal deficit, an increase (two or more points) in NIH Stroke Scale or a decrease (two or more points) in Glasgow Coma Scale score in the absence of medication administration, fever, seizure, rebleed, increased intracranial pressure, or hydrocephalus. Delayed cerebral ischemia was defined as the presence of CND accompanied by evidence of impaired CBF (simultaneously or within 12 hours predetermination or postdetermination of CND). Impaired CBF was determined using surrogate markers of blood flow including angiography (⩾25% cerebral vessel narrowing) and elevated transcranial Doppler flow velocities (⩾200 cm/s or Lindegaard ratio ⩾3) or CT/magnetic resonance perfusion scans (impaired perfusion or new cerebral infarction). Long-term outcomes were determined by global functional recovery at 3 and 12 months using the Modified Rankin Scale (MRS) obtained during a face-to-face interview or phone call with the patient or their surrogate.
Statistical Analysis
For the purpose of analysis, HH scores were dichotomized into high (3 to 5) and low (1 to 2) groups and MRS scores were dichotomized into favorable (MRS 0 to 2) and unfavorable (MRS 3 to 6) groups. Cytochrome P450 eicosanoid CSF concentrations below the LLQ (lower limit of quantitation) were reported as LLQ/2. Genetic power analysis was calculated based on association between key genotype CYP2C9 and outcome of MRS at 3 months by using web-based software of Genetic Power Calculator developed by Purcell et al.21 Given the sample size of 304 in our study, we are able to achieve 78% power to detect association for SNP of Rs1799853 with 13% risk allele frequencies and genotype relative risk (2.01 for C/T and 4.46 for T/T) with a chi-square test of allelic association at the significant level of 0.05.
Acute and long-term outcomes were compared using Pearson correlation coefficient (r). The mean and maximum CYP eicosanoid levels for each patient were calculated and were used to compare the mean±standard error of the mean (s.e.m.) in the genotype and outcome groups using t-test (with Levene's test for equality of variances) or ANOVA with Bonferroni's post hoc test. To examine homogeneous latent trajectory classes of DHET CSF levels after aSAH, group-based trajectory analysis was performed with the PROC TRAJ macro in SAS version 9.4 as previously described.22 The time range of 2 to 11 days was selected to minimize missing data and the time from hemorrhage was rounded up to the nearest day. Log transformation was applied to reduce sample variation and skewness for better model fitting. The Bayesian Information Criterion and the substantive utility of the classes (e.g., distinctiveness of the trajectories, proportion assigned to a given class) were used to determine the optimal solution for the number of trajectory groups. Mean concentration values from the trajectory analysis were reported as the geometric mean±95% confidence interval (CI). Dihydroxyeicosatetraenoic acid trajectory groups and genotype/allele frequencies were compared with acute outcomes, long-term outcomes, and covariate groups using chi-square analysis or t-test. Also, the relationship between genotype, CYP eicosanoid concentrations/trajectory groups, and outcomes was determined using logistic regression after controlling for covariates such as age, sex, race (for comparison of CYP eicosanoid levels and outcomes), and either Fisher grade (for analyses involving acute outcomes) or HH score (for analyses involving long-term outcomes). Previous studies report that Fisher grade and HH score are strongly associated with acute and long-term outcomes, respectively.23 The cumulative incidence of acute outcomes during the inpatient stay in each genotype group was compared using Kaplan–Meier log-rank analysis and the model was adjusted for covariates using Cox regression. Multivariable Cox proportional hazards regression was used to evaluate associations between CYP2C8*4 allele and survival outcomes controlling for age, gender, and fisher grade. We tested the proportional-hazards assumption using Schoenfeld residuals and the assumptions were satisfied for both models. In addition, CYP eicosanoid levels and outcomes were compared in genetic haplotype groups before and after adjustment for covariates using the SNPstats software.19 Haplotypes with frequencies <2% were excluded from the analysis. Cox models were run in Stata (College Station, TX, USA) version 11. All other statistical analysis was performed using the Statistical Package for the Social Sciences (SPSS) version 21.0 for Windows (SPSS Inc., Chicago, IL, USA). Statistical significance was determined at P values <0.05.
Results
Acute and Long-Term Outcomes
The presence of CND and DCI was detected in 55.1% (n=332) and 37.2% (n=333), respectively. Impaired CBF was detected via angiography, transcranial Doppler flow velocities, transcranial Doppler Lindegaard ratio, and CT/magnetic resonance perfusion scans in 44.3% (n=203), 51.9% (n=337), 81.9% (n=337), and 57.7% (n=196), respectively. The presence of CND was significantly correlated with DCI and unfavorable MRS at 3 and 12 months (r=0.70, r=0.34, r=0.29; P<0.001), respectively. In addition, the presence of DCI was significantly correlated with unfavorable MRS at 3 and 12 months (r=0.22, r=0.29; P<0.001), respectively. Also, unfavorable MRS scores at 3 months were significantly correlated with unfavorable MRS scores at 12 months (r=0.72; P<0.001).
Covariates and Outcomes
Results comparing covariates with acute and long-term outcomes are shown in Table 1. As expected, the severity of injury measured by Fisher grade and HH scores were associated with worse acute and long-term outcomes (P<0.001), respectively. Increased age was associated with the presence of CND (P=0.001) but not DCI and long-term outcomes. Race was associated with better outcomes at 3 months, but not at 12 months, in Caucasians compared with Non-Caucasians (P=0.003). Sex was not associated with acute and long-term outcomes.
Table 1. Covariates and outcomes.
| Covariate | Group | CND (+) N (%) | CND (−) N (%) | OR (95% CI) | P value | DCI (+) N (%) | DCI (−) N (%) | OR (95% CI) | P value |
|---|---|---|---|---|---|---|---|---|---|
| Race | Non-Caucasian | 21 (11.5) | 15 (10.1) | 1.16 (0.57–2.33) | 0.681 | 16 (12.9) | 20 (9.6) | 1.40 (0.70–2.82) | 0.344 |
| Caucasian | 162 (88.5) | 134 (89.9) | 108 (87.1) | 189 (90.4) | |||||
| Gender | Male | 53 (29) | 40 (26.8) | 1.11 (0.69–1.80) | 0.669 | 37 (29.8) | 56 (26.8) | 1.16 (0.71–1.90) | 0.549 |
| Female | 130 (71) | 109 (73.2) | 87 (70.2) | 153 (73.2) | |||||
| Age | Years | 54.4 (11.2) | 50.5 (10.2) | NA | 0.001a | 52.3 (10.9) | 53.3 (11.1) | NA | 0.434 |
| Fisher grade | 2 | 39 (21.3) | 68 (45.9) | NA | <0.001a | 26 (21) | 81 (38.9) | NA | <0.001a |
| 3 | 98 (53.6) | 71 (48) | 67 (54) | 102 (49) | |||||
| 4 | 46 (25.1) | 9 (6.1) | 31 (25) | 25 (12) |
| Covariate | Group | MRS3 (3–6) N (%) | MRS3 (0–2) N (%) | OR (95% CI) | P value | MRS12 (3–6) N (%) | MRS12 (0–2) N (%) | OR (95% CI) | P value |
|---|---|---|---|---|---|---|---|---|---|
| Race | Non-Caucasian | 14 (17.7) | 10 (5.8) | 3.51 (1.48–8.30) | 0.003a | 8 (11.8) | 12 (6.8) | 1.83 (0.71–4.70) | 0.202 |
| Caucasian | 65 (82.3) | 163 (94.2) | 60 (88.2) | 165 (93.2) | |||||
| Gender | Male | 24 (30.4) | 47 (27.2) | 1.17 (0.65–2.10) | 0.599 | 16 (23.5) | 50 (28.2) | 0.78 (0.41–1.50) | 0.456 |
| Female | 55 (69.6) | 126 (72.8) | 52 (76.5) | 127 (71.8) | |||||
| Age | Years | 53.0 (10.9) | 55.0 (10.9) | NA | 0.175 | 53.9 (11.1) | 55.5 (11.6) | NA | 0.326 |
| Hunt and Hess | Favorable (0–2) | 17 (21.5) | 94 (54.3) | 4.34 (2.35–8.02) | <0.001a | 13 (19.1) | 96 (54.2) | 5.01 (2.56–9.83) | <0.001a |
| Unfavorable (3–5) | 62 (78.5) | 79 (45.7) | 55 (80.9) | 81 (45.8) |
CI, confidence interval; CND, clinical neurologic deterioration; DCI, delayed cerebral ischemia; MRS, Modified Rankin Scale score at 3 and 12 months: Unfavorable (3 to 6), Favorable (0 to 2); OR, odds ratio.
