Abstract
Background
Numerous experimental studies suggest that B-type natriuretic peptide (BNP) is cardioprotective, yet in clinical studies, higher plasma BNP concentrations have been associated with incident cardiovascular disease and higher left ventricular mass (LVM). Genetic association studies may allow us to determine the true causal directions without confounding by compensatory mechanisms.
Methods and Results
We performed meta-analysis of two genome-wide association (GWA) results from a total of 2,790 African Americans. We assumed an additive genetic model in association analysis of imputed 2.5 million SNP dosages with residuals generated from multivariable-adjusted logarithmically-transformed BNP controlling for relevant covariates and population stratification. Two loci were genome-wide significant, a candidate gene locus NPPB (rs198389, p-value=1.18×10−09) and novel missense variant in the KLKB1 locus (rs3733402, p-value=1.75×10−11) that explained 0.4% and 1.9% of variation in log BNP concentration, respectively. The observed increase in BNP concentration was proportional to the number of effect allele copies, an average of 8.1 pg/dl increase associated with two allele copies. SNPs in this loci were subsequently cross-checked with GWA results for the aldosterone-to-renin ratio in individuals of European ancestry, and only rs3733402 was genome-wide significant (p<5.0×10−8), suggesting possible shared genetic architecture for these two pathways. Other statistically significant relations for these SNPs included: rs198389 with systolic blood pressure in blacks (COGENT consortium) rs198389 and rs3733402 with LVM in whites (EchoGEN consortium).
Conclusions
These findings improve our knowledge of the genetic basis of BNP variation in African Americans, demonstrate possible shared allelic architecture for BNP with ARR and motivate further studies of underlying mechanisms.
Keywords: genetic association, genetics, association studies, Genome Wide Association Study, genotype
Introduction
B-type natriuretic peptide (BNP) is a member of the natriuretic peptide hormone family, characterized by a common 17 amino-acid ring and shared biological function (i.e., their effect on the cardiovascular system). BNP is secreted from the cardiac ventricles in response to volume expansion and pressure overload.1 It acts on various tissues e.g. the blood vessels and the kidneys to induce vasodilatation, natriuresis and diuresis.2 Paradoxically, plasma BNP concentrations have been associated with hypertension, diastolic dysfunction and/or renal dysfunction and congestive heart failure3 in clinical and population-based studies. These observations appear to result from compensatory mechanisms.
Plasma BNP concentration is moderately heritable4, which suggests a multifactorial inheritance. From candidate genetic studies, common variants in the Natriuretic Peptide Precursor Gene B (NPPB) have been reported to be associated, not only with plasma BNP concentrations, but also with BP and hypertension5,6 type 2 diabetes7, 8, echocardiographic indices and diastolic dysfunction and congestive heart failure.9 Thus, in elucidating the genetic architecture of BNP concentration, we are likely to understand the biological basis of these associations. Recently, Del Greco FM et al10 in the first ever GWA study discovered and replicated a genome-wide significant signal in the MTHFR-CLCN6-NPPA-NPPB gene cluster associated with N-Terminal-proBNP (the inactive form of plasma BNP that is also secreted by cardiac myocytes)11, 12 in populations of European ancestry. Previously, Benjamin et al13 reported a genome-wide significant locus associated with circulating BNP concentration in the Framingham Heart Study, although that finding has not been replicated. Overall, there is compelling evidence that genetic factors play a key role in the regulation of BNP concentrations. Except for one study which found that corin serine protease cleaves proANP (pro-atrial natriuretic peptide) and proBNP converting them into biologically active peptide hormones in populations of African ancestry,14 most genetic studies have focused on populations of European ancestry. We have carried out the first meta-analysis of GWA studies for BNP concentration in African Americans. We aimed to identify novel BNP-associated variants/loci and evaluate the independence of the associations for SNPs at nearby loci. We further characterized the novel loci by evaluating evidence of their association with aldosterone-renin ratio (ARR), blood pressure, renal traits and left ventricular mass (LVM) in African Americans, Hispanics and Europeans.
Materials and Methods
Data and Recruitment of Subjects
The study population was recruited between September 2000 and March 2004 from the Jackson, MS metropolitan statistical area (MSA) i.e., Hinds, Madison and Rankin Counties using four strategies; (i) all living Jackson MS participants enrolled in the Atherosclerosis Risk in Communities (ARIC) study aged 35–84 years at initiation of the study were invited to participate. They constitute 31 percent of the JHS participants and we hereby refer to this group of participants as the ARIC and JHS overlap (ARIC-JHS), and was used as a replication sample in the present investigation; (ii) a random sample (17%) recruited from a commercially available list (AccuData Integrated Marketing, Fort Myers, FL) of all of community residents 35–84 years in the tri-county area; (iii) a volunteer sample (30%) aged between 35–84 years who responded to targeted advertisements: radio, newspaper, local churches, and civic/social organizations. These volunteers were representative of the Jackson MSA African American population in terms of age, sex and socioeconomic characteristics; (iv) family members (22%) mainly family members of enumerated households including those < 35 years old for purposes of genetic studies. Participants recruited in strategies ii-iv are hereby referred to as the JHS-specific sample. The final study sample consisted of 5,301 men and women between the ages of 21 and 94 years and has been reported to be geographically representative of the age-eligible African Americans in the Jackson MSA.15 Of the total recruited participants, 3,029 or 57.1 percent (892 ARIC-JHS overlap and 2,136 JHS-specific) gave consent for genetic analyses and thus were genotyped separately in the CARe consortium16 using Affymetrix 6.0 platform (See Supplemental Material). The ARIC-JHS overlap samples were genotyped together with the larger ARIC subset and JHS specific with the larger JHS subset. In recognition of the separate genotyping, we performed two separate GWA and then meta-analyzed the results. The study protocol was approved by the University of Mississippi Medical Center Internal Review Board committee on human subjects.
Relevance of BNP GWA results to parallel GWA of Aldosterone and Renin in samples of European ancestry
Due to the paucity of cohorts of African ancestry with available BNP concentration measures and GWA data, we sought to assess our results by conducting cross-check of the identified BNP-associated novel loci in the GWA results of ARR in a large European consortium available to us through our collaborators. This large consortium included four studies: Framingham Heart Study, Study of Health in Pomerania, Cooperative Health Research in the Augsburg Region, and Supplementation en Vitamines et Mineraux Antioxiydants Study. This step was motivated by the known reciprocal action of BNP and ARR. Together with atrial natriuretic peptide, BNP blocks cardiac sympathetic nervous system activity and inhibits the renin-angiotensin-aldosterone axis.17 In clinical studies, when heart failure patients are treated aggressively with angiotensin-II receptor antagonists, such treatment often results in reduced BNP concentrations.18, 19 Evidence from animal studies indicates that BNP inhibits renin synthesis, angiotensin-II generation and angiotensin-associated aldosterone synthesis in the adrenal glands.20, 21Thus, there is evidence that links BNP and the ARR pathways, thereby providing an incentive for assessing commonality in GWA hits for prototype marker of the pathways.
Inclusion and Exclusion Criteria
Participants with self-reported congestive heart failure or with BNP > 100 pg/mL (N=84); serum creatinine levels >2.0 mg/dL i.e. representing renal insufficiency (N=35) and missing BNP measurements (N=26) were excluded from the present analysis. In total, we retained 2,036 participants in JHS-specific sample and 754 in ARIC-JHS sample (Table 1).
