Summary
Stimulation of Escherichia coli with acetate elevates the acetylation level of the chemotaxis response regulator CheY. This elevation, in an unknown mechanism, activates CheY to generate clockwise rotation. Here, using quantitative selective reaction monitoring mass spectrometry and high-resolution targeted mass spectrometry, we identified K91 and K109 as the major sites whose acetylation level in vivo increases in response to acetate. Employing single and multiple lysine replacements in CheY, we found that K91 and K109 are also the sites mainly responsible for acetate-dependent clockwise generation. Furthermore, we showed that clockwise rotation is repressed when residue K91 is non-modified, as evidenced by an increased ability of CheY to generate clockwise rotation when K91 was acetylated or replaced by specific amino acids. Using molecular dynamics simulations we show that K91 repression is manifested in the conformational dynamics of the β4α4 loop, shifted towards an active state upon mutation. Removal of β4α4 loop repression may represent a general activation mechanism in CheY, pertaining also to the canonical phosphorylation activation pathway as suggested by crystal structures of active and inactive CheY from Thermotoga maritima. By way of elimination we further suggest that K109 acetylation is actively involved in generating clockwise rotation.
Keywords: CheY acetylation, response regulator activation, lysine acetylation, bacterial chemotaxis, flagellar rotation, molecular dynamics
Introduction
Acetylation of proteins on lysine residues (N-acetylation), a reversible post-translational covalent modification that adds an acetyl group to the ε-amino group of a lysine residue, is widespread in nature, from bacteria to mammals (Kim and Yang, 2011; Thao and Escalante-Semerena, 2011; J D Jones and O'Connor, 2011). In Escherichia coli alone, more than one hundred proteins undergo N-acetylation. One of them, among the first to be identified as acetylatable, is the excitatory response regulator of chemotaxis, CheY (Barak et al., 1992; Barak et al., 2004; Barak et al., 2006; Yan et al., 2008). This protein undergoes acetylation by at least two mechanisms: acetylation by the enzyme acetyl-CoA synthetase (Acs) with acetate as an acetyl donor (Barak et al., 1992; Barak et al., 2004) and autoacetylation with AcCoA as an acetyl donor (Barak et al., 2006). CheY was found in vitro to be acetylated by each of these mechanisms at six acetylation sites — lysine residues 91, 92, 109, 119, 122 and 126 (Ramakrishnan et al., 1998; Barak et al., 2004; Barak et al., 2006; R Li et al., 2010). The main function, shown thus far for CheY acetylation by Acs, is shifting the direction of flagellar rotation from the default, counterclockwise, to clockwise. This function, termed hereafter ‘acetate-dependent clockwise generation’, is observed even in the absence of the chemotaxis machinery. Thus, acetate addition to gutted cells expressing CheY from a plasmid but lacking all other chemotaxis proteins and some of the receptors (Wolfe et al., 1988; Barak et al., 1998; Ramakrishnan et al., 1998) or to cytoplasm-free cell envelopes containing Acs (Barak et al., 1992) resulted in prolonged clockwise rotation. Indeed, the acetylation level of CheY in vivo is high (Yan et al., 2008), but it is not known which of the sites are affected by acetate and, accordingly, which of them is likely to be responsible for clockwise generation. Here we identified that these sites are K91 and K109. Furthermore, the use of gutted cells fully devoid of the phosphorelay machinery uniquely enabled us to study, without perturbations of phosphorylation-dependent signaling, how subtle modifications in CheY activate the protein to generate clockwise rotation.
Results
To identify the acetylation site(s) responsible for clockwise generation we first determined which of the acetylation sites is more often acetylated in vivo and influenced by acetate, and then examined whether this site is, indeed, involved in clockwise generation.
Acetylation sites of CheY in vivo
To identify the acetylation sites of CheY in vivo we exposed extracts of E. coli ΔcheY cells expressing 6xHis-CheY from a plasmid to SDS polyacrylamide gel electrophoresis. We studied two expression levels of CheY. One without induction, producing a relatively low level of CheY as judged by both Western blots (supplementary Fig. S1) and functional assays of expanding ring formation on semi-solid agar (Adler, 1966) (Fig. S2). The other was with induction by isopropyl β-D-thiogalactopyranoside (IPTG; 1 mM), making the detection easier (Fig. S1). We subjected the CheY gel bands to shotgun proteomics using tandem mass spectrometry (MS/MS). We performed in-gel digestion using trypsin and chymotrypsin, which yielded a mixture of peptides for each gel band. We analyzed these mixtures by high-resolution tandem mass spectrometry, which facilitates identification of the CheY peptides through collision-induced dissociation in the instrument. This resulted in peptide fragmentation spectra also used for identification of acetylation sites on the peptide sequences. The analysis identified essentially all the sites known to be acetylated in vitro and, in addition, lysine residues 4, 7, and 70 (Table 1, Fig. S3). Yet, it only provided a qualitative view of CheY acetylation in vivo.
Table 1.
Acetylation sites of CheY in vivo revealed from MS/MS analyses
| Peptide Sequence | Acetylation site on CheYa |
MH+ [Da]b | Xcorrc |
|---|---|---|---|
| Wild-type level of CheY (no IPTG) | |||
| ATLEEKLNacKIFEacKLGMGLCG | K122; K126 | 2351.21573 | 2.2 |
| Wild-type level of CheY (no IPTG) + acetate | |||
| HHTDPALRAADacKELacK | K4; K7 | 1785.91252 | 2.36 |
| DALNKLQAGGYGFVISDWNMPNMDGLELLacKTIR | K70 | 3724.82656 | 3.16 |
| AacKKENIIAAAQAG | K91 | 1328.71623 | 2.06 |
| LMVTAEAKacKENIIAAAQAGASG | K92 | 2202.14408 | 2.11 |
| LNacKIFEK | K122 | 933.53984 | 2.75 |
| Elevated levels of CheY (IPTG) | |||
| VVacKPFTAATLEEKLNK | K109 | 1830.02666 | 3.21 |
| LEEacKLNKI | K119 | 1028.59380 | 1.91 |
| TAATLEEacKLNKIFEacKL | K119; K126 | 1933.04265 | 1.86 |
| Elevated levels of CheY (IPTG) + acetate | |||
| ALPVLMVTAEAacKKENIIAAAQ | K91 | 2240.21514 | 2.17 |
| VVacKPFTAATLEEKLNK | K109 | 1830.02666 | 3.07 |
| LEEacKLNKI | K119 | 1028.59380 | 1.98 |
| TAATLEEacKLNKIFEacKL | K119; K126 | 1933.0265 | 1.92 |
According to protein accession P0AE67. The sites were determined according to acetylated peptides obtained by proteolytic digestion with trypsin and chymotrypsin.
MH+ is the mass of the protonated form of the peptide.
Xcorr is a cross-correlation score function calculated to assess the quality of the match between a tandem mass spectrum and amino acid sequence information in a database (Eng et al., 1994). The peptides were identified using the Sequest search engine.
K91 and K109 are the sites most affected by acetate in vivo
To determine the quantitative differences between the acetylation levels in the various sites and to establish the effect of acetate on these levels, we used synthetic acetylated standard peptides combined with quantitative, selective reaction monitoring (SRM) mass spectrometry — an established approach for quantification of peptides and proteins in complex biological samples (Addona et al., 2009). We first separated proteins from each of the samples by SDS-PAGE and then subjected the relevant bands to SRM. Using this method in conjunction with synthetic, acetylated peptide standards (Table S1), we obtained absolute concentrations of the targeted acetylated peptides. We mainly relied on peptides obtained from IPTG-induced CheY (Table 2) because more peptides at higher levels of expression could be detected. Excluding the K45-containing peptide (see below), whenever peptides from non-induced CheY could be detected (Table S2), they were those already obtained from induced CheY (Table 2). Although we found lysine residues 26, 45, 91, 92, 122 and 126 to be acetylated, there were some batch-to-batch differences. For example, K126 was only detected in batch 3 (Table 2), and K45 was only detected in samples without IPTG (Table S2).
Table 2.
