Abstract
Vinculin is a cytoplasmic actin-binding protein enriched in focal adhesions and adherens junctions that is essential for embryonic development. Much is now known regarding the role of vinculin in governing cell–matrix adhesion. In the past decade that the crystal structure of vinculin and the molecular details for how vinculin regulates adhesion events have emerged. The recent data suggests a critical function for vinculin in regulating integrin clustering, force generation, and strength of adhesion. In addition to an important role in cell– matrix adhesion, vinculin is also emerging as a regulator of apoptosis, Shigella entry into host cells, and cadherin-based cell–cell adhesion. A close inspection of this work reveals that there are similarities between vinculin’s role in focal adhesions and these processes and also some intriguing differences.
Keywords: Vinculin, Integrins, Cadherins, Cell adhesion, Force generation, Cell migration and Actin
1. Introduction
Vinculin was originally isolated from chicken gizzard smooth muscle in 1979 as a molecule that copurified with α-actinin (Geiger, 1979). Initial characterization revealed that this protein was localized at regions where the ends of actin bundles terminated in membrane attachment sites (Burridge and Feramisco, 1980; Geiger et al., 1980). Thus, the protein was named vinculin from the Latin word vinculum—meaning a bond signifying a union or unity. The subsequent cloning of full-length chicken vinculin in 1988 paved the way for numerous experiments aimed at better understanding this protein’s function. We now know that vinculin is a 116-kDa cytoplasmic protein that is a component of the membrane-associated adhesion complexes that tether cells to the extracellular matrix (cell–matrix adhesions or focal adhesions) and adjacent cells (cell–cell adhesions or adherens junctions). Vinculin has no enzymatic activity; consequently, its function is governed by protein–protein interactions that are highly regulated, in part by conformational changes. The nature of these conformational changes and how they are regulated is emerging. These insights, together with information from other functional assays, have established a role for vinculin in regulating many aspects of cell–matrix adhesion, including the assembly, turnover, and strength of focal adhesions, as well as the transmission of force by these cellular structures. In addition to its more established role in cell–matrix adhesion, vinculin is also emerging as a functional regulator of cadherin-based cell–cell adhesions. Indeed, recent studies have shown that vinculin is important for surface expression and the mechanosensing of cadherins. These observations are corroborated by in vivo studies illustrating vinculin’s vital roles in embryogenesis and diseased states. This chapter highlights recent progress and the emerging models for how vinculin function and regulation are governed.
2. Vinculin Structure
2.1. Global structure
For many years, vinculin has been known to be a compact globular protein (Isenberg et al., 1982) that undergoes a conformational change that exposes a 90-kDa N-terminal globular head (residues 1–811), a short flexible proline-rich linker (residues 811–881), and a 27-kDa C-terminal, extended, rod-shaped tail (residues 881–1066) (Coutu and Craig, 1988; Eimer et al., 1993; Milam, 1985; Molony and Burridge, 1985). The vinculin head domain is further divided into three globular protein masses of similar size, organized in a trilobed planar arrangement associated with a short stem (Winkler and Jockusch, 2001; Winkler et al., 1996). Similar to the head, the tail is also globular. It possesses four distinct masses arranged like pearls on a string (Winkler and Jockusch, 2001; Winkler et al., 1996). The proline-rich linker between the head and tail domains of vinculin is flexible, allowing for sharp kinks in the molecule (Winkler and Jockusch, 2001; Winkler et al., 1996).
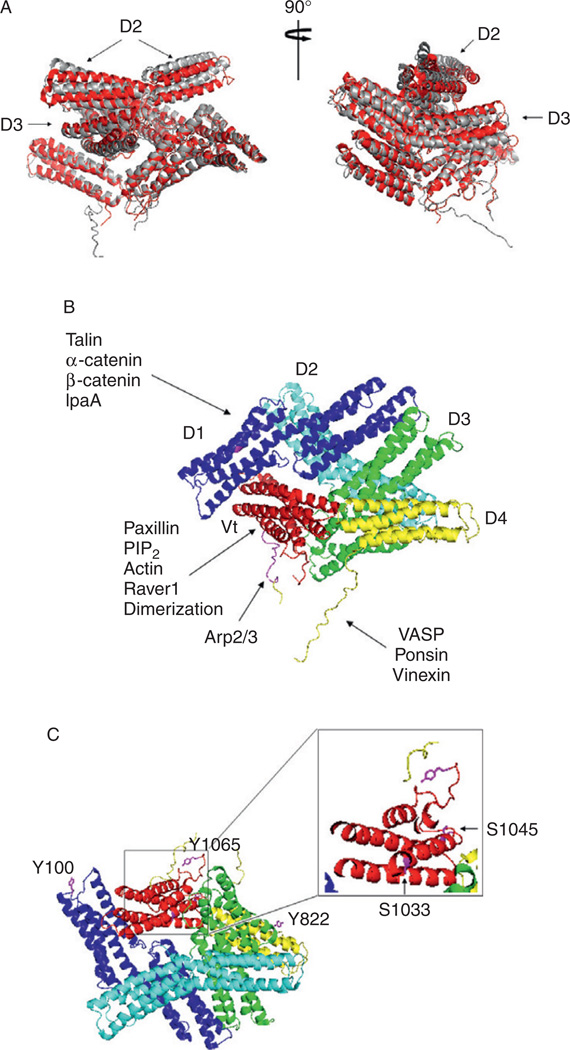
More recently, the crystal structures of full-length chicken vinculin (Bakolitsa et al., 2004), full-length human vinculin (Borgon et al., 2004), and a fragment of the human vinculin head (1–258) in complex with a fragment encompassing residues 879–1066 of the tail (Izard et al., 2004) have been solved. All three structures share striking similarities with the original structure deduced from electron micrograph images described above (Winkler and Jockusch, 2001; Winkler et al., 1996). They reveal that vinculin is comprised of eight, antiparallel, α-helical bundles organized into five distinct domains that are denoted domains 1–5 (D1–D5) by Bakolitsa et al. (2004), and vinculin head 1–3 (Vh1–3) and vinculin tail 1– 2 (Vt1–2) by Izard et al. (Borgon et al., 2004). For the remainder of this chapter, we will use the D1–D5 nomenclature to discuss the vinculin structure. With the exception of small differences in vinculin D2 and D3, the two full-length crystal structures are very similar (Fig. 5.1A).
