Abstract
MicroRNAs (miRNAs) play a key role in cancer progression by coordinately repressing target genes involved in cell proliferation, migration, and invasion. miRNAs regulate gene expression by repressing translation or directing sequence-specific degradation of complementary mRNA. Here, we report that expression of miR-1280 is significantly suppressed in human melanoma specimens when compared with nevi, and in human melanoma cell lines when compared with cultured normal human melanocytes. The proto-oncogene Src was identified as a target of miR-1280 action. Levels of Src expression were significantly higher in melanoma samples and cell lines than in nevi and normal melanocytes. miR-1280 overexpression significantly suppressed the luciferase activity of reporter plasmids containing the full-length 3′ untranslated region of Src. miR-1280-mediated suppression of Src led to substantial decreases in melanoma cell proliferation, cell cycle progression, invasion, as well as induced melanoma cell apoptosis. The effects of miR-1280 overexpression on melanoma cell proliferation and growth were reversed by Src overexpression. Intratumoral delivery of miR-1280 significantly suppressed melanoma cell growth in vivo. Our results demonstrate a novel role for miR-1280 as a tumor suppressor in melanoma, identify the Src signaling pathway as a target of miR-1280 action, and suggest a potential therapeutic role for miR-1280 in melanoma.
Introduction
Melanoma is the sixth most common cancer in the United States, and one of the leading causes of death from skin cancer. Each year, more than 70,000 patients are diagnosed with melanoma in the United States, and around 9,500 die from the disease.1 Melanomas are genetically complex malignancies characterized by dysregulation of multiple signaling and tumor suppressor/oncogene pathways, including B-Raf proto-oncogen, serine/threonine kinase, neuroblastoma RAS viral oncogene homolog, and phosphatase and tensin homolog.2,3 The global incidence of melanoma continues to rise faster than any other malignancy, and despite considerable research efforts, no curative therapy is available for advanced metastatic melanoma.
MicroRNAs (miRNAs) are small, ~18–24 nt, endogenously synthesized nonprotein-coding sequences thought to regulate >90% of human genes.4 miRNAs regulate gene expression by complementary base pairing with the 3′ untranslated region (UTR) of target mRNAs, causing their degradation,5 or by directly mediating mRNA degradation.6 miRNAs are expressed in a tissue-specific manner and are central regulators of gene expression. They can act either as oncomirs by targeting tumor suppressors, or as tumor suppressors by targeting oncogenes.7,8 Inactivation of oncogenic miRNAs9,10 or restoration of tumor-suppressor miRNAs11,12,13 may have great potential for cancer treatment. Due to their tremendous regulatory potential and tissue-specific and disease-specific expression patterns, there is increasing evidence that miRNA expression profiles could be indicative of disease risk or burden.14,15
Src family kinases are nonreceptor cytoplasmic protein mediators of signal transduction which act as proto-oncogenes by mediating tumor cell proliferation, adhesion, angiogenesis, as well as invasion and metastasis.16 Src kinase activity is elevated in a variety of cancers17,18 and has been linked with the progression of different human cancers,17,19,20 including melanoma.21 Src inhibition has preclinical activity in melanoma cell lines,22 and inhibition of Src-kinase decreases levels of phosphorylated Stat3 and promotes melanoma cell apoptosis.23
This report describes, for the first time, a functional role for miR-1280 in melanoma progression, identifies the proto-oncogene Src as a target of miR-1280 action, and suggests its potential therapeutic role.
Results
miR-1280 expression in melanoma tissues and cell lines
miRNA qRT-PCR of nevus (n = 24) and melanoma (n = 37) samples indicated that miR-1280 expression is significantly (P < 0.005) downregulated in melanomas when compared with nevus samples (Figure 1a). The clinicopathological characteristics of the melanoma and nevus cohort are presented in Supplementary Table S1. In addition, miR-1280 expression was substantially downregulated in a panel of melanoma cell lines when compared with normal human melanocytes (Figure 1b). This analysis demonstrated the downregulation of miR-1280 in melanoma specimens and cell lines, thereby indicating a possible tumor suppressor role .
Figure 1.
