Abstract
We synthesized and characterized two novel fluorescent sigma-2 receptor selective ligands, SW120 and SW116, and evaluated these ligands as potential probes for imaging cell proliferation. Both ligands are highly selective for sigma-2 receptors versus sigma-1 receptors. SW120 and SW116 were internalized into MDA-MB-435 cells, and 50% of the maximum fluorescent intensity was reached in 11 and 24 minutes, respectively. In vitro studies showed that 50% of SW120 or SW116 washed out of cells in 1 hour. The internalization of SW120 was reduced ≈30% by phenylarsine oxide, an inhibitor of endocytosis, suggesting that sigma-2 ligands are internalized, in part, by an endocytotic pathway. Subcellular localization studies using confocal and two-photon microscopy showed that SW120 and SW116 partially colocalized with fluorescent markers of mitochondria, endoplasmic reticulum, lysosomes, and the plasma membrane, suggesting that sigma-2 receptors localized to the cytoplasmic organelles and plasma membrane. SW120 did not colocalize with the nuclear dye 4′,6-diamidino-2-phenylindole. In vivo studies showed that the uptake of SW120 in solid tumors and peripheral blood mononuclear cells of mice positively correlated with the expression level of the cell proliferation marker Ki-67, suggesting that sigma-2 fluorescent probes may be used to image cell proliferation in mice.
Sigma receptors are a class of proteins that were originally thought to be a subtype of the opiate receptors.1 Subsequent studies revealed that sigma binding sites represent a distinct class of receptors.2–5 Two sigma binding site subtypes were distinguished based on differences in their drug binding profiles and molecular weight. The two binding sites are known as sigma-1 and sigma-2 receptors. The sigma-1 receptor has a molecular weight of ≈25 kDa, whereas the sigma-2 receptor has a molecular weight of ≈21.5 kDa. The sigma-1 receptor gene has been cloned from guinea pig liver, human placental choriocarcinoma, rat brain, and mouse kidney.6–8 The sigma-2 receptor has not yet been cloned.
Sigma-2 receptors are overexpressed in a variety of human and rodent tumors9–11 and have been validated as a biomarker of the proliferative status of solid tumors.12,13 These studies suggest that sigma-2 receptors can serve as a potential target for diagnostic tumor imaging agents and cancer chemotherapeutic drugs. Our group has developed sigma-2 receptor selective radioligands that have shown promise in imaging solid tumors with positron emission tomography (PET) in rodent models of cancer.14–17 A number of laboratories have explored the anticancer effects of sigma-2 ligands; proposed mechanisms of cell death include caspase-independent apoptosis,18 lysosomal leakage,19 Ca2+ release,20,21 oxidative stress,19 ceramide production,22 and autophagy.23
We previously synthesized and characterized two fluorescent sigma-2 receptor probes, SW107 and K05-138, which contain the sigma-2 selective ligand SV119 and a fluorophore, either dansyl chloride or 4-chloro-7-nitrobenzo-2-oxa-1,3-diazole (NBD chloride).24 The confocal and two-photon microscopy studies with these fluorescent compounds suggest that sigma-2 receptors are localized in the mitochondria, endoplasmic reticulum, lysosomes, and plasma membrane. Although very useful, these sigma-2 receptor fluorescent probes possess relatively low binding affinities for sigma-2 receptors (145 nM for SW107 and 45 nM for K05-138) compared to their precursor compound SV119 (5 nM).25 In the current study, we synthesized two novel fluorescent sigma-2 ligands, SW120 and SW116. Both compounds show improved sigma-2 receptor binding affinities. SW120 also shows greater selectivity for sigma-2 versus sigma-1 receptors. We have extensively characterized these two compounds in cell culture and evaluated SW120 as a proliferation probe in an animal model of breast cancer and melanoma. The data suggest that sigma-2 fluorescent ligands are useful reagents to study the biologic functions of sigma-2 receptors and can be potentially used as proliferation probes in vivo.
Materials and Methods
Materials
MitoTracker Red CMXRos, ER-Tracker Red dye, Lyso-Tracker Red DND-99, FM1-43FX, and FM4-64FX were purchased from Invitrogen Corporation (Carlsbad, CA). [3H]-Pentazocine (31.6 Ci/mmol) was purchased from PerkinElmer (Boston, MA). [3H]RHM-1 (80 Ci/mmol) was synthesized by American Radiolabeled Chemicals, Inc. (St. Louis, MO) via O-alkylation of the corresponding phenol precursor.14 Phenylarsine oxide (PAO) was purchased from Sigma Chemical Company (St. Louis, MO). Cell media were purchased from the Washington University Tissue Culture Center (St. Louis, MO). All other chemicals were purchased from Aldrich Chemical Company, Inc. (Milwaukee, WI) or Sigma Chemical Company.
Chemical Synthesis of N-(9-(10-(5-Dimethylamino-1-naphthalensulfonamido))decyl)-9-azabicyclo[3.3.1]-nonan-3α-yl)-N-(2-methoxy-5-methylphenyl)carbamate Oxalate (SW116)
A solution of dansyl chloride (590 mg, 2.18 mmol) in CH3CN (6 mL) was added dropwise to a mixture of the amine precursor SW43 (500 mg, 1.09 mmol) and K2CO3 (452 mg, 3.27 mmol) in CH3CN (5 mL). The reaction mixture was stirred at room temperature for 24 hours. The mixture was filtered, and volatiles were removed in vacuo. The product was purified by column chromatography (CH3OH: CH2Cl2: NH4OH 10:90:0.5) to give 734 mg of SW116 (97% yield) as a yellow oil. The oxalate salt was made for analysis; 1H NMR (free base, CDCl3) δ: 8.54 (d, J = 8.6 Hz, 1H), 8.29 (d, J = 8.6 Hz, 1H), 8.25 (dd, J = 1.2 and 7.8 Hz, 1H), 7.94 (br s, 1H), 7.50 to 7.59 (m, 2H), 7.18 (d, J = 7.4 Hz, 1H), 7.14 (s, 1H), 6.73 to 6.81 (m, 2H), 5.13 (q, J = 6.8 Hz, 1H), 4.65 (br s, 1H), 3.84 (s, 3H), 3.10 to 3.18 (m, 2H), 2.89 (s, 6H), 2.84 to 2.84 (m, 2H), 2.44 to 2.68 (m, 4H), 2.30 (s, 3H), 1.10 to 1.98 (m, 24H); MS (FAB+) exact mass calculated for C39H56N4O5S [M+H]+ 693.4050, found 693.4019.
Chemical Synthesis of N-9-(10-(7-nitrobenzo-2-oxa-1,3-diazol-4-ylamino)decyl)-9-azabicyclo[3.3.1]nonan-3α-yl-N-(2-methoxy-5-methylphenyl)carbamate (SW120)
A solution of 4-chloro-7-nitrobenzofurazan (100 mg, 0.5 mmol) in CH3OH (5 mL) was added dropwise to a mixture of the amine precursor SW43 (230 mg, 0.5 mmol) and NaHCO3 (50 mg, 0.6 mmol) in CH3OH (3 mL). The reaction mixture was stirred at room temperature for 3 hours. The mixture was filtered, and volatiles were removed in vacuo. The product was purified by column chromatography (CH3OH: CH2Cl2 8:92) to give 260 mg of SW120 (83% yield) as a dark red solid. 1H NMR (free base, CDCl3) δ: 8.48 (d, J = 8.6 Hz, 1H), 7.89 (s, 1H), 7.14 (s, 1H), 6.72 to 6.80 (m, 2H), 6.17 (d, J = 8.6 Hz, 1H), 5.11 to 5.19 (m, 1H), 3.85 (s, 3H), 3.48 to 3.54 (m, 2H), 3.30 to 3.36 (m, 2H), 2.60 to 2.84 (m, 4H), 2.28 (s, 3H), 1.30 to 2.20 (m, 25H); MS (electrospray) exact mass calculated for C33H46N6O6 [M+H]+ 623.3557, found 623.3560.