Statistical significance established at P<0.05.
Cytochrome P450 Eicosanoid Quantitation
Results showing the percentage of patients and samples with detectable CYP eicosanoid levels and the mean and maximum CYP eicosanoid levels in all patients are shown in Supplementary Table 2. Detectable EET and DHET levels were measured in 64.3% and 98.9% of patients (n=269) and 13.1% and 97.9% of samples (n=3151), respectively. Mean and maximum EET levels were 0.073±0.007 ng/mL and 0.153±0.018 ng/mL, respectively. Mean and maximum DHET levels were 1.271±0.069 ng/mL and 2.462±0.140 ng/mL, respectively.
Genetics Hardy–Weinberg Equilibrium and Hapmap Comparison
Our genetic analysis was limited to Caucasians which represented 89% of patients genotyped. Results of the HWE test and the observed versus Hapmap frequency comparison are shown in Table 2. The genotype frequencies for CYP2C9 g.18470G>A were not in HWE (P=0.003). Also, our aSAH Caucasian population showed different genotype frequencies from those reported in the Hapmap CEU (Caucasian) population for CYP2C8 g.24879A>T (P=0.041), CYP2C9 g.18470G>A (P=0.009), and EPHX2 g.54788T>C (P=0.031). Linkage disquilibrium data are presented in Supplementary Table 3.
Table 2. HWE and observed versus hapmap frequencies.
| Gene SNP (rs#) | Genotype | Observed N (%) | Hapmap N (%) | P value |
|
|---|---|---|---|---|---|
| HWE | Obs-Hap | ||||
| CYP2C8 | A/A | 148 (50.5) | 135 (46.0) | 0.067 | 0.041a |
| g.24879A>Tb | T/A | 111 (37.9) | 137 (46.9) | ||
| (rs11572133) | T/T | 34 (11.6) | 21 (7.1) | ||
| CYP2C9 | G/G | 157 (52.9) | 122 (41.1) | 0.003a | 0.009a |
| g.18470G>Ab | G/A | 103 (34.7) | 138 (46.4) | ||
| (rs4918766) | A/A | 37 (12.5) | 37 (12.5) | ||
| EPHX2 | T/T | 149 (49.3) | 150 (49.6) | 0.666 | 0.031a |
| g.54788T>Cb | C/T | 124 (41.1) | 139 (46) | ||
| (rs2071575) | C/C | 29 (9.6) | 13 (4.4) | ||
| CYP2J2 | T/T | 160 (53.5) | 183 (61.1) | 0.466 | 0.053 |
| g.30354600T>Gb | G/T | 114 (38.1) | 103 (34.5) | ||
| (rs7515289) | G/G | 25 (8.4) | 13 (4.4) | ||
CYP, cytochrome P450; HWE, Hardy–Weinberg Equilibrium; Obs-Hap, observed versus Hapmap frequency comparison; SNP, single-nucleotide polymorphism.
Statistical significance established at P<0.05.
Tagging SNP.
Cytochrome P450 Eicosanoid Levels in Genetic Groups
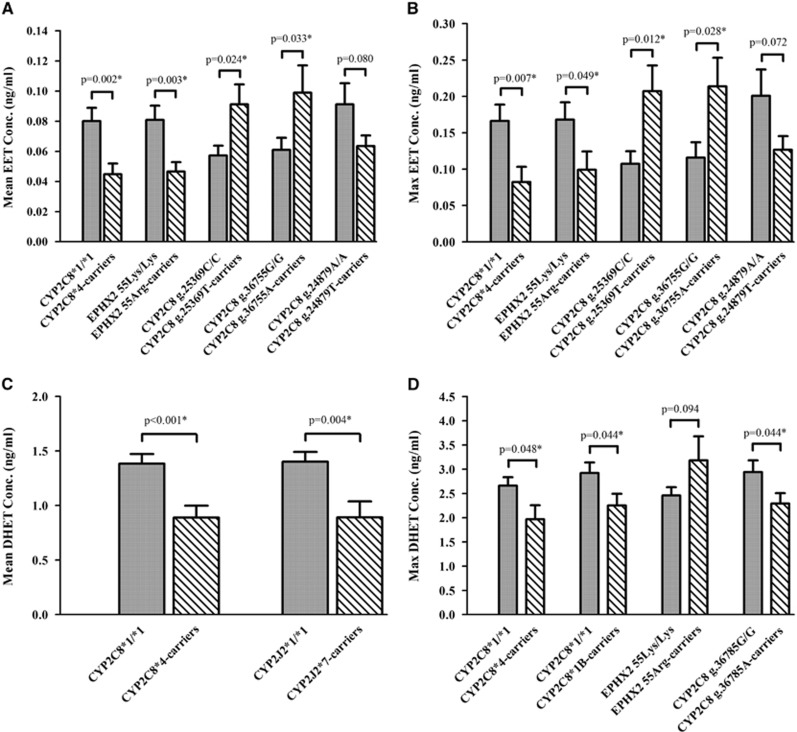
Results comparing CYP eicosanoid CSF levels in genetic groups are shown in Figure 1 and Supplementary Table 4. CYP2C8*4 (g.16136G) allele carriers (n=24) had 44% and 36% lower mean EET and DHET levels when compared with CYP2C8*1/*1 carriers (n=197) (P=0.002 and P<0.001), respectively. Similar results were observed when comparing maximum EET and DHET levels in these genotype groups. EPHX2 55Arg (g.14861G) allele carriers (n=38) had 42% and 41% lower mean and maximum EET levels (P=0.003 and P=0.049, respectively) when compared with EPHX2 55Lys/Lys carriers (n=180). CYP2C8*1B (g.4825A) allele carriers (n=95) had 23% reduction in maximum DHET levels when compared with CYP2C8*1/*1 carriers (n=123) (P=0.044). Also, CYP2J2*7 (g.4930T) allele carriers (n=28) had 36% lower mean DHET levels when compared with CYP2J2*1/*1 carriers (n=193) (P=0.004). In the tSNP analysis, CYP2C8 g.25369T allele carriers (n=119) had 60% and 93% higher mean and maximum EET levels when compared with CYP2C8 g.25369C/C carriers (n=98) (P=0.024 and P=0.012), respectively. CYP2C8 g.36755A allele carriers (n=104) had 49% and 84% higher mean and maximum EET levels when compared with CYP2C8 g.36755G/G carriers (n=71) (P=0.033 and P=0.028), respectively. CYP2C8 g.36785G allele carriers had 22% lower maximum DHET levels compared with CYP2C8 g.36785A/A carriers (P=0.044). When comparing individual genotype groups, CYP2C8 g.36785A/G carriers (n=97) had 29% lower maximum DHET levels compared with CYP2C8 g.36785A/A carriers (n=100) (P=0.023) (Supplementary Figure 1).