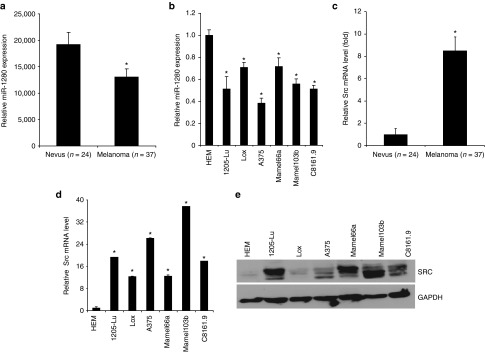
Suppression of miR-1280 expression in melanoma is accompanied by Src overexpression. (a, b) miRNA-qRT-PCR analysis showing expression of miR-1280 expression in a cohort of melanomas (n = 37) and nevi (n = 24) and in a panel of melanoma cell lines and normal human melanocytes (HEM). (c, d) Src mRNA expression in a cohort of melanomas (n = 37) and nevi (n = 24), and in a panel of melanoma cell lines and normal human melanocytes (HEM). (e) Src protein expression in melanoma cell lines and normal human melanocyte. (b, d) Data presented reflect mean ± SEM of three replicates. *P < 0.05.
Src oncogene as a target of miR-1280
To identify potential effectors of miR-1280, we used in silico algorithm and sequence alignments to predict its mRNA targets. This analysis identified Src as a putative target, as the seed sequence of miR-1280 was complementary to the 3′UTR of Src (Figure 2a). To investigate the association between expression of miR-1280 and of Src, we determined Src expression in the same tissue samples and panel of cell lines. Src expression was significantly higher in melanoma tissues when compared with nevus samples (Figure 1c). Similarly, Src expression at the mRNA and protein levels was higher in melanoma cells when compared with the normal human melanocyte line, although the absolute level of expression varied among different melanoma cell lines (Figure 1d,e). Thus, the downregulation of miR-1280 in melanoma is accompanied by overexpression of Src, which is known to play an important oncogenic role with pleiotropic effects on multiple signaling pathways involved in tumor cell proliferation, migration and invasion.16,24
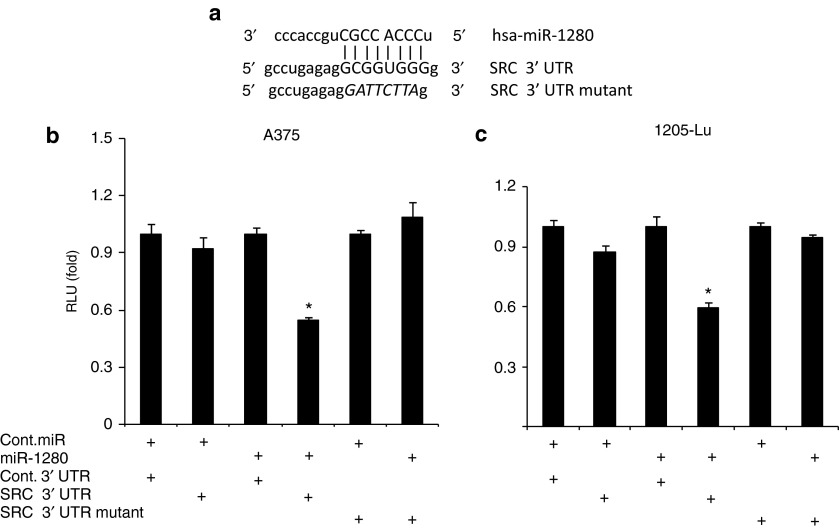
Figure 2.
Src as a target of miR-1280. (a) The miR-1280 seed sequence is complementary to the 3′UTR of Src. (b, c) Luciferase assays showing reporter activity after cotransfection of Src-3′UTR or Src- 3′UTR Mutant with miR-1280 in A375 and 1205-Lu cells, respectively. (b, c) Data presented reflect mean ± SEM of three independent experiments. *P < 0.05.
To assess Src as a functional target of miR-1280, we cotransfected a full-length Src-3′UTR-luciferase expression vector along with miR-1280 into A375 and 1205-Lu human melanoma cells. This treatment resulted in a statistically significant decrease in reporter gene expression when compared with the control 3′UTR vector (Figure 2b,c). However, cotransfection with a vector containing a mutated Src-3′UTR site had no effect on reporter gene activity (Figure 2b,c). These results indicate that the conserved nucleotides in the 3′UTR of Src are responsible for miR-1280 targeting in vitro.