Receptor Binding Assays
The sigma-1 and sigma-2 receptor binding affinities of SW120 and SW116 were determined as previously described.26 Briefly, guinea pig brain (sigma-1 assay) or rat liver (sigma-2 assay) membrane homogenates (≈300 μg protein) were diluted with 50 mM Tris-HCl, pH 8.0, and incubated with either ≈5 nM [3H](+)-pentazocine (34.9 Ci/mmol; sigma-1 assay) or 1 nM [3H]RHM-1 (80 Ci/mmol; sigma-2 assay) in a total volume of 150 μL in 96-well plates at 25°C. The concentrations of SW120 and SW116 ranged from 0.1 nM to 10 μM. After incubating for 60 minutes, the reactions were terminated by the addition of 150 μL of cold wash buffer (10 mM Tris-HCl, 150 mM NaCl, pH 7.4) using a 96-channel transfer pipette (Fisher Scientific, Pittsburgh, PA), and the samples were harvested and filtered rapidly into a 96-well fiberglass filter plate (Millipore, Billerica, MA) that had been presoaked with 100 μL of 50 mM Tris-HCl at pH 8.0 for 1 hour. Each filter was washed three times with 200 μL of ice-cold wash buffer, and the bound radioactivity was quantified using a Wallac 1450 MicroBeta liquid scintillation counter (PerkinElmer). Nonspecific binding was determined in the presence of 10 μM cold haloperidol.
Absorption, Excitation, Emission Spectra, and Molar Extinction Coefficients (ε) of SW120 and SW116
The absorbance spectra of the tested compounds dissolved in methanol (10 μM) were collected using ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) with an optical path length of 1 mm. The excitation and emission spectra were then collected in standard quartz cuvettes using a LS 50B spectrofluorometer (PerkinElmer, Waltham, MA) with excitation and emission slits set to 3 nm. To determine the excitation spectra, SW120 was illuminated at wavelengths ranging from 300 to 500 nm, and the fluorescent emission intensity was collected at 520 nm. To determine the emission spectra of SW120, the excitation wavelengths were set to 335 and 465 nm, and the emission spectra were recorded. The excitation spectra of SW116 were recorded by varying the illumination wavelength between 300 and 500 nm and collecting the fluorescence at 506 nm. To determine the emission spectra of SW116, the excitation wavelengths were set to 333 nm, and the emission spectra were recorded.
To determine the molar extinction coefficients (ε), stock solutions of the tested compounds were diluted with methanol to the desired concentrations (1–20 μM) and absorbance spectra (300–700 nm) were collected using Beckman DU650 spectrophotometer (Beckman Coulter, Fullerton, CA) in standard quartz cuvettes. The absorbance values (A) at absorption maxima were then plotted against the compound concentration (c), and the molar extinction coefficients (ε) were determined by fitting the Beer law:
where d was the optical path length (d = 1 cm) to the experimental data using the linear regression method (Sigma Plot, Systat, San Jose, CA). The measurements were repeated three to four times and presented as mean ± SE.
Cell Culture Conditions
Human melanoma MDA-MB-435 cells were grown in Minimum Essential Medium (MEM) containing 10% fetal bovine serum, 2 mM l-glutamine, 1 mM sodium pyruvate, 1× non-essential amino acids, 2% MEM vitamins, and 1× penicillin-streptomycin solution. The cells were maintained at 37°C in a humidified incubator with a 5% CO2/95% air atmosphere. MDA-MB-435 cells were seeded on 35 mm glass-bottomed dishes at 2 × 105 cells/dish for 24 hours prior to initiating any treatment. For studying subcellular localization of SW120, cells were incubated with 30 nM SW120 and one of the subcellular organelle fluorescent markers (50 nM MitoTracker, 0.5 μM ERTracker, 50 nM LysoTracker, 300 nM 4′,6-diamidino-2-phenylindole [DAPI]) at 37°C for 2 hours or incubated with 50 nM SW120 and a plasma membrane marker (FM4-64FX) in Hank's Buffered Salt Solution (HBSS) buffer, which does not contain Ca2+ and Mg2+, at 0°C for 15 minutes. Cells were then imaged using a confocal laser scanning microscope. For studying the subcellular localization of SW116, cells were incubated with 100 nM SW116 and one of the subcellular organelle fluorescent markers (20 nM MitoTracker, 0.5 μM ERTracker, 100 nM LysoTracker) at 37°C for 2 hours or incubated with 100 nM SW116 and 5 μg/mL plasma membrane marker (FM1-43FX) in HBSS buffer, which does not contain Ca2+ and Mg2+, at 0°C for 15 minutes. Cells were then imaged using a two-photon microscope.
Two-Photon Microscopy
Cells loaded with respective compounds were imaged using a multiphoton microscope (LSM510 META NLO, Carl Zeiss Microimaging, Thornwood, NY) with a tunable Ti-Sapphire Chameleon XR laser (Coherent, Santa Clara, CA). The in situ emission spectra were collected at fixed excitation wavelengths (735 and 800 nm for SW116 and SW120, respectively) by acquiring individual images at emission wavelengths ranging from 400 to 600 nm at 10 nm intervals using the META head. To determine the in situ excitation spectra, images excited at 720 to 900 nm were collected at 10 nm intervals through 500 to 550 nm emission filter, a range corresponding to maximum emission of the compounds. The images of SW116 and SW120 in cells were collected using an emission fingerprinting technique27 to separate the actual compound fluorescence from spectrally similar autofluorescence of cells. The emission spectra of compounds in buffers and unstained cells (autofluorescence) acquired on the same system were used as references.
For colocalization experiments, the excitation wavelength was set at 740 nm for SW116, and the emission was collected using a 480 to 520 nm bandpass filter. MitoTracker, ER-Tracker, LysoTracker, and FM1-43FX were excited using the 543 nm line from a helium-neon laser, and the emission was collected using a 558 to 665 nm bandpass filter. The cells were viewed with a 40×/0.8 NA water immersion lens. To reduce interchannel crosstalk, a multitracking technique was used. Images were taken at a resolution of 1,024 × 1,024 pixels. Two-photon scanning parameters were set up so that the cells in the well without the compounds had no fluorescent signal.
Confocal Microscopy
A confocal laser scanning microscope (LSM5 Pascal, Carl Zeiss Microimaging) was used. SW120 was excited using the 488 nm line from an argon laser, and the emission was collected through a 505 to 530 nm bandpass filter. MitoTracker, ER-Tracker, LysoTracker, or membrane marker (FM4-64FX) was excited using the 543 nm line from a helium-neon laser, and the emission was collected with a 560 nm long-pass filter. The nuclear dye (DAPI) was excited using the 405 nm line from a diode laser, and the emission was collected with a 420 to 480 nm bandpass filter. A multitracking technique was used to reduce interchannel crosstalk. Image acquisition was performed using a 40×/1.20 NA water immersion lens. Images were taken at a resolution of 1,024 × 1,024 pixels. Confocal scanning parameters were set up so that the cells in the well without the compounds had no fluorescent signal.
Flow Cytometry
Flow cytometric analysis was performed using a FACScan (Becton Dickinson, Franklin Lakes, NJ) equipped with an air-cooled argon laser. SW120 was excited at a wavelength of 488 nm, and its emission was collected at a wavelength of 550 nm. Phycoerythrin (PE)-conjugated Ki-67 antibody (BD Pharmingen, San Jose, CA) was excited at 488 nm, and its emission was collected with a 570 nm filter.