Figure 5.1. Structure of full-length vinculin in the autoinhibited state.
(A) Overlay of two published full-length vinculin structures. Chicken vinculin structure as solved by Bakolitsa et al. (2004) is colored in light gray, and the human vinculin structure reported by Borgon et al. (2004) is colored in dark gray. These two structures overlap in most domains except for domain 2 (D2) and part of the domain 3 (D3). Two different views are shown. (B) Binding partners to different regions of vinculin. Vinculin domains are labeled: D1, residues 1–252; D2, residues 253–485; D3, residues 486–717; D4, 718–837; and Vt 891–1066. The proline region is partly disordered and contains a strap (residues 878–890). Binding partners to the vinculin head, linker, and tail region are indicated. This figure is adapted from the paper published by Bakolitsa et al. (2004). (C) Vinculin phosphorylation sites. Vinculin phosphorylation sites Y100, Y822, S1033, S1045, and Y1065 are labeled.
The observation that vinculin comprises almost exclusively helical bundles is not surprising, given the prevalence of helical bundles in cytoskeletal proteins, with examples including (but not limited to) talin (Fillingham et al., 2005; Papagrigoriou et al., 2004), α-catenin (Pokutta et al., 2002; Yang et al., 2001), and focal adhesion kinase (Ceccarelli et al., 2006; Prutzman et al., 2004). However, unlike most helical bundles, which are structurally rigid and retain their conformation upon ligand binding, vinculin’s helical bundle, D1, undergoes remarkable conformational changes upon ligand binding (a process termed helical-bundle conversion; Borgon et al., 2004; Izard et al., 2004). Hence, despite being structurally similar to other cytoskeletal proteins, vinculin possesses some dynamic properties.
2.2. Vinculin head domain
The full-length crystal structures of vinculin reveal that the region previously known as the vinculin head (residues 1–811) comprises D1–D3 and a small portion of D4. The D1–D3 region contains two four-helix bundles connected by a long, centrally shared α-helix (Bakolitsa et al., 2004; Borgon et al., 2004). This region resembles the amino-terminal fragment of α-catenin; consequently, it has been referred to as a “vinculin/α-catenin repeat” (Bakolitsa et al., 2004). Several proteins bind to the vinculin head domain. These include talin (Burridge and Mangeat, 1984), α-catenin (Weiss et al., 1998), β-catenin (Hazan et al., 1997), α-actinin (Geiger, 1979), and IpaA (Tran Van Nhieu et al., 1997) (Fig. 5.1B). Among these, only talin binds to vinculin exclusively in cell–matrix adhesions (Burridge and Mangeat, 1984), both α- and β-catenins bind vinculin exclusively in adherens junctions (Drenckhahn and Franz, 1986; Geiger, 1979; Hazan et al., 1997; Watabe-Uchida et al., 1998). The crystal structures of the talin:vinculin D1 and α-actinin:vinculin D1 complexes reveal that both of these binding partners interact with vinculin by inserting an α-helix between helices 1 and 2 of the N-terminal helical bundle of vinculin D1 (Bois et al., 2005; Izard and Vonrhein, 2004; Izard et al., 2004). However, they insert their α-helices in opposite orientations suggesting that D1 has remarkable flexibility and takes on distinct conformations when bound to different ligands (Bois et al., 2005; Izard and Vonrhein, 2004; Izard et al., 2004).
2.3. Proline-rich linker
Based on the full-length crystal structure of vinculin, the proline-rich linker has been redefined as residues 838–878 (Bakolitsa et al., 2004). Following the linker is a strap (residues 878–890) that packs against the first two helices of the vinculin tail. Ligands that interact with the proline-rich region include vasodilator-stimulated phosphoprotein (VASP) (Brindle et al., 1996), the Arp2/3 complex (DeMali et al., 2002), vinexin (Kioka et al., 1999), and ponsin (Mandai et al., 1999; Fig. 5.1B). At the N-terminus of the proline-rich linker is an FPPPPP motif (residues 842–847), which is essential for vinculin binding to VASP, vinexin, and ponsin. The C-terminal end of the proline-rich linker contains a PPPP motif (residues 875–878), which mediates Arp2/3 complex recruitment to vinculin (DeMali et al., 2002).
2.4. Vinculin tail domain
Like the head domain, the vinculin tail (Vt, D5) is mostly α-helical. It contains five antiparallel α-helices (H1–H5) (Bakolitsa et al., 1999, 2004; Borgon et al., 2004) and features two basic elements on the domain surface: a basic ladder formed primarily by the H3 side chains and a basic collar that surrounds a C-terminal, five-residue hairpin. A number of proteins bind to the vinculin tail, including paxillin (Turner et al., 1990; Wood et al., 1994), F-actin (Johnson and Craig, 1995b), phosphatidylinositol-4,5-bisphosphate (PIP2) (Johnson and Craig, 1995a), protein kinase Cα (PKCα) (Weekes et al., 1996), and raver1 (Huttelmaier et al., 2001; Fig. 5.1B). The tail domain also contains two serine residues, S1033 and S1045, that are highly phosphorylated by protein kinase C (Fig. 5.1C) (Schwienbacher et al., 1996; Weekes et al., 1996; Ziegler et al., 2002).
3. Autoinhibited Conformation and Vinculin Activation
3.1. Autoinhibited conformation
The isolated vinculin head domain readily binds to several ligands. However, binding to two of these, talin and α-actinin, is significantly reduced in full-length vinculin (Chen et al., 2006; Groesch and Otto, 1990; Johnson and Craig, 1994; Kroemker et al., 1994). Similarly, the ability of the isolated vinculin tail domain to bind paxillin (Turner et al., 1990), bind PIP2 (Johnson and Craig, 1995a), or cosediment and cross-link actin filaments are lost in the full-length molecule. Binding of a number of proteins to the proline-rich linker, including vinexin (Takahashi et al., 2005), VASP (Huttelmaier et al., 1998), and the Arp2/3 complex (DeMali et al., 2002) are also blocked in full length vinculin. Moreover, the recruitment of some of these binding partners, including talin and actin, could be promoted by addition of phospholipids (Gilmore and Burridge, 1996). These observations led to a model whereby vinculin exists in two conformations: an autoinhibited or inactive conformation in which the binding sites for a number of ligands are masked and an open or active conformation in which the binding sites are exposed (Bakolitsa et al., 2004). It is important to note that although the open conformation is often referred to as the “active conformation,” vinculin has no enzymatic activity.