Effects of miR-1280 overexpression on melanoma progression
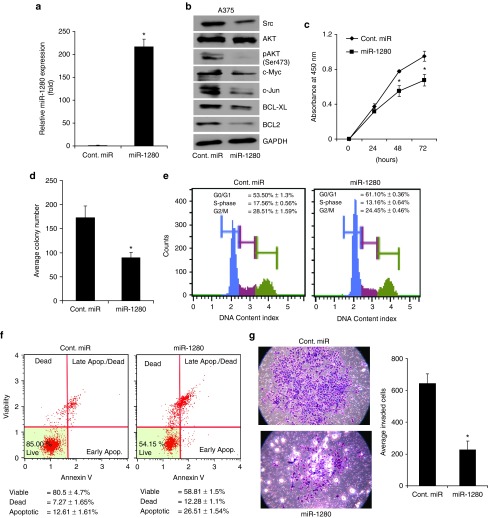
Transient overexpression of miR-1280 (Figure 3a) suppressed Src at the protein level, with a concomitant suppression of downstream genes in the Src signaling pathway (Figure 3b). Thus, we observed a substantial decrease in the expression of AKT, MYC, JUN, BCL-XL, and BCL2 in A375 melanoma cells following miR-1280 overexpression (Figure 3b). Suppression of endogenous miR-1280 in normal human melanocytes by anti-miR-1280 resulted in induction of Src expression (Supplementary Figure S1), confirming miR-1280-mediated regulation of Src in both melanocytes and melanoma cells. Overexpression of miR-1280 in A375 cells substantially suppressed tumor cell proliferation (Figure 3c) when compared with cells expressing a control miRNA (cont. miR). The miR-1280-transfected A375 cells showed lower colony formation ability, as both the size and number of foci were reduced when compared to control miR-expressing cells (Figure 3d). Cell cycle analysis revealed a statistically significant increase in the G0/G1 phase and a decrease in the S-phase of A375 cells overexpressing miR-1280 when compared with control miR (Figure 3e). miR-1280 overexpression induced apoptosis in A375 cells when compared with control miR (Figure 3f). Furthermore, miR-1280 overexpression suppressed the invasive capability of A375 cells (Figure 3g). To confirm the effects of miR-1280 overexpression, miR-1280 was transfected into 1205-Lu human melanoma cells. As shown in Supplementary Figure S2a–g, similar decreases in cell proliferation, colony formation and S-phase, together with an increase in apoptosis, were observed in 1205-Lu cells transfected with miR-1280. These results confirm the phenotypic effects of miR-1280 overexpression in human melanoma cells.
Figure 3.
miR-1280 suppresses Src and inhibits A375 melanoma cell proliferation, colony formation, and invasion, and induces apoptosis. (a) Relative miR-1280 expression levels in A375 cells following transfection with miR-1280 as determined by miR qRT-PCR. (b) Western blot analysis showing miR-1280-mediated effect on Src expression and its downstream targets. (c) The proliferative ability of A375 cells after miR-1280 transfection (d) miR-1280 overexpression significantly inhibits the colony formation ability of A375 melanoma cells. (e) Cell cycle analysis of A375 cells after transfection of miR-1280. (f) Apoptotic index of A375 cells after miR-1280 overexpression when compared with cont.miR-expressing cells. (g) miR-1280 overexpression significantly suppressed the invasive ability of A375 melanoma cell lines. (c–g) Data presented reflect mean ± SEM of three independent experiments. *P < 0.05.
The role of Src in mediating the effects of miR-1280
To further explore the role of Src as a target of miR-1280, we cotransfected A375 cells with miR-1280 and a Src expression vector, and examined their effects on melanoma cell survival. Cotransfection of miR-1280 and an empty vector control resulted in suppression of Src and its downstream targets pAKT(Ser473), BCL2 and BCL-XL, accompanied by suppression in A375 melanoma cell survival and colony formation ability (Figure 4a–c). These effects were largely reversed following cotransfection of miR-1280 and a vector expressing Src cDNA (Figure 4a–c). These results indicate that the effects of miR-1280 on melanoma proliferation appear to be mediated largely by its inhibition of Src expression.
Figure 4.
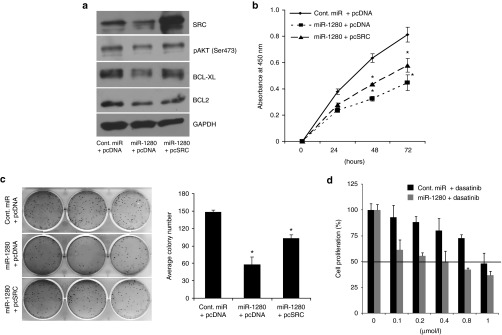
Src overexpression reverses miR-1280-mediated effects. Effects of cotransfection of Src, along with miR-1280 on gene expression (a), cell proliferation (b), and colony formation (c) ability of A375 melanoma cells. (d) Effects of miR-1280 in combination with the Src inhibitor dasatinib on melanoma cell proliferation. (b, d) Data presented reflect mean ± SEM of three independent experiments. *P < 0.05.