Kinetics Studies of the Internalization and Efflux of SW120 and SW116 in MDA-MB-435 Cells
MDA-MB-435 cells were plated in 35 mm diameter glass-bottomed dishes at 2 × 105 cells/dish for 24 hours. For the internalization kinetics study, 10 nM SW120 or 100 nM SW116 was added to the cell culture dishes, and images were taken at the time intervals indicated in Figure 1. For the efflux kinetics study on washout, the cells were incubated with 10 nM SW120 or 100 nM SW116 for 2 hours. The medium with SW120 or SW116 was then removed, and the cells were incubated with fresh medium without SW120 or SW116 for 1, 2, 3, 4, 5, and 6 hours. At each time point of incubation, images were taken. The fluorescent intensity for each cell was determined using either an inverted confocal microscope for SW120 or a multiphoton microscope for SW116. About 20 cells (from at least three fields) were analyzed in each dish. For internalization studies of SW120 or SW116, the average intensity of the cells versus time was fitted by equation A using PRISM software (GraphPad Software, Inc., San Diego, CA):
where I is the fluorescent intensity at time point, T, Imax is the maximum fluorescent intensity, and T1/2 is the time at which the intensity, I, equals one half of Imax.
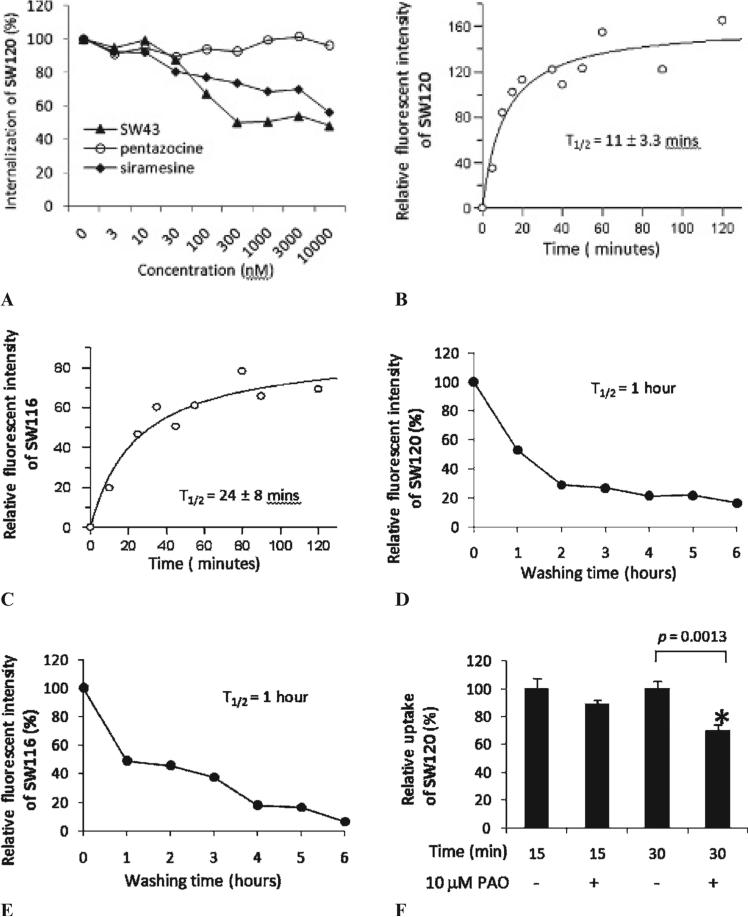
Figure 1.
Characterization of SW120. A, Flow cytometric determination of the internalization of SW120 in MDAMB-435 cells with and without blocking by SW43 (solid triangles), sirame-sine (solid diamonds), or (+)-pentazocine (open circles). B, Kinetics for the internalization of SW120 in MDA-MB-435 cells. C, Kinetics for the internalization of SW116 in MDAMB-435 cells. D, Kinetics for the efflux of SW120 in MDA-MB-435 cells. E, Kinetics for the efflux of SW116 in MDA-MB-435 cells. F, The inhibition of SW120 internalization by phenyl-arsine oxide (PAO). MDA-MB-435 cells were preincubated with or without 10 μM PAO for 30 minutes at 37°C and then incubated with 10 nM SW120 for an additional 15 and 30 minutes. The cells were analyzed by flow cytometry. The internalization of SW120 was significantly reduced by 10 μM PAO (*p < .005).
Tumor Implantation and Tissue Harvesting
All animal experiments were conducted in compliance with the National Institutes of Health (NIH) guidelines for the care and use of laboratory animals, with the approval of Washington University's Animal Studies Committee. Approximately 1.5 × 106 exponentially growing murine mammary adenocarcinoma 66 or 1 × 107 exponentially growing human melanoma MDA-MB-435 cells were injected subcutaneously in the axial region of adult (20– 25 g) female nude mice. Similarly, approximately 1.5 × 105 exponentially growing mouse mammary carcinoma cell line EMT6 were implanted in adult female BALB/c mice. The tumors were allowed to develop for 2 to 3 weeks before fluorescent probe administration. The tumor sizes were ≈0.4 g for murine 66 tumors, ≈0.7 g for human MDA-MB-435 tumors, and ≈0.4 g for murine EMT6 tumors. Mice were euthanized 1 hour after intravenous injection of 50 μg SW120 in 100 μL phosphate-buffered saline (PBS). Blood was taken by cardiac puncture under isoflurane/oxygen anesthesia for preparation of peripheral blood mononuclear cells (PBMCs) as a control tissue, whereas tumors were removed, minced with fine scissors, and then placed in a flask with 25 to 30 mL of an enzyme cocktail consisting of 0.04% collagenase, 0.04% pronase, and 0.05% DNAase I in Waymouth's medium. After incubating for 45 minutes at 37°C with continuous stirring, the cell suspension was filtered to remove large pieces of tissue. The filtrate was centrifuged for 5 minutes at 225g at 4°C, and the pellet was resuspended in Waymouth's medium containing 10% serum for flow cytometry analysis.
Preparation of the PBMCs
Mouse blood samples were collected in heparinized syringes. Blood was diluted 1:1 with PBS, layered onto Histopaque-1083 (Sigma Chemical Company) at a volume ratio of 1:1, and centrifuged at 300g for 15 minutes. The PBMC layer was removed and washed once with PBS and used as nonproliferative control cells.
Statistical Analyses
The results are expressed as the mean ± SD of two or three independent experiments performed in triplicate. Comparisons between two experimental groups were performed using a two-tailed Student t-test. A p value < .05 was considered significant.
Results
Chemical Synthesis and Characterization of the Sigma-2 Selective Fluorescent Ligands
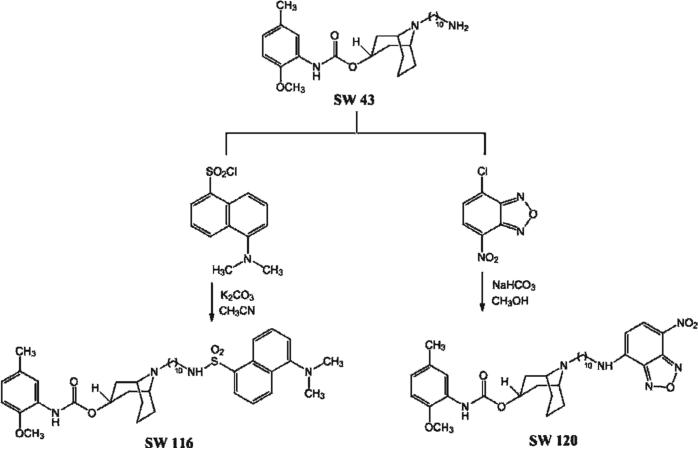
We prepared two novel sigma-2 fluorescent probes that can be used with confocal microscopy or two-photon microscopy. The synthesis of the precursor SW43 and its binding affinities for sigma-1 and sigma-2 receptors have been reported previously (Kiσ1 = 134 nM and Kiσ2 = 7 nM).25 The primary amine SW43 was condensed with the fluorophore NBD chloride in the presence of sodium bicarbonate to form the sigma-2 selective fluorescent ligand SW120 in 83% yield. The reaction between SW43 and dansyl chloride in the presence of potassium carbonate gave compound SW116 in 97% yield (Figure 2).
Figure 2.
The synthetic scheme for generating SW120 and SW116.