The recent descriptions of the crystal structures of full-length vinculin in its autoinhibited and talin-bound active states revealed the intricacies of the intramolecular interaction (Bakolitsa et al., 2004; Borgon et al., 2004): the head domains D1–D3 form a pair of pincers, which together with D4, make contact and hold the vinculin tail in an autoinhibited state. The crystal structure of the autoinhibited state also revealed why PIP2, the Arp2/3 complex, and F-actin bind only to the open conformation (Bakolitsa et al., 2004). In the case of PIP2, this is because it binds to the two basic surfaces on the vinculin tail (the above-described basic ladder and basic collar) (Bakolitsa et al., 1999), but in the autoinhibited conformation the collar is partially obscured by the strap. Binding of the Arp2/3 complex is inhibited because the proline residue (P878) that is critical for its recruitment is present in the strap; P878 packs against the vinculin tail, consistent with the low affinity of autoinhibited vinculin for the Arp2/3 complex (DeMali et al., 2002). Binding by F-actin to vinculin depends on two regions in the vinculin tail, one of which is partially blocked by the vinculin head in the closed conformation.
How the head–tail intramolecular interaction is maintained has been the subject of intense scrutiny. Early biochemical analyses showed that the strong intramolecular interaction can be partially explained by the complementary pIs for the two domains: the head domain has a pI of 5.5 and the tail has a pI of 9.6 (Miller et al., 2001). The crystal structures also revealed that the vinculin head–tail interaction is maintained by (1) hydrophobic interactions and hydrogen bonds formed at the D1–D5 interface, (2) polar interactions between D3 and D5, and (3) hydrogen bonds and electrostatic interactions between D4 and D5. Of these, the D1–D5 interface is thought to be the most critical for maintaining the autoinhibited state (Miller et al., 2001). Biochemical analyses tested which of these interactions maintain the autoinhibited conformation (Cohen et al., 2005). These studies established that D5 interacts with D1–D4 with a higher affinity (10−7M) than with D1 alone (10−5 M) (Cohen et al., 2005); this finding not only underscored the importance of the D1–D5 interaction but also suggested that other domains play a role in promoting the autoinhibited conformation. Mutagenesis of charged residues to alanines revealed a series of D5 mutants that disrupt the D4–D5, but not the D1–D5 interface (Cohen et al., 2005). The importance of the D4 interface was verified by reciprocal mutation of residues in D4 that contact D5. In these experiments, the head–tail interaction was reduced by about 100-fold and talin binding was permitted (Cohen et al., 2005). Moreover, these effects do not appear to be limited to interactions in vitro. In cells expressing mutant versions of vinculin with a disrupted D4–D5 interaction, the number and size of focal adhesions were increased and residency times were longer (Cohen et al., 2006). Collectively, these data suggest that not only a D1–D5 interaction but also a D4–D5 interaction is required to maintain vinculin in an autoinhibited state. The contribution of the D3–D5 interaction to maintaining the autoinhibited conformation of vinculin has not been tested by biochemical assays.
3.2. Activation of vinculin
3.2.1. Combinatorial activation
As vinculin lacks enzymatic activity and functions by interacting with other proteins, it is important to understand how the autoinhibited conformation is disrupted. From the crystal structures and biochemical analyses, we have learned that vinculin is held in the autoinhibited conformation by at least two head–tail interfaces and that the conformational changes in the vinculin head, tail, and linker are structurally and thermodynamically linked (Bakolitsa et al., 2004; Cohen et al., 2005). Based on the fact that no known vinculin ligands bind to full-length vinculin with an affinity comparable to that of the head–tail interaction (<1 nM), it has been proposed that two ligands are required to disrupt both interfaces leading to vinculin activation (Bakolitsa et al., 2004). To directly test this hypothesis, the Craig laboratory developed a Fo¨rster resonance energy transfer (FRET) system that reports on vinculin conformational changes (Chen et al., 2005, 2006). The FRET probe is a vinculin construct tagged with CFP at the N-terminus and a YFP inserted immediately proceed D5. When vinculin is in the autoinhibited conformation, the two fluorophores are in close proximity and produce a high FRET signal; upon vinculin activation, the two fluorophores are separated and the FRET signal is reduced. This system has been used extensively to study conformational changes in vinculin (Chen et al., 2005, 2006; Grashoff et al., 2010), as well as to test the ability of talin to activate vinculin (Chen et al., 2006). Neither the talin rod (which contains at least three vinculin-binding sites) nor a short talin peptide containing a single vinculin-binding site induced a change in FRET (Chen et al., 2006). These findings were consistent with the notion that two or more ligands are required for vinculin activation. Indeed, when F-actin was applied alongside either the talin rod or the peptide, vinculin was activated in a dose-dependent manner (Chen et al., 2006). These data support a mechanism whereby at least two proteins, talin and F-actin, are required to activate vinculin (Fig. 5.2).
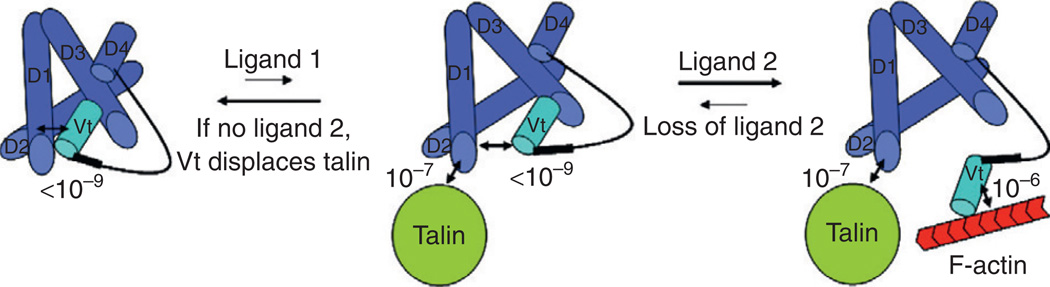
Figure 5.2. A proposed model for combinatory activation of vinculin by talin and F-actin.