Combined effects of a Src inhibitor and miR-1280
Next, we determined if overexpression of miR-1280 in combination with a Src inhibitor (dasatinib) resulted in increased antiproliferative effects. As shown in Figure 4d, A375 cells transfected with miR-1280 were 2.5-fold more sensitive to dasatinib treatment than cont.-miR-transfected cells, indicating that miR-1280 sensitizes melanoma cell lines to dasatinib treatment.
Intratumoral delivery of miR-1280 suppresses tumor growth in vivo
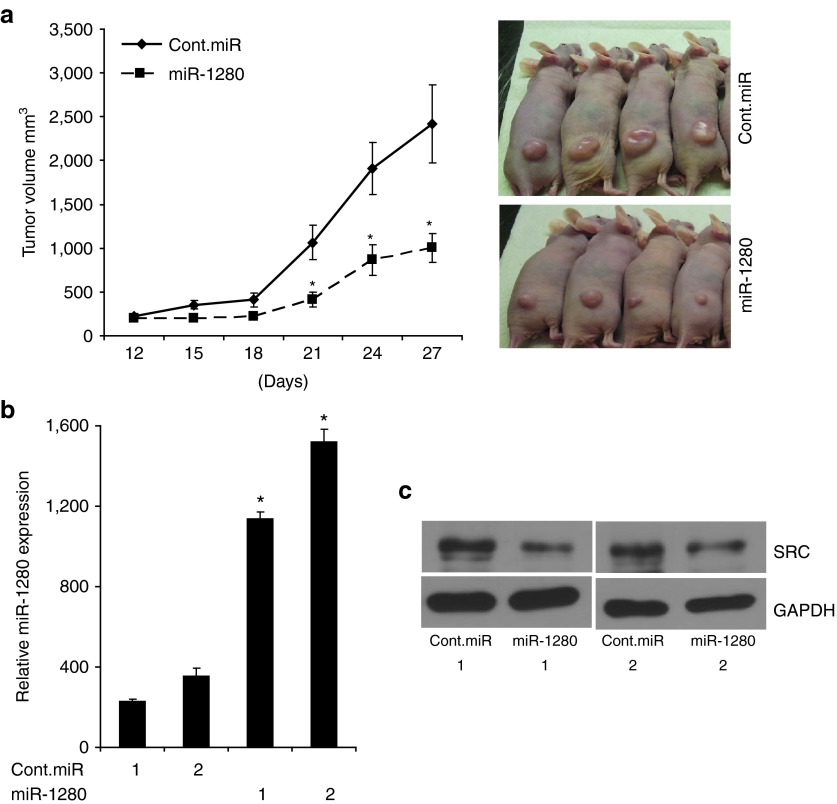
To study the effects of miR-1280 on melanoma progression in vivo, we used an intratumoral delivery approach.25 A375 cells (1 × 106) were injected subcutaneously into nude mice, and miR-1280 or control miRNA, complexed with the siPORT transfection reagent, was injected intratumorally every 2 days once palpable tumors were detected (usually on day 12) until day 27, after which the mice were killed due to the presence of large tumors in the control group. Intratumoral injection of miR-1280 significantly suppressed tumor cell growth when compared with control miR (Figure 5a). We then determined the expression of miR-1280 and Src in harvested tumors. A significant increase in the expression of miR-1280 was observed in miR-1280-treated tumors when compared with controls, with a corresponding decrease in Src expression (Figure 5b,c). These findings further confirm the tumor suppressor effects of miR-1280 in a xenograft model of melanoma.
Figure 5.
Intratumoral delivery of miR-1280 suppresses melanoma cell growth. (a) Tumor volume following intratumoral delivery of miR-1280 or cont. miRNA into palpable tumors generated by subcutaneous injection of A375 cells. (b) Expression of miR-1280 in tumors injected with cont. miRNA or miR-1280. (c) Western blot analysis of Src expression in tumors injected with cont. miRNA or miR-1280. In panel A data presented reflect mean ± SEM of 10 mice in each group. *P < 0.05.
Discussion
The Src nonreceptor tyrosine kinase and the structurally homologous Src family kinases are known to promote cell proliferation, survival, motility, and angiogenic potential, all of which support the neoplastic phenotype.17,26 Elevated kinase activity of c-Src proto-oncogenes is found in a number of tumors, and Src inhibition has recently emerged as a target of drug therapy. Increased Src expression and activity has been reported in melanoma cell lines and in melanoma tumors in vivo.21,27 Src expression has been shown to be regulated by small molecule inhibitors such as dasatinib and bosutinib. However, the regulation of Src by miRNAs in melanoma has not been explored in detail.