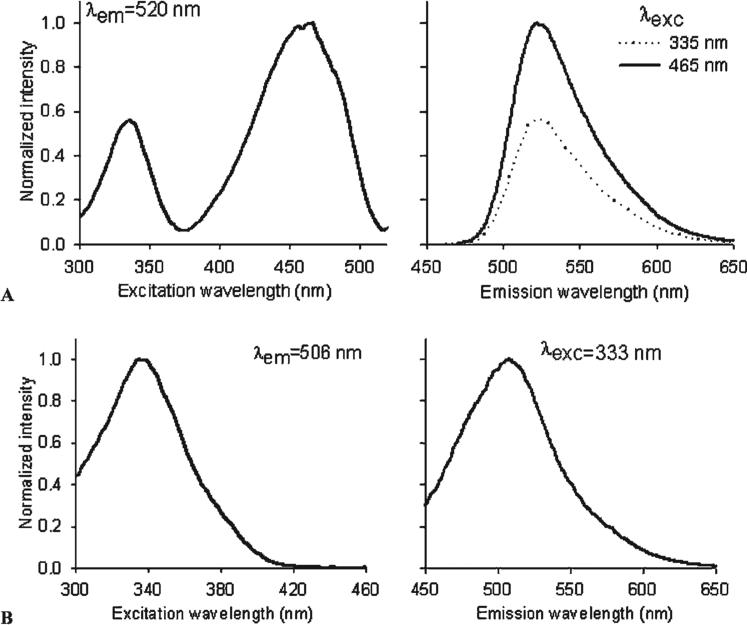
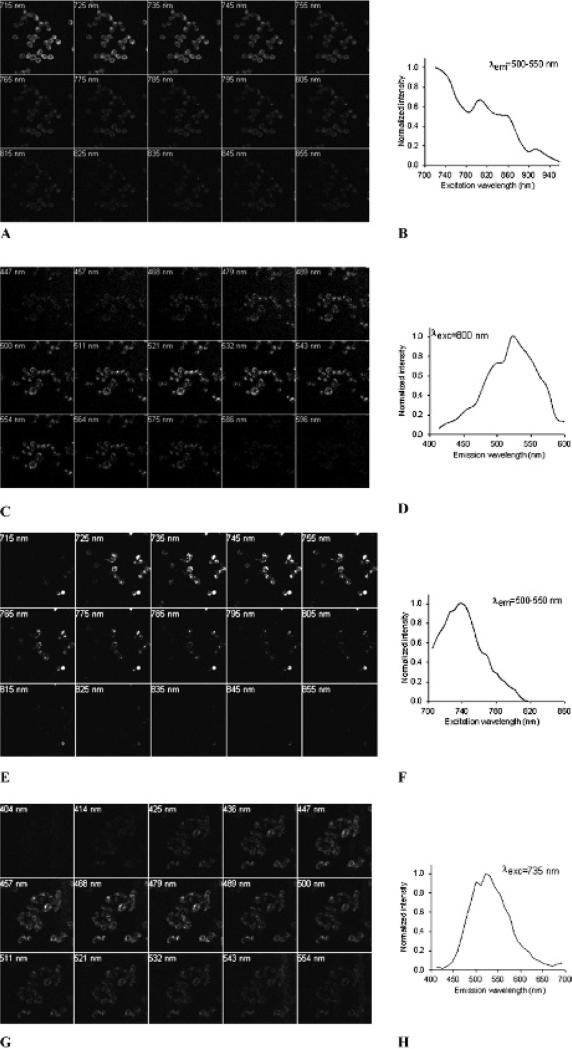
In vitro binding studies were conducted to determine the affinities of SW120 and SW116 for sigma-1 and sigma-2 receptors. Both SW120 and SW116 were found to be highly selective for sigma-2 receptors versus sigma-1 receptors. The sigma-2 receptor inhibition constants (Kiσ2) for SW120 and SW116 were 11 nM and 14 nM, respectively, as determined by inhibiting the binding of [3H]RHM-1 to rat liver membrane homogenates. SW120 and SW116 both had low affinities for sigma-1 receptors (Kiσ1 = 450 nM for SW120 and Kiσ1 = 1,055 for SW116). The excitation and emission spectra of SW120 and SW116 in methanol were obtained using a spectrofluorometer. SW120 displayed excitation peaks at two different wavelengths, 335 and 465 nm. The maximum emission wavelength for both peak excitation wavelengths was 525 nm (Figure 3A). SW116 had a peak excitation wavelength of 333 nm and a maximal emission wavelength of 506 nm (Figure 3B). We also acquired in situ excitation and emission spectra for both SW120 (Figure 4, A–D) and SW116 (Figure 4, E–H) using multiphoton microscopy when these ligands were internalized in live cells (MDAMB-435). Both compounds share similar emission wavelengths between 500 and 550 nm. The data indicate that the SW120 can be excited in situ using either the 488 nm standard argon laser line or a 800 nm multiphoton excitation, whereas SW116 is most efficiently imaged using an excitation wavelength of 730 to 740 nm.
Figure 3.
Excitation and emission spectra of SW120 (A) and SW116 (B) in methanol. The concentration of SW120 or SW116 is 10 μM.
Figure 4.
In situ excitation and emission spectra of SW120 (A–D) and SW116 (E–H) in MDA-MB-435 cells determined with two-photon microscopy. A, Images of excitation spectra of SW120. B, An excitation spectra chart of SW120 displaying the normalized fluorescent intensity versus excitation wavelengths. C, Images of emission spectra of SW120. D, An emission spectra chart of SW120 displaying the normalized fluorescent intensity versus emission wavelengths. E, Images of excitation spectra of SW116. F, An excitation spectra chart of SW116 showing the normalized fluorescent intensity versus excitation wavelengths. G, Images of emission spectra of SW116. H, An emission spectra chart of SW116 showing the normalized fluorescent intensity versus emission wavelengths.
Molar extinction coefficients (ε) of SW120 and SW116 were determined. The molar extinction coefficient (ε) of SW120 is 29,516 ± 482 at 465 nm, whereas the molar extinction coefficient (ε) of SW116 is 7,226 ± 119 at 333 nm. These data indicate that the fluorescent intensity of SW120 is stronger than that of SW116.
To study whether these sigma-2 receptor fluorescent ligands selectively bind to sigma-2 receptors and not sigma-1 receptors in live tumor cells, a series of blocking experiments were performed using a sigma-1 selective ligand, (+)-pentazocine, and the sigma-2 selective ligands SW43 and siramesine. MDA-MB-435 cells were preincubated with the sigma-1 or sigma-2 selective ligand for 1 hour at 37°C at concentrations ranging from 3 to 10,000 nM. The cells were then treated with 10 nM of SW120 for 30 minutes, and the fluorescent intensity of the labeled cells was analyzed by flow cytometry. The data indicate that SW43 and siramesine blocked the binding of SW120 in a concentration-dependent manner (Figure 1A). Approximately 52% and 44% of the binding of SW120 was blocked by SW43 or siramesine at a concentration of 10 μM, respectively. In contrast, (+)-pentazocine did not block the binding of SW120 at 10 μM, the highest concentration used in this study. These data suggest that SW120 enters the cell by both receptor-mediated and passive diffusion mechanisms.
Kinetic Studies of the Internalization and Efflux of SW120 or SW116 in MDA-MB-435 Cells
The kinetic internalization of SW120 and SW116 into MDA-MB-435 cells was studied using confocal microscopy and two-photon microscopy, respectively. MDA-MB-435 cells were incubated with 10 nM SW120 or 100 nM SW116 for varying lengths of time as indicated in Figure 1, B and C, fluorescent images were taken at each time point, and the fluorescent intensity of SW120 or SW116 in the cells was determined. After fitting the data using the equation described in the Materials and Method section, the length of time for SW120 or SW116 to reach 50% of the maximal fluorescent intensity (T1/2) was 11 and 24 minutes, respectively.