Low-resolution targeted mass spectrometry measurements of lysine acetylated residues on CheYa with and without acetate.
| Peptide sequence | Acetylation site on CheY |
Acetylated peptides [pM] | |
|---|---|---|---|
| Experiment I |
Absence of acetate |
Presence of acetate |
|
| NLLacKELGFNNVEEAEDGVDALNK | K26 | 28.9 | 68.2 |
| TIRADGAMSALPVLMVTAEAacK | K91 | 0 | 52.2 |
| ADGAMSALPVLMVTAEAacKacK | K91, K92 | 0.03 | 0.52 |
| LNacKIFEK | K122 | 0.4 | 13.35 |
| Experiment II | |||
| NLLacKELGFNNVEEAEDGVDALNK | K26 | 5.4 | 6.4 |
| TIRADGAMSALPVLMVTAEAacK | K91 | 0 | 137.05 |
| LNacKIFEK | K122 | 0.42 | 13.35 |
| Experiment III | |||
| IVRNLLacK | K26 | 3.2 | 1.7 |
| NLLacKELGFNNVEEAEDGVDALNK | K26 | 2.5 | 0.3 |
| TIRADGAMSALPVLMVTAEAacK | K91 | 142.2 | 322.3 |
| LNacKIFEK | K122 | 5942.3 | 2793.3 |
| LNKIFEacK | K126 | 3940.2 | 4692.3 |
| LNacKIFEacK | K122; K126 | 128.5 | 261.3 |
IPTG-induced CheY.
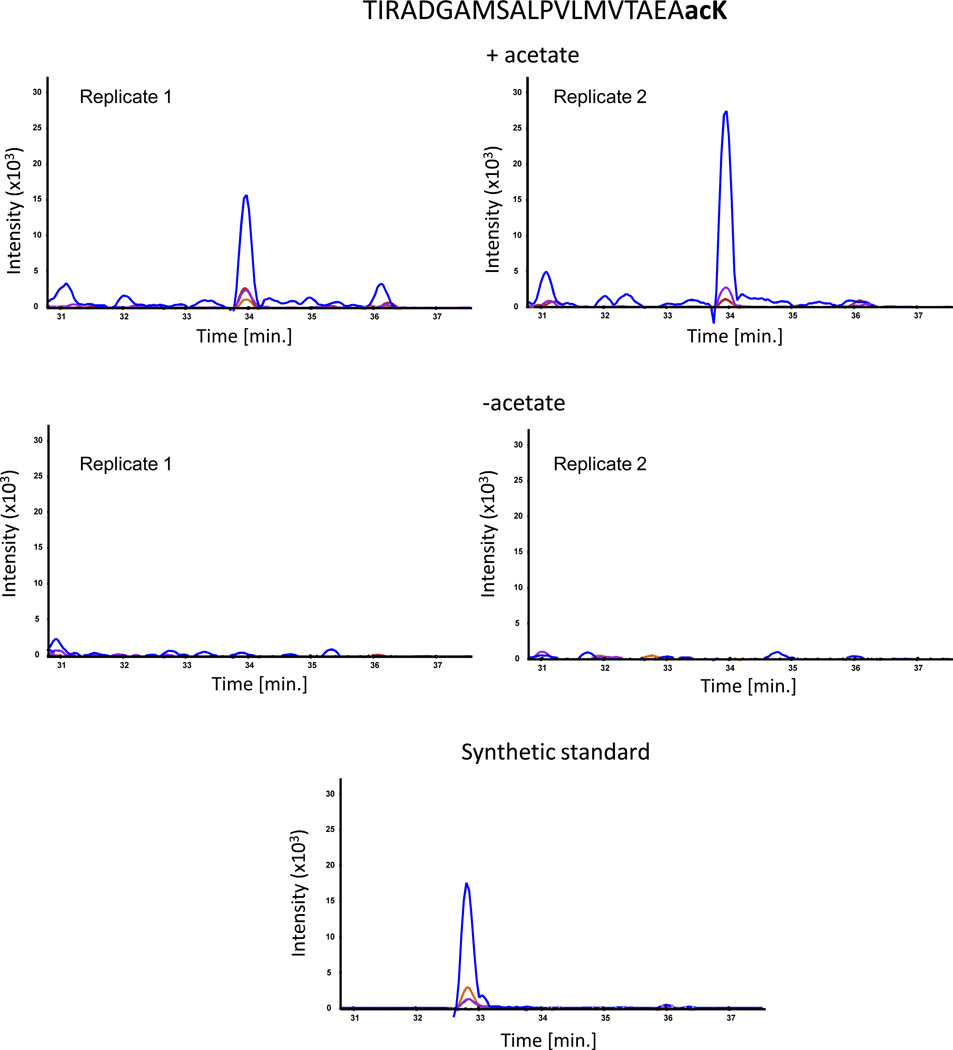
To determine the effect of acetate on the acetylation sites, we supplemented portions of the same cultures with acetate for 1 h growth prior to harvesting. The only acetylation site that showed a consistent change in the acetylation level in response to acetate was K91, whose level consistently increased (Fig. 1 and Table 2). However, these low-resolution targeted mass spectrometry measurements could not detect acetylation on residues K109 and K119, if occurred. This is because there is no trypsin digestion site between them (i.e., unmodified Lys or Arg), meaning that the resulting peptides after protein digestion were too long for analysis by mass spectrometry.
Fig. 1.
SRM mass spectrometry analysis of the acetylated peptide TIRADGAMSALPVLMVTAEAacK. The plots show overlaid chromatograms of the measured SRM transitions (product ions): b11 (blue), y10 (orange), b14 (purple) and b15 (red). The synthetic standard was external (not spiked directly into the sample), hence the slightly different retention time from the native peptide.
To circumvent this difficulty, we digested the IPTG-induced samples by both trypsin and chymotrypsin, and then employed high-resolution targeted mass spectrometry measurements, which enabled relative quantification of the acetylation ratios prior and subsequent to treatment with acetate. With this approach we identified four different peptides containing acetylated K109 (Table 3). The acetylation levels of all of them were consistently elevated (doubled on average) in response to acetate (Fig. 2 and Table 3). Even though acetylation of K119 was detected as well, its level did not change in response to acetate treatment (Table 3). Thus, in vivo, acetate treatment predominantly results in K91 and K109 acetylation, suggesting that these two sites are responsible for the acetate-dependent clockwise generation.
Table 3.
High-resolution targeted mass spectrometry measurements of K109 and K119 on CheY with and without acetate
| Peptide sequence | Site on CheY |
Acetate (+/−) ratio |
|---|---|---|
| KENIIAAAQAGASGYVVacK | K109 | 1.55 |
| KENIIAAAQAGASGYVVacKPF | K109 | 1.51 |
| ENIIAAAQAGASGYVVacKPFTAATL | K109 | 2.56 |
| VVacKPFTAATLEEK | K109 | 2.46 |
| TAATLEEacKLNK | K119 | 1.01 |
Fig. 2.
Targeted Parallel Reaction Monitoring analysis of acetylated peptides using high-resolution mass spectrometry. Presented are overlaid chromatograms of the product ions from peptide KENIIAAAQAGASGYVVacK: b5 (blue), b6 (purple), b7 (red), b9 (orange), b11 (turquois), and b12 (green). Unlike the SRM experiments, in this analysis there were no synthetic standards, thus only relative quantification was performed.
K91 and K109 are responsible for the acetate-dependent clockwise generation
To examine this possibility we prepared by site-directed mutagenesis arginine-for-lysine replacement mutants at sites 91 and 109. As controls, we similarly prepared KR replacements at the other previously identified acetylation sites — K92, K119, K122, and K126. [A KR replacement conserves the positive electric charge but does not undergo acetylation (M Li et al., 2002; Kim and Yang, 2011).] We expressed the CheY mutant proteins to a low level (without IPTG) in a gutted background (mentioned under Experimental Procedures), thus avoiding a repellent response to acetate (Wolfe et al., 1988). Furthermore, to prevent the generation of acetyl phosphate, which can generate clockwise rotation by phosphorylating CheY, all the gutted strains were also deleted for both ack and pta (encoding acetate kinase and phosphotransacetylase, respectively) (Barak et al., 1998). To examine the ability of acetate to generate clockwise rotation in the mutants, we tethered the cells to glass slides via their flagella and looked for changes in the direction of flagellar rotation in response to acetate addition.