Figure 1.
Cytochrome P450 (CYP) eicosanoid levels in genetic groups. Mean epoxyeicosatrienoic acid (EET) (A), maximum EET (B), mean dihydroxyeicosatetraenoic acid (DHET) (C), and maximum DHET (D) levels in cerebrospinal fluid (CSF) from patients with aneurysmal subarachnoid hemorrhage (aSAH) are compared in genotype groups. The mean and maximum EET and DHET CSF levels for each patient were calculated and were used to compare the mean±s.e.m. of the CYP eicosanoid levels for each patient in the variant allele carrier (striped bars) and wild-type (WT) genotype (solid bars) groups using t-test with Welch's correction as appropriate. Genotype groups were compared using ANOVA in Supplementary Figure 1. Statistical significance established at *P<0.05.
The most common haplotype (Haplotype 1) for CYP2C8, CYP2C9, CYP2J2, and EPHX2 was GTCAG (rs7909236, rs11572133, rs11572139, rs1934953, and rs1934952), ATGCGT (rs12772884, rs1934967, rs2253635, rs4086116, rs4918766, and rs9332104), CT (rs1155002 and rs7515289), and TA (rs2071575 and rs7816586) seen in 30.2%, 22.7%, 47.0%, and 39.0% of patients, respectively. CYP2C9 Haplotype 4 (ACGCGT) was present in 14.8% of patients and was associated with 0.44 and 1.06 ng/mL higher mean and maximum DHET levels (P=0.028 and P=0.008), respectively. CYP2C8 Haplotype 2 (GATAA) was present in 29.1% of patients and was associated with 0.03 and 0.1 ng/mL higher mean and maximum EET levels (P=0.019 and P=0.006), respectively. CYP2C8 Haplotype 4 (GACGG) was present in 8.3% of patients and was associated with 0.54 ng/mL and 1.0 ng/mL higher mean and maximum DHET levels (P=0.010 and P=0.018), respectively. EPHX2 Haplotype 4 (CA) present in 2.1% of patients was associated with 2.09 and 3.84 ng/mL higher mean and maximum DHET levels (P<0.001). Similar relationships between CYP eicosanoids and haplotypes were observed after controlling for age, sex, and Fisher grade. There were no differences in CYP eicosanoid levels in haplotype groups when compared with the most common haplotype for CYP2J2.
Genetics and Outcomes
Results comparing the genotype and allele frequencies with acute outcomes are shown in Table 3. Carriers of the CYP2C8*4 (g.16136G) allele were ~2.2- and 2.5-fold more likely to develop DCI (P=0.041) and CND (P=0.039), respectively. Conversely, patients with CYP2J2 g.30345693C/T genotype were ~1.9-fold less likely to develop CND (P=0.025). Supplementary Figure 2 shows patients with the variant CYP2C8*4 (g.16136G) allele had a greater cumulative incidence of DCI (53.1%) and a trend for greater cumulative incidence of CND (71.8%) over 14 days compared with those with CYP2C8*1/*1 genotype (35.3% and 54%, P=0.032 and P=0.052, respectively). These relationships did not change after controlling for clinical covariates. Additionally, multivariable Cox proportional hazards regression was used to evaluate associations between independent variables of interest and survival outcomes. We tested the proportional-hazards assumption using Schoenfeld residuals plot and the assumptions were satisfied for both models (Supplementary Table 5). Table 4 shows the results comparing the genotype and allele frequencies with long-term outcomes. Results show that patients with the EPHX2 404Thr (g.50690-50691insGTC) and CYP2C8*2C (g.8633T) alleles were 2.6- and 2.1-fold more likely to have unfavorable outcomes at 3 months (P=0.040 and P=0.036), respectively. Conversely, patients with CYP2C9 g.816A/G, CYP2J2*1/*7 (g.4930G/T), and EPHX2 287Arg/Gln (g.30221G/A) genotypes were ~2.1-, 3.5-, and 2.6-fold less likely to develop unfavorable outcomes at 12 months (P=0.048, P=0.048, and P=0.027), respectively. Our haplotype analysis showed no differences in outcomes among haplotype groups when compared with the most common haplotype for each candidate gene. Supplementary Table 6 shows a summary of findings for our genetic studies.
Table 3. Genetics and acute outcomes.
| Gene SNP (rs#) | Genotype | CND (+) N (%) | CND (−) N (%) | Unadjusted P value |
Adjusteda |
|
|---|---|---|---|---|---|---|
| OR (95% CI) | P value | |||||
| CYP2C8 | C/C | 137 (85.6) | 117 (92.9) | 0.054 | Reference | — |
| g.16136C>Gb | C/G | 23 (14.4) | 9 (7.1) | 2.48 (1.05–5.89) | 0.039c | |
| Ile264Met [c4] | G/G | 0 (0) | 0 (0) | — | — | |
| (rs1058930) | G-carriers | 23 (14.4) | 9 (7.1) | 0.054 | 2.48 (1.05–5.89) | 0.039c |
| CYP2J2 | C/C | 68 (42.8) | 41 (32.8) | 0.135 | Reference | — |
| g.30345693C>Td | C/T | 69 (43.4) | 69 (55.2) | 0.53 (0.31–0.92) | 0.025c | |
| (rs1155002) | T/T | 22 (13.8) | 15 (12) | 0.96 (0.42–2.19) | 0.922 | |
| T carriers | 91 (57.2) | 84 (67.2) | 0.086 | 0.60 (0.36–1.02) | 0.057 | |
| Gene SNP (rs#) | Genotype | DCI (+) N (%) | DCI (−) N (%) | Unadjusted P value |
Adjusteda |
|
|---|---|---|---|---|---|---|
| OR (95% CI) | P value | |||||
| CYP2C8 | C/C | 90 (81.4) | 165 (91.7) | 0.049c | Reference | — |
| g.16136C>Gb | C/G | 17 (15.9) | 15 (8.3) | 2.21 (1.03–4.73) | 0.041c | |
| Ile264Met [c4] | G/G | 0 (0) | 0 (0) | — | — | |
| (rs1058930) | G-carriers | 17 (15.9) | 15 (8.3) | 0.049c | 2.21 (1.03–4.73) | 0.041c |
CI, confidence interval; CND, clinical neurologic deterioration; CYP, cytochrome P450; DCI, delayed cerebral ischemia; OR, odds ratio; SNP, single-nucleotide polymorphism.