In this study, we examined both the regulation of the Src pathway by miR-1280 in melanoma, as well as its functional significance. miR-1280 is a largely unstudied miRNA, other than a previous study in bladder cancer.28 As a result, little is known of its potential function in cancer. Here, we report suppression of miR-1280 and demonstrate its role as a tumor suppressor in melanoma. We observed miR-1280 to be downregulated in melanomas when compared with nevi, as well as in melanoma cell lines when compared with cultured melanocytes. In silico algorithm and sequence alignment identified the Src proto-oncogene as a possible target. Our results demonstrated that miR-1280 directly targets the 3′UTR of Src, as its overexpression was associated with suppression of luciferase activity in a reporter plasmid driven by the Src-3′UTR.
In addition, a significant downregulation of Src protein levels was observed following miR-1280 overexpression, indicating the post-transcriptional regulation of Src via targeting its 3′UTR. Src has been reported to be expressed at high levels in malignant melanoma,29 whereas its expression is lower in benign melanocytic lesions or normal skin,30 suggesting a role for Src in the development and/or progression of melanoma. Src is a proto-oncogene with demonstrated functions in tumor formation and progression.16,24 Our results suggest silencing of miR-1280 as a possible mechanism for Src overexpression in melanoma. Recently, Src was also reported to be targeted by miR-31 in melanoma.31 Src expression is frequently elevated in a variety of cancers, including breast, pancreatic, and lung cancer, as well as melanoma,21,32,33,34 and may therefore represent a promising molecular target for anticancer therapy. Due to increased activity of Src in melanoma, a phase II study using the Src inhibitor saracatinib (AZD0530) was performed in melanoma, although no objective clinical responses were observed.35 Other Src inhibitors including dasatinib and bosutinib have also been used to target Src. Suppression of Src by tyrosine kinase inhibitors caused retardation of melanoma cell growth and reduced Stat3 activation.36 However, they are multitargeted inhibitors, and higher concentrations are needed to produce antiproliferative effects in melanoma.30 miRNA-mediated targeting of Src alone or in combination with Src inhibitors provides a novel alternative approach to regulate its activity.
Src has been reported to be expressed at higher levels in malignant melanoma than nonmelanoma skin cancers such as basal cell carcinoma and squamous cell carcinoma.29 Accordingly, we observed Src to be overexpressed in melanoma cell lines when compared with melanocytes, and in melanomas when compared with nevi. Additional studies identified Src as a direct functional target of miR-1280. miR-1280-mediated suppression of Src was accompanied by suppression of downstream targets of Src and induction of antiproliferative and proapoptotic effects. Mechanistic studies showed that the tumor suppressor effects of miR-1280 were mediated, in large part, by inhibition of Src expression. Src inhibitor-mediated effects were phenocopied by miR-1280 overexpression, consonant with prior studies documenting Src-mediated regulation of cell motility and invasion.16 miR-1280 overexpression also sensitized melanoma cell lines to the Src inhibitor dasatinib. The in vivo intratumoral delivery of miR-1280 into subcutaneous xenografts significantly reduced tumor burden. The tumors receiving miR-1280 showed significantly higher (>fivefold) expression of miR-1280, accompanied by a >60% reduction in Src expression when compared to the control miRNA group, thus, highlighting the role of miR-1280 in Src-mediated tumor progression in vivo. The intratumoral delivery of miR-1280 also reduced the tumor burden by >60%, underscoring the therapeutic potential of miR-1280 in melanoma. Given the propensity of melanoma to recur locoregionally as satellite or intransit metastases, intralesional approaches to melanoma therapy, including gene therapy approaches, have been actively pursued in the management of melanoma patients with locally recurrent disease.37,38,39 To date, one other study has described a role for intralesional therapy of a miRNA (miR-205)40 for melanoma therapy.
In conclusion, our study demonstrates that miR-1280 is significantly suppressed in melanoma. miR-1280 overexpression results in downregulation of Src, suppression of the proliferative and invasive ability of melanoma cells, and induction of melanoma cell apoptosis. Overall, these studies describe a promising therapeutic role for overexpressing miR-1280 in melanoma, which appears to act at least in part by mimicking pharmacological inhibitors of Src.