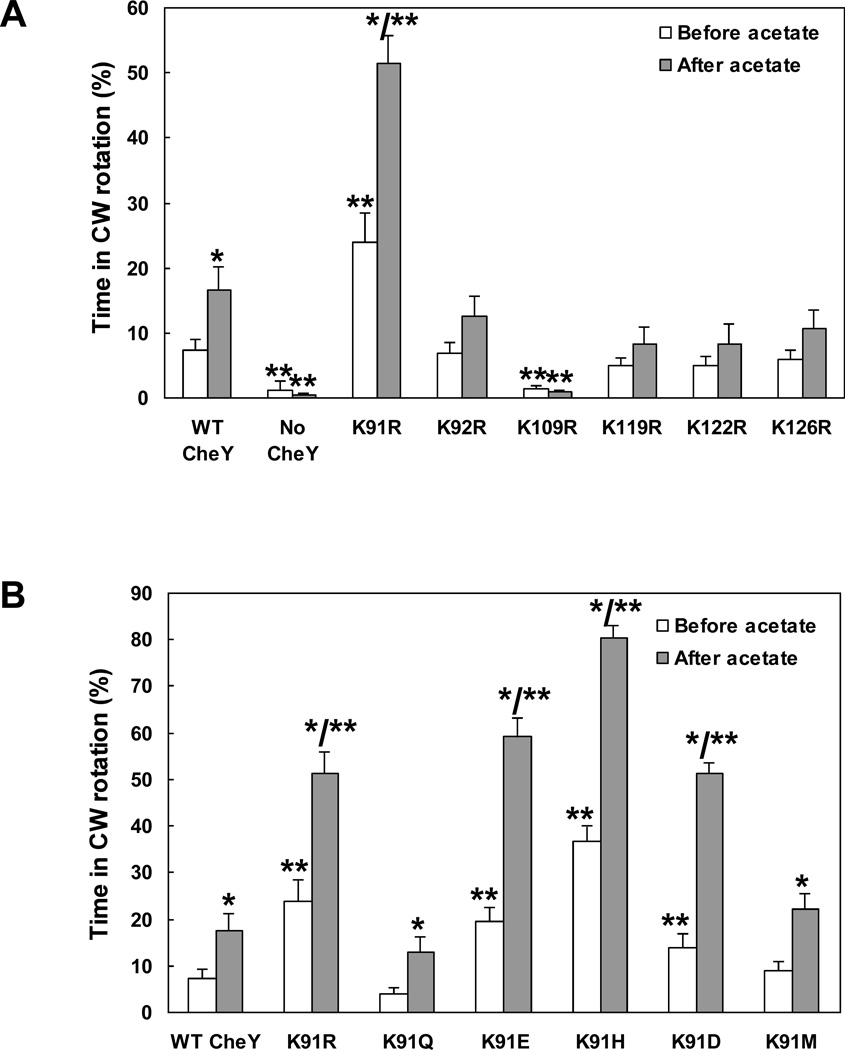
Cells expressing CheY(K91R) and CheY(K109R) were significantly different from cells expressing wild-type CheY or CheY with arginine replacements at other lysine positions. Cells expressing CheY(K91R) spent twice as much time in clockwise rotation as did cells expressing wild-type CheY. Cells expressing CheY(K109R), consistent with earlier observations (Lukat et al., 1991; Bourret et al., 1993), exhibited almost no clockwise rotation (Fig. 3A). All strains used in this experiment were verified to express CheY to similar levels (Fig. S1). Cells expressing CheY(K91R) and cells expressing CheY(K109R) were also exceptional when flagellar rotation was measured after acetate addition: Cells expressing CheY(K91R) doubled the time spent in clockwise rotation, and cells expressing CheY(K109R) continued rotating counterclockwise almost exclusively (Fig. 3A). The other single KR replacements did not significantly affect rotation behavior. These observations, taken together with the mass spectrometry results pointing to K91 and K109 as the major sites that undergo acetylation in response to acetate, suggest that K91 and K109 are the main players in generating clockwise rotation.
Fig. 3.
Acetate-dependent clockwise generation by CheY containing single replacements. A, KR replacements at the known acetylation sites. B, Various replacements of K91. All plasmids were expressed (without IPTG) in gutted RBB1097 background to a similar level (Fig. S1C). The direction of flagellar rotation was determined for 1 min prior to, and 10 s subsequent to, addition of acetate (100 mM, pH 7.0). The results are the mean ± SEM of at least three independent experiments (the number of experiments was determined on the basis of the criterion that additional experiments did not change the mean). *P < 0.05 for the difference between before and after acetate treatment, determined by paired t-test. **P < 0.05 for the difference between the mutants and the wild-type control, determined by ANOVA with Dunnet Multiple Comparison Test as posttest.
K91 likely represses CheY activation
To determine whether the observed clockwise-rotation enhancement in cells expressing CheY(K91R) (Fig. 3A) is specific for K replacement by R, we studied additional K replacements at position 91. Clearly, K replacements by H, E, and D were at least as effective as R in enhancing clockwise rotation (Fig. 3B). In contrast, K replacements by Q and M were not effective and the strains behaved like cells expressing wild-type CheY. Notably, the effect of K91R mutation could only be seen in gutted cells expressing CheY, not in wild-type, ΔcheY cells expressing CheY (Fig. S4). One possible explanation is that the repressive function of K91R can only be observed when most of CheY is not CheA-bound at the receptor complex. [In wild-type cells CheY localizes to the cell poles in a CheA-dependent manner (Sourjik and Berg, 2000).]
Two major generalizations can be made from these results. One is that the differences between the K91 replacement mutants are in the basal level of clockwise rotation, i.e., prior to acetate addition. Acetate roughly doubled the clockwise level in all these mutants. The other generalization is that none of the mutations was inhibitory. They either activated CheY to generate more clockwise rotation or had no effect compared with wild-type CheY. This suggests that non-modified K91 represses to some extent the clockwise generating activity of the protein, and acetylation of K91 or its replacement by specific amino acids removes this repression, resulting in enhanced clockwise rotation.
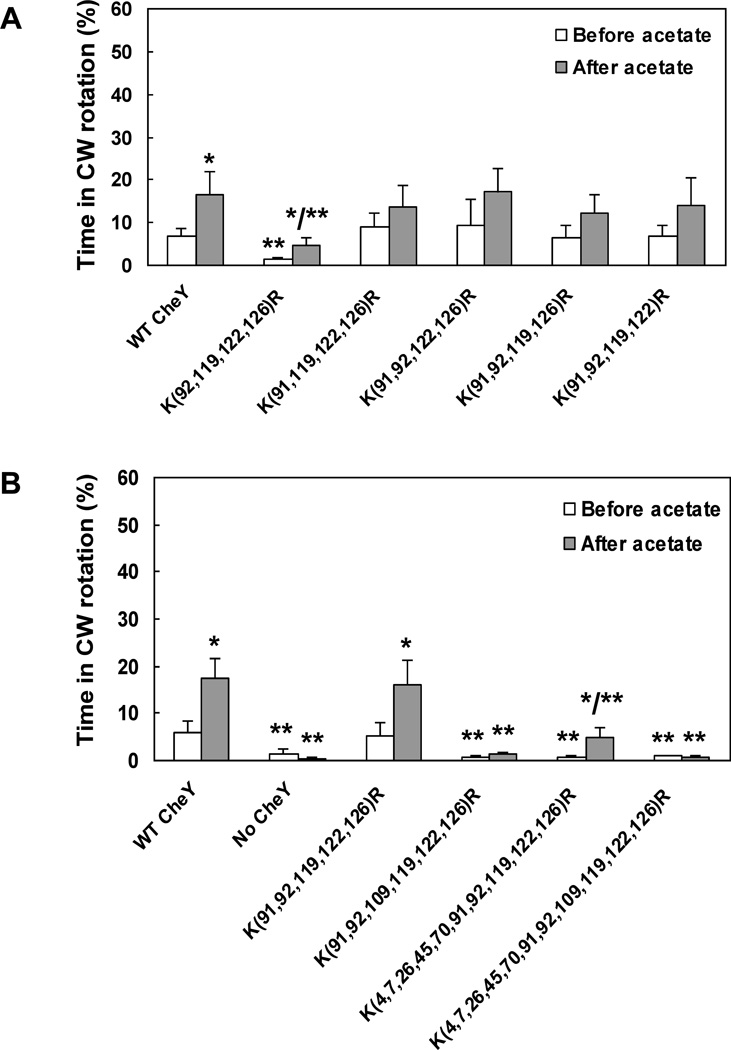
To test this conclusion we constructed a set of multiple mutants in which one of the six acetylation sites was not modified (additional to K109, which we did not modify to enable clockwise generation), while the other four sites were KR replaced. Evidently, only when K91 was not replaced, clockwise rotation was significantly lower than in the wild-type control both before and after acetate addition (Fig. 4A).