Multivariate analysis included correction for age, sex, and Fisher grade.
Functional SNP.
Statistical significance established at P<0.05.
Tagging SNP.
Table 4. Genetics and long-term outcomes.
| Gene SNP (rs#) | Genotype | MRS3 (3–6) N (%) | MRS3 (0–2) N (%) | Unadjusted P value |
Adjusteda |
|
|---|---|---|---|---|---|---|
| OR (95% CI) | P value | |||||
| CYP2C9 | C/C | 39 (62.9) | 117 (75) | 0.186 | Reference | — |
| g.8633C>Tb | C/T | 22 (35.5) | 38 (24.4) | 2.01 (1.01–3.97) | 0.045c | |
| Arg144Cys [c2C] | T/T | 1 (1.6) | 1 (0.6) | 4.46 (0.23–84.97) | 0.320 | |
| (rs1799853) | T carriers | 23 (37.1) | 39 (25) | 0.074 | 2.06 (1.05–4.04) | 0.036c |
| EXPH2 | DEL/DEL | 51 (81) | 145 (91.8) | 0.021c | Reference | — |
| g. 50690-50691insGTCb | DEL/GTC | 12 (19) | 11 (7) | 2.81 (1.12–7.05) | 0.028c | |
| Thr404del | GTC/GTC | 0 (0) | 2 (1.3) | — | 0.999 | |
| (rs71553864) | GTC carriers | 12 (19) | 13 (8.2) | 0.022c | 2.55 (1.04–6.24) | 0.040c |
| Gene SNP (rs#) | Genotype | MRS12 (3–6) N (%) | MRS12 (0–2) N (%) | Unadjusted P value |
Adjusteda |
|
|---|---|---|---|---|---|---|
| OR (95% CI) | P value | |||||
| CYP2C9 | A/A | 28 (50.9) | 58 (37.9) | 0.034c | Reference | — |
| g.816A>Gd | A/G | 17 (30.9) | 78 (51) | 0.48 (0.23–0.99) | 0.048c | |
| (rs2253635) | G/G | 10 (18.2) | 17 (11.1) | 1.15 (0.44–2.97) | 0.780 | |
| G carriers | 27 (49.1) | 95 (62.1) | 0.093 | 0.61 (0.32–1.18) | 0.140 | |
| CYP2J2 | G/G | 55 (93.2) | 133 (84.7) | 0.037c | Reference | — |
| g.4930G>Tb | G/T | 3 (5.1) | 24 (15.3) | 0.28 (0.08–0.99) | 0.048c | |
| c.-76G>T[c7] | T/T | 1 (1.7) | 0 (0) | — | 1.000 | |
| (rs890293) | T carriers | 4 (6.8) | 24 (15.3) | 0.115 | 0.36 (0.12–1.14) | 0.082 |
| EXPH2 | C/C | 51 (86.4) | 109 (70.3) | 0.047c | Reference | — |
| g.30221G>Ab | T/C | 8 (13.6) | 44 (28.4) | 0.38 (0.16–0.89) | 0.027c | |
| Arg287Gln | T/T | 0 (0) | 2 (1.3) | — | 0.999 | |
| (rs751141) | T carriers | 8 (13.6) | 46 (29.7) | 0.015c | 0.36 (0.15–0.84) | 0.019c |
CI, confidence interval; CYP, cytochrome P450; MRS, Modified Rankin Scale score at 3 and 12 months: Unfavorable (3 to 6), Favorable (0 to 2); OR, odds ratio; SNP, single-nucleotide polymorphism.
Multivariate analysis included correction for age, sex, and Hunt & Hess Score.
Functional SNP.
Statistical significance established at P<0.05.
Tagging SNP.
Cytochrome P450 Eicosanoid Levels in Outcome Groups
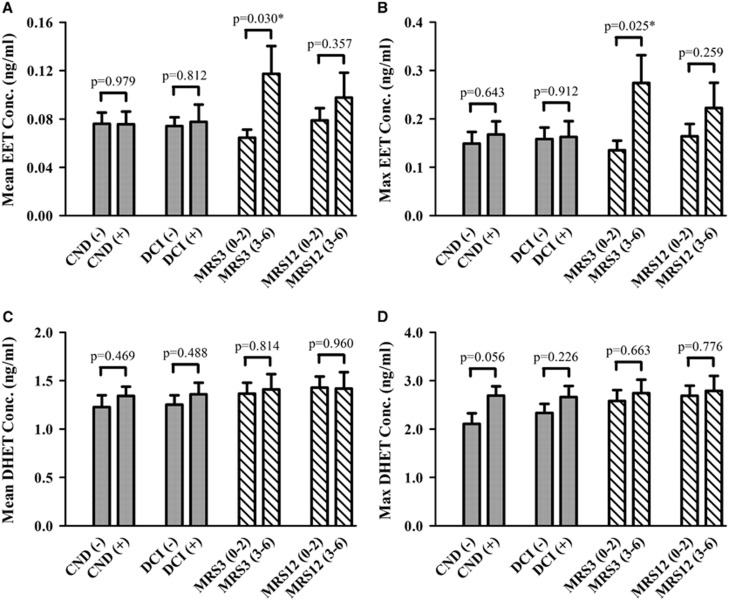
Results comparing CYP eicosanoid CSF levels in outcome groups are shown in Figure 2 and Supplementary Table 7. Mean and maximum EET CSF levels were ~2-fold higher in patients with unfavorable outcomes at 3 months (n=67) when compared with those with favorable outcomes (n=115) (P=0.030 and P=0.025), respectively. Similar relationships were observed in the multivariate analysis. No relationship was observed between mean and maximum CSF levels of individual DHET isomers and outcomes (data not shown). Cytochrome P450 eicosanoid levels in Fisher grade groups are shown in Supplementary Figure 3. Fisher grade was associated with mean EET levels (P=0.014) and maximum EET levels (P=0.012). Patients classified as Fisher grade 4 had higher mean and maximum EET levels (0.104±0.021 ng/mL and 0.227±0.047 ng/mL) compared with those in Fisher grade 2 (0.043±0.003 ng/mL and 0.071±0.010 ng/mL, P=0.010 and P=0.012, respectively).
Figure 2.
Cytochrome P450 (CYP) eicosanoid levels in outcome groups. Mean epoxyeicosatrienoic acid (EET) (A), maximum EET (B), mean dihydroxyeicosatetraenoic acid (DHET) (C), and maximum DHET (D) levels in cerebrospinal fluid (CSF) from patients with aneurysmal subarachnoid hemorrhage (aSAH) are compared in outcomes groups. Acute outcomes (solid bars) included the presence or absence of delayed cerebral ischemia (DCI) and clinical neurologic deterioration (CND) up to 14 days after the hemorrhage. Long-term outcomes (stripped bars) were determined by global functional recovery at 3 and 12 months using the Modified Rankin Scale (MRS) and were dichotomized into favorable (MRS 0 to 2) and unfavorable (MRS 3 to 6) groups. The mean and maximum EET and DHET CSF levels for each patient were calculated and were used to compare the mean±s.e.m. of the CYP eicosanoid levels in outcome groups using t-test with Welch's correction as appropriate. Statistical significance established at *P<0.05.