Table 1.
Characteristics of the Study Samples
| Study Name | JHS | ARIC-JHS Overlap |
|---|---|---|
| Samples, n (%) | 2036 (100) | 754 (100) |
| Females, n (%) | 1236 (61) | 492 (65) |
| Age (years) | 50±12 | 66±5 |
| BNP (pg/mL) Median (25%, 75%) | 5.8 (1.9, 13.5) | 10.7 (4.6, 24.3) |
| BNP below limit of detection n (%) | 509 (25) | 105 (14) |
| Body Mass Index (kg/m2) | 32±8 | 31±6 |
| Systolic BP (mm Hg) | 129±19 | 131±17 |
| Diastolic BP (mm Hg) | 82±11 | 77±10 |
| Hypertension, (%) | 56 | 78 |
| Antihypertensive therapy, (%) | 44 | 70 |
| History of Myocardial Infarction, (%) | 5 | 5 |
| Prevalent stroke, (%) | 3 | 7 |
| Atrial fibrillation, (%) | 0.2 | 0.6 |
| Diabetes, (%) | 16 | 26 |
| Creatinine, mg/dL | 1.0±0.2 | 1.0±0.2 |
BNP Measurement
Of the participants that attended Jackson Heart examination cycle 1, a total of 4,093 (77%) from the entire sample were assayed for plasma BNP using chemiluminescent immunoassay performed on the Siemens Advia Centaur (Siemens) with the assay’s limit of detection of 2.0 pg/mL. The intra-assay coefficients of variation are reported elsewhere.22 Genotyping, quality control and imputation are presented are presented in the online supplemental materials.
Statistical Analysis
We log-transformed BNP concentration before using Tobit regression models23 to generate sex specific residuals due to the skewed and left censored distribution (caused by the assay’s limit of detection) of BNP concentration. The diagnostic assessment had a limit of detection of 2.0 pg/mL and affected 25% and 14% participants in the JHS-specific and ARIC-JHS overlap samples, respectively. Tobit regression permits analysis of all data unlike conventional regression methods.6, 10 We adjusted for age, sex, body mass index, systolic and diastolic BP, anti-hypertensive therapy, history of myocardial infarction, diabetes, atrial fibrillation, serum creatinine, and 10 principal components to account for population stratification. The natriuretic peptide residuals were then standardized within sex (mean=0 and variance=1) and tested for association with imputed SNPs using an additive genetic model. We used a linear regression framework implemented in the MACH2QTL version 1.08 software24 for both samples. The individual sample GWA results were then combined through meta-analysis using METAL software,25 which employs inverse-variance weighted fixed-effects models to combine beta coefficients and standard errors from sample level regression results for each SNP to derive pooled estimates. The meta-analyzed results were corrected for λ by multiplying the standard errors (SE) of the regression coefficients by the square-root of the study-specific λ. To correct for multiple testing, we declared a significance threshold level a priori of p-value ≤ 2.5×10−8 due to the small LD blocks in populations of African ancestry. We used LocusZoom, a web-based tool developed by Pruim and colleagues26 to inspect regions showing association to determine the extent of the association signal and the position relative to nearby genes.
Conditional analysis was conducted in the individual sample GWA by adding the dosage of the SNP with the strongest association signal at the locus of interest into the regression model as a covariate and testing the residual association with all remaining SNPs within ±500 kb flanking region. We used ProbABEL software package version 0.1.327 to run GWA after adjusting for rs198389 and rs3733402 and 10 principal components. We used Wald test to identify SNPs with strong association signals after accounting for the leading SNP.
In the reciprocal ‘look-up’ (i.e. cross-checking the association of variant related to BNP levels with another trait) of the ARR GWA results (Figure 1), we restricted analyses to the top 12 SNPs around the loci of the two leading variants observed in our meta-analysis results. We set threshold for declaring significance at 0.05/12 SNPs = 4.16 × 10−3. Further, we also attempted to characterize the potential pleiotropic effects of the novel loci by evaluating their association with traits that are physiologically related to BNP concentration (Spearman correlations are provided on Supplemental Materials Table 1), such as systolic blood pressure (BP), left ventricular mass (LVM), chronic kidney disease (CKD), glomerular filtration rate (GFR) and microalbuminuria (MA) in African Americans (COGENT, GENOA), European Americans (EchoGEN, CKDGen) and Hispanic Americans (NOMAS).We used a nominal p-value (p≤0.05) to declare significant association for these latter look-ups. Additionally, we estimated the genetic effects of BNP-associated novel SNPs on plasma BNP concentration. We adjusted for age, sex, body mass index, blood pressure medications, systolic BP, creatinine, type II diabetes and significant principal components.
Figure 1.
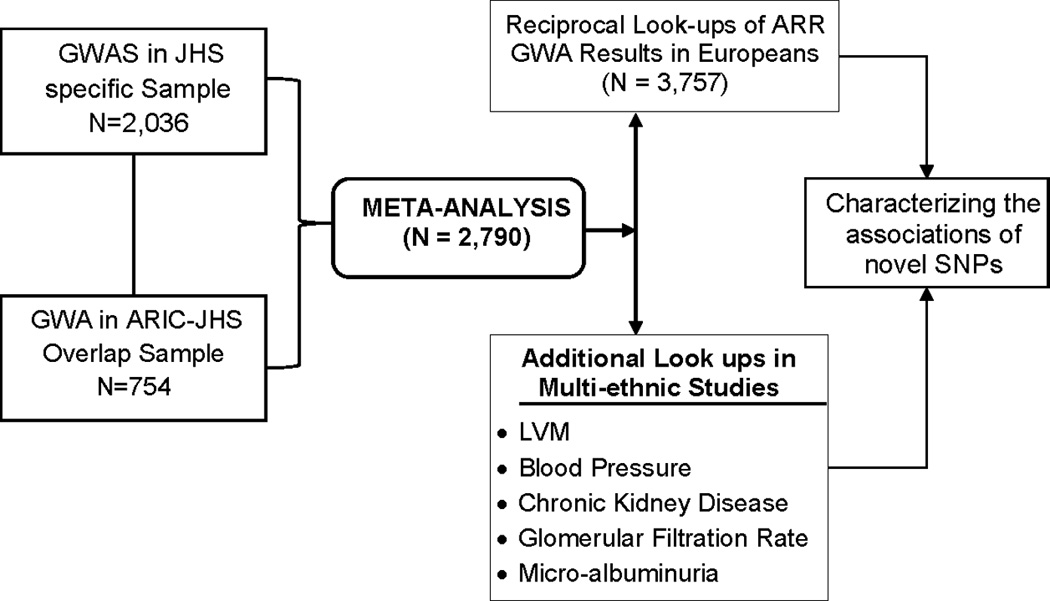
Study Design for Plasma BNP GWA in AA
Results
Characteristics of Study Participants
Table 1 summarizes the characteristics of the two study samples (JHS specific and ARIC-JHS overlap) we meta-analyzed. Compared to the JHS specific, the ARIC-JHS overlap sample was older (>17 years) and had a higher prevalence of hypertension, stroke and diabetes. The median plasma BNP concentration was also higher in the ARIC-JHS sample (10.7 pg/mL) than in the JHS specific (5.8 pg/mL), and 14 and 25 percent, respectively had plasma BNP concentrations less than the limit of detection.