Fig. 4.
Acetate-dependent clockwise generation by CheY containing multiple replacements. A, Mutants expressing CheY protein with four KR replacements each. B, Multiple KR replacements. All CheY proteins were expressed (without IPTG) in gutted RBB1097 background to a similar level, excluding the proteins with 10 and 11 KR replacements (see text for details). The direction of flagellar rotation was determined before and after (1 min and 10 s, respectively) the addition of acetate (100 mM, pH 7.0). The results are the mean ± SEM of at least three independent experiments (the number of experiments was determined on the basis of the criterion that additional experiments did not change the mean). *P < 0.05 for the difference between before and after acetate treatment, determined by paired t-test. **P < 0.05 for the difference between the mutants and the wild-type control, determined by ANOVA with Dunnet Multiple Comparison Test as posttest.
By way of elimination — K109 acetylation likely activates CheY
The observations that, in all the cells containing CheY with K91 replacements, addition of acetate roughly doubled the fraction of time spent in clockwise rotation (Fig. 3B) indicate that acetylation of a site other than K91 generates clockwise rotation in these cells. The best candidate for this function is K109. This is for two reasons. First, it is the only site, other than K91, identified to undergo acetate-dependent acetylation in vivo (Fig. 2). And second, a mutation in a clockwise-generating site is expected to reduce the clockwise level. From all the single KR replacement mutants, only the one expressing CheY(K109R) exhibited this phenotype (Fig. 3A). To examine the validity of this possibility, we constructed a set of multiple mutants having KR replacements additional to the one in site 91. Because the mass spectrometry results demonstrated that lysine residues other than those identified earlier can be occasionally acetylated as well (Tables 1, 2), we also constructed mutants with all 11 lysine residues, or all of them but K109, replaced by arginine (termed hereafter x11KR and x10KR mutants, respectively). Clearly, presence of K109 (i.e., avoiding replacement at this site) enabled the response to acetate, irrespective of which other sites were replaced (Fig. 4B). These observations and the generation of clockwise rotation by acetate even in the x10KR mutant endorses the possibility that acetylation of K109 is the cause of clockwise generation.
A possible role for position 91 in the repression of CheY activation
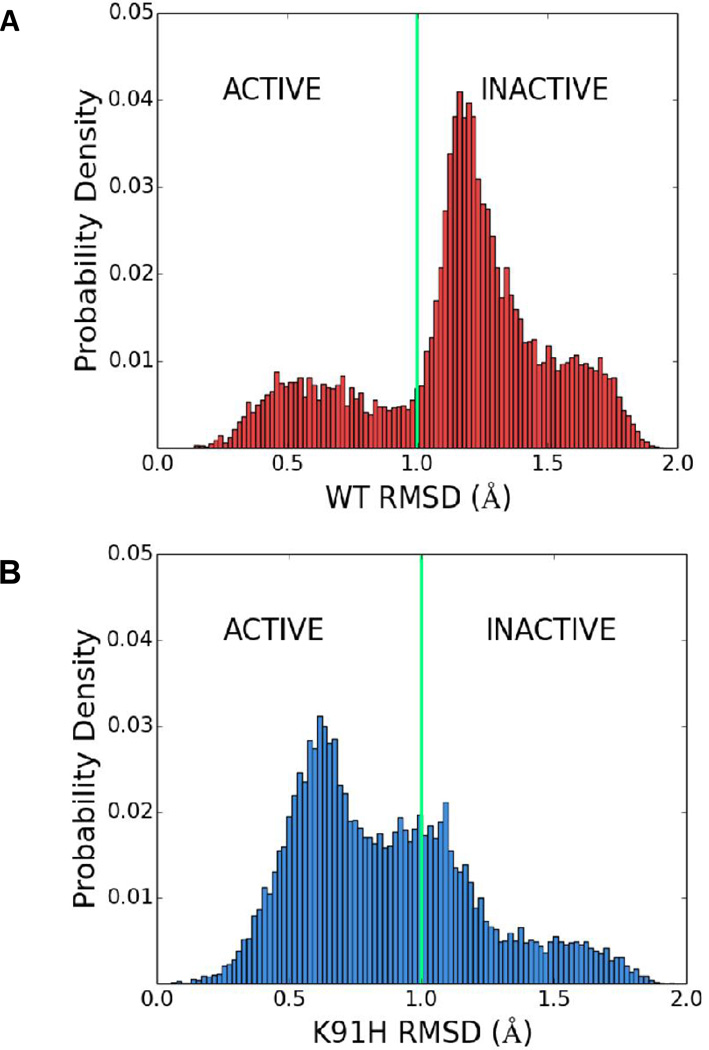
Generally speaking, because the K91-activating mutations were different from each other in their side chains (including volume and charge), it is likely that their activating effect is produced by the exclusion of lysine from position 91. To examine how lysine exclusion from position 91 can affect CheY activation at the molecular level, we used all-atom molecular dynamics (MD) simulations for studying the effect of the K91H mutation (shown above to give rise to the most hyperactive phenotype, i.e., the least repressed) on activation, and compared it with wild-type CheY. Initial atomic coordinates for both the K91H (with histidine protonated at the δ position) and wild-type CheY models were derived from the crystal structure of E. coli CheY (PDB 3CHY) (Volz and Matsumura, 1991). Each system was minimized along with solvent and equilibrated for 250 ns. An ensemble of ten, statistically independent, 250 ns production simulations were subsequently carried out on both the K91H and wild-type systems, resulting in a total of 2.75 µs of sampling for each model. These simulations revealed a higher level of flexibility in the protein regions surrounding position 91 of the K91H system. In particular, residues 88–102, corresponding to the β4α4 loop and α4 helix, showed, on average, an increase in their root mean squared fluctuations (RMSF) as compared to the corresponding regions in the ensemble of wild-type simulations (Fig. 5). Based on this result, we hypothesized that the K91H mutation might give rise to a hyperactive phenotype by altering the conformational dynamics of the β4α4 loop, a region previously shown to correlate with CheY activation (Knaggs et al., 2007; Ma and Cui, 2007; Mottonen et al., 2010).
Fig. 5.
K91H exhibits high flexibility of β4α4 loop. RMSF versus residue number for wild-type (WT; A) and K91H (B) ensemble simulations. Individual RMSF traces for the ten, 250 ns simulations in each ensemble (gray lines) were averaged to produce a mean RMSF value for all 129 CheY residues (colored lines). Overlaying the averaged RMSF traces from the wild-type and K91H ensembles (C) highlights regions of differing stiffness. Vertical dotted lines emphasize the increased flexibility in regions of CheY surrounding the K91 mutation (residues 88–105), including the β4α4 loop (residues 88–91).
