Abstract
Motivation: Nonlinear dose–response models are primary tools for estimating the potency [e.g. half-maximum inhibitory concentration (IC) known as IC50] of anti-cancer drugs. We present drexplorer software, which enables biologists to evaluate replicate reproducibility, detect outlier data points, fit different models, select the best model, estimate IC values at different percentiles and assess drug–drug interactions. drexplorer serves as a computation engine within the R environment and a graphical interface for users who do not have programming backgrounds.
Availability and implementation: The drexplorer R package is freely available from GitHub at https://github.com/nickytong/drexplorer. A graphical user interface is shipped with the package.
Contact: jingwang@mdanderson.org
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Understanding the dose–response relationship of therapeutic compounds is a major focus of clinical oncology. Large-scale drug screening in cancer cell lines has demonstrated that different genomic characteristics (mutations) confer distinct drug sensitivity, supporting patient stratification into treatment regimens on the basis of genomic markers (Barretina et al., 2012; Yang et al., 2013). Cell lines resistant to single-drug treatment may respond to synergistic drug combinations (Lehár, et al., 2009). As cell viability assays become widely used, it is important to provide easily accessible tools to analyze drug profiling data.
We developed drexplorer software in the R environment (R Core Team, 2014) to encompass several aspects of dose–response analysis: assess reproducibility of replicated experiments, detect outlier data points, fit different models, identify the best model, estimate inhibitory concentration (IC) values and evaluate drug–drug interactions.
2 Algorithms
A typical drug screening experiment exposes cells to a range of concentrations (doses) of a drug and quantifies cell viability (response). To account for experimental variations, multiple measurements are obtained for each dose. We denote the jth repeat of the ith dose by and the corresponding response by. For anti-cancer drug screening, response is further scaled by the mean response of the controls (i.e. under) and is called relative viability.
2.1 Outlier detection
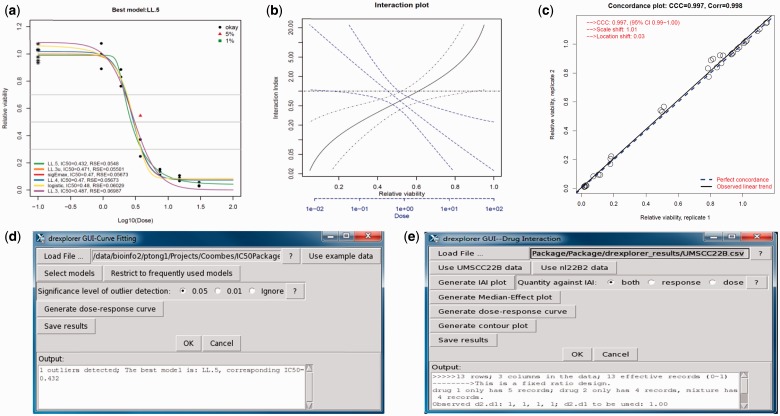
Outlier values in response will affect dose–response curve fitting and IC estimation. We implement the Newman test (Newman, 1939) to detect outliers, defining the test statistic as, where is the maximum difference among the response vector at the ith dose and is the standard deviation of the response in untreated controls. Once calculated, the test statistic can be compared with the threshold value from the null distribution under the specified type I error (). We apply this test to identify outlier data points at each dose separately. Figure 1a shows outlier detection using the ryegrass data (Ritz and Streibig, 2005).
Fig. 1.
Example of drexplorer output and GUI. (a) Outlier data at type I error = 0.01 and 0.05 are, respectively, indicated by the green square and red triangle. Various dose–response curves are fitted; RSEs are indicated; title indicates model selected by RSE. (b) Interaction index estimated at different doses (blue line) and relative viability (black line); 95% confidence interval also shown. (c) Reproducibility assessment using concordance. Responses from two replicates are highly concordant showing good reproducibility. (d) GUI screenshot for dose–response estimation. (e) GUI screenshot for interaction index estimation.