Vinculin has a strong head–talin interaction with a Kd less than 10−9 M. A small fraction of vinculin in the open conformation binds talin. As talin’s affinity for vinculin is ~10−7 M, talin alone cannot effectively compete with the vinculin tail and little vinculin–talin complex is formed. F-actin binding to vinculin–talin complexes captures vinculin in an active conformation. This figure is adapted from the paper published by Chen et al. (2006).
3.2.2. Activation by a single ligand
Notwithstanding the data described above, several lines of evidence support the notion that a single ligand can activate vinculin (Bois et al., 2005, 2006; Izard and Vonrhein, 2004; Izard et al., 2004). First, upon binding of vinculin D1 to either a talin or an α-actinin peptide containing a single vinculin-binding site, D1 undergoes helical-bundle conversion (Bois et al., 2005; Izard and Vonrhein, 2004; Izard et al., 2004), an event that could in theory produce tail dissociation (Bois et al., 2005; Izard and Vonrhein, 2004; Izard et al., 2004). In further support of the helical bundle conversion, the addition of head ligands to full length vinculin increases its sensitivity to proteases and perturbs its NMR chemical shift patterns. (Bois et al., 2006). However, it remains to be determined if these conformational changes are similar to those that occur when these proteins are present at physiological concentrations in a cell. The second line of evidence supporting the ability of a single ligand to activate vinculin is that the binding sites for talin and α-actinin are not occluded by the vinculin head–tail interaction (Bakolitsa et al., 2004; Borgon et al., 2004). However, unless the affinity of these ligands for the vinculin head is greater than that of the vinculin tail, it is unlikely that this interaction alone is sufficient to unfurl the molecule. This possibility was tested by adding a peptide containing a single vinculin-binding site to preformed D1:D5 or D1–D4:D5 vinculin complexes and measuring its ability to interact with these complexes. Both peptides were able to compete with the vinculin tail for vinculin head binding supporting the notion that single ligand can activate vinculin (Bois et al., 2006; Izard and Vonrhein, 2004). However, these experiments employed vinculin fragments which have a 10,000-fold (D1:D5, lacks the D4–V5 interface) or 100-fold (D1–D4:D5) weaker affinity than the full-length molecule, due to the lack of a covalent linkage between the vinculin head and tail. Further studies tested if a peptide containing a binding site can activate the full-length molecule (Bois et al., 2005, 2006). Based on surface plasmon resonance analysis, the affinity of vinculin-binding site peptides for the full-length molecule was calculated to be ~ 70–80 nM, which is comparable to the affinity of the head for the tail in the absence of linker (Bois et al., 2006). The α-actinin peptide binds to full-length vinculin or D1 with a Kd of ~ 2 nM, suggesting that this vinculin-binding site peptide can sever the interaction between the vinculin head and talin (Bois et al., 2005). However, these effects may potentially be accounted for by artificial vinculin activation as a consequence of its immobilization to a solid surface. Consistent with this notion, full-length vinculin absorbed to nitrocellulose, but not vinculin in solution, bound a vinculin-binding site peptide (Adey and Kay, 1997; Steimle et al., 1999). Similarly, in solution-phase binding studies, a talin vinculin-binding-site peptide failed to bind the full-length molecule but readily bound the isolated vinculin head domain (Chen et al., 2006).
3.3. Activation by IpaA
One emerging concept regarding the mechanism employed by talin to activate vinculin is that it may not apply to all ligands. In FRET studies using a full-length vinculin probe, Chen et al. (2006) showed that application of the Shigella invasin, IpaA, is sufficient to decrease FRET. This finding suggests that unlike talin, IpaA has the ability to activate vinculin independently of F-actin. However, adding F-actin to the IpaA-treated FRET probe further increased vinculin activation, suggesting that F-actin also plays a role (Chen et al., 2006). It remains unclear how IpaA coordinately disrupts both the D1:D5 and D4:D5 α-actinin, whose vinculin-binding sites are buried inside helical bundles (Bois et al., 2005; Fillingham et al., 2005; Gingras et al., 2006; Papagrigoriou et al., 2004; Ylanne et al., 2001), those of IpaA are exposed and accessible for binding (Hamiaux et al., 2006). Moreover, each of these vinculin-binding sites simultaneously binds two vinculin D1 domains. Hence, it is possible that the accessibility of these sites contributes to IpaA activation of vinculin (Hamiaux et al., 2006).
3.4. Activation in vivo
The in vitro biochemical studies that have been carried out on vinculin have significantly advanced our understanding of its activation. However, the relevance of these findings to cellular events has not been evident until recently. The Craig group has pioneered the study of vinculin conformation at focal adhesions in live cells, using the FRET-based probes described above (Chen et al., 2005). Their initial examination of these probes confirmed the original model with regard to vinculin at focal adhesions being in the open conformation, and vinculin in the cytoplasm being in the closed conformation. Strikingly, however, their data also revealed that vinculin conformations in vivo are much more complicated than predicted in this simplified model (Chen et al., 2005). While most of the vinculin at focal adhesions exists in an open conformation, there is a fraction in the closed conformation: immediately following its recruitment to focal adhesions and later as the adhesions disassemble and the membrane retracts (Chen et al., 2005). These data indicate that vinculin recruitment to focal adhesions is distinct from its activation and that vinculin is inactivated preceding the disassembly of focal adhesions. Additional work is needed to establish which physiological ligands other than talin activate vinculin and to identify the signal transduction pathways that regulate these events in vivo.
4. Biological Functions
4.1. Modulation of cell adhesion and migration
Vinculin’s function is best understood at sites of cell–matrix adhesion. Cell adhesion to the extracellular matrix regulates numerous basic biological processes, including cell growth, migration, differentiation, and survival (Berrier and Yamada, 2007; Huveneers and Danen, 2009; Schwartz and DeSimone, 2008). The most pronounced adhesion complexes in cultured cells are focal adhesions; they are flat, elongated structures rich in heterodimeric (α and β) cell-surface adhesion receptors known as integrins. The cytoplasmic tails of integrins bind with a wide array of proteins that regulate the activation of integrins and their interaction with the extracellular matrix. These focal adhesion proteins also send and respond to extracellular cues and physical properties of the extracellular matrix and provide physical linkage between integrins and the actin cytoskeletal network. Vinculin is recruited to the cytoplasmic tails of β integrin proteins, via its interaction with talin (Burridge and Mangeat, 1984; Horwitz et al., 1986).