The kinetic washout of SW120 or SW116 from the cells was also studied by confocal or two-photon microscopy, respectively. MDA-MB-435 cells were incubated with 10 nM SW120 or 100 nM SW116 at 37°C for 2 hours. Then the SW120 or SW116 was removed and the cells were incubated in medium without the fluorescent ligand for varying lengths of time (1–6 hours). The fluorescent images were taken at each time point, and the fluorescent intensity of the labeled cells was determined. Figure 1, D and E, shows that the fluorescence intensity of labeled cells decreased with time, indicating that SW120 and SW116 were washed out from the cells. After 1 hour, 50% of SW120 or SW116 had washed out of the cells. This time course was also confirmed by flow cytometry analysis for SW120 (data not shown). It appears that washout occurs in two phases. The initial phase (0–2 hours for SW120, 0–1 hour for SW116) is more rapid than the later phase (2–6 hours for SW120, 1–6 hours for SW116). It is possible that the initial phase is due to the efflux of free ligand from the cells, whereas the later phase may reflect the efflux of sigma-2 receptor-bound ligand, which may take a longer time to dissociate from the receptor before it can be washed out.
To study whether the internalization of sigma-2 receptors is mediated by endocytosis, we examined the effect of PAO, a well-characterized endocytosis inhibitor,28 on the internalization of SW120. MDA-MB-435 cells were pretreated with 10 μM PAO for 30 minutes and then treated with 10 nM SW120 in the absence or presence of PAO for an additional 30 minutes. Flow cytometry analysis (Figure 1F) showed that 10 μM PAO significantly (p = .0013) blocked internalization of SW120 by 30%. These data demonstrate that 30% of the sigma-2 receptor ligand was internalized by an endocytosis-mediated mechanism, whereas the remaining 70% was internalized by other mechanisms, such as passive diffusion.
Colocalization of SW120 and Subcellular Organelle Markers by Confocal Microscopy
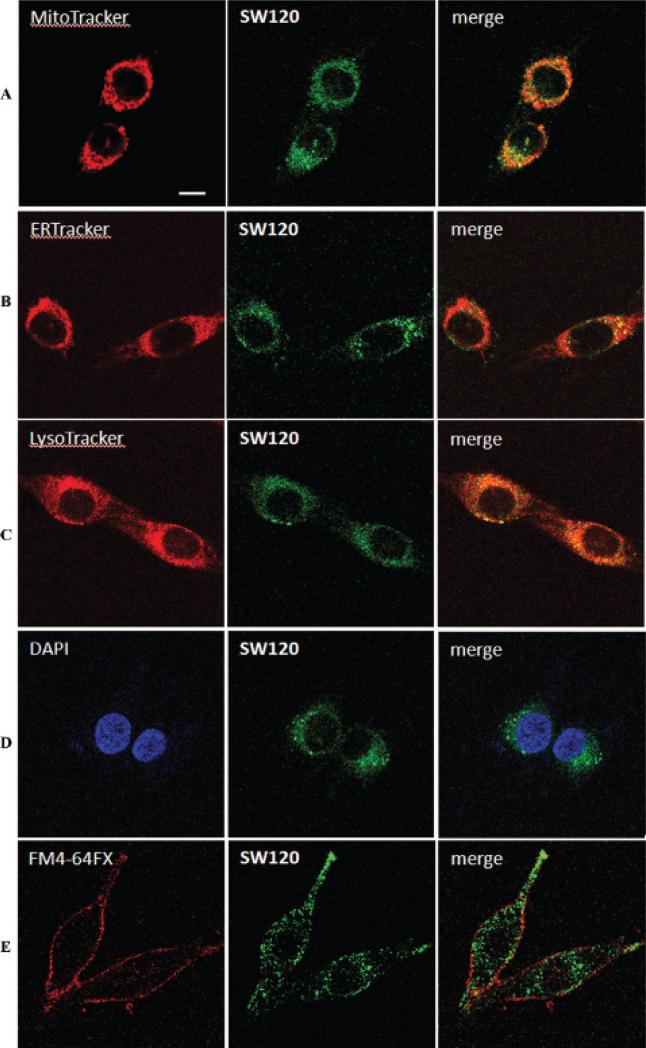
The subcellular localization of sigma-2 receptors was studied using SW120 and subcellular organelle markers. MDA-MB-435 cells were incubated with 30 nM SW120 and each of the four subcellular organelle markers at 37°C for 2 hours: the mitochondria tracker MitoTracker Red CMXRos (20 nM), the endoplasmic reticulum tracker ER-Tracker Red (500 nM), the lysosome tracker LysoTracker Red DND-99 (50 nM), or the nuclear marker DAPI (300 nM). The results show that SW120 distributes in the cytoplasm of cells in two forms: an evenly distributed form and a punctate form. SW120 appears to partially colocalize with MitoTracker, ER-Tracker, and LysoTracker, suggesting that sigma-2 receptors may localize in mitochondria, endoplasmic reticulum, and lysosomes (Figure 5). The data also showed that SW120 does not colocalize with the nuclear marker DAPI, suggesting that either the sigma-2 receptor does not exist in the nucleus or SW120 does not enter the nucleus. MDA-MB-435 cells were also incubated with 50 nM SW120 and the plasma membrane tracker, FM 4-64FX (5 μg/mL), for 15 minutes at 0°C. The results showed that SW120 is also colocalized with the plasma membrane tracker, suggesting that sigma-2 receptor may partially localize in the plasma membrane.
Figure 5.
Determination of the intra-cellular distribution of SW120 in MDA-MB-435 cells with and without MitoTracker (A), ER-Tracker (B), LysoTracker (C), a nuclear marker, DAPI (D), or a membrane tracker, FM4-64FX (E), using confocal microscopy. MDA-MB-435 cells were incubated with 30 nM SW120 and either 20 nM MitoTracker, 500 nM ER-Tracker, 50 nM LysoTracker, or 300 nM DAPI. After incubating for 2 hours at 37°C, live cells were imaged by confocal microscopy. MDA-MB-435 cells were also incubated with 50 nM SW120 and 5 μg/mL of the membrane tracker, FM4-64FX, for 15 minutes at 0°C. The live cells were imaged by confocal microscopy. Scale bar = 10 μm.
Colocalization of SW116 and Subcellular Organelle Markers by Two-Photon Microscopy
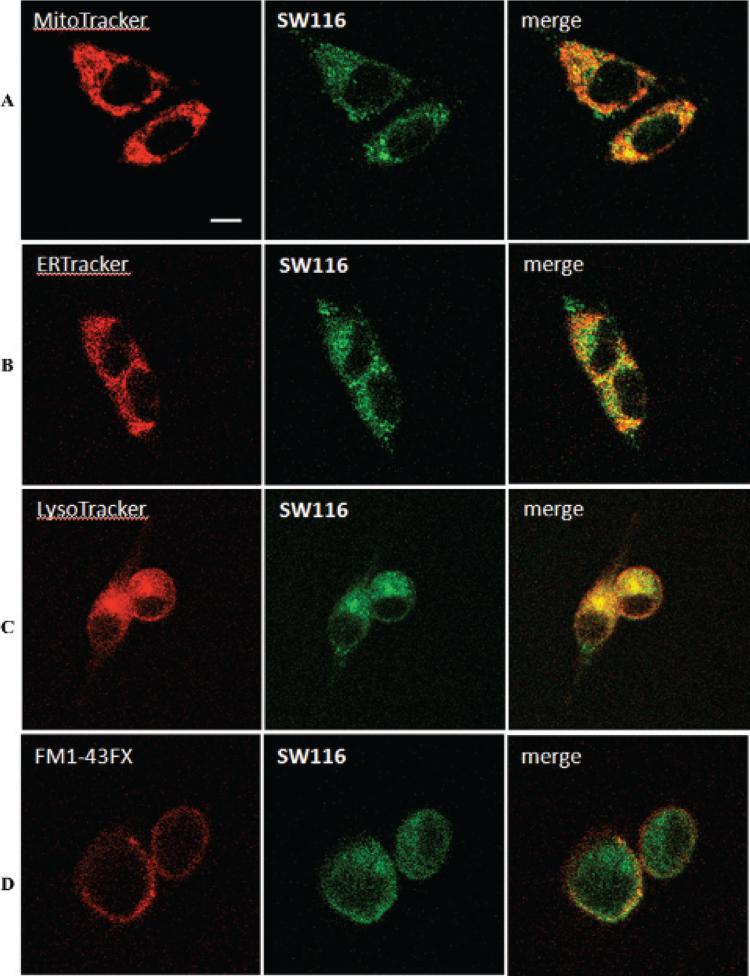
To study the subcellular localization of sigma-2 receptors, MDA-MB-435 cells were incubated with 100 nM SW116 and one of the four subcellular organelle markers as described above for confocal microscopy: the mitochondria tracker MitoTracker Red CMXRos (20 nM), the endoplasmic reticulum tracker ER-Tracker Red (500 nM), the lysosome tracker LysoTracker Red DND-99 (100 nM), or the plasma membrane tracker FM 1-43FX (5 μg/mL). The live cells were then imaged by two-photon microscopy. Our results show that SW116 is distributed throughout the cytoplasm of the cells (Figure 6). The SW116 staining was highly punctate, suggesting that the label was sequestered in small, membrane-bound compartments. The data demonstrate that SW116 partially colocalizes with the Mito-Tracker, ER-Tracker, LysoTracker, and plasma membrane tracker, suggesting that sigma-2 receptors are localized in the mitochondria, lysosomes, endoplasmic reticulum, and plasma membrane.