Meta-Analysis of Association Results of ARIC-JHS and JHS specific samples
The meta-analyzed GWA results of plasma BNP concentration are summarized in the Manhattan plot (Figure 2A) and QQ plot (Figure 2B). The inflation factors (λ) for ARIC-JHS and JHS specific GWAS analyses were 0.998 and 1.029 respectively, while the meta-analysis λ was 1.02, suggesting controlled cryptic relatedness in all analyses. We present regions showing association signals across the genome in regional association plots shown in Figure 3 panels A (rs198389) and B (rs3733402). These SNPs are located NPPB and KLKB1 loci, respectively.
Figure 2.
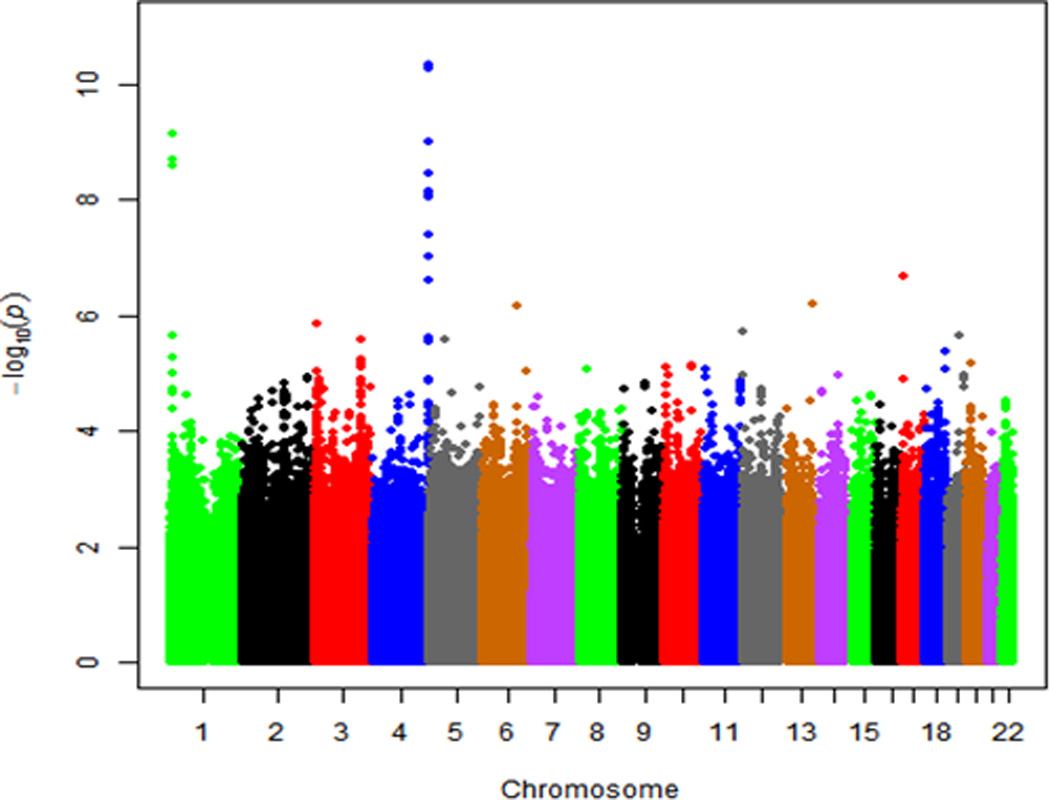
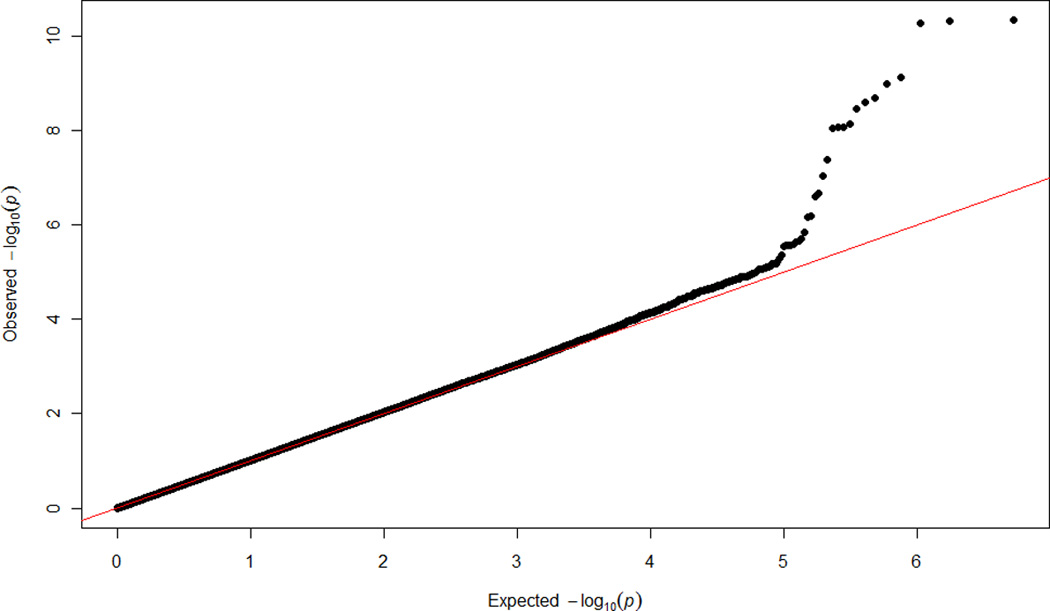
Plots showing the genome-wide –log10 p values for interrogated SNPs across autosomal chromosomes in meta-analyzed samples: (A) Manhattan Plot, (B) QQ Plot.
Figure 3.