A significant body of evidence suggests that vinculin’s presence in focal adhesions is critical for integrin-mediated cell adhesion and migration. For example, cells devoid of vinculin have fewer and smaller adhesions and close a wound more rapidly than vinculin-positive cells (Coll et al., 1995; Saunders et al., 2006; Volberg et al., 1995; Xu et al., 1998b). Conversely, in vinculin-overexpressing cells, the number and size of focal adhesions are increased and cell motility is reduced (Rodriguez Fernandez et al., 1992). Hence, vinculin negatively regulates cell motility via its effects on cell adhesion.
4.1.1. Clustering of integrins and turnover of focal adhesions
Despite its well-established role in modulating focal adhesion stability and cell migration, the molecular mechanisms underlying vinculin’s effects remained elusive until recently. Cohen et al. (2006) showed that the residency of both vinculin and talin in focal adhesions is increased in cells expressing a mutant vinculin in which head–tail interactions are reduced. When the talin-binding site is ablated, this mutant vinculin exhibits enhanced dynamics in focal adhesions (Cohen et al., 2006). Hence, confor-mational changes in vinculin regulate the formation and lifetime of talin– vinculin complexes in cells. This observation combined with knowledge that talin regulates integrin activation led to the notion that vinculin could regulate integrin dynamics (Calderwood et al., 1999). The effect of vinculin on integrin dynamics was directly examined using real-time interference reflection microscopy (Saunders et al., 2006). Focal adhesions are smaller and less abundant in vinculin-null cells than in their wild-type counterparts and turnover more rapidly (Saunders et al., 2006). Using mutant versions of vinculin that lack the tail domain to mimic the open vinculin conformation, Humphries et al. (2007) showed that vinculin regulates integrin dynamics and clustering. Importantly, talin must bind vinculin to exert these effects (Humphries et al., 2007). These findings have led to a new model for the mechanism, whereby vinculin regulates cell–matrix adhesions (Carisey and Ballestrem, 2010; Humphries et al., 2007). According to this model, the stability of adhesion complexes is dependent on vinculin activation; when vinculin is activated, the vinculin–talin complexes stabilize the focal adhesions and enhance integrin clustering (Fig. 5.3A). Further support of a role for vinculin in regulating integrin clustering comes from a more recent study showing that small hairpin RNA-based inhibition of vinculin decreases the number of active integrins on the cell surface (Ohmori et al., 2010).
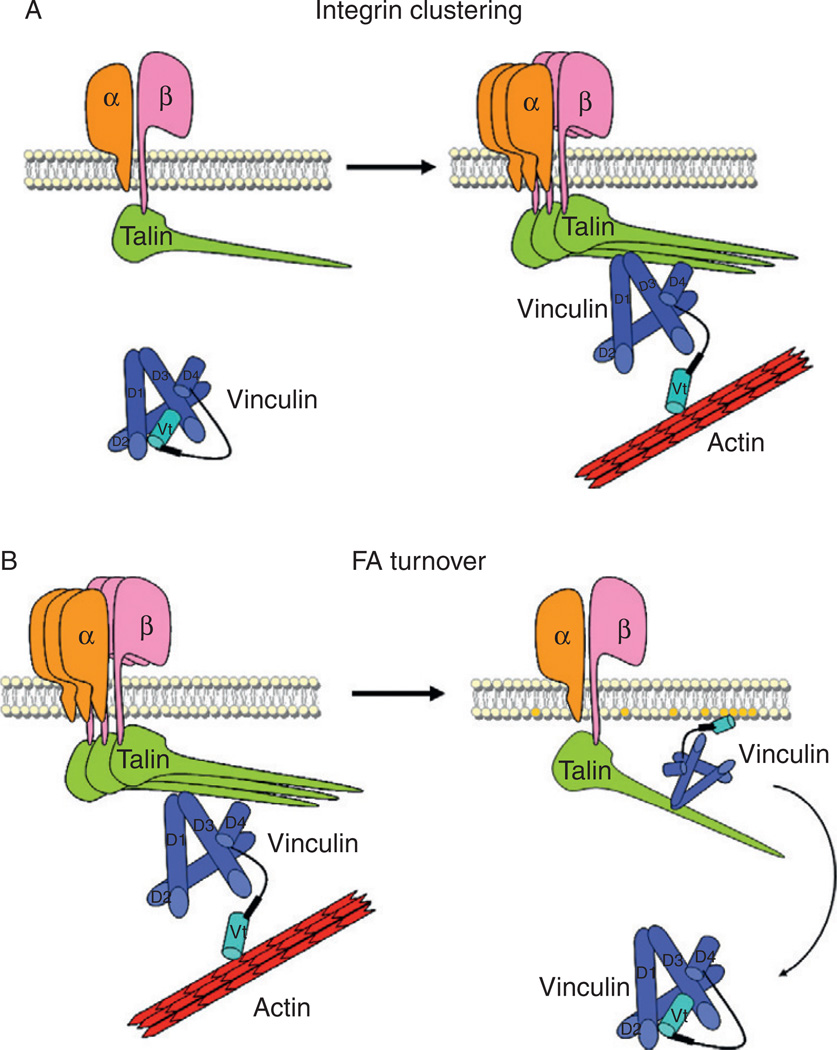
Figure 5.3. Vinculin regulates integrin clustering and focal adhesion turnover.
(A) Vinculin–talin interaction regulates integrin clustering. In the absence of vinculin (left panel), talin bound-active integrins form small focal adhesions with few clustered integrins. Upon activation by talin and F-actin (right panel), vinculin regulates integrin dynamics by clustering integrins in an active conformation. (B) The vinculin–PIP2 interaction regulates focal adhesion turnover. Recent data suggest that stable adhesions (left panel) disassemble when vinculin is lost from focal adhesions when increased phosphoinositide levels causes PIP2 to bind and displace F-actin from vinculin (right panel) (Chandrasekar et al., 2005; Saunders et al., 2006).