Figure 6.
Determination of the intra-cellular distribution of SW116 in MDA-MB-435 cells with and without MitoTracker (A), ER-Tracker (B), LysoTracker (C), or a membrane tracker, FM1-43FX (D), using two-photon microscopy. MDA-MB-435 cells were incubated with 100 nM SW116 and either 20 nM Mito-Tracker, 500 nM ER-Tracker, or 100 nM LysoTracker for 2 hours at 37°C. MDA-MB-435 cells were also incubated with 100 nM SW116 and 5 μg/mL of the membrane tracker, FM1-43FX, for 15 minutes at 0°C. The live cells were imaged by two-photon microscopy.
SW120 Preferentially Labels Murine and Human Tumors rather than PBMCs In Vivo
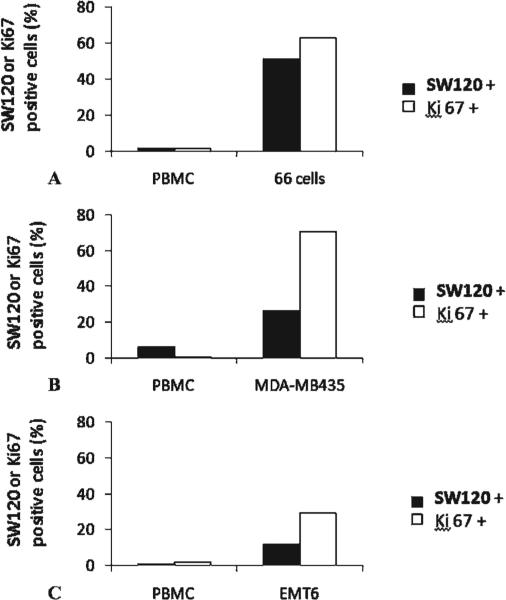
The sigma-2 receptor has been validated as a proliferation marker in cell culture and in solid tumors. We tested the hypothesis that sigma-2 selective ligands preferentially label proliferating cells versus nonproliferating cells in vivo. Nude mice implanted with either murine mammary adenocarcinoma 66 cells or human melanoma MDA-MB-435 cells received an injection of 50 μg SW120 in 100 μL PBS. BALB/c mice implanted with mouse mammary carcinoma cell line EMT6 were also treated with SW120 (50 μg in 100 μL PBS). One hour later, the mice were euthanized. Blood was taken, and the PBMCs, which are commonly used as controls for nonproliferative cells, were prepared. Tumors were removed, and the tumor cells were dissociated. The dissociated tumor cells and the isolated PBMCs were analyzed for SW120-labeled cells by flow cytometry. These cells were also analyzed by flow cytometry for Ki-67 expression, a commonly used proliferation marker. Our data showed that PBMCs were Ki-67 negative, whereas a large portion of the dissociated tumor cells were Ki-67 positive in all three tumor models (Figure 7). The data also showed that PBMCs were not labeled by SW120, whereas a portion of the tumor cells were labeled with SW120. The observation that not all Ki-67-positive cells were labeled with SW120 may be due to the relatively high molecular weight (622.76 kDa) and lipophilicity (ClogP = 8.56) of the fluorescent probe, which may prevent the labeling of Ki-67-positive cells in poorly perfused regions of the tumor. The trend of the positive correlation between Ki-67 expression and SW120 labeling implies that the fluorescent sigma-2 ligand may possess in vivo selectivity toward proliferating cells versus nonproliferative cells.
Figure 7.
Ki-67 expression and SW120 fluorescent intensity in solid tumors and peripheral blood mononuclear cells (PBMCs) of mice by flow cytometric analysis. The female nude mice implanted with either murine mammary adenocarcinoma 66 cells (A) or human melanoma MDA-MB-435 (B) or BALB/C mice implanted with mouse mammary carcinoma cell line EMT6 (C) were intravenously injected with SW120 (50 μg/mouse), and tissues were harvested after 1 hour. The 66 cells, EMT6 cells, or MDA-MB-435 cells were dissociated from the solid tumors. PBMCs were prepared from the blood of mice. The cells were analyzed for the fluorescent intensity of SW120, and the Ki-67 expression was determined by Ki-67 immunostaining using a flow cytometer.
Discussion
The development of fluorescent ligands with high affinity and selectivity for sigma-2 receptors is important for studying the biologic functions of the sigma-2 receptor. These two novel sigma-2 fluorescent ligands can be imaged by confocal and two-photon microscopy, permitting flow cytometry studies and the use of optical imaging systems in vitro and in vivo, and can thus provide both temporal and spatial information regarding the interactions between sigma-2 ligands and sigma-2 receptors. Such information will be helpful for understanding the pharmacokinetics of sigma-2 ligands as imaging and chemotherapeutic reagents in animal models and in future human studies. In the current study, we synthesized and characterized two new sigma-2 fluorescent ligands, SW120 and SW116. Both SW120 and SW116 are highly selective for sigma-2 receptors versus sigma-1 receptors. Studies of their subcellular localization have shown that both ligands partially colocalize with the mitochondria, endoplasmic reticulum, lysosome, and plasma membrane trackers. SW120 does not colocalize with the nuclear marker DAPI. Kinetic studies of the internalization and efflux of the fluorescent ligands have shown that SW120 and SW116 were each internalized into cells, reaching half of the maximal fluorescent intensity in 11 or 24 minutes, respectively, and subsequently washed out of cells, decreasing their fluorescent intensities to half of the maximum value in 1 hour. We also explored the possibility that the fluorescent sigma-2 ligands can be used to image proliferating cells in vivo by correlating the uptake of SW120 with Ki-67 expression in solid tumors and PBMCs in mice. Our data suggest that fluorescent sigma-2 ligands are useful for studying biologic functions of the sigma-2 receptor and could be used to image solid tumors in vivo by optical imaging methods.
In the current study, SW120 and SW116 were synthesized by treating the primary amine SW43 with the fluorophores NBD chloride and dansyl chloride, respectively. In our previous study, we prepared K05-138 and SW107 by reacting the primary amine SV119 with the fluorophores NBD chloride and dansyl chloride, respectively.24 The difference between SW43 and SV119 is that SW43 has a 10-methylene group between the primary amino group and the bridgehead nitrogen atom, whereas SV119 has a six-methylene linker group. SW120 showed a fourfold increase in the binding affinity at sigma-2 receptors (Kiσ2 = 11 nM) and a 1.7-fold increase in the selectivity at sigma-2 versus sigma-1 receptors compared to K05-138. SW116 showed a 10-fold increase in the binding affinity at sigma-2 receptors (Kiσ2 = 14 nM), with no significant difference in selectivity at sigma-2 versus sigma-1 receptors compared to SW107. The increase in binding affinities is likely due to the longer 10-carbon spacer, which further separates the sigma-2 receptor binding moiety from the fluorophore and thus reduces the interference of the fluorophore with the binding of the probe to the sigma-2 receptor. The increase in binding affinity and selectivity allows the use of lower concentrations of these new sigma-2 ligands and subsequently decreases nonspecific binding of the sigma-2 ligands to other proteins.