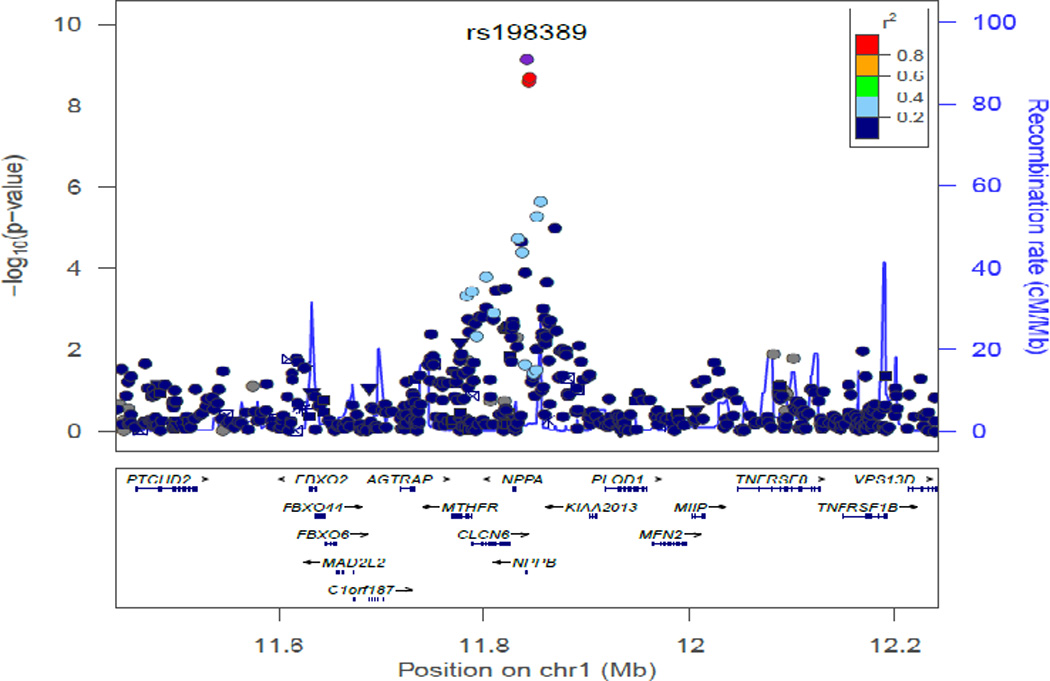
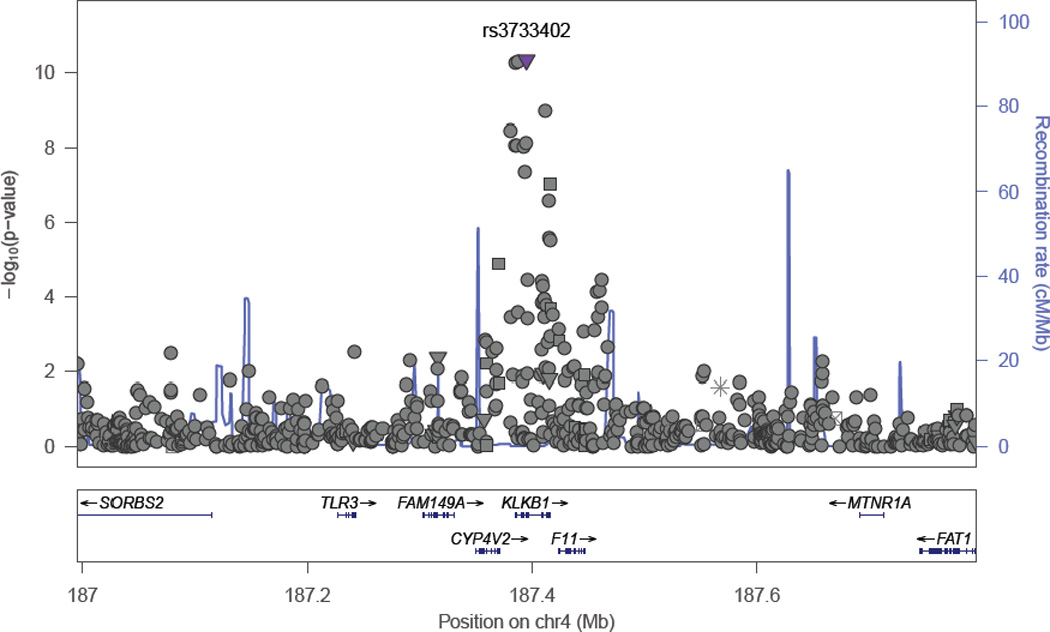
Regional Association Plots obtained using LocusZoom web-based tool for SNPs genome-wide significantly associated with plasma BNP concentration in African Americans for (A) NPPB locus (rs198389) (B) KLKB1 locus (rs3733402)
We identified a total of 12 SNPs that were genome-wide significantly associated with BNP concentration; three SNPs at the1p36.22 locus and 9 SNPs at the KLKB1 (4q35.2) locus (Table 2). We show in Table 2, their base-pair positions, and distance from the nearest gene and type. The three genome-wide significant (rs198389, MAF=0.40, p-value=1.18×10−09; rs12406089, MAF=0.39, p-value=3.67×10−09; and rs6668659, MAF=0.37, p-value=3.08×10−09) SNPs in chromosome 1p36.22 were clustered around the promoter region of the NPPB gene. The other 9 SNPs were clustered around the Kallikrein plasma factor (KLKB1) gene. Rs3733402 is a missense mutation; rs1511801, rs2048 and rs4253238 were upstream of KLKB1 and the rest were intronic. The G allele of the leading SNP rs3733402 (MAF=0.27) is associated with increased plasma BNP concentrations, [β(SE) 0.20(0.03); p-value = 1.75×10−11].
Table 2.
Genome-wide significant SNPs associated with Log plasma BNP concentrations
| SNP | CHR | Position* | EA | Imputation quality |
Imputed [Yes, No] |
EAF | Nearest Gene |
Distance gene (bp) |
Function | Beta (SE) | P-value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs198389 | 1 | 11841858 | G | 0.88 | Yes | 0.40 | NPPB | 279 | Promoter | 0.18(.03) | 1.18×10−09 |
| rs6668659 | 1 | 11844885 | G | … | No | 0.39 | NPPB | 3306 | Unknown | −0.18(.03) | 3.08×10−09 |
| rs12406089 | 1 | 11843768 | C | 0.89 | Yes | 0.37 | NPPB | 2189 | unknown | 0.19(.03) | 3.67×10−09 |
| rs3733402 | 4 | 187395028 | G | 0.99 | Yes | 0.26 | KLKB1 | 9363 | Missense | 0.20(.03) | 1.75×10−11 |
| rs1511801 | 4 | 187387704 | T | 0.96 | Yes | 0.73 | KLKB1 | 2039 | Upstream | −0.21(.03) | 2.19×10−11 |
| rs4253238 | 4 | 187385381 | C | 0.98 | Yes | 0.26 | KLKB1 | 285 | Upstream | 0.20(.03) | 3.08×10−11 |
| rs4253311 | 4 | 187411677 | G | 0.99 | Yes | 0.61 | KLKB1 | 4942 | Intronic | −0.17(.03) | 3.22×10−10 |
| rs11132382 | 4 | 187380796 | G | 0.97 | Yes | 0.65 | KLKB1 | 4870 | NA | −0.17(.03) | 1.53×10−09 |
| rs4253252 | 4 | 187394452 | G | … | No | 0.64 | KLKB1 | 8786 | Intronic | −0.16(.02) | 2.27×10−09 |
| rs2048 | 4 | 187385127 | G | 0.98 | Yes | 0.35 | KLKB1 | 538 | Upstream | 0.17(.03) | 3.36×10−09 |
| rs1912826 | 4 | 187386534 | G | 0.98 | Yes | 0.35 | KLKB1 | 868 | intronic | 0.17(.03) | 3.55×10−09 |
| rs4253248 | 4 | 187392482 | G | 0.99 | Yes | 0.35 | KLKB1 | 6816 | intronic | 0.17(.03) | 3.76×10−09 |
Base pair position (bp) according to NCBI36/hg18; CHR: Chromosome; EA: Effect Allele, EAF: Effect Allele Frequency; ARR: Aldosterone-Renin Ratio
To explore the presence of additional signals at the BNP-associated loci, we performed conditional analyses at NPPB and KLKB1 loci separately by conditioning on the lead SNPs rs198389 and rs3733402, respectively, and tested the residual association with all the remaining SNPs within ±500 kb flanking regions of these lead SNPs. The conditional p-values for the top SNPs are presented in Supplemental Material Table 2 none remained genome-wide significantly associated with BNP concentration. The drop in the strength of the association signal implied that rs198389 and rs3733402 were likely the sole drivers behind most of the association signal at NPPB and KLKB1 loci, respectively.