To characterize the effect of increased flexibility in the protein regions surrounding K91 on the conformational sampling of the β4α4 loop, we used principal components analysis (PCA) (Bishop, 2006) to provide a concise description of the β4α4 loop conformations witnessed by our MD simulations. The utility of the reduced PCA description to capture the salient features of the dynamics of the β4α4 loop was demonstrated by the relatively small number of principal components (PCs) required to describe a significant degree of the conformational variability within the simulation trajectories (Table S3) (Skjaerven et al., 2011). In the present application, the cumulative fractional variance of the top three PCs accounted for over 75% of the total trajectory variance, providing a practical, low-dimensional subspace on to which to project the individual β4α4 loop conformations observed in the K91H and wild-type ensemble simulations. The resulting projections, along with projections of the analogous portions of existing activated CheY structures (PDBs 1FQW, 1ZDM, 1F4V, 1DJM) and the equilibrated, inactive CheY structure revealed two well-defined clusters of β4α4 loop conformations (Fig. S5). Notably, β4α4 loop conformations from both the K91H and wild-type simulations inhabited either cluster while the active and inactive β4α4 loop conformations localized to single clusters opposite one another (Fig. S5). This suggests that both the K91H and wild-type systems sampled distinct functional states corresponding to the active and inactive β4α4 loop conformations in our MD simulations.
To characterize the organization of projected β4α4 loop conformations in three-dimensional principal component space, we used the K-means clustering algorithm (Bishop, 2006) to systematically assign loop conformations to two clusters (Fig. S6). Strikingly, K-means revealed that over 80% of the K91H loop conformations were assigned to the same cluster as the activated loop structures whereas only <30% of wild-type loop conformations were assigned to this cluster (Table S4). To further survey the distribution of the conformations in 3D PC space, the root mean squared deviation (RMSD) between projections of the activated β4α4 loop conformations and those arising within the K91H mutant and wild-type ensemble simulations was calculated (see Experimental Procedures). The resulting cumulative RMSD distributions (Fig. 6) clearly demonstrated an asymmetry in the occupation of the active and inactive β4α4 loop states by the K91H and wild-type CheY systems in agreement with the K-means result. Taken together the results from our MD simulations and accompanying analysis suggest that the increased flexibility in the β4α4 loop and α4 helix associated with the K91H mutation gives rise to a structural population shift in which the β4α4 loop fluctuates more often near the conformation typical of activated loop structures. It is highly likely that K91 acetylation has an equivalent function in CheY activation. Whether or not this is the case will be the subject of a future study.
Fig. 6.
K91 represses the active state of CheY’s β4α4 loop. Distributions of root mean squared deviations (RMSDs) between projections of an activated β4α4 loop structure (PDB 1FQW) and conformations of the β4α4 loop sampled in the wild-type (WT; A) and K91H (B) ensemble simulations. Vertical green lines indicate the rough boundary between clusters in principle component space computed using K-means and highlight the population shift incurred by the K91H mutation. See Experimental Procedures for more details.
Discussion
In this study — the first to identify the acetylation sites on CheY in vivo and the first to quantify their acetylation levels — we addressed the question, which of these sites are responsible for acetate-dependent clockwise generation. The main findings made are that K91 and K109 are the predominant sites that undergo acetylation in response to acetate, and that these are the sites mainly responsible for the acetate-dependent clockwise generation. The study was carried out with gutted cells in order to avoid perturbations from the phosphorylation system. This means that the results and conclusions are solely relevant to the mechanism of acetylation-dependent clockwise generation.
K91 and K109 are the major sites responsible for acetate-dependent acetylation and clockwise generation
Using SRM mass spectrometry in conjunction with synthetic, acetylated peptide standards, we were able to obtain absolute quantification of the targeted acetylated peptides. One potential drawback of the approach was the fact that peptide concentrations were obtained from in-gel digestion and, therefore, did not reflect the concentration in the sample. Nevertheless, the concentrations still provided an accurate account of the acetylation sites that responded to acetate stimulation. This information combined with high-resolution targeted mass-spectrometry measurements pointed to K91 and K109 as the major sites that undergo acetate-stimulated acetylation in vivo. These two sites are also the ones found to be autoacetylated in vitro (R Li et al., 2010). In addition, the responses of tethered cells expressing CheY proteins with a variety of KR replacements (Fig. 3) indicated that K91 and K109 are the sites responsible for acetate-dependent clockwise generation. The rules that emerge from these studies are that K91 must be replaced (by specific amino acids) and K109 must not be replaced for obtaining higher clockwise rotation before or after acetate stimulation (Fig. 3A, Fig 4B).
K91 function in acetate-dependent clockwise generation
The observations that clockwise rotation is low whenever K91 is not replaced suggest that K91 represses clockwise rotation. Removal of this repression can apparently be done by two means. One, a physiological mean, is acetylation of K91. The other, an artificial mean, is elimination of K91 by a specific replacement. Obviously, the replacement is more effective than acetylation [see, for example, the much lower clockwise generation by acetate when K91 is not in CheY[K(92,119,122,126)R compared to when K is replaced at position 91 in CheY[K(91,92,119,122,126)R — Fig. 4A and 4B, respectively] because the replacement is on every CheY molecule, whereas acetylation only occurs in a fraction of the CheY molecules (Yan et al., 2008). The effective replacements were K91R, K91H, K91E, and K91D. K91Q and K91M replacements were not effective (Fig. 3B).
MD simulations demonstrated that the β4α4 loop of wild-type CheY co-exists in active and inactive states, spending more time in the inactive state (Fig. 6A). The simulations further demonstrated that when K91 is replaced by histidine the loop spends much more time in the active state (Fig. 6B). Since the K91-activating mutations were different from each other in their side chains and since K91 acetylation is phenotypically similar to K91H replacement, we suggest that the effect of K91 acetylation is mechanistically similar to K91H replacement. This possibility as well as the role of β4α4 loop in the generation of clockwise rotation and the exact regulatory mechanism of loop flexibility should still be studied.
We further suggest that the mechanism of β4α4 repression has a fundamental role in the regulation of CheY activity. A similar mechanism can apparently be observed in the crystal structures of the active and inactive forms of CheY of Thermotoga maritima, in which the β4α4 loop was reported to have a mechanism of activation similar to that of CheY in E. coli (Mottonen et al., 2010). The equivalence of E. coli K91 (K91EC) in T. maritima is Q86 (Q86TM; compare Fig. 7A and Fig. 7B). In the inactive form of the protein, Q86TM blocks the space that, in the active form, is occupied by Y84™ (the equivalent of E89EC) (Fig. 7B). Such an interaction should enhance the stability of the β4α4 loop in its inactive conformation and inhibit the conformational shift to the active form by preventing the burial of Y84™ near the phosphorylation pocket. We interpret the clear interactions of Q86™ in the inactive state versus the likely weaker interactions involved in the function of K91EC (Fig. 7A) as an evolutionary temperature adaptation of CheY in T. maritima. This adaptation possibly allows the regulation mechanism of CheY activation to be sustained at higher temperatures. In E. coli phosphorylation-dependent signaling, a possible role for K91 acetylation may be prolongation of the lifespan of CheY’s active state in a similar manner to the role of E89EC in CheY’s autophosphorylation reaction (Thomas et al., 2013).
Fig. 7.
The equivalent of position 91 plays a role in β4α4 loop repression in CheY from Thermotoga Maritima. Aligned structures of active (blue) and inactive (green) conformations of CheY from E. coli (A; PDBs 2BIJ and 3CHY) and Thermotoga Maritima (B; PDBs 4IGA and 4TMY). The structures were visualized and aligned by PyMOL (The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC). See text for details.
K109 function in acetate-dependent clockwise generation
Earlier studies, in which any tested replacement of K109 by another amino acid resulted in loss of clockwise rotation (Lukat et al., 1991; Bourret et al., 1993), taken together with our observation that all K109R–replacement mutants failed to support clockwise rotation, indicate that K109 is essential for clockwise rotation. Conversely, the observations that the x10KR mutant was capable of exhibiting acetate-dependent clockwise generation (Fig. 4B) and that K109 is one of the two acetylation sites that undergo acetate-stimulated enhancement of the acetylation level in vivo (Fig. 2) suggest that acetylation of K109 generates CW rotation. As discussed above, this clockwise generation can mainly be observed when K91 is replaced or acetylated.