How is the stabilizing effect of vinculin on cell–matrix adhesions relieved? The evidence to date suggests that vinculin binding to PIP2 regulates the release of vinculin from focal adhesions. Evidence of a role for PIP2 comes from a vinculin mutant defective for lipid binding, that is, the putative vinculin–PIP2 interaction surfaces on D5 are disrupted (K952Q, K956Q, R963Q, K966Q, R1060Q, K1061Q) (Chandrasekar et al., 2005). This mutant ablates the majority of PIP2 binding but preserves the vinculin–actin and the D1:D5 interaction. When expressed in embryonic fibroblasts derived from vinculin-null mice, this mutant localized to focal adhesions at wild-type levels, but did not rescue the spreading defects characteristic of these cells. The lack of spreading suggested that perturbing the vinculin–PIP2 interaction may lead to more stable focal adhesions. Indeed, the assembly, breakdown, and retrograde transport of focal adhesions in cells expressing the vinculin lipid-binding mutant are all greatly impaired compared to the same processes in their wild-type counterparts (Saunders et al., 2006). Consistent with the reduction in focal adhesion turnover, cells expressing the lipid-binding mutant also exhibit increased adhesion and reduced cell migration (Chandrasekar et al., 2005). These data have led to a model in which the vinculin–PIP2 interaction regulates the turnover of focal adhesions (Chandrasekar et al., 2005; Saunders et al., 2006; Ziegler et al., 2006). According to this model, cues for focal adhesion disassembly increase phosphoinositide levels, resulting in competition of PIP2 with F-actin for binding to vinculin (Fig. 5.3B). The observation that overexpression of PtdInsP 5-kinase α (but not a kinase-dead mutant) increases PIP2 levels and stimulates the loss of focal adhesions lends further support to a role of vinculin binding to PIP2 in the disassembly of focal adhesions (Chandrasekar et al., 2005).
4.1.2. The strength of cell–matrix adhesion
In addition to regulating the dynamics of focal adhesions, vinculin is important for adhesive strength. Adhesive strength is established by the initial integrin–ligand binding step, and is then rapidly enhanced by the clustering of integrins and the recruitment of focal adhesions proteins (Lotz et al., 1989). The latter contributes to adhesive strength by distributing bond forces along the cell–substrate interface (Gallant et al., 2005; Lotz et al., 1989). The role of vinculin in regulating adhesion strength is illustrated by the observation that its recruitment correlates with the formation of focal complexes, and by the fact that its loss results in the weakening of adhesion (Galbraith et al., 2002). Recently, Gallant et al. systematically analyzed the effects of vinculin in adhesion strength. Vinculin-deficient F9 cells exhibit adhesive strength values 20% lower than parental F9 cells (Gallant et al., 2005). These effects are not limited to the F9 cells, as the silencing of vinculin in NIH3T3 cells produced a 25% loss in adhesive strength (Gallant et al., 2005). Thus, vinculin makes minor yet significant contributions to the overall strength of adhesion.
4.1.3. Force transmission
The transmission of force requires focal adhesion complexes that link the extracellular matrix on the outside of the cell to the actin cytoskeleton on the inside of the cell. In motile cells, at least two distinct types of force must be generated (Ridley et al., 2003; Stricker et al., 2010). One is provided by actin polymerization and is the protrusive force needed to extend lamellipodia or filopodia (Pantaloni et al., 2001; Pollard and Borisy, 2003). The other is myosin dependent and leads to rearrangement of the actin cytoskeleton (Ridley et al., 2003). This second force was originally thought to generate the contraction needed to move the cell body forward (Maciver, 1996; Verkhovsky et al., 1999), but more recent work suggests that it is not required for migration (Lombardi et al., 2007).
Focal adhesions assemble and enlarge when force increases and shrink or disassemble when force decreases. This unique property allows cells to respond appropriately to external and internal forces. Several lines of evidence indicate that vinculin regulates the transmission of force between the extracellular matrix and the actin cytoskeleton. For example, cells lacking vinculin exert lower traction forces and are less stiff than their wild-type counterparts (Alenghat et al., 2000; Mierke et al., 2008), and the application of external or internal force increases vinculin recruitment to cell–matrix adhesion sites (Galbraith et al., 2002; Gallant et al., 2005) in a myosin II-dependent manner (le Duc et al., 2010). These findings led to the idea that vinculin recruitment to focal adhesions is force dependent. Using a biosensor that measures force across vinculin, the Schwartz lab showed that vinculin is recruited to focal adhesions and remains in the open conformation under conditions that reduce myosin-dependent contractility (i.e., treatment with Rho kinase inhibitors or reduction of myosin II expression) (Grashoff et al., 2010). Moreover, in migrating cells, the force across vinculin was found to be high in nascent adhesions and to decrease toward moderate levels as the focal adhesions enlarge, with force across vinculin remaining low or decreasing slightly as focal adhesions disassemble (Grashoff et al., 2010). Taken together, these findings indicate that vinculin recruitment and force bearing are independent and that the ability of vinculin to bear force determines whether adhesions assemble or disassemble under force.
How might vinculin regulate the transmission of force? The connection between the actin cytoskeleton and cell adhesions is not rigid, but rather involves points where slippage occurs. Consequently, cell adhesion has been likened to a molecular clutch (Hu et al., 2007). When the clutch is engaged, there is no slippage between adhesion and the cytoskeleton, and movement is productive. When the clutch is disengaged, however, polymerization pressure at the membrane and myosin-dependent contraction cause actin to slip backward, a process known as retrograde flow. Vinculin is a key component of this molecular clutch, and its interaction with F-actin is required for its effects in this context (Hu et al., 2007; Humphries et al., 2007). Moreover, the vinculin-containing clutch colocalizes with areas of high force during lamellipodia protrusion ( Ji et al., 2008). These data indicate that in protruding regions, transient binding of vinculin to F-actin is important for force transduction.
4.1.4. Linkages between the extracellular matrix and actin cytoskeleton
4.1.4.1. Requirement of talin in linking vinculin to mechanical stimuli
One means by which vinculin could potentially link the integrins to extracellular matrix complexes to strengthen adhesion is by binding to talin. Recent biochemical and structural studies have provided new insights into the molecular details of the vinculin–talin interaction. Talin is an elongated and flexible antiparallel dimer, whose small globular head is connected to an extended rod (Isenberg and Goldmann, 1998; Winkler et al., 1997). Talin uses an N-terminal FERM domain (residues 86–400) to bind to the NPxY motif in the β-integrin cytoplasmic tail (Garcia-Alvarez et al., 2003). The talin rod contains a second integrin-binding site (residues 1984–2541) of lower affinity (Tremuth et al., 2004; Xing et al., 2001), a C-terminal actin-binding site (residues 2345–2541) (Hemmings et al., 1996; McCann and Craig, 1997), and at least 11 vinculin-binding sites (Gingras et al., 2005; Hemmings et al., 1996).