Reciprocal Look-ups of Genome-wide Results
The characteristics of the study population used for reciprocal look-up are provided in the companion manuscript by [Wolfgang and colleagues]. We looked up the lead SNPs near NPPB and KLKB1 loci for evidence of association with ARR in European discovery data. All the 12 SNPs we identified on chromosome 4q35.2 were significantly (p≤4.16×10−3) associated with ARR in Europeans (Table 3), although the effects of the index alleles were all in the opposite direction of the effect on BNP concentrations. Alleles associated with increased BNP concentration were associated with low ARR levels in the European samples. Further, the allele frequencies of the effect alleles were always higher than in our study. The SNPs around the NPPB locus were not significantly associated with ARR although the direction of effect was consistent.
Table 3.
Results of reciprocal look-ups of ARR GWA results
| SNP | CHR | NCBI36 / hg18 Position |
EA | EAF | Meta-analysis of BNP GWAS Results (N=2790) |
Reciprocal ARR GWA Look-up Results (N=3757) |
||
|---|---|---|---|---|---|---|---|---|
| Beta(SE) | P-value | Beta (SE) | P-value | |||||
| rs198389 | 1 | 11841858 | G | 0.40 | 0.18 (0.03) | 1.18×10−09 | 0.02±0.02 | 0.42 |
| rs12406089 | 1 | 11844885 | C | 0.37 | 0.19 (0.03) | 3.08×10−09 | 0.03±0.02 | 0.15 |
| rs6668659 | 1 | 11843768 | G | 0.39 | −0.18 (0.03) | 3.67×10−09 | −0.02±0.02 | 0.24 |
| rs3733402 | 4 | 187395028 | G | 0.26 | 0.20 (0.03) | 1.75×10−11 | −0.10±0.02 | 5.00×10−08 |
| rs1511801 | 4 | 187387304 | T | 0.27 | −0.21 (0.03) | 2.19×10−11 | 0.10±0.02 | 5.32×10−08 |
| rs4253238 | 4 | 187385381 | C | 0.74 | 0.20 (0.03) | 3.08×10−11 | −0.10±0.02 | 5.42×10−08 |
| rs4253311 | 4 | 187411677 | G | 0.40 | −0.17 (0.03) | 3.22×10−10 | 0.10±0.02 | 3.54×10−08 |
| rs11132382 | 4 | 187380796 | G | 0.36 | −0.17 (0.03) | 1.53×10−09 | 0.10±0.02 | 5.60×10−08 |
| rs4253252 | 4 | 187394452 | G | 0.36 | −0.16 (0.03) | 2.27×10−09 | 0.10±0.02 | 5.03×10−08 |
| rs2048 | 4 | 187385127 | G | 0.65 | 0.17 (0.03) | 3.36×10−09 | −0.10±0.02 | 5.44×10−08 |
| rs1912826 | 4 | 187386534 | G | 0.65 | 0.17 (0.03) | 3.55×10−09 | −0.10±0.02 | 5.34×10−08 |
| rs4253248 | 4 | 187392482 | G | 0.65 | 0.17 (0.03) | 3.76×10−09 | −0.10±0.02 | 5.14×10−08 |
CHR: Chromosome; EA: Effect Allele, EAF: Effect Allele Frequency; ARR: Aldosterone-Renin Ratio; NCBI36/hg18: Human genomes build 18
In additional look-ups, we characterized the association of the identified novel loci with physiologically correlated traits across multi-ethnic cohorts. Initially, we confirmed correlations of the traits with BNP concentration in our study population, and as presented in Supplemental Materials Table 1, all Spearman correlations were strong and highly significant (p<0.001).
The index allele of rs198389 (NBBP locus) was significantly associated with systolic BP in African Americans (p=0.019) and LVM in Europeans (p=0.05), while the index allele for rs3733402 was associated with LVM (p=0.048) in Europeans (Table 4). We observed borderline significant association of rs198389 (p=0.073) and rs3733402 (p=0.071) with MA and CKD, respectively in Europeans only (Table 4). Associations with the remainder of the correlated traits were not significant.
Table 4.
Characterization of BNP associated novel loci for association with physiologically related traits
| NPPB – rs198389 | KLKB1-rs3733402 | |||||||
|---|---|---|---|---|---|---|---|---|
| Trait | Consortium | Ethnicity | Direction† | P-value | N | Direction† | P-value | N |
| LVM | CARe | Blacks | + | 0.69 | 6,765 | + | 0.53 | 6,765 |
| LVM | GENOA | Blacks | + | 0.31 | 651 | + | 0.57 | 651 |
| LVM | NOMAS | Hispanics | + | 0.29 | 802 | + | 0.21 | 802 |
| LVM | EchoGEN | Europeans | − | 0.050 | 12,612 | − | 0.048 | 12,612 |
| Systolic BP | COGENT | Blacks | − | 0.019 | 7,473 | + | 0.96 | 7,473 |
| eGFR | CKDGen | Europeans | − | 0.86 | 11,606 | − | 0.11 | 9,093 |
| CKD | CKDGen | Europeans | − | 0.93 | 13,187 | + | 0.07 | 6,087 |
| MA | CKDGen | Europeans | + | 0.073 | 9,238 | − | 0.67 | 7,302 |
The directions of effect are based on the alleles (rs198389-A; rs3733402-G) associated with increased BNP concentration in this study.
LVM = Left Ventricular Mass; BP=Blood Pressure; eGFR=Estimated Glomerular Filtration Rate; MA=Microalbuminuria; CARe: NHLBI Candidate Gene Resource Consortium;
We also characterized the association of the novel loci with BNP concentrations in its untransformed units. We found that a unit increase in the dosage of the effect allele at the NPPB and KLKB1 loci was significantly associated with 8.1±2.1 pg/dL (p=1.38×10−4) and 8.3±2.2 pg/dL (p=1.38×10−4) of plasma BNP concentration, respectively (Table 5). We also observed that subjects with two copies of the effective allele showed 15.3 pg/dl and 14.2 pg/dl higher concentrations than subjects with none (Table 5).
Table 5.
Average increase in BNP concentration and odds ratios (95% confidence interval) of risk of hypertension according to the number of copies of the effect allele of novel loci
| BNP Concentration | ||
|---|---|---|
| SNPs | Beta (SE) | p-value |
| NPPB-rs198389 | ||
| 0 | Reference | … |
| 1 | 2.3 (3.0) | 0.43 |
| 2 | 15.7 (4.0) | 1.02×10−4 |
| Trend | 8.1 (2.1) | 1.38×10−4 |
| KLKB1-rs3733402 | ||
| 0 | Reference | |
| 1 | 9.2 (2.8) | 1.26×10−3 |
| 2 | 14.3 (5.4) | 7.73×10−3 |
| Trend | 8.3 (2.2) | 1.39×10−4 |
0: zero G alleles; 1, 2 one and two alleles of the effect allele (G); CI: confidence interval
Discussion
In our first ever GWA analysis of plasma BNP concentration in a sample population of African American ancestry, we identified two loci NPPB (rs198389) and KLKB1 (rs3733402) that were genome-wide significantly associated with BNP concentration. We confirmed previously reported NPPB locus and provided convincing evidence of a novel BNP-associated locus near KLKB1 using meta-analysis and reciprocal look-ups of GWA results of ARR levels in persons of European ancestry. Each additional copy of effective allele for both variants were associated with increased plasma BNP concentration in this community based African American population. Through look-up of GWA results of traits related to plasma BNP concentration in multi-ethnic populations, we found evidence that these genes probably have pleiotropic effects. Conditional analyses did not reveal any additional SNPs at the loci.