We have determined the excitation and emission spectra for SW120 and SW116 in both methanol and live cells. When the fluorescent ligands are internalized into cells, they can interact with the cytoplasmic environment and may have different excitation and emission spectra from those determined in methanol. Therefore, the excitation/emission wavelength for confocal and two-photon images based on the spectra in methanol may not be suitable for imaging the sigma-2 ligand in live cells. In the current study, we measured the excitation and emission spectra of the sigma-2 ligands when they are in live cells with multiphoton laser scanning confocal microscopy system. The data showed that the maximal emission wavelengths of the two ligands are similar to those in methanol, suggesting that the cellular environment had little effect on the fluorescence emission wavelengths of the two ligands. On the other hand, we found that fluorescence intensity of SW120 and SW116 dramatically increased as they entered cells. It is possible that SW120 and SW116 interact with the hydrophobic membrane-rich environment in the cell and thereby increase the fluorescent intensity. It is also possible that SW120 and SW116 accumulate in the cells and that their concentrations inside cells may be higher than those in the media.
To study the selectivity of fluorescent sigma-2 ligands in live cells, in vitro blocking studies were performed. The internalization of SW120 was blocked by sigma-2 ligands SW43 and siramesine by approximately 52% and 44%, respectively, but was not affected by (+)-pentazocine, a sigma-1 receptor ligand. The data suggested that SW120 internalization is mediated, in part, by sigma-2 receptors. The remainder of the uptake of SW120 is believed to occur via a passive diffusion mechanism, which is not blocked by the sigma-2 receptor ligands.
We studied the subcellular localization of SW120 and SW116. Our data showed that SW120 and SW116 largely colocalized with lysosome trackers. There are at least three explanations for the lysosomal localization: (1) sigma-2 receptors localize in lysosomes; (2) SW120 and SW116 are internalized into cells by endocytosis-mediated mechanisms, and then the endosomes are fused to lysosomes29; and (3) SW120 and SW116 are basic molecules and are trapped into acidic organelles such as lysosomes. The lysosomal localization of SW120 and SW116 is consistent with the report that siramesine, a sigma-2 ligand, induces cell death by destabilizing lysosomes.19 We have also shown that SW120 and SW116 partially colocalize with the trackers for mitochondria and endoplasmic reticulum. These data suggest that sigma-2 receptors may exist in mitochondria and endoplasmic reticulum, consistent with the previous studies that sigma-2 ligands trigger apoptosis in tumor cells by acting on mitochondria and modulating Ca2+ release from endoplasmic reticulum.18,20,21,24 The data are also consistent with our observation by transmission electron microscopy that the sigma-2 ligand siramesine induces distortion of mitochondria (unpublished data). The partial colocalization of SW120 and SW116 with plasma membrane markers suggested that sigma-2 receptor localized in the plasma membrane. It is possible that SW120 and SW116 bind to functional structures of the membrane such as lipid rafts for initiating signal transduction pathways. The data are consistent with the report that sigma-2 receptors exist in lipid rafts.30 Costaining cells with SW120 and a nuclear marker, DAPI, showed that SW120 localized in organelles in the cytoplasm but not in the nucleus. The observation suggested that either sigma-2 receptor does not exist in the nucleus or that SW120 cannot cross the nuclear membrane. The sigma-2 receptor binding assay on the isolated nuclei should be performed to distinguish these two possibilities. In addition, although the data suggest that the sigma-2 receptors may localize in mitochondria, endoplasmic reticulum, lysosomes, and plasma membrane, these data are solely based on the confocal and two-photon microscopy studies. Owing to the resolution limitation of this method, the subcellular localization of the sigma-2 receptors needs to be further studied using different experimental methods. For example, the sigma-2 receptor densities should be determined on the isolated mitochondria.
Our previous data showed that sigma-2 receptor is a biomarker of cell proliferation. The sigma-2 receptor density in proliferating mouse mammary adenocarcinoma cells is 10 times higher than that in the quiescent cells.12,13 We questioned whether sigma-2 ligands selectively label proliferating cells versus nonproliferating cells. The current in vivo studies showed that SW120 labels tumor cells but not PBMCs. Our data also showed that a large portion of tumor cells are Ki-67 positive, whereas the PBMCs are Ki-67 negative. The positive correlation between SW120-positive cell numbers and Ki-67-positive cell numbers suggests that SW120, to a certain degree, has selectivity for proliferating cells versus nonproliferating cells. These data suggest that fluorescent sigma-2 selective ligands may be used as optical imaging probes for cell proliferation.
The current studies suggest that sigma-2 selective ligands may be useful in various applications, including noninvasive diagnosis and chemotherapy of cancer. For example, SW43 or SV119 can be used as a scaffold for a PET tracer to image proliferating tumor cells in cancer patients. With respect to chemotherapy, sigma-2 selective ligands may be used as antitumor drugs to selectively target proliferating tumors and reduce toxicity to non-proliferating normal cells. In fact, several groups have reported that the sigma-2 selective ligands show antitumor efficacy in cell culture and in animal models.18,19,31
Conclusion
We synthesized and characterized two novel fluorescent sigma-2 receptor selective ligands, SW120 and SW116. These ligands may serve as useful tools to study the function of sigma-2 receptors and to image cell proliferation.
Acknowledgments
Financial disclosure of authors: This research was funded by NIH grant CA 102869 and the Washington University Molecular Imaging Center P50 seed grant. This work is supported by the Alafi Neuroimaging Laboratory, the Hope Center for Neurological Disorders, and NIH Neuroscience Blueprint Center Core Grant P30 NS057105 to Washington University. This work is also partly supported by NIH P01 NS032636 and NIH R01 NS036265 to Dr. Mark P. Goldberg.
Footnotes
Financial disclosure of reviewers: None reported.