Yet, the gutted strain expressing CheY x10KR mutant protein spent less time in clockwise rotation than that expressing wild-type CheY (Fig. 4B). The reason for this is, at least in part, the much-reduced expression levels of the x10KR and x11KR CheY mutant proteins relative to all other strains (right panel in Fig. S1C). This reduced expression was probably due to the lower solubility of the x10KR and x11KR proteins, evident from the formation of inclusion bodies when they are produced. Consistent with this explanation, the mutant CheY[K(91,92,119,122,126)R], which lacks all the acetylation sites except for K109, had wild-type-like rotation.
Earlier studies that observed the inability of cells expressing CheY(K109R) to generate clockwise rotation attributed this flaw to breaking presumably important intra-molecular interactions of K109 (Bourret et al., 1993). Thus, it was suggested that K109 may form a salt bridge with Asp-57 of non-phosphorylated CheY (Silverman and Simon, 1974; Volz and Matsumura, 1991) and that it may be involved in coordinating one of the oxygen atoms of the phosphoryl group on CheY (Robinson et al., 2000). Yet, the observations that CheY(K109R) is hyperphosphorylated (Lukat et al., 1991) and that K109 is apparently uninvolved in phosphotransfer catalysis (Silversmith et al., 1997) taken together with the fact that all our results were obtained in gutted cells lacking the phosphorylation system and the enzymes that convert acetate to acetyl phosphate, strongly argue that the ability of K109 acetylation to generate clockwise rotation is phosphorylation-independent. The mechanism by which K109 activates CheY is still largely unknown. Once the function of K109 in activating CheY is fully understood, the mechanism by which K109 acetylation activates the protein can be resolved.
Experimental procedures
Bacterial strains and plasmids
The E. coli strains, plasmids, and primers used in this study are listed in Tables 4 and S5. All mutants were generated by site-directed mutagenesis derived from a polymerase chain reaction, using an appropriate set of primers and, as a template, a pCA24N vector carrying wild-type cheY with N-terminal His-tag from the ASKA library (Kitagawa et al., 2005). The resulting recombinant plasmids were introduced into E. coli XL-1 Blue cells and then isolated using a plasmid purification kit (QIAGEN) and sequenced. All DNA manipulations and standard molecular biology protocols were performed as described (Sambrook and Russell, 2001).
Table 4.
Strains and plasmids used in this study
| Relevant genotype | Source/reference | |
|---|---|---|
| A. Strains | ||
| AG1 | recA1 endA1 gyrA96 thi-1 hsdR17(rK−mK+) supE44 relA1 | Stratagene |
| JW1871 | AG1+ pCA24N | (Kitagawa et al., 2005) |
| XL-1 Blue |
recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F´ proAB lacIqZΔM15 Tn10 (TetR)] |
Stratagene |
| UU1631 | Δ(cheY) Δthr(Am)-1 leuB6 his-4 metF(Am)159 eda-50 rpsL136 StrepR [thi-1 ara-14 lacY1 mtl-1 xyl-5 tonA31 tsx-78] |
VS100 in (Sourjik & Berg, 2000) |
| RBB1097 | CP875 Δ(cheA - cheZ)::ZeocinR, Δ(ackA pta hisJ hisP dhu) zej- 223::Tn10 |
(Barak et al., 1998) |
| EW289 | UU1631 + pCA24N (cheY) | This study |
| EW330 | RBB1097 + pCA24N (cheY) | This study |
| EW333 | RBB1097 + pCA24N [cheY(K92,119,122,126R)] | This study |
| EW334 | RBB1097 + pCA24N [cheY(K91,92,119,122,126R)] | This study |
| EW335 | RBB1097 + pCA24N [cheY(K91R)] | This study |
| EW336 | RBB1097 + pCA24N [cheY(K92R)] | This study |
| EW337 | RBB1097 + pCA24N [cheY(K109R)] | This study |
| EW338 | RBB1097 + pCA24N [cheY(K119R)] | This study |
| EW339 | RBB1097 + pCA24N [cheY(K122R)] | This study |
| EW340 | RBB1097 + pCA24N [cheY(K126R)] | This study |
| EW411 | RBB1097 + pCA24N [cheY(K4,7,26,45,70,91,92,119,122,126R)] | This study |
| EW415 | RBB1097 + pCA24N [cheY(K4,7,26,45,70,91,92,109,119,122,126R)] | This study |
| EW418 | RBB1097 + pCA24N [cheY(K91,92,109,119,122,126R)] | This study |
| EW451 | RBB1097 + pCA24N [cheY(K91,92,122,126R)] | This study |
| EW455 | RBB1097 + pCA24N [cheY(K91,92,119,122R)] | This study |
| EW465 | RBB1097 + pCA24N [cheY(K91,119,122,126R)] | This study |
| EW466 | RBB1097 + pCA24N [cheY(K91,92,119,126R)] | This study |
| EW547 | RBB1097 + pCA24N [cheY(K91Q)] | This study |
| EW588 | RBB1097 + pCA24N [cheY(K91E)] | This study |
| EW590 | RBB1097 + pCA24N [cheY(K91H)] | This study |
| EW595 | RBB1097 + pCA24N [cheY(K91M)] | This study |
| EW596 | RBB1097 + pCA24N [cheY(K91D)] | This study |
| B. Plasmid | ||
| pCA24N | CheY-His6 overexpression, chloramphenicol resistance | (Kitagawa et al., 2005) |
Quantitative LCMS analyses
Gutted E. coli cells of strain UU1631 expressing 6xHis-CheY from a plasmid, were grown at 37°C on tryptone broth supplemented with chloramphenicol (Sigma, Israel; 34 µg/ml) to O.D590 = 0.9 with or without induction of IPTG (1 mM). One hour before harvesting, the culture was divided into two equal portions, one incubated with sodium acetate (100 mM; pH 7.0; Merck) and the other without acetate. The harvested cells were washed twice with motility buffer [10 mM potassium phosphate buffer (pH 7.0), 0.1 mM EDTA and 0.1 mM L-methionine (Adler, 1973)], lysed by BugBuster (Novagen), and subjected to SDS-PAGE 12%. The CheY bands were cut from the gel and destained. Proteins were then reduced by incubation with dithiothreitol (5 mM; Sigma) for 30 min at 60°C, and alkylated with 10 mM iodoacetamide (Sigma) in the dark for 30 min at 21 °C. Proteins were then subjected to digestion with trypsin (Promega; Madison, WI, USA) or to digestion with chymotrypsin (Sigma Aldrich, St. Louise MO, USA) for 6 h followed by trypsin for 16 h at 37°C. The digestions were stopped by trifluroacetic acid (1%). The samples were stored in −80°C.
Liquid Chromatography
ULC/MS grade solvents were used for all chromatographic steps. Each sample was loaded using split-less nano-Ultra Performance Liquid Chromatography (10 kpsi nanoAcquity; Waters, Milford, MA, USA). The mobile phase was: (A) H2O + 0.1% formic acid and (B) acetonitrile + 0.1% formic acid. Desalting of the samples was performed online using a reversed-phase C18 trapping column (180 µm internal diameter, 20 mm length, 5 µm particle size; Waters). The peptides in the samples were separated using a C18 T3 HSS nano-column (75 µm internal diameter, 150 mm length, 1.8 µm particle size; Waters) at 0.4 µl/min. Peptides were eluted from the column into the mass spectrometer using the following gradient: 3% to 30%B in 30 min, 30% to 95%B in 5 min, maintained at 95% for 7 min and then back to initial conditions.
Low-resolution mass spectrometry
The nanoLC was coupled online through a nanoESI emitter (7 cm length, 10 mm tip; New Objective; Woburn, MA, USA) to a tandem quadrupole mass spectrometer (Xevo TQ-S, Waters Corp.). Data were acquired in SRM using Masslynx 4.1. In the high-resolution targeted analysis, the nanoLC was coupled to a quadrupole Orbitrap mass spectrometer (Thermo Scientific) and operated in ‘targeted MS2’ mode.