The genome-wide significant association of rs198389 with BNP concentration is the first evidence from GWA showing the association in African Americans, although several previous candidate gene studies have investigated the NPPA/NPPB for association with BNP concentrations6, 28, 29 and with traits physiologically related to BNP such as ventricular dysfunction,9 blood pressure/hypertension,5, 6 type 2 diabetes,8, 30 NT-proBNP,5, 10 and echocardiographic indices.5 In the majority of these studies, rs198389 is used as a genetic variant to represent variation at the NPPB locus. A recent GWA on serum NT-proBNP in Europeans supplied additional evidence on the genetic role of NPPB locus.10
This association of the NPPB locus with plasma BNP concentration may be a reflection of the function of NPPB in the synthesis of circulating BNP. The NPPB gene has been previously described as having a role in encoding for the pre-proBNP precursor. After removal of a 26-amino acid signal peptide from this protein, it becomes a 108-amino acid proBNP polypeptide. Further processing of this polypeptide results in a physiologically active 32-amino acid carboxyl-terminal BNP molecule (BNP32) and an inactive N-terminal fragment (NT-BNP).11 Meirhaeghe and colleagues recently demonstrated that there is an increased promoter activity in vitro of the C allele of rs198389 SNP in type 2 diabetes patients.30 They also observed that COS7 cells transfected with C-allele showed higher BNP gene transcription compared to T-allele.10, 30 Distribution of the C allele in the general adult population has been reported to be common but has no effect on cardiovascular outcome.31
To refine and thus better explain the association signal at the NPPB locus in our study, we generated LD plots centered on rs198389 but within 25 kb across YRI and CEU (Supplemental Materials Figure 1 panels A and B). LD around rs198389 extends along a 9 kb region in HapMap CEU (release 3) and contains all SNPs previously associated with plasma BNP and serum proBNP in populations of European ancestry. However, the interval is only 3 kb in YRI and contains rs6668659 only, which has previously been associated with plasma BNP in EA. This difference in the LD block between CEU and YRI may explain the difference in the polymorphisms with strongest association signal at the NPPB locus in our study as compared to previous studies in Whites.6
Our finding that a missense SNP (rs3733402) on human KLKB1 is genome-wide significantly associated with plasma BNP concentration is novel and has not been previously reported. Rs4253238, which is in complete LD (r2=1.00) with rs3733402, was recently found to be associated with mid regional proadrenomedullin in individuals of European ancestry.32 KLKB1 is known to encode plasma kallikrein, a serine protease synthesized in the liver as plasma prekallikrein, and when secreted into the blood it is converted into plasma kallikrein.33 The plasma kallikrein controls the release of bradykinin and activates conversion of prorenin to renin. Previous research work to identify polymorphisms of KLKB1 and their linkage analysis to end-stage renal disease among African Americans yielded inconclusive results.34 Despite these findings, the role of plasma kallikrein in the regulation of blood pressure and renal perfusion through secretion of kinins and vasopeptides35 qualifies as a candidate gene for essential hypertension and chronic renal failure (conditions that are both associated with plasma BNP concentration through its biological effects on peripheral and renal vasculature).36 In candidate gene studies, Lu and colleagues found that genetic variation at the KLKB1 is associated with increased risk of essential hypertension in Han Chinese.37 Thus, it is in this regard that we pursued reciprocal look-up of ARR GWA results for evidence of pleiotropic effects of the KLKB1 locus. Plasma BNP concentration is linked to RAAS through its role as a “counter-regulatory” peptide that is protective in conditions of over activation of RAAS (such as in heart failure). Natriuretic peptides lower renin and aldosterone concentrations and decrease angiotensin II stimulated aldosterone secretion. These actions result in a natriuretic effect that is compensatory though limited in volume overload conditions.38 Our results corroborate this inverse relation in that alleles responsible for higher plasma BNP are associated with decrease in ARR (Table 3). The lack of a similar significant association between the ARR and rs198389 SNP in the NPPB gene underscores possible complex pleotropic relationship between rs198389 SNP on NPPB gene and ARR traits. Pleoitropy can either be direct (i.e., a gene influence single-gene traits) or indirect39 (i.e., a single gene influences traits in multiple pathways).
Statistically significant associations in additional look-ups of rs198389 and rs3733402 with systolic BP in participants of African ancestry, and with LVM in individuals of European ancestry (Table 4) are all supported by biological data. For instance, BNP is known to promote vasodilation, inhibit vasoconstriction and lower sympathetic tone and thus lowers BP.2 Findings from animal studies using transgenic mice that overexpress BNP show that the systolic BP is 20 to 30 mmHg lower in affected mice compared to their normal counterparts.40 Also, the link between BNP and LVM is well established. BNP is primarily synthesized, stored and released by the ventricular myocardium in response to increased wall stress.2, 41 As a result of its relation to the ventricular myocardium, BNP concentrations are higher in patients with heart failure. Currently BNP concentrations are used clinically as part of the diagnostic workup for heart failure.42 In the Jackson Heart Study cohort, higher BNP concentrations are significantly associated with higher echocardiographic LVM. Furthermore, the natriuretic peptide system is strongly related to kidney function through both renal artery hemodynamics and through direct tubular effects. Natriuretic peptide increases blood flow to the kidneys through vasodilation of the renal arteries promoting diuresis.2, 43, 44 Natriuretic peptide more directly causes dilatation of afferent arterioles and constriction of efferent arterioles in the tubules leading to increased glomerular filtration.2
The nominally significant associations of BNP GWA SNPs with both plasma BNP and LVM are consistent with the notion that genetic variation in BNP concentrations may impact target organ damage such as LVH. BNP knock-out mice develop cardiac fibrosis and LVH.45 Since human knock-outs do not exist, the present GWA allows us to characterize their association with LVM in the community.
We also examined the interaction between the top GWAS loci through pathway analysis, which would provide another potential route to investigate the collective effects of genetic variants on biological systems. Multiple pathways were found to be enriched with BNP-related genes, but the evidence was inconclusive and warrants further investigation (Supplemental Material text) to understand the definite mechanism underlying these associations.
Strengths and Limitations
This is so far, the largest GWA study of BNP concentration in a community-based sample of African Americans. Previous candidate gene-based analyses were limited to specific genes suspected to influence genetic variation of plasma BNP concentration. This current study allowed us to test several other regions in the genome for possible association with BNP concentration. Our results confirm findings on the NPPB locus and reports one novel SNP in the KLKB1 locus. The lack of an appropriate replication sample is a major limitation of the study. Although, we exploited possible reciprocal relation between BNP and ARR to cross-check the relevance of our GWA results and look-ups of association results from physiologically related traits to characterize the effects of our findings, replication is usually the standard procedure of ensuring credibility of genotype-phenotype associations. Further, the unavailability of genotype data on the missense corin variants barred us from checking the widely known association with natriuretic peptides in African Americans.