References
- 1.Martin WR, Eades CG, Thompson JA, et al. The effects of morphine- and nalorphine-like drugs in the nondependent and morphine-dependent chronic spinal dog. J Pharmacol Exp Ther. 1976;197:517–32. [PubMed] [Google Scholar]
- 2.Walker JM, Bowen WD, Walker FO, et al. Sigma receptors: biology and function. Pharmacol Rev. 1990;42:355–402. [PubMed] [Google Scholar]
- 3.Hellewell SB, Bowen WD. A sigma-like binding site in rat pheochromocytoma (PC12) cells: decreased affinity for (+)-benzomorphans and lower molecular weight suggest a different sigma receptor form from that of guinea pig brain. Brain Res. 1990;527:244–53. doi: 10.1016/0006-8993(90)91143-5. doi:10.1016/0006-8993(90)91143-5. [DOI] [PubMed] [Google Scholar]
- 4.Rothman RB, Reid A, Mahboubi A, et al. Labeling by [3H]1, 3-di(2-tolyl)guanidine of two high affinity binding sites in guinea pig brain: evidence for allosteric regulation by calcium channel antagonists and pseudoallosteric modulation by sigma ligands. Mol Pharmacol. 1991;39:222–32. [PubMed] [Google Scholar]
- 5.Quirion R, Bowen WD, Itzhak Y, et al. A proposal for the classification of sigma binding sites. Trends. Pharmacol Sci. 1992;13:85–6. doi: 10.1016/0165-6147(92)90030-a. doi:10.1016/0165-6147(92)90030-A. [DOI] [PubMed] [Google Scholar]
- 6.Hanner M, Moebius FF, Flandorfer A, et al. Purification, molecular cloning, and expression of the mammalian sigma1-binding site. Proc Natl Acad Sci U S A. 1996;93:8072–7. doi: 10.1073/pnas.93.15.8072. doi:10.1073/pnas.93.15.8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kekuda R, Prasad PD, Fei YJ, et al. Cloning and functional expression of the human type 1 sigma receptor (hSigmaR1). Biochem Biophys Res Commun. 1996;229:553–8. doi: 10.1006/bbrc.1996.1842. doi:10.1006/bbrc.1996.1842. [DOI] [PubMed] [Google Scholar]
- 8.Seth P, Leibach FH, Ganapathy V. Cloning and structural analysis of the cDNA and the gene encoding the murine type 1 sigma receptor. Biochem Biophys Res Commun. 1997;241:535–40. doi: 10.1006/bbrc.1997.7840. doi:10.1006/bbrc.1997.7840. [DOI] [PubMed] [Google Scholar]
- 9.Bem WT, Thomas GE, Mamone JY, et al. Overexpression of sigma receptors in nonneural human tumors. Cancer Res. 1991;51:6558–62. [PubMed] [Google Scholar]
- 10.Vilner BJ, Bowen WD. Sigma receptor-active neuroleptics are cytotoxic to C6 glioma cells in culture. Eur J Pharmacol. 1993;244:199–201. doi: 10.1016/0922-4106(93)90029-9. doi:10.1016/0922-4106(93)90029-9. [DOI] [PubMed] [Google Scholar]
- 11.Vilner BJ, de Costa BR, Bowen WD. Cytotoxic effects of sigma ligands: sigma receptor-mediated alterations in cellular morphology and viability. J Neurosci. 1995;15:117–34. doi: 10.1523/JNEUROSCI.15-01-00117.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mach RH, Smith CR, al-Nabulsi I, et al. Sigma 2 receptors as potential biomarkers of proliferation in breast cancer. Cancer Res. 1997;57:156–61. [PubMed] [Google Scholar]
- 13.Wheeler KT, Wang LM, Wallen CA, et al. Sigma-2 receptors as a biomarker of proliferation in solid tumours. Br J Cancer. 2000;82:1223–32. doi: 10.1054/bjoc.1999.1067. doi:10.1054/bjoc.1999.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tu Z, Dence CS, Ponde DE, et al. Carbon-11 labeled sigma2 receptor ligands for imaging breast cancer. Nucl Med Biol. 2005;32:423–30. doi: 10.1016/j.nucmedbio.2005.03.008. doi:10.1016/j.nucmedbio.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 15.Tu Z, Xu J, Jones LA, et al. Fluorine-18-labeled benzamide analogues for imaging the sigma2 receptor status of solid tumors with positron emission tomography. J Med Chem. 2007;50:3194–204. doi: 10.1021/jm0614883. doi:10.1021/jm0614883. [DOI] [PubMed] [Google Scholar]
- 16.Hou C, Tu Z, Mach R, et al. Characterization of a novel iodinated sigma-2 receptor ligand as a cell proliferation marker. Nucl Med Biol. 2006;33:203–9. doi: 10.1016/j.nucmedbio.2005.10.001. doi:10.1016/j.nucmedbio.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Rowland DJ, Tu Z, Xu J, et al. Synthesis and in vivo evaluation of 2 high-affinity 76Br-labeled sigma2-receptor ligands. J Nucl Med. 2006;47:1041–8. [PubMed] [Google Scholar]
- 18.Crawford KW, Bowen WD. Sigma-2 receptor agonists activate a novel apoptotic pathway and potentiate antineoplastic drugs in breast tumor cell lines. Cancer Res. 2002;62:313–22. [PubMed] [Google Scholar]
- 19.Ostenfeld MS, Fehrenbacher N, Hoyer-Hansen M, et al. Effective tumor cell death by sigma-2 receptor ligand siramesine involves lysosomal leakage and oxidative stress. Cancer Res. 2005;65:8975–83. doi: 10.1158/0008-5472.CAN-05-0269. doi:10.1158/0008-5472.CAN-05-0269. [DOI] [PubMed] [Google Scholar]
- 20.Vilner BJ, Bowen WD. Modulation of cellular calcium by sigma-2 receptors: release from intracellular stores in human SK-N-SH neuroblastoma cells. J Pharmacol Exp Ther. 2000;292:900–11. [PubMed] [Google Scholar]
- 21.Cassano G, Gasparre G, Niso M, et al. F281, synthetic agonist of the sigma-2 receptor, induces Ca2+ efflux from the endoplasmic reticulum and mitochondria in SK-N-SH cells. Cell Calcium. 2009;45:340–5. doi: 10.1016/j.ceca.2008.12.005. doi:10.1016/j.ceca.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 22.Crawford KW, Coop A, Bowen WD. Sigma(2) receptors regulate changes in sphingolipid levels in breast tumor cells. Eur J Pharmacol. 2002;443:207–9. doi: 10.1016/s0014-2999(02)01581-9. doi:10.1016/S0014-2999(02) 01581-9. [DOI] [PubMed] [Google Scholar]
- 23.Ostenfeld MS, Hoyer-Hansen M, Bastholm L, et al. Anti-cancer agent siramesine is a lysosomotropic detergent that induces cytoprotective autophagosome accumulation. Autophagy. 2008;4:487–99. doi: 10.4161/auto.5774. [DOI] [PubMed] [Google Scholar]
- 24.Zeng C, Vangveravong S, Xu J, et al. Subcellular localization of sigma-2 receptors in breast cancer cells using two-photon and confocal microscopy. Cancer Res. 2007;67:6708–16. doi: 10.1158/0008-5472.CAN-06-3803. doi:10.1158/ 0008-5472.CAN-06-3803. [DOI] [PubMed] [Google Scholar]
- 25.Vangveravong S, Xu J, Zeng C, et al. Synthesis of N-substituted 9-azabicyclo[3.3.1]nonan-3alpha-yl carbamate analogs as sigma2 receptor ligands. Bioorg Med Chem. 2006;14:6988–97. doi: 10.1016/j.bmc.2006.06.028. doi:10.1016/ j.bmc.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 26.Xu J, Tu Z, Jones LA, et al. [3H]N-[4-(3, 4-dihydro-6, 7-dimethoxyisoquinolin-2(1H)-yl)butyl]-2-methoxy-5-methylbenzamide: a novel sigma-2 receptor probe. Eur J Pharmacol. 2005;525:8–17. doi: 10.1016/j.ejphar.2005.09.063. doi:10.1016/j.ejphar.2005.09.063. [DOI] [PubMed] [Google Scholar]
- 27.Zimmermann T, Rietdorf J, Pepperkok R. Spectral imaging and its applications in live cell microscopy. FEBS Lett. 2003;546:87–92. doi: 10.1016/s0014-5793(03)00521-0. doi:10.1016/S0014-5793(03)00521-0. [DOI] [PubMed] [Google Scholar]
- 28.Hertel C, Coulter SJ, Perkins JP. A comparison of catecholamine-induced internalization of beta-adrenergic receptors and receptor-mediated endocytosis of epidermal growth factor in human astrocytoma cells. Inhibition by phenylarsine oxide. J Biol Chem. 1985;260:12547–53. [PubMed] [Google Scholar]
- 29.Luzio JP, Parkinson MD, Gray SR, et al. The delivery of endocytosed cargo to lysosomes. Biochem Soc Trans. 2009;37:1019–21. doi: 10.1042/BST0371019. doi:10.1042/BST0371019. [DOI] [PubMed] [Google Scholar]
- 30.Gebreselassie D, Bowen WD. Sigma-2 receptors are specifically localized to lipid rafts in rat liver membranes. Eur J Pharmacol. 2004;493:19–28. doi: 10.1016/j.ejphar.2004.04.005. doi:10.1016/j.ejphar.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 31.Kashiwagi H, McDunn JE, Simon PO, Jr, et al. Selective sigma-2 ligands preferentially bind to pancreatic adenocarcinomas: applications in diagnostic imaging and therapy. Mol Cancer. 2007;6:48. doi: 10.1186/1476-4598-6-48. doi:10.1186/1476-4598-6-48. [DOI] [PMC free article] [PubMed] [Google Scholar]