Trajectory Analysis of Cytochrome P450 Eicosanoid Levels
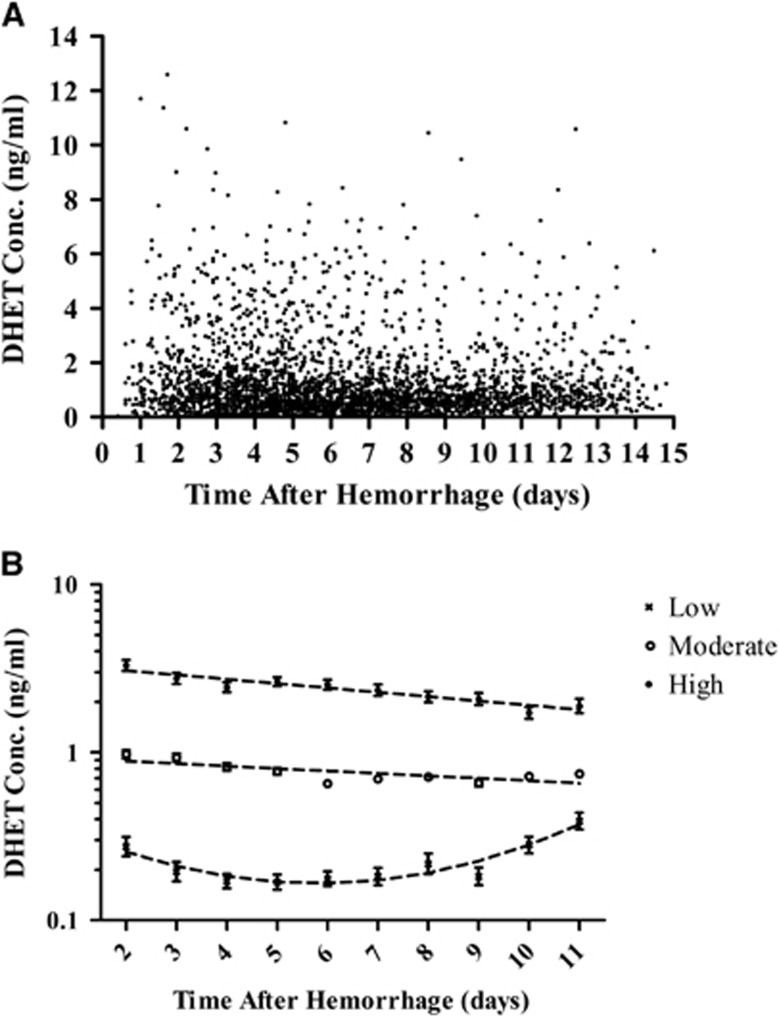
Dihydroxyeicosatetraenoic acid temporal concentration profiles and their relationship with outcomes are shown in Figure 3 and Supplementary Table 8. The trajectory model evaluating DHET CSF levels show three groups of patients with significantly different concentration profiles from days 2 to 11 after aSAH (P<0.001). Dihydroxyeicosatetraenoic acid trajectory groups were not associated with acute or long-term outcomes. Concentration values in trajectory groups are presented as geometric mean with 95% CI. Patients in the ‘low' group (n=45, 16.7%) have relatively low DHET levels that slightly increase over time (day 2: 0.27±1.30 ng/mL; day 11: 0.39±1.26 ng/ml). Patients in the ‘moderate' group (n=159, 59.1%) have relatively moderate DHET levels that slightly decrease over time (day 2: 0.98±1.12 ng/mL; day 11: 0.75±1.09 ng/mL). Patients in the ‘high' group (n=65, 24.2%) have relatively high DHET levels that decrease over time (day 2: 3.30±1.16 ng/mL; day 11: 1.89±1.21 ng/mL). Dihydroxyeicosatetraenoic acid trajectory groups were not associated with acute or long-term outcomes before and after adjusting for clinical covariates. When trajectory patterns of individual DHET isomer concentrations were evaluated, no relationship was observed between the trajectory groups and outcomes (data not shown).
Figure 3.
Cytochrome P450 (CYP) eicosanoid temporal concentration profiles and trajectory patterns. (A) Raw population values of dihydroxyeicosatetraenoic acid (DHET) cerebrospinal fluid (CSF) concentrations (ng/mL) from 269 patients up to 14 days after hemorrhage are shown. (B) DHET CSF concentration versus time from hemorrhage (days) in high (filled circles, n=65 (24.2%)), moderate (open circles, n=159 (59.1%)), and low (X, n=45 (16.7%)) concentration groups as identified by trajectory analysis is shown. Concentration data are presented as geometric mean with 95% confidence interval.
Although statistically significant associations (P<0.05) were noted in this article, none of these associations passed Bonferroni's multiple testing correction criteria.
Discussion
This clinical study is the first to investigate the impact of gene variants in the EET metabolic pathway on CYP eicosanoid CSF levels, risk for aSAH, and subsequent acute/long-term outcomes in aSAH patients. We report that patients with CYP2C8*4 showed lowered EET and DHET levels in CSF and had a greater likelihood of DCI and CND during the inpatient stay. Also, patients with the loss-of-function variants CYP2J2*7 and CYP2C8*1B showed lower EET and DHET levels, respectively, while those with the CYP2C8 g.25369T and 2C8 g.36755A allele had higher EET levels. Patients with the gain-of-function EPHX2 55Arg variant had lower EET levels. Patients classified as Fisher grade 4 (ventricular bleed) had increased EET levels compared with those in the Fisher grade 2 group (bleed <1 mm thick). Dihydroxyeicosatetraenoic acid CSF levels and trajectory patterns were not associated with outcomes. Patients with EPHX2 404del and CYP2C8*2C variants were more likely to have unfavorable long-term outcomes. Conversely, patients with CYP2J2*7, EPHX2 287Gln, and CYP2C9 g.816G>A variants were less likely to develop unfavorable long-term outcomes. Furthermore, we identified tSNPs that were not in HWE and showed genotype frequencies in our aSAH population that were different from those in Hapmap CEU database which may indicate putative genetic markers for aSAH risk.
Our observed relationships between CND, DCI, and MRS scores are similar to previous reports.16 Likewise, we observed previously established relationships between HH score, Fisher grade, race, and increasing age with acute and/or long-term outcomes.24 Caucasians had better 3-month outcomes compared with non-Caucasians as previously reported in patients with aSAH and other stroke subtypes.25 Collectively, these data suggest that the impact of race and age on EET formation/metabolism and outcomes after aSAH may warrant further investigation.
Our genetic analysis showed that multiple gene variants were associated with altered EET and DHET CSF levels. Patients with the CYP2C8*4 allele (11%) have reduced EET and DHET levels in CSF and are more likely to develop DCI and CND. Previous studies report that expression of CYP2C8*4 in vitro results in a ~5-fold reduction in protein levels and enzymatic activity toward arachidonic acid to form 14,15-EET.26, 27 CYP2C8*4 enzymes showed reduced carbon monoxide binding to the heme moiety and were more sensitive to proteinase K digestion indicating improper heme insertion and protein folding.28 Furthermore, human liver samples and microsomes from Caucasians harboring CYP2C8*4 or a haplotype containing CYP2C8*4 showed reduced protein expression.29, 30 Collectively, these studies suggest that CYP2C8*4 is a loss-of-function SNP associated with reduced EET and DHET levels and worse outcomes.