Materials and Methods
Cell culture, plasmids, and transfection. Melanoma cell lines Lox (provided by Dr. Oystein Fodstad), A375 (ATCC, Manassas, VA), Mamel66a and Mamel103b (provided by Dr. Dirk Schadendorf) were grown in RPMI with 5% fetal bovine serum. 1205-Lu (Coriell Institute, Camden, NJ) were grown in TU-2% media. C8161.9 cells (obtained from Dr. Danny Welch) were grown in DMEM/F12 with 5% fetal bovine serum (Life Technologies, Carlsbad, CA). Normal human melanocytes were grown in LL-0027 media (Lifeline cell technology, Walkersville MD). All cells were cultivated at 37 °C in an atmosphere containing 5% CO2. Plasmids pcDNA3.1 (Life Technologies), pcDNA-SRC (Addgene, Cambridge, MA), pEZX-MT01 miRNA 3′-UTR target expression clones for Src (HmiT017696-MT01) and miRNA target clone control vector for pEZX-MT01 (CmiT000001-MT01) (GeneCopoeia, Rockville, MD) were purchased. Mutated Src 3′UTR sequences complementary to miR-1280 were cloned in the pmirGLO-dual luciferase vector (Promega, Madison, WI). miRNA precursors and anti-miRNA for miR-1280 and controls were purchased from Life Technologies. Transient transfections were carried out by Lipofectamine-2000 (Life Technologies) according to the manufacturer's protocol. Src inhibitor dasatinib was purchased from Chemietek (Indianapolis, IN).
Quantitative real-time PCR. Mature miRNAs and mRNAs were assayed using the TaqMan MicroRNA Assays and Gene Expression Assays, respectively, in accordance with the manufacturer's instructions (Life Technologies). All RT reactions, including no-template controls and RT minus controls, were run in a 7500 Fast real-time PCR system (Life Technologies). RNA concentrations were determined with a NanoDrop (Thermo Scientific, Rockford, IL). Samples were normalized to RNU48 for miRNAs or HPRT1 for mRNAs (Life Technologies) as indicated. Gene expression levels were quantified using the 7500 Fast real-time sequence detection system software (Life Technologies). Comparative real-time PCR was performed in triplicate, including no-template controls. Relative expression was calculated using the comparative Ct method.
RNA and miRNA extraction from tissue samples and cell lines. Samples from melanoma patients (n = 37) and benign nevi (n = 24) were obtained under a protocol approved by the California Pacific Medical Center Institutional Review Board (CPMC IRB# 2009.112EXP, Approved on April 2014, Expires April 2015) Informed consent was obtained from each patient for this study. RNA was extracted by using RNeasy Mini Kit (Qiagen, Valencia, CA) following the manufacturer's protocol. miRNAs were extracted by using the mirVana miRNA extraction kit (Life Technologies) from tissues and cell lines following the manufacturer's instructions.
Cell viability, colony formation, and cell cycle analysis. Cells were plated in 96-well plates at a density of 3 × 103 cells per well. Cell viability was assessed at 24, 48, and 72 hours post-transfection using Cell Counting Kit-8 (Dojindo, Rockville, MD) following the manufacturer's protocol. For the colony-formation assay, 200 cells were plated in a six-well plate in triplicates and allowed to grow till visible colonies appeared. Colonies were stained with Giemsa and counted. Cell cycle analysis was performed as described previously.41
Western blot analysis. Cell lysates were prepared in phosphate-buffered saline containing 1× Halt protease inhibitor cocktail and 1× Halt phosphatase inhibitor cocktail (Pierce, Rockford, IL) centrifuged at 3500 rpm for 10 minutes at 4 °C. Proteins (10–15 ug) from each sample were subjected to sodium dodecyl sulfate/polyacrylamide gel electrophoresis (PAGE) and transferred onto a nitrocellulose membrane. Target proteins were detected by using specific antibodies against SRC, AKT, pAKT (Ser473), c-Myc, c-JUN, BCL-XL, GAPDH (Cell Signaling Technology, Danvers, MA), and BCL2 (Santa Cruz Biotechnology, Santa Cruz, CA).
Luciferase assays. For reporter assays, cells were transiently transfected with Src-3′UTR or cont. 3′UTR along with miR-1280. Firefly luciferase activities were measured by using the Dual Luciferase Assay (Promega) 24 hour after transfection and the results were normalized with Renilla luciferase. Each reporter plasmid was transfected at least three times and each sample was assayed in triplicate.
In vivo intratumoral delivery of miR-1280. The antitumor effect of miR-1280 was determined by local administration of miR-1280 precursor in established tumors as described previously.25 Each mouse was injected subcutaneously with 1.0 × 106 A375 melanoma cells. Once palpable tumors developed (average volume of 200 mm3), 6.25 μg of synthetic miRNA complexed with 1.6 μl siPORT Amine transfection reagent (Ambion) in 20 μl phosphate-buffered saline was delivered intratumorally at 2-day intervals. All animal care was in accordance with institutional guidelines.
Statistical analysis. All quantified data represent an average of at least triplicate samples or as indicated. Error bars represent standard error of the mean. Statistical significance was determined by the Student's t-test and two-tailed P values < 0.05 were considered significant.