2.2 Assessing reproducibility
When replicate experiments have been performed, it is necessary to assess the quality of the reproducibility. We use the concordance correlation coefficient (CCC) to capture both the location shift and the scale shift between replicates (Lawrence and Lin, 1989). CCC ranges between −1 and 1, where CCC = 1 means perfect agreement. Figure 1c illustrates the reproducibility assessment for two technical replicates run on different dates.
2.3 The dose–response model
The dose–response model can be expressed as a nonlinear function:
where is the vector of unknown parameters, is the measurement error for the response , is the total number of doses administered and is the number of repeats under the ith dose. The error term is assumed to be identically independently distributed as. In the case of the sigmoid Emax model, has the following form:
Here, is the vector of parameters to be estimated.
The dose–response models in drexplorer include all the models in the drc (Ritz and Streibig, 2005) and DoseFinding (Bornkamp, et al., 2010) packages and can be easily used through a uniform interface.
Parameters in the dose–response models can be estimated by minimizing the sum of the squared residuals (SSR):
After fitting the model, we estimate the IC at percentile by solving in equation where is the vector of the fitted parameters.
Different models can be fit, so we identify the best model as that with minimum residual standard error, calculated as SSR divided by degrees of freedom. Approaches for parametric dose–response models may use the Akaike information criterion or Bayesian information criterion.
2.4 The Interaction Index for Drug Combinations
The interaction index (IAI) is useful to quantify the degree of interaction in drug combinations. For a 2-drug combination, IAI can be defined as
Here, Dy,1 and Dy,2 and are the doses needed to induce response y for drugs 1 and 2 when administered alone; and are the doses in the mixture that produce the same response y. We adopt the source code used by Lee, et al. (2007) and Lee and Kong (2009) to estimate the IAI and 95% confidence interval (see Fig. 1b using UMSCC22B data; Lee and Kong, 2009). Graphical user interfaces (GUIs) help users without advanced programming skills to analyze dose–response data (Fig. 1d and e).
3 Conclusion
Drexplorer is a versatile R package encompassing several aspects of dose–response and drug–drug interaction analysis. The GUI enables biologists without programming skills to analyze their data.
Supplementary Material
Acknowledgements
This project was partially supported by the NCI/NIH through the Lung SPORE (P50 CA070907) and Cancer Center Support Grant (CA016672), and by the Mary K. Chapman Foundation.
Conflict of Interest: none declared.
References
- Barretina J., et al. (2012) The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature, 483, 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornkamp B., et al. (2010) DoseFinding: planning and analyzing dose finding experiments, R package version 0.4-1. http://www.icesi.edu.co/CRAN/web/packages/DoseFinding/DoseFinding.pdf. [Google Scholar]
- Lawrence I., Lin K. (1989) A concordance correlation coefficient to evaluate reproducibility. Biometrics, 255–268. [PubMed] [Google Scholar]
- Lee J.J., Kong M. (2009) Confidence intervals of interaction index for assessing multiple drug interaction. Stat. Biopharm. Res., 1, 4–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.J., et al. (2007) Interaction index and different methods for determining drug interaction in combination therapy. J. Biopharm. Stat., 17, 461–480. [DOI] [PubMed] [Google Scholar]
- Lehár J., et al. (2009) Synergistic drug combinations tend to improve therapeutically relevant selectivity. Nat. Biotechnol., 27, 659–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman D. (1939) The distribution of range in samples from a normal population, expressed in terms of an independent estimate of standard deviation. Biometrika, 31, 20–30. [Google Scholar]
- R Core Team. (2014) R: A Language and Environment for Statistical Computing. The R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org.
- Ritz C., Streibig J.C. (2005) Bioassay analysis using R. J. Stat. Softw., 12, 1–22. [Google Scholar]
- Yang W., et al. (2013) Genomics of drug sensitivity in cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res., 41, D955–D961. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.