Conclusion
We conducted first genome-wide association analysis of plasma BNP concentrations in African Americans and confirmed that the genetic variants with the strongest effects are all within and around the NPPB and KLKB1 genes, both determinants of blood pressure control and hypertension. Although we were constrained by a relatively small sample size, and, therefore a potentially lower statistical power to detect weak effect sizes, we were able to successfully characterize the association of these loci with other traits through look-ups. Larger sample sizes of African American samples will likely confirm these results and facilitate discovery of more loci regulating plasma BNP concentrations.
Supplementary Material
Acknowledgments
The authors thank the Jackson Heart Study team (University of Mississippi Medical Center, Jackson State University and Tougaloo College) and participants for their long-term commitment that continues to improve our understanding of the genetic epidemiology of cardiovascular and other chronic diseases.
Funding Sources: The Jackson Heart Study is supported by contracts HSN268201300046C, HHSN268201300047C, HHSN268201300048C, HHSN268201300049C, HHSN268201300050C from the National Heart, Lung, and Blood Institute on Minority Health and Health Disparities.
Footnotes
Conflict of Interest Disclosures: None
References
- 1.Cheung BM, Kumana CR. Natriuretic peptides--relevance in cardiovascular disease. JAMA. 1998;280:1983–1984. doi: 10.1001/jama.280.23.1983. [DOI] [PubMed] [Google Scholar]
- 2.Levin ER, Gardner DG, Samson WK. Natriuretic peptides. N Engl J Med. 1998;339:321–328. doi: 10.1056/NEJM199807303390507. [DOI] [PubMed] [Google Scholar]
- 3.Fox ER, Musani SK, Singh P, Bidulescu A, Nagarajarao HS, Samdarshi TE, et al. Association of plasma B-type natriuretic peptide concentrations with longitudinal blood pressure tracking in African Americans: Findings from the Jackson Heart Study. Hypertension. 2013;1:48–54. doi: 10.1161/HYPERTENSIONAHA.112.197657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang TJ, Larson MG, Levy D, Benjamin EJ, Corey D, Leip EP, Vasan RS. Heritability and genetic linkage of plasma natriuretic peptide levels. Circulation. 2003;108:13–16. doi: 10.1161/01.CIR.0000081657.83724.A7. [DOI] [PubMed] [Google Scholar]
- 5.Ellis KL, Newton-Cheh C, Wang TJ, Frampton CM, Doughty RN, Whalley GA, et al. Association of genetic variation in the natriuretic peptide system with cardiovascular outcomes. J Mol Cell Cardiol. 2011;50:695–701. doi: 10.1016/j.yjmcc.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 6.Newton-Cheh C, Larson MG, Vasan RS, Levy D, Bloch KD, Surti A, et al. Association of common variants in NPPA and NPPB with circulating natriuretic peptides and blood pressure. Nat Genet. 2009;41:348–353. doi: 10.1038/ng.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choquet H, Cavalcanti-Proenca C, Lecoeur C, Dina C, Cauchi S, Vaxillaire M, et al. The T-381C SNP in BNP gene may be modestly associated with type 2 diabetes: An updated meta-analysis in 49 279 subjects. Hum Mol Genet. 2009;18:2495–2501. doi: 10.1093/hmg/ddp169. [DOI] [PubMed] [Google Scholar]
- 8.Pfister R, Sharp S, Luben R, Welsh P, Barroso I, Salomaa V, et al. Mendelian randomization study of B-type natriuretic peptide and type 2 diabetes: Evidence of causal association from population studies. PLoS Med. 2011;8:e1001112. doi: 10.1371/journal.pmed.1001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox AA, Collard CD, Shernan SK, Seidman CE, Seidman JG, Liu KY, et al. Natriuretic peptide system gene variants are associated with ventricular dysfunction after coronary artery bypass grafting. Anesthesiology. 2009;110:738–747. doi: 10.1097/aln.0b013e31819c7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del Greco MF, Pattaro C, Luchner A, Pichler I, Winkler T, Hicks AA, et al. Genome-wide association analysis and fine mapping of NT-proBNP level provide novel insight into the role of the MTHFR-CLCN6-NPPA-NPPB gene cluster. Hum Mol Genet. 2011;20:1660–1671. doi: 10.1093/hmg/ddr035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sudoh T, Maekawa K, Kojima M, Minamino N, Kangawa K, Matsuo H. Cloning and sequence analysis of cDNA encoding a precursor for human brain natriuretic peptide. Biochem Biophys Res Commun. 1989;159:1427–1434. doi: 10.1016/0006-291x(89)92269-9. [DOI] [PubMed] [Google Scholar]
- 12.Yandle TG. Biochemistry of natriuretic peptides. J.Intern.Med. 1994;235:561–576. doi: 10.1111/j.1365-2796.1994.tb01263.x. [DOI] [PubMed] [Google Scholar]
- 13.Benjamin EJ, Dupuis J, Larson MG, Lunetta KL, Booth SL, Govindaraju DR, et al. Genome-wide association with select biomarker traits in the framingham heart study. BMC Med Genet. 2007;8(Suppl 1):S11. doi: 10.1186/1471-2350-8-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dries DL, Victor RG, Rame JE, Cooper RS, Wu X, Zhu X, et al. Corin gene minor allele defined by 2 missense mutations is common in blacks and associated with high blood pressure and hypertension. Circulation. 2005;112:2403–2410. doi: 10.1161/CIRCULATIONAHA.105.568881. [DOI] [PubMed] [Google Scholar]
- 15.Fuqua SR, Wyatt SB, Andrew ME, Sarpong DF, Henderson FR, Cunningham MF, Taylor HA., Jr Recruiting african-american research participation in the Jackson Heart Study: Methods, response rates, and sample description. Ethn Dis. 2005;15:S6–S29. [PubMed] [Google Scholar]
- 16.Musunuru K, Lettre G, Young T, Farlow DN, Pirruccello JP, Ejebe KG, et al. Candidate gene association resource (CARe): Design, methods, and proof of concept. Circ Cardiovasc Genet. 2010;3:267–275. doi: 10.1161/CIRCGENETICS.109.882696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakagawa O, Ogawa Y, Itoh H, Suga S, Komatsu Y, Kishimoto I, et al. Rapid transcriptional activation and early mrna turnover of brain natriuretic peptide in cardiocyte hypertrophy. Evidence for brain natriuretic peptide as an "emergency" cardiac hormone against ventricular overload. J Clin Invest. 1995;96:1280–1287. doi: 10.1172/JCI118162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsutamoto T, Wada A, Maeda K, Mabuchi N, Hayashi M, Tsutsui T, et al. Effect of spironolactone on plasma brain natriuretic peptide and left ventricular remodeling in patients with congestive heart failure. J Am Coll Cardiol. 2001;37:1228–1233. doi: 10.1016/s0735-1097(01)01116-0. [DOI] [PubMed] [Google Scholar]