SUPPLEMENTARY MATERIAL Figure S1. Anti-miR-1280 induces Src expression in normal human melanocytes. Figure S2. Src-mediated effects of miR-1280 on 1205-Lu melanoma cell proliferation, apoptosis, and invasion. Table S1. Clinico-pathologic characteristics of primary melanoma patient cohort and nevus cohort.
Acknowledgments
The authors declare no conflict of interest. This work was supported by United States Public Health Service Grants CA114337 and CA122947, by the Mary R. and Joseph R. Payden Foundation, and by the T. Robert and Katherine Burke Fund.
Supplementary Material
References
- Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- Sharpless E, Chin L. The INK4a/ARF locus and melanoma. Oncogene. 2003;22:3092–3098. doi: 10.1038/sj.onc.1206461. [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Ambros V, Chen X. The regulation of genes and genomes by small RNAs. Development. 2007;134:1635–1641. doi: 10.1242/dev.002006. [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43:371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setién F, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- Lujambio A, Calin GA, Villanueva A, Ropero S, Sánchez-Céspedes M, Blanco D, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Fuchs E. MicroRNA-mediated control in the skin. Cell Death Differ. 2010;17:229–235. doi: 10.1038/cdd.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels CL, Tsongalis GJ. MicroRNAs: novel biomarkers for human cancer. Clin Chem. 2009;55:623–631. doi: 10.1373/clinchem.2008.112805. [DOI] [PubMed] [Google Scholar]
- Kim LC, Song L, Haura EB. Src kinases as therapeutic targets for cancer. Nat Rev Clin Oncol. 2009;6:587–595. doi: 10.1038/nrclinonc.2009.129. [DOI] [PubMed] [Google Scholar]
- Irby RB, Yeatman TJ. Role of Src expression and activation in human cancer. Oncogene. 2000;19:5636–5642. doi: 10.1038/sj.onc.1203912. [DOI] [PubMed] [Google Scholar]
- Kumble S, Omary MB, Cartwright CA, Triadafilopoulos G. Src activation in malignant and premalignant epithelia of Barrett's esophagus. Gastroenterology. 1997;112:348–356. doi: 10.1053/gast.1997.v112.pm9024288. [DOI] [PubMed] [Google Scholar]
- Aligayer H, Boyd DD, Heiss MM, Abdalla EK, Curley SA, Gallick GE. Activation of Src kinase in primary colorectal carcinoma: an indicator of poor clinical prognosis. Cancer. 2002;94:344–351. doi: 10.1002/cncr.10221. [DOI] [PubMed] [Google Scholar]
- Trevino JG, Summy JM, Lesslie DP, Parikh NU, Hong DS, Lee FY, et al. Inhibition of SRC expression and activity inhibits tumor progression and metastasis of human pancreatic adenocarcinoma cells in an orthotopic nude mouse model. Am J Pathol. 2006;168:962–972. doi: 10.2353/ajpath.2006.050570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homsi J, Cubitt C, Daud A. The Src signaling pathway: a potential target in melanoma and other malignancies. Expert Opin Ther Targets. 2007;11:91–100. doi: 10.1517/14728222.11.1.91. [DOI] [PubMed] [Google Scholar]
- Buettner R, Mesa T, Vultur A, Lee F, Jove R. Inhibition of Src family kinases with dasatinib blocks migration and invasion of human melanoma cells. Mol Cancer Res. 2008;6:1766–1774. doi: 10.1158/1541-7786.MCR-08-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu G, Bowman T, Huang M, Shivers S, Reintgen D, Daud A, et al. Roles of activated Src and Stat3 signaling in melanoma tumor cell growth. Oncogene. 2002;21:7001–7010. doi: 10.1038/sj.onc.1205859. [DOI] [PubMed] [Google Scholar]
- Wheeler DL, Iida M, Dunn EF. The role of Src in solid tumors. Oncologist. 2009;14:667–678. doi: 10.1634/theoncologist.2009-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majid S, Dar AA, Saini S, Arora S, Shahryari V, Zaman MS, et al. miR-23b represses proto-oncogene Src kinase and functions as methylation-silenced tumor suppressor with diagnostic and prognostic significance in prostate cancer. Cancer Res. 2012;72:6435–6446. doi: 10.1158/0008-5472.CAN-12-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtneidge SA, Fumagalli S, Koegl M, Superti-Furga G, Twamley-Stein GM. The Src family of protein tyrosine kinases: regulation and functions. Development (Cambridge, England) 1993. pp. 57–64. [PubMed]