High-resolution mass spectrometry
The nanoLC was coupled online through a FlexIon nanospray using a nanoESI emitter (7 cm length, 10 mm tip; New Objective; Woburn, MA, USA), to a quadrupole orbitrap mass spectrometer (Q Exactive, Thermo Scientific). The mass spectrometer was operated in Parallel Reaction Monitoring mode (Peterson et al., 2012). Orbitrap resolution was set to 35,000, automatic gain control target was set to 2e5, and injection time to 250 ms.
Data analysis
Raw data were then imported into Skyline software (MacLean et al., 2010) for final processing and evaluation. Quantification was based on the area under the curve of extracted ion chromatogram from the most intense transition per peptide. At least three overlapping SRM transitions were acquired per peptide. Acetylated peptides were synthesized (JPT, Germany) and ran prior to the samples for use as external standards (Table S1). The signal from the native peptides was referenced to the standards to obtain absolute concentrations.
Acetate-dependent clockwise generation
For studying acetate-dependent clockwise generation, plasmids expressing wild-type or mutant CheY proteins (listed in Table 4) were introduced into the gutted strain RBB1097 lacking all the genes from cheA to cheZ and also deleted for ack and pta (Table 4). These strains, lacking all the chemotaxis proteins, some of the chemotaxis receptors, and the enzymes that convert acetate to AcCoA via acetyl phosphate and vice versa, were grown at 37°C on tryptone broth supplemented with chloramphenicol (34 µg/ml) to O.D590 = 0.7. These cells expressed the plasmids listed in Table 4 even though they were grown without IPTG induction due to leakiness of the plasmids. To determine the direction of flagellar rotation and the response to acetate, we used the tethering assay (Silverman and Simon, 1974). Cells were washed twice and resuspended in a motility buffer containing 10 mM potassium phosphate (pH 7.0), 0.1 mM EDTA and 0.1 mM L-methionine (Adler, 1973). The cells were tethered through their flagella to glass slides in a flow chamber (Berg and Block, 1984) as described earlier (Ravid and Eisenbach, 1983), and the rotation of the tethered cells (at room temperature, 23–25°C) was recorded and analyzed using homemade computer software. When needed, acetate (100 mM; pH 7.0) was added in the motility buffer to the flow chamber.
Estimation of CheY expression levels in Δ cheY strains
Cells of each strain were grown to mid-exponential phase (OD590 = 0.6) and divided into two portions. IPTG (1 mM) was added to one of the portions, and both portions were further incubated at 37°C for 3 h. Cell-free extracts were prepared by BugBuster according to manufacturer instructions and subjected to SDS-PAGE (12% polyacrylamide) followed by Western blots with an anti-6xHis antibody.
Statistical analysis
All statistical analyses were carried out using InStat 3 software package (Graph Pad Software, USA).
Molecular dynamics simulations
System Setup
Atomic coordinates for the initial wild-type and K91H CheY protein models were taken from the crystallographically derived PDB 3CHY (Volz and Matsumura, 1991). For residues in the PDB in which multiple orientations of the side-chains were present, the conformation with the highest reported occupancy was taken. Water molecules present in the PDB were also retained. In the case of the K91H CheY model, lysine 91 was replaced with a histidine protonated at the δ position using the psfgen module in VMD (Humphrey et al., 1996). Models were then hydrated with TIP3P water molecules using VMD’s solvate plugin, resulting in a simulation box of size 64 by 59 by 60 Å. Using VMD’s autoionize plugin the wild-type and K91H systems were then neutralized by adding four and five sodium ions, respectively, and subsequently ionized with sodium and chloride ions to the physiological concentration of 150 mM. The resulting wild-type and K91H systems contained 21,191 and 21,193 atoms, respectively, including protein, water, and ions.
Simulation Protocol
All molecular dynamics simulations were performed using the parallel molecular dynamics code, NAMD 2.9 (Phillips et al., 2005; Mei et al., 2011), and CHARMM22 force field (MacKerell et al., 1998). Simulations were conducted in the NPT ensemble with isobaric and isothermal conditions maintained at 1 atm and 310°K using the Nosé-Hoover Langevin piston with period 200 fs and decay 50 fs and the Langevin thermostat with a temperature coupling of 5 ps−1. The r-RESPA integrator scheme (Phillips et al., 2005) with an integration time step of 2 fs was used in all simulations. SHAKE constraints were applied to all hydrogen atoms (Ryckaert et al., 1977). Short-range, non-bonded interactions, calculated every 2 fs with a cutoff of 12 Å and long-range electrostatics, were evaluated every 6 fs using the particle mesh Ewald (PME) method (Darden et al., 1993) with a grid size of 1 Å. Periodic boundary conditions were used in all simulations. Both wild-type and K91H models were subjected to a series of conjugant gradient energy minimizations: 100,000 steps with all protein atoms excluding hydrogen restrained, 100,000 steps with backbone atoms restrained, and 100,000 steps with all atoms unrestrained. Before proceeding with unrestrained equilibration simulations, each model underwent a 2 ns NPT simulation in which backbone atoms were harmonically restrained with spring constant = 0.5 kcal/mol x nm2.
Simulation analysis
Visualization and extraction of raw trajectory data for analysis was performed using VMD (Humphrey et al., 1996). Root mean squared fluctuation (RMSF) calculations were performed using the measure function in VMD (Humphrey et al., 1996). Clustering analysis was performed using the implementation of the K-means algorithm from the python scientific computing library, Scipy (E Jones et al., 2001). Principal components analysis (PCA) was performed using the protein dynamics analysis package, ProDy (Bakan et al., 2011). Before conducting PCA, the backbone atomic coordinates of the β4α4 loop (residues 88 to 91) were extracted from the concatenated K91H and wild-type ensemble simulations and aligned using root mean square deviation fitting in VMD. A total of 25,000 conformations (12,500 per model), sampled over 5 µs of simulation, were analyzed. The top three resulting principal components were used as a basis on which the backbone structures of the β4α4 loop were projected for further analysis. The root mean squared deviation values depicted in Fig. 6 were calculated in the three-dimensional principal component space described above and are equivalent to the Euclidean distances between projections of a representative activated loop conformation (PDB 1FQW) and those arising in the K91H and wild-type ensemble simulation trajectories. Illustrations of K-means and PCA results were produced using the python plotting library, Matplotlib (Hunter, 2007).
Supplementary Material
Acknowledgments
We thank the Smoler Proteomics Research Center of the Technion, Haifa, Israel for carrying out tandem mass spectrometry for the identification of CheY acetylation sites in vivo. We also thank Yulian Gavrilov and Dr. Yaakov Levy (Weizmann Institute of Science) for their help with the production and analysis of the molecular dynamics simulations at the preliminary stage, Olga Khersonsky (Weizmann Institute) for her advice in site-directed mutagenesis, Dr. Leah Armon (Weizmann Institute) for statistical advice, and Dr. Juan R. Perilla and Dr. Wei Han (University of Illinois at Urbana-Champaign) for insightful discussions and advice regarding the computational aspects of this manuscript. M.E. is an incumbent of the Jack and Simon Djanogly Professorial Chair in Biochemistry. This study was supported by grant no. 534/10 from the Israel Science Foundation and by the Minerva Foundation with funding from the Federal German Ministry for Education and Research. Funding was also provided by National Institutes of Health grant No. 9P41GM104601 and National Science Foundation grant No. PHY-1430124. Molecular dynamics simulations were performed on the Blue Waters supercomputer as part of the Petascale Computational Resource (PRAC) grant "The Computational Microscope", which is supported by the National Science Foundation (award number ACI-1440026).