Patients with at least one copy of the variant EPHX2 55Arg had lower mean and maximum EET levels. This reduction in EET concentration is consistent with the findings of Przybyla-Zawislak et al31 who showed that this variant increases sEH enzyme activity in vitro. Similarly, studies report that CYP2J2*7 results in lower mRNA transcription in vitro.32 In this study, we did not observe lower EET levels, but we report lower mean DHET levels in patients with the variant CYP2J2*7 allele. These results are expected since DHETs are considered as a surrogate marker for EETs. We expect worse outcomes in patients with CYP2J2*7, but we observed the opposite trend suggesting that other mediators may be involved. Since in vitro expression of CYP2C8*1B results in increased mRNA transcription compared with CYP2C8*1, we expect that patients with CYP2C8*1B may have higher EET and DHET levels.30 However, CYP2C8*1B was associated with reduced maximum DHET levels in our study possibly due to differences in gene regulation in various species.30 Also, CYP2C8 tSNPs and CYP2C8, CYP2C9, and EPHX2 haplotypes were associated with altered CYP eicosanoid levels. These data suggest that there are multiple SNPs on our candidate genes, potentially including SNPs that were not investigated in this study, which may affect EET synthesis and metabolism.
Results revealed that patients heterozygous for CYP2C9*2 (21%) are 2-fold more likely to develop unfavorable outcomes at 3 months. Lundbad et al33 reported that expression of CYP2C9*4 in vitro result in a 33% reduction in EET formation and that incubations of human liver microsomes from patients homozygous for CYP2C9*2 and CYP2C8*3 led to a 34% decrease in EET formation.33 Moreover, human liver samples from two patients heterozygous for CYP2C9*2 showed 5- to 10-fold greater mRNA expression of 144Cys over 144Arg.34 Collectively, these studies suggest that CYP2C9*2 is a loss-of-function SNP associated with reduced EET and DHET levels and worse outcomes.
We report that patients with the EPXH2 287Gln variant (25%) are ~2.6-fold less likely to develop unfavorable outcomes at 12 months. Previous studies report that in vitro expression of the EPHX2 (sEH) 287Gln enzyme resulted in ~5-fold reduction in 14,15-EET hydrolysis, higher sEH monomer:dimer ratio, and reduced enzyme stability.31, 35, 36 Rat neuronal cell cultures and cardiomyocytes from sEH knockout mice transduced with the EPHX2 287Gln enzyme reduced 14,15-DHET levels to 34% and ~40% of WT values after administration of excess 14,15-EET and reduced ischemic cell death to 80% and 78% of values for untreated cells, respectively.12, 37 Moreover, clinical studies report that the EPHX2 287Gln variant was associated with reduced epoxyoctadecenoic acid (EpOME) metabolism and higher cholesterol and triglyceride levels in plasma.38, 39 These findings suggest that the EPHX2 287Gln variant is a loss-of-function SNP associated with reduced EET metabolism and ischemic cell death. Therefore, it is expected that patients with EPHX2 287Gln would have increased EET levels, reduced DHET levels, and favorable outcomes. This hypothesis is consistent with our results regarding long-term outcomes.
Although multiple fSNPs and tSNPs were associated with acute and long-term outcomes, candidate gene haplotypes were not associated with outcomes. This discrepancy may be due to reduced power when using haplotypes or because the causative variant shows different linkage disequilibrium with haplotypes compared with tSNPs. These data suggest that additional studies are needed using a more focused genotype approach and improved characterization of candidate gene haplotypes.
Hardy–Weinberg Equilibrium tests show that CYP2C9 g.18470G>A is not in HWE and, similar to other tSNPs in our candidate genes, showed genotype frequencies in our aSAH population that were different from those in the Hapmap CEU population. Given that our population is enriched for individuals with aSAH, these results suggest that genetic variation in the EET metabolic pathway may contribute to the formation and rupture of intracranial aneurysms. Support for these results includes numerous in vivo studies which show that EETs exhibit diverse physiologic functions including vasodilation of the cerebrovasculature and antihypertensive effects.6, 40 In vitro studies report that EETs exhibit proliferative, migratory, angiogenic, fibrinolytic, antiapoptotic, antiinflammatory, and antiplatelet aggregation effects in vascular endothelial cells and inhibit migration and apoptosis in vascular smooth muscle cells.4 On the basis of these reported mechanisms of action, it is expected that EETs have an important role in vascular homeostasis and remodeling and may impact intracranial aneurysm formation, rupture, or recovery after aSAH.
We showed the ability to measure DHET, and to a lesser extent EET, levels in CSF from patients with aSAH. Our previous studies showed room temperature stability of EET and DHET analytes during the collection period of 12 hours14 and studies in our laboratory have showed reproducibility of sample quantification over years of −80°C freezer storage (data not shown). Although EET CSF levels in patients with aSAH have not been previously reported in clinical studies, our DHET levels were consistent with our previous studies.18, 41 Epoxyeicosatrienoic acid and DHET CSF levels measured in this study are comparable to EET levels reported to show physiologic effects on cerebrovascular tissues. Epoxyeicosatrienoic acids have been shown to relax isolated cerebral arteries and increase the activity of large conductance calcium-dependent potassium channels (BKCa) in cerebral vascular muscle cells at 1 nmol/L (0.32 ng/mL).42 It is expected that the CYP eicosanoid levels in the brain will be higher than those reported in this study because these compounds act locally in an autocrine/paracrine manner4 and then are diluted in CSF. In addition, any spikes in concentration will be diluted in the CSF drainage bags during the collection period.
Our analysis of CYP eicosanoids showed that DHET CSF levels and trajectory patterns were not associated with acute or long-term outcomes, but increased EET CSF levels were associated with unfavorable 3-month outcomes. This relationship was not expected due to the antiinflammatory, vasodilatory, and neuroprotective properties of EETs.4 However, increased EET levels were associated with Fisher grade 4, which is reported to be strongly associated with worse acute and long-term outcomes.24 These data suggest that the degree and location of the hemorrhage may affect EET formation and release in the brain and possibly long-term outcomes. Red blood cells can release EETs directly from phospholipid membranes or synthesize EETs through the release and subsequent metabolism of arachidonic acid from phospholipid membranes.43 However, it is unknown whether an increase in EET CSF levels is due to EET release or synthesis directly from the blood in the CSF or from brain tissues. Also, it is important to note that this relationship was not observed in acute outcomes and 12-month outcomes and that the analysis did not include the measurement of vasoconstrictors, such as 20-HETE (20-hydroxyeicosatetraenoic acid), which may counteract the effects of EETs in the brain. The role of the EET metabolic pathway in aSAH has not been previously investigated in clinical studies and therefore warrants further study.