Crystal (Izard and Vonrhein, 2004; Izard et al., 2004) and NMR structures (Fillingham et al., 2005) of vinculin D1 complexed with various talin peptides that contain one or more vinculin-binding sites have been solved, revealing that both molecules must undergo a conformational change to interact. Izard et al. first demonstrated that vinculin D1 must undergo a conformational change before talin can bind to it (Izard and Vonrhein, 2004; Izard et al., 2004). Specifically, all four α-helices in the N-terminal bundle of D1 rearrange into a five-helix bundle to accommodate the vinculin-binding-site peptide. Interestingly, the structural changes to D1 that are induced by different vinculin-binding sites in talin appear to be quite similar (Izard and Vonrhein, 2004; Izard et al., 2004). Of the vinculin-binding sites in talin, only one is exposed on its surface in the basal state; the others become accessible only when structural rearrangements occur. In support of a requirement for a talin conformation change, studies show that although isolated talin peptides bind vinculin with high affinity, full-length talin binds only weakly (Bass et al., 2002; Izard and Vonrhein, 2004; Johnson and Craig, 1994; Patel et al., 2006). Also, the stability of the talin helical bundles negatively correlates with their ability to bind vinculin, a finding consistent with a conformational change (Patel et al., 2006). Most recently, NMR and electron paramagnetic resonance spectroscopy have enabled direct visualization of unfurling of the talin α-helical bundles (Gingras et al., 2006; Papagrigoriou et al., 2004).
As talin must undergo a conformational change to bind vinculin, it is critical to determine how its vinculin-binding sites become exposed. The application of force using magnetic tweezers leads to unfolding of the talin rod and exposes cryptic binding sites for vinculin (del Rio et al., 2009). The number of vinculin molecules bound to talin in this context depends on the amount of force applied. Interestingly, findings from this analysis were consistent with the presence of three intermediate talin transition states (del Rio et al., 2009), as previously suggested based on a molecular simulation study (Hytonen and Vogel, 2008). Collectively, these studies establish mechanical force as an important determinant of vinculin binding to talin and in mechanosignaling transduction in vivo.
4.1.4.2. Direct binding between vinculin and actin
The vinculin–actin interaction has long been studied from the perspective of vinculin. These works revealed vinculin is an actin-bundling protein. However, little else is known about the effects of vinculin on actin dynamics and filament structure. Many early studies that attempted to examine vinculin’s effects on actin were performed on vinculin preparations later found to be contaminated with actin and/or other proteins (Otto, 1986; Rosenfeld et al., 1985; Wilkins and Lin, 1982, 1986). We also now know that vinculin purified under the original conditions is in a closed autoinhibited conformation that binds actin with only very low affinity, probably accounting for the negative results obtained from the vast majority of these studies.
The advent of DNA technology allowed for the production of vinculin free of protein contaminants and facilitated exploration of vinculin’s effects on actin. Although work as far back as the 1990s showed that vinculin binds actin directly ( Johnson and Craig, 1995b), precisely where the actin-binding site lies remains a matter of debate (Table 5.1). Analysis of the vinculin protein sequence revealed putative consensus binding sites between helix 1 and helix 3 (H1–H3), and helix 1 and helix 4 (H1–H4) of D5. Although a more C-terminally located region (1016–1066) was also reported to be an actin-binding site (Huttelmaier et al., 1997; Johnson and Craig, 1995b), other binding studies showed that a fragment containing this region does not bind F-actin (Goldmann et al., 1998; Janssen et al., 2006). These differences may be explained in part by the fact that some of these residues are important for maintaining the vinculin head–tail interaction and by the finding that perturbing this interaction may impact actin binding.
Table 5.1.
Putative actin-binding sites in vinculin
| Actin-binding sites | Methods | References |
|---|---|---|
| 940–1012, 1012–1066 | In vitro binding to GST fusion Proteins | Johnson and Craig (2000) |
| 893–1016 | Maltose-binding protein fused with vinculin fragments to cosediment with F-actin, and colocalization analysis in cells | Menkel et al. (1994) |
| 893–985, 1016–1066 | Maltose-binding protein fused with vinculin fragments to cosediment with F-actin | Huttelmaier et al. (1997) |
| 894–1012, no binding to 1012–1066 | Stopped-flow and dynamic light scattering analysis | Goldmann et al. (1998) |
| 883–890, base of H3 and a stretch of residues in C-terminus | EM reconstruction | Janssen et al. (2006) |
It is well established that upon binding to actin vinculin dimerizes and assembles actin filaments into bundles ( Janssen et al., 2006; Jockusch and Isenberg, 1981; Johnson and Craig, 1995b, 2000; Pardo et al., 1983a,b). However, less is known with respect to the effects of vinculin on actin filament formation and structure. The earliest detectable actin-associated adhesions are “dots or doublets of actin dots.” These plaques are highly enriched for integrins, paxillin, and vinculin (Zimerman et al., 2004), suggesting that one of these proteins may have the capability to initiate actin filament formation. This possibility stimulated renewed interest in studying the effects of vinculin on actin assembly and/or dynamics. Our study of the association of the vinculin tail with monomeric and filamentous actin has defined two novel effects of vinculin on actin (Wen et al., 2009). The vinculin tail binds to monomeric G-actin, in which case it forms a nucleus from which genuine actin filament bundles assemble; the characteristics of filaments formed under these conditions are similar to those of filaments assembled under physiological conditions. The vinculin tail also binds to existing filaments and alters their structure. Hence, vinculin has two novel effects on actin: it recruits G-actin to form a nucleus for actin polymerization and it modifies the structure of existing actin filaments.