References
- Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol. 2009;27:633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler J. A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli . J Gen Microbiol. 1973;74:77–91. doi: 10.1099/00221287-74-1-77. [DOI] [PubMed] [Google Scholar]
- Bakan A, Meireles LM, Bahar I. ProDy: protein dynamics inferred from theory and experiments. Bioinformatics. 2011;27:1575–1577. doi: 10.1093/bioinformatics/btr168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak R, Abouhamad WN, Eisenbach M. Both acetate kinase and acetyl Coenzyme A synthetase are involved in acetate-stimulated change in the direction of flagellar rotation in Escherichia coli . J Bacteriol. 1998;180:985–988. doi: 10.1128/jb.180.4.985-988.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak R, Prasad K, Shainskaya A, Wolfe AJ, Eisenbach M. Acetylation of the chemotaxis response regulator CheY by acetyl-CoA synthetase from Escherichia coli . J Mol Biol. 2004;342:383–401. doi: 10.1016/j.jmb.2004.07.020. [DOI] [PubMed] [Google Scholar]
- Barak R, Welch M, Yanovsky A, Oosawa K, Eisenbach M. Acetyladenylate or its derivative acetylates the chemotaxis protein CheY in vitro and increases its activity at the flagellar switch. Biochemistry (USA) 1992;31:10099–10107. doi: 10.1021/bi00156a033. [DOI] [PubMed] [Google Scholar]
- Barak R, Yan J, Shainskaya A, Eisenbach M. The chemotaxis response regulator CheY can catalyze its own acetylation. J Mol Biol. 2006;359:251–265. doi: 10.1016/j.jmb.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Berg HC, Block SM. A miniature flow cell designed for rapid exchange of media under high-power microscope objectives. J Gen Microbiol. 1984;130:2915–2920. doi: 10.1099/00221287-130-11-2915. [DOI] [PubMed] [Google Scholar]
- Bishop CM. Pattern Recognition and Machine Learning. Springer Verlag; 2006. [Google Scholar]
- Bourret RB, Drake SK, Chervitz SA, Simon MI, Falke JJ. Activation of the phosphosignaling protein CheY .II. Analysis of activated mutants by19F NMR and protein engineering. J Biol Chem. 1993;268:13089–13096. [PMC free article] [PubMed] [Google Scholar]
- Darden T, York D, Pedersen L. Particle mesh Ewald: An N⋅ log (N) method for Ewald sums in large systems. The Journal of chemical physics. 1993 [Google Scholar]
- Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14 doi: 10.1016/0263-7855(96)00018-5. 33-8-27-8. [DOI] [PubMed] [Google Scholar]
- Hunter JD. Matplotlib: A 2D graphics environment. Computing in Science & Engineering. 2007 [Google Scholar]
- Jones E, Oliphant T, Peterson P. SciPy: Open source scientific tools for Python. 2001 http://www scipy org/
- Jones JD, O’Connor CD. Protein acetylation in prokaryotes. Proteomics. 2011;11:3012–3022. doi: 10.1002/pmic.201000812. [DOI] [PubMed] [Google Scholar]
- Kim GW, Yang XJ. Comprehensive lysine acetylomes emerging from bacteria to humans. Trends Biochem Sci. 2011;36:211–220. doi: 10.1016/j.tibs.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- Knaggs MH, Salsbury FRJ, Edgell MH, Fetrow JS. Insights into correlated motions and long-range interactions in CheY derived from molecular dynamics simulations. Biophys J. 2007;92:2062–2079. doi: 10.1529/biophysj.106.081950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Luo J, Brooks CL, Gu W. Acetylation of p53 inhibits its ubiquitination by Mdm2. J Biol Chem. 2002;277:50607–50611. doi: 10.1074/jbc.C200578200. [DOI] [PubMed] [Google Scholar]
- Li R, Gu J, Chen YY, Xiao CL, Wang LW, Zhang ZP, et al. CobB regulates Escherichia coli chemotaxis by deacetylating the response regulator CheY. Mol Microbiol. 2010;76:1162–1174. doi: 10.1111/j.1365-2958.2010.07125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukat GS, Lee BH, Mottonen JM, Stock A, Stock JB. Roles of the highly conserved aspartate and lysine residues in the response regulator of bacterial chemotaxis. J Biol Chem. 1991;266:8348–8354. [PubMed] [Google Scholar]
- Ma L, Cui Q. Activation mechanism of a signaling protein at atomic resolution from advanced computations. J Am Chem Soc. 2007;129:10261–10268. doi: 10.1021/ja073059f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei C, Sun Y, Zheng G, Bohm EJ, Kale LV, Phillips CJ, Harrison C. Enabling and scaling biomolecular simulations of 100 million atoms on petascale machines with a multicore-optimized message-driven runtime; 2011 International conference for High performance computing, networking, storage and analysis (SC); 2011. pp. 1–11. [Google Scholar]
- Mottonen JM, Jacobs DJ, Livesay DR. Allosteric response is both conserved and variable across three CheY orthologs. Biophys J. 2010;99:2245–2254. doi: 10.1016/j.bpj.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson AC, Russell JD, Bailey DJ, Westphall MS, Coon JJ. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol Cell Proteomics. 2012;11:1475–1488. doi: 10.1074/mcp.O112.020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan R, Schuster M, Bourret RB. Acetylation at Lys-92 enhances signaling by the chemotaxis response regulator protein CheY. Proc Natl Acad Sci USA. 1998;95:4918–4923. doi: 10.1073/pnas.95.9.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid S, Eisenbach M. Correlation between bacteriophage chi adsorption and mode of flagellar rotation of Escherichia coli chemotaxis mutants. J Bacteriol. 1983;154:604–611. doi: 10.1128/jb.154.2.604-611.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson VL, Buckler DR, Stock AM. A tale of two components: a novel kinase and a regulatory switch. Nat Struct Biol. 2000;7:626–633. doi: 10.1038/77915. [DOI] [PubMed] [Google Scholar]
- Ryckaert J-P, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. Journal of Computational Physics. 1977;23:327–341. [Google Scholar]
- Sambrook J, Russell D. Molecular cloning: a laboratory manual. 3rd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Silverman M, Simon M. Flagellar rotation and the mechanism of bacterial motility. Nature. 1974;249:73–74. doi: 10.1038/249073a0. [DOI] [PubMed] [Google Scholar]
- Silversmith RE, Appleby JL, Bourret RB. Catalytic mechanism of phosphorylation and dephosphorylation of CheY: Kinetic characterization of imidazole phosphates as phosphodonors and the role of acid catalysis. Biochemistry (USA) 1997;36:14965–14974. doi: 10.1021/bi9715573. [DOI] [PubMed] [Google Scholar]
- Skjaerven L, Martinez A, Reuter N. Principal component and normal mode analysis of proteins; a quantitative comparison using the GroEL subunit. Proteins. 2011;79:232–243. doi: 10.1002/prot.22875. [DOI] [PubMed] [Google Scholar]
- Sourjik V, Berg HC. Localization of components of the chemotaxis machinery ofEscherichia coliusing fluorescent protein fusions. Mol Microbiol. 2000;37:740–751. doi: 10.1046/j.1365-2958.2000.02044.x. [DOI] [PubMed] [Google Scholar]
- Thao S, Escalante-Semerena JC. Control of protein function by reversible Nε-lysine acetylation in bacteria. Curr Opin Microbiol. 2011;14:200–204. doi: 10.1016/j.mib.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SA, Immormino RM, Bourret RB, Silversmith RE. Nonconserved active site residues modulate CheY autophosphorylation kinetics and phosphodonor preference. Biochemistry (USA) 2013;52:2262–2273. doi: 10.1021/bi301654m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz K, Matsumura P. Crystal structure of Escherichia coli CheY refined at 1.7-Å resolution. J Biol Chem. 1991;266:15511–15519. doi: 10.2210/pdb3chy/pdb. [DOI] [PubMed] [Google Scholar]
- Wolfe AJ, Conley MP, Berg HC. Acetyladenylate plays a role in controlling the direction of flagellar rotation. Proc Natl Acad Sci USA. 1988;85:6711–6715. doi: 10.1073/pnas.85.18.6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Barak R, Liarzi O, Shainskaya A, Eisenbach M. In vivo acetylation of CheY, a response regulator in chemotaxis of Escherichia coli. J Mol Biol. 2008;376:1260–1271. doi: 10.1016/j.jmb.2007.12.070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.