In spite of these novel findings, several limitations of this study should be noted. Although this work is the first to show a relationship between genetic polymorphisms, CYP eicosanoid levels, and outcomes in aSAH patients, it is important to note that these findings did not pass correction criteria for multiple testing. Therefore, these results will be important for validation by inclusion in either future meta-analyses or design of multicenter trial to thoroughly evaluate this relationship. The analysis of CYP eicosanoid CSF levels was not compared to control values due to lack of access to control CSF. Since the CSF samples were taken from drainage bags that collected CSF over a 12-hour time period, reported CYP eicosanoid levels represent time-averaged values which may be lower than those at the site of action. Also, there was a significantly lower number of CSF samples with EET levels above the quantitation limit when compared with samples with detectable DHET levels possibly due to EET conversion to DHETs or reduced recovery from the collection bag. In our genetic analysis, some functional genetic variants in the EET metabolic pathway were not genotyped. Many SNPs had allele frequencies that were too low for informative analysis. Our analyses were limited to single fSNP analyses, thus the effects of multiple fSNPs simultaneously were not assessed. It is not possible to determine whether our putative genetic markers were causative or were in linkage disequilibrium with the causative SNP. Furthermore, this study represents one of the first reports of a large population of aSAH patients for genotype/CYP eicosanoid analysis, therefore, a validation cohort is not readily available for confirmation of these genotype findings. Future studies that include a focused genotype approach, validation cohort are needed.
Conclusions
In summary, EETs have been shown to have an important role in the regulation of cerebrovascular tone and vessel remodeling in vitro and significantly affect CBF and cerebral ischemic injury in vivo. Other studies report that SNPs in the genes responsible for the formation or metabolism of EETs alter enzyme expression or activity. This study, involving one of the largest aSAH cohorts to date, suggests that gene variants involved in EET formation/metabolism are associated with the risk for complications after aSAH and subsequent long-term outcomes. These results are important to help elucidate the mechanisms involved in the pathogenesis and pathophysiology of aSAH and possibly identify patients at high risk for unfavorable outcomes so that intervention strategies may be implemented earlier or more aggressively in these patients.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Journal of Cerebral Blood Flow & Metabolism website (http://www.nature.com/jcbfm)
Author Contributions
All authors contributed to multiple aspects of the this manuscript including study design, sample collection, data collection, data analysis, interpretation of data, writing of the manuscript, and the decision to submit the paper for publication. All authors have approved the final article is true.
Sources of research support for this study include NIH National Institute of Nursing Research (NINR) for grants R01NR0044339-05 and F31NR012608-01 and the National Center of Research Resources for grant S10RR023461.
Supplementary Material
References
- Ferro JM, Canhao P, Peralta R. Update on subarachnoid haemorrhage. J Neurol. 2008;255:465–479. doi: 10.1007/s00415-008-0606-3. [DOI] [PubMed] [Google Scholar]
- Rose MJ. Aneurysmal subarachnoid hemorrhage: an update on the medical complications and treatments strategies seen in these patients. Curr Opin Anaesthesiol. 2011;24:500–507. doi: 10.1097/ACO.0b013e32834ad45b. [DOI] [PubMed] [Google Scholar]
- Dorsch NW, King MT. A review of cerebral vasospasm in aneurysmal subarachnoid haemorrhage Part I: Incidence and effects. J Clin Neurosci. 1994;1:19–26. doi: 10.1016/0967-5868(94)90005-1. [DOI] [PubMed] [Google Scholar]
- Imig JD, Simpkins AN, Renic M, Harder DR. Cytochrome P450 eicosanoids and cerebral vascular function. Exp Rev Mol Med. 2011;13:e7. doi: 10.1017/S1462399411001773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer RM, Stephenson DT, Roberts EF, Clemens JA. Cytosolic phospholipase A2 (cPLA2) and lipid mediator release in the brain. J Lipid Mediat Cell Signal. 1996;14:3–7. doi: 10.1016/0929-7855(96)01501-5. [DOI] [PubMed] [Google Scholar]
- Fleming I. Cytochrome p450 and vascular homeostasis. Circ Res. 2001;89:753–762. doi: 10.1161/hh2101.099268. [DOI] [PubMed] [Google Scholar]
- Sura P, Sura R, Enayetallah AE, Grant DF. Distribution and expression of soluble epoxide hydrolase in human brain. J Histochem Cytochem. 2008;56:551–559. doi: 10.1369/jhc.2008.950659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkayed NJ, Birks EK, Hudetz AG, Roman RJ, Henderson L, Harder DR. Inhibition of brain P-450 arachidonic acid epoxygenase decreases baseline cerebral blood flow. Am J Physiol. 1996;271:H1541–H1546. doi: 10.1152/ajpheart.1996.271.4.H1541. [DOI] [PubMed] [Google Scholar]
- Zhang W, Koerner IP, Noppens R, Grafe M, Tsai HJ, Morisseau C, et al. Soluble epoxide hydrolase: a novel therapeutic target in stroke. J Cereb Blood Flow Metab. 2007;27:1931–1940. doi: 10.1038/sj.jcbfm.9600494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Otsuka T, Sugo N, Ardeshiri A, Alhadid YK, Iliff JJ, et al. Soluble epoxide hydrolase gene deletion is protective against experimental cerebral ischemia. Stroke. 2008;39:2073–2078. doi: 10.1161/STROKEAHA.107.508325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Alkayed NJ. Hypoxic preconditioning and tolerance via hypoxia inducible factor (HIF) 1alpha-linked induction of P450 2C11 epoxygenase in astrocytes. J Cereb Blood Flow Metab. 2005;25:939–948. doi: 10.1038/sj.jcbfm.9600085. [DOI] [PubMed] [Google Scholar]
- Koerner IP, Jacks R, DeBarber AE, Koop D, Mao P, Grant DF, et al. Polymorphisms in the human soluble epoxide hydrolase gene EPHX2 linked to neuronal survival after ischemic injury. J Neurosci. 2007;27:4642–4649. doi: 10.1523/JNEUROSCI.0056-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallek M, Alexander S, Crago E, Sherwood P, Horowitz M, Poloyac S, et al. Endothelin-1 and endothelin receptor gene variants and their association with negative outcomes following aneurysmal subarachnoid hemorrhage. Biol Res Nurs. 2012;15:390–397. doi: 10.1177/1099800412459674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher CM, Kistler JP, Davis JM. Relation of cerebral vasospasm to subarachnoid hemorrhage visualized by computerized tomographic scanning. Neurosurgery. 1980;6:1–9. doi: 10.1227/00006123-198001000-00001. [DOI] [PubMed] [Google Scholar]
- Hunt WE, Hess RM. Surgical risk as related to time of intervention in the repair of intracranial aneurysms. J Neurosurg. 1968;28:14–20. doi: 10.3171/jns.1968.28.1.0014. [DOI] [PubMed] [Google Scholar]