- Barnekow A, Paul E, Schartl M. Expression of the c-src protooncogene in human skin tumors. Cancer Res. 1987;47:235–240. [PubMed] [Google Scholar]
- Majid S, Dar AA, Saini S, Shahryari V, Arora S, Zaman MS, et al. MicroRNA-1280 inhibits invasion and metastasis by targeting ROCK1 in bladder cancer. PLoS One. 2012;7:e46743. doi: 10.1371/journal.pone.0046743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Pyon JK, Kim DW, Lee SH, Nam HS, Kim CH, et al. Elevated c-Src and c-Yes expression in malignant skin cancers. J Exp Clin Cancer Res. 2010;29:116. doi: 10.1186/1756-9966-29-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homsi J, Cubitt CL, Zhang S, Munster PN, Yu H, Sullivan DM, et al. Src activation in melanoma and Src inhibitors as therapeutic agents in melanoma. Melanoma Res. 2009;19:167–175. doi: 10.1097/CMR.0b013e328304974c. [DOI] [PubMed] [Google Scholar]
- Asangani IA, Harms PW, Dodson L, Pandhi M, Kunju LP, Maher CA, et al. Genetic and epigenetic loss of microRNA-31 leads to feed-forward expression of EZH2 in melanoma. Oncotarget. 2012;3:1011–1025. doi: 10.18632/oncotarget.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxbury MS, Ito H, Zinner MJ, Ashley SW, Whang EE. Inhibition of SRC tyrosine kinase impairs inherent and acquired gemcitabine resistance in human pancreatic adenocarcinoma cells. Clin Cancer Res. 2004;10:2307–2318. doi: 10.1158/1078-0432.ccr-1183-3. [DOI] [PubMed] [Google Scholar]
- Jallal H, Valentino ML, Chen G, Boschelli F, Ali S, Rabbani SA. A Src/Abl kinase inhibitor, SKI-606, blocks breast cancer invasion, growth, and metastasis in vitro and in vivo. Cancer Res. 2007;67:1580–1588. doi: 10.1158/0008-5472.CAN-06-2027. [DOI] [PubMed] [Google Scholar]
- Masaki T, Igarashi K, Tokuda M, Yukimasa S, Han F, Jin YJ, et al. pp60c-src activation in lung adenocarcinoma. Eur J Cancer. 2003;39:1447–1455. doi: 10.1016/s0959-8049(03)00276-4. [DOI] [PubMed] [Google Scholar]
- Gangadhar TC, Clark JI, Karrison T, Gajewski TF. Phase II study of the Src kinase inhibitor saracatinib (AZD0530) in metastatic melanoma. Invest New Drugs. 2013;31:769–773. doi: 10.1007/s10637-012-9897-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortylewski M, Jove R, Yu H. Targeting STAT3 affects melanoma on multiple fronts. Cancer Metast Rev. 2005;24:315–327. doi: 10.1007/s10555-005-1580-1. [DOI] [PubMed] [Google Scholar]
- Dummer R, Rochlitz C, Velu T, Acres B, Limacher JM, Bleuzen P, et al. Intralesional adenovirus-mediated interleukin-2 gene transfer for advanced solid cancers and melanoma. Mol Ther. 2008;16:985–994. doi: 10.1038/mt.2008.32. [DOI] [PubMed] [Google Scholar]
- Kaufman HL, Kim DW, DeRaffele G, Mitcham J, Coffin RS, Kim-Schulze S. Local and distant immunity induced by intralesional vaccination with an oncolytic herpes virus encoding GM-CSF in patients with stage IIIc and IV melanoma. Ann Surg Oncol. 2010;17:718–730. doi: 10.1245/s10434-009-0809-6. [DOI] [PubMed] [Google Scholar]
- Weide B, Derhovanessian E, Pflugfelder A, Eigentler TK, Radny P, Zelba H, et al. High response rate after intratumoral treatment with interleukin-2: results from a phase 2 study in 51 patients with metastasized melanoma. Cancer. 2010;116:4139–4146. doi: 10.1002/cncr.25156. [DOI] [PubMed] [Google Scholar]
- Noguchi S, Iwasaki J, Kumazaki M, Mori T, Maruo K, Sakai H, et al. Chemically modified synthetic microRNA-205 inhibits the growth of melanoma cells in vitro and in vivo. Mol Ther. 2013;21:1204–1211. doi: 10.1038/mt.2013.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar AA, Zaika A, Piazuelo MB, Correa P, Koyama T, Belkhiri A, et al. Frequent overexpression of Aurora Kinase A in upper gastrointestinal adenocarcinomas correlates with potent antiapoptotic functions. Cancer. 2008;112:1688–1698. doi: 10.1002/cncr.23371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.