- 19.Latini R, Masson S, Anand I, Judd D, Maggioni AP, Chiang YT, et al. Valsartan Heart Failure Trial I. Effects of valsartan on circulating brain natriuretic peptide and norepinephrine in symptomatic chronic heart failure: The valsartan heart failure trial (val-heft) Circulation. 2002;106:2454–2458. doi: 10.1161/01.cir.0000036747.68104.ac. [DOI] [PubMed] [Google Scholar]
- 20.Nawata H, Ohashi M, Haji M, Takayanagi R, Higuchi K, Fujio N, et al. Atrial and brain natriuretic peptide in adrenal steroidogenesis. J Steroid Biochem Mol Biol. 1991;40:367–379. doi: 10.1016/0960-0760(91)90204-i. [DOI] [PubMed] [Google Scholar]
- 21.Akabane S, Matsushima Y, Matsuo H, Kawamura M, Imanishi M, Omae T. Effects of brain natriuretic peptide on renin secretion in normal and hypertonic saline-infused kidney. Eur J Pharmacol. 1991;198:143–148. doi: 10.1016/0014-2999(91)90613-u. [DOI] [PubMed] [Google Scholar]
- 22.Fox ER, Musani SK, Bidulescu A, Nagarajarao HS, Samdarshi TE, Gebreab SY, et al. Relation of obesity to circulating B-type natriuretic peptide concentrations in blacks: The Jackson Heart Study. Circulation. 2011;124:1021–1027. doi: 10.1161/CIRCULATIONAHA.110.991943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tobit J. Estimation of relationship for limited dependent variables. Econometrica. 1958;26:24–36. [Google Scholar]
- 24.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willer CJ, Li Y, Abecasis GR. Metal: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. Locuszoom: Regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aulchenko YS, Struchalin MV, van Duijn CM. Probabel package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lanfear DE, Stolker JM, Marsh S, Rich MW, McLeod HL. Genetic variation in the b-type natiuretic peptide pathway affects BNP levels. Cardiovasc Drugs Ther. 2007;21:55–62. doi: 10.1007/s10557-007-6007-5. [DOI] [PubMed] [Google Scholar]
- 29.Takeishi Y, Toriyama S, Takabatake N, Shibata Y, Konta T, Emi M, et al. Linkage disequilibrium analyses of natriuretic peptide precursor B locus reveal risk haplotype conferring high plasma bnp levels. Biochem Biophys Res Commun. 2007;362:480–484. doi: 10.1016/j.bbrc.2007.08.028. [DOI] [PubMed] [Google Scholar]
- 30.Meirhaeghe A, Sandhu MS, McCarthy MI, de GP, Cottel D, Arveiler D, Ferrieres J, et al. Association between the T-381C polymorphism of the brain natriuretic peptide gene and risk of type 2 diabetes in human populations. Hum Mol Genet. 2007;16:1343–1350. doi: 10.1093/hmg/ddm084. [DOI] [PubMed] [Google Scholar]
- 31.Costello-Boerrigter LC, Boerrigter G, Ameenuddin S, Mahoney DW, Slusser JP, Heublein DM, et al. The effect of the brain-type natriuretic peptide single-nucleotide polymorphism rs198389 on test characteristics of common assays. Mayo Clin Proc. 2011;86:210–218. doi: 10.4065/mcp.2010.0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verweij N, Mahmud H, Mateo Leach I, de Boer RA, Brouwers FP, et al. Genome-wide association study on plasma levels of midregional-proadrenomedullin and c-terminal-pro-endothelin-1. Hypertension. 2013;61:602–608. doi: 10.1161/HYPERTENSIONAHA.111.203117. [DOI] [PubMed] [Google Scholar]
- 33.Chung DW, Fujikawa K, McMullen BA, Davie EW. Human plasma prekallikrein, a zymogen to a serine protease that contains four tandem repeats. Biochemistry. 1986;25:2410–2417. doi: 10.1021/bi00357a017. [DOI] [PubMed] [Google Scholar]
- 34.Yu H, Anderson PJ, Freedman BI, Rich SS, Bowden DW. Genomic structure of the human plasma prekallikrein gene, identification of allelic variants, and analysis in end-stage renal disease. Genomics. 2000;69:225–234. doi: 10.1006/geno.2000.6330. [DOI] [PubMed] [Google Scholar]
- 35.Pravenec M, Kren V, Kunes J, Scicli AG, Carretero OA, Simonet L, Kurtz TW. Cosegregation of blood pressure with a kallikrein gene family polymorphism. Hypertension. 1991;17:242–246. doi: 10.1161/01.hyp.17.2.242. [DOI] [PubMed] [Google Scholar]
- 36.Wang AY, Lai KN. Use of cardiac biomarkers in end-stage renal disease. J Am Soc Nephrol. 2008;19:1643–1652. doi: 10.1681/ASN.2008010012. [DOI] [PubMed] [Google Scholar]
- 37.Lu X, Zhao W, Huang J, Li H, Yang W, Wang L, et al. Common variation in KLKB1 and essential hypertension risk: Tagging-SNP haplotype analysis in a case-control study. Hum Genet. 2007;121:327–335. doi: 10.1007/s00439-007-0340-4. [DOI] [PubMed] [Google Scholar]
- 38.Richards AM. Natriuretic peptides: Update on peptide release, bioactivity, and clinical use. Hypertension. 2007;50:25–30. doi: 10.1161/HYPERTENSIONAHA.106.069153. [DOI] [PubMed] [Google Scholar]
- 39.Hodgkin J. Seven types of pleiotropy. Int J Dev Biol. 1998;42:501–505. [PubMed] [Google Scholar]
- 40.Kuroski de Bold ML. Atrial natriuretic factor and brain natriuretic peptide gene expression in the spontaneous hypertensive rat during postnatal development. Am J Hypertens. 1998;11:1006–1018. doi: 10.1016/s0895-7061(98)00116-2. [DOI] [PubMed] [Google Scholar]
- 41.Houben AJ, Willemsen RT, van de, Ven H, de Leeuw PW. Microvascular adaptation to changes in dietary sodium is disturbed in patients with essential hypertension. J Hypertens. 2005;23:127–132. doi: 10.1097/00004872-200501000-00022. [DOI] [PubMed] [Google Scholar]
- 42.McCullough PA, Nowak RM, McCord J, Hollander JE, Herrmann HC, Steg PG, et al. B-type natriuretic peptide and clinical judgment in emergency diagnosis of heart failure: Analysis from breathing not properly (BNP) multinational study. Circulation. 2002;106:416–422. doi: 10.1161/01.cir.0000025242.79963.4c. [DOI] [PubMed] [Google Scholar]
- 43.Yamada Y, Yokota M. Production of C-type natriuretic peptide in human aortic endothelial cells induced by activation of protein kinase C. Am J Hypertens. 1996;9:924–929. doi: 10.1016/s0895-7061(96)00107-0. [DOI] [PubMed] [Google Scholar]
- 44.Yamamoto K, Burnett JC, Jr, Jougasaki M, Nishimura RA, Bailey KR, Saito Y, et al. Superiority of brain natriuretic peptide as a hormonal marker of ventricular systolic and diastolic dysfunction and ventricular hypertrophy. Hypertension. 1996;28:988–994. doi: 10.1161/01.hyp.28.6.988. [DOI] [PubMed] [Google Scholar]
- 45.Tamura N, Ogawa Y, Chusho H, Nakamura K, Nakao K, Suda M, et al. Cardiac fibrosis in mice lacking brain natriuretic peptide. Proc Natl Acad Sci U S A. 2000;97:4239–4244. doi: 10.1073/pnas.070371497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