- Frontera JA, Fernandez A, Schmidt JM, Claassen J, Wartenberg KE, Badjatia N, et al. Defining vasospasm after subarachnoid hemorrhage: what is the most clinically relevant definition. Stroke. 2009;40:1963–1968. doi: 10.1161/STROKEAHA.108.544700. [DOI] [PubMed] [Google Scholar]
- Hong MG, Pawitan Y, Magnusson PK, Prince JA. Strategies and issues in the detection of pathway enrichment in genome-wide association studies. Hum Genet. 2009;126:289–301. doi: 10.1007/s00439-009-0676-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller TM, Donnelly MK, Crago EA, Roman DM, Sherwood PR, Horowitz MB, et al. Rapid, simultaneous quantitation of mono and dioxygenated metabolites of arachidonic acid in human CSF and rat brain. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:3991–4000. doi: 10.1016/j.jchromb.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sole X, Guino E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- Crago EA, Thampatty BP, Sherwood PR, Kuo CW, Bender C, Balzer J, et al. Cerebrospinal fluid 20-HETE is associated with delayed cerebral ischemia and poor outcomes after aneurysmal subarachnoid hemorrhage. Stroke. 2011;42:1872–1877. doi: 10.1161/STROKEAHA.110.605816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- Nagin DS, Tremblay RE. Analyzing developmental trajectories of distinct but related behaviors: a group-based method. Psychol Methods. 2001;6:18–34. doi: 10.1037/1082-989x.6.1.18. [DOI] [PubMed] [Google Scholar]
- Rosen DS, Macdonald RL. Subarachnoid hemorrhage grading scales: a systematic review. Neurocrit Care. 2005;2:110–118. doi: 10.1385/NCC:2:2:110. [DOI] [PubMed] [Google Scholar]
- de Rooij NK, Greving JP, Rinkel GJ, Frijns CJ. Early prediction of delayed cerebral ischemia after subarachnoid hemorrhage: development and validation of a practical risk chart. Stroke. 2013;44:1288–1294. doi: 10.1161/STROKEAHA.113.001125. [DOI] [PubMed] [Google Scholar]
- Ayala C, Greenlund KJ, Croft JB, Keenan NL, Donehoo RS, Giles WH, et al. Racial/ethnic disparities in mortality by stroke subtype in the United States, 1995-1998. Am J Epidemiol. 2001;154:1057–1063. doi: 10.1093/aje/154.11.1057. [DOI] [PubMed] [Google Scholar]
- Gao Y, Liu D, Wang H, Zhu J, Chen C. Functional characterization of five CYP2C8 variants and prediction of CYP2C8 genotype-dependent effects on in vitro and in vivo drug-drug interactions. Xenobiotica. 2010;40:467–475. doi: 10.3109/00498254.2010.487163. [DOI] [PubMed] [Google Scholar]
- Smith HE, Jones JP, 3rd, Kalhorn TF, Farin FM, Stapleton PL, Davis CL, et al. Role of cytochrome P450 2C8 and 2J2 genotypes in calcineurin inhibitor-induced chronic kidney disease. Pharmacogenet Genomics. 2008;18:943–953. doi: 10.1097/FPC.0b013e32830e1e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Ting JG, Pan Y, Teh LK, Ismail R, Ong CE. Functional role of Ile264 in CYP2C8: mutations affect haem incorporation and catalytic activity. Drug Metab Pharmacokinet. 2008;23:165–174. doi: 10.2133/dmpk.23.165. [DOI] [PubMed] [Google Scholar]
- Naraharisetti SB, Lin YS, Rieder MJ, Marciante KD, Psaty BM, Thummel KE, et al. Human liver expression of CYP2C8: gender, age, and genotype effects. Drug Metab Dispos. 2010;38:889–893. doi: 10.1124/dmd.109.031542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Antona C, Niemi M, Backman JT, Kajosaari LI, Neuvonen PJ, Robledo M, et al. Characterization of novel CYP2C8 haplotypes and their contribution to paclitaxel and repaglinide metabolism. Pharmacogenomics J. 2008;8:268–277. doi: 10.1038/sj.tpj.6500482. [DOI] [PubMed] [Google Scholar]
- Przybyla-Zawislak BD, Srivastava PK, Vazquez-Matias J, Mohrenweiser HW, Maxwell JE, Hammock BD, et al. Polymorphisms in human soluble epoxide hydrolase. Mol Pharmacol. 2003;64:482–490. doi: 10.1124/mol.64.2.482. [DOI] [PubMed] [Google Scholar]
- Spiecker M, Darius H, Hankeln T, Soufi M, Sattler AM, Schaefer JR, et al. Risk of coronary artery disease associated with polymorphism of the cytochrome P450 epoxygenase CYP2J2. Circulation. 2004;110:2132–2136. doi: 10.1161/01.CIR.0000143832.91812.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad MS, Stark K, Eliasson E, Oliw E, Rane A. Biosynthesis of epoxyeicosatrienoic acids varies between polymorphic CYP2C enzymes. Biochem Biophys Res Commun. 2005;327:1052–1057. doi: 10.1016/j.bbrc.2004.12.116. [DOI] [PubMed] [Google Scholar]
- Bhasker CR, Miners JO, Coulter S, Birkett DJ. Allelic and functional variability of cytochrome P4502C9. Pharmacogenetics. 1997;7:51–58. doi: 10.1097/00008571-199702000-00007. [DOI] [PubMed] [Google Scholar]
- Srivastava PK, Sharma VK, Kalonia DS, Grant DF. Polymorphisms in human soluble epoxide hydrolase: effects on enzyme activity, enzyme stability, and quaternary structure. Arch Biochem Biophys. 2004;427:164–169. doi: 10.1016/j.abb.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Sandberg M, Hassett C, Adman ET, Meijer J, Omiecinski CJ. Identification and functional characterization of human soluble epoxide hydrolase genetic polymorphisms. J Biol Chem. 2000;275:28873–28881. doi: 10.1074/jbc.M001153200. [DOI] [PubMed] [Google Scholar]
- Merkel MJ, Liu L, Cao Z, Packwood W, Young J, Alkayed NJ, et al. Inhibition of soluble epoxide hydrolase preserves cardiomyocytes: role of STAT3 signaling. American journal of physiology. Heart Circ Physiol. 2010;298:H679–H687. doi: 10.1152/ajpheart.00533.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JP, Yang SH, Kim DK, Lee H, Kim B, Cho JY, et al. In vivo activity of epoxide hydrolase according to sequence variation affects the progression of human IgA nephropathy. Am J Physiol. 2011;300:F1283–F1290. doi: 10.1152/ajprenal.00733.2010. [DOI] [PubMed] [Google Scholar]
- Sato K, Emi M, Ezura Y, Fujita Y, Takada D, Ishigami T, et al. Soluble epoxide hydrolase variant (Glu287Arg) modifies plasma total cholesterol and triglyceride phenotype in familial hypercholesterolemia: intrafamilial association study in an eight-generation hyperlipidemic kindred. J Hum Genet. 2004;49:29–34. doi: 10.1007/s10038-003-0103-6. [DOI] [PubMed] [Google Scholar]
- Pratt PF, Medhora M, Harder DR. Mechanisms regulating cerebral blood flow as therapeutic targets. Curr Opin Investig Drugs. 2004;5:952–956. [PubMed] [Google Scholar]
- Poloyac SM, Reynolds RB, Yonas H, Kerr ME. Identification and quantification of the hydroxyeicosatetraenoic acids, 20-HETE and 12-HETE, in the cerebrospinal fluid after subarachnoid hemorrhage. J Neurosci Methods. 2005;144:257–263. doi: 10.1016/j.jneumeth.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Campbell WB, Fleming I. Epoxyeicosatrienoic acids and endothelium-dependent responses. Pflugers Arch. 2010;459:881–895. doi: 10.1007/s00424-010-0804-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H. Erythrocyte-derived epoxyeicosatrienoic acids. Prostaglandins Other Lipid Mediat. 2007;82:4–10. doi: 10.1016/j.prostaglandins.2006.05.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.