4.1.4.3. Linkage of vinculin and actin via the Arp2/3 complex
We (DeMali et al., 2002) and others (Moese et al., 2007; Nolz et al., 2007; Schindeler et al., 2005) have shown that vinculin binds and recruits the Arp2/3 complex. In migrating cells, the interaction between vinculin and the Arp2/3 complex occurs transiently in response to signals that trigger membrane protrusion. The interaction is regulated by phosphatidylinositol-3-kinase and Rac1 activity and is sufficient to recruit the Arp2/3 complex to new sites of integrin clustering (DeMali et al., 2002). Vinculin with point mutations that disrupt the Arp2/3 complex binding site fail to restore cell spreading and lamellipodia formation to the same extent as wild-type vinculin when transfected into vinculinnull cells, in spite of the fact that the encoded proteins are targeted to focal adhesions and mediate adhesion (DeMali et al., 2002). These observations indicate that the interaction between the Arp2/3 complex and vinculin promotes the extension of lamellipodia and cell spreading. Further, they support a model whereby nascent adhesion receptors are linked to the existing actin cytoskeleton via vinculin and the Arp2/3 complex.
The current data support the existence of two vinculin-dependent mechanisms for linking nascent adhesions to the actin cytoskeleton: (1) vinculin itself triggers filament formation and (2) vinculin recruits the Arp2/3 complex, which in turn nucleates filament formation. It is not readily apparent why there is a need for two vinculin-dependent mechanisms that trigger filament formation, but the most straightforward explanation is that vinculin might act alone when physiological conditions dictate the need for a bundled actin filament, but operate in concert with the Arp2/3 complex when a branched actin network, such as those that are present in lamellipodia, is needed. The other possibility is that vinculin itself may nucleate polymerization to link nascent adhesions to the actin cytoskeleton, and the vinculin:Arp2/3 complex interaction might be required to allow for further protrusion for these adhesion sites (DeMali and Burridge, 2003). More work is needed to establish the physiological conditions under which each mechanism operates and to fully understand how nascent adhesions form during cell migration, as well as how they are linked to the actin cytoskeletal network that underlies this process.
4.2. Modulation of apoptosis
Loss of vinculin has been correlated with tumorigenesis (Kawahara et al., 1999; Lifschitz-Mercer et al., 1997; Meyer and Brinck, 1997; Sadano et al., 1992; Somiari et al., 2003). This observation can be explained, in part, by vinculin’s role in regulating cell adhesion, migration, and invasion (Mierke et al., 2010; Ziegler et al., 2006). However, emerging evidence suggests that vinculin also plays a key role in regulating apoptosis. For example, vinculinnull F9 cells are resistant to caspase-3 activation under conditions that would normally trigger a massive apoptotic response (Subauste et al., 2004). It appears that this modulation of the apoptotic response by vinculin is due to its ability to compete with FAK for paxillin binding. Evidence for this comes from the finding that in cells lacking vinculin, more paxillin than usual is available to bind FAK and to regulate downstream cell-survival signaling. In contrast, when vinculin, or a vinculin fragment containing residues 811–1066, is expressed in vinculin null cells, the paxillin–FAK interaction is inhibited and the apoptotic response is rescued (Subauste et al., 2004). These results suggest that vinculin may regulate cell survival by modulating interactions between paxillin and FAK. Interestingly, a vinculin phosphorylation mutant, Y822F, does not restore the apoptotic response in vinculin-null cells. How this mutation affects vinculin function has not been explored.
4.3. Regulation of bacterial entry
Vinculin has been implicated in facilitating the invasion of pathogenic bacteria and their propulsion through the host–cell cytoplasm. This phenomenon is best characterized in the case of the gram-negative bacterium Shigella. Upon contact with the intestinal epithelium, Shigella secretes at least four invasins (IpaA, IpaB, IpaC, and IpaD) and induces robust rearrangements in the host cytoskeleton that produce a phagocytic cup that engulfs the bacterium. Entry is abolished by deletion of the genes encoding ipaB, ipaC, or ipaD and is inhibited tenfold by inactivation of ipaA (Menard et al., 1993; Tran Van Nhieu et al., 1997). This ability to reduce invasion reflects a requirement for IpaB-D in formation of the phagocytic cup; IpaA, in contrast, induces the rearrangement of actin networks present in and around the cup.
The mechanism underlying IpaA-induced host cell rearrangements involves vinculin. The C-terminus of IpaA contains two bona fide binding sites and a third, putative, binding site (Izard et al., 2006). The first two of these sites are required for vinculin binding and recruitment to the site of bacterial entry. Once IpaA is bound to the vinculin head, the vinculin tail is free to associate with F-actin and to trigger a loss of filamentous actin (Bourdet-Sicard et al., 1999; Tran Van Nhieu et al., 1997).
There is some controversy as to how IpaA–vinculin binding triggers rearrangement of actin filaments at the site of Shigella entry. Polymerization experiments showed that IpaA binding uncovers a site in vinculin that allows it to partially cap the barbed ends of actin filaments, resulting in an overall net loss of filamentous structures (Ramarao et al., 2007). While this mechanism has received the most attention, it is unlikely that partial capping of the filament ends alone accounts for the instantaneous loss of actin filaments observed in the host cell upon contact with Shigella. In further support of this notion, the deletion of a single vinculin-binding site in IpaA completely ablated the partial capping activity of vinculin, but only slightly reduced the invasive potential of Shigella. Hence, it appears that the IpaA C-terminus (which contains the vinculin-binding sites) modestly contributes to bacterial invasion (Izard et al., 2006).
We have examined the mechanism for the effects of IpaA on the actin cytoskeleton and found other regions of importance. The IpaA-induced loss of actin stress fibers and cell rounding in cells exposed to IpaA does not require an intact vinculin-binding site on IpaA or vinculin (DeMali et al., 2006). Rather, we find that cells expressing IpaA exhibit elevated Rho activity and increased myosin light chain phosphorylation. In addition, IpaA decreases integrin’s affinity for extracellular matrix ligands by interfering with talin recruitment to the integrin cytoplasmic tail. The combination of these two effects, namely weakened adhesion and increased contractility, accounts for the loss of actin stress fibers and cell rounding observed in cells exposed to IpaA. Taken together, these results and previous work suggest a model whereby IpaA binding to vinculin localizes IpaA to adhesion sites. In this locale, IpaA can interfere with the actin cytoskeleton by unmasking the barbed-end capping activity of vinculin and by perturbing talin’s interaction with vinculin and integrins. These combined effects would result in a massive loss of F-actin at the site of Shigella entry.
Vinculin has also been implicated in the pathogenesis of Helicobacter pylori, but less is known about its role in this context. Like Shigella, H. pylori initiates pathogenesis by translocating bacterial proteins to the host cell cytoplasm. The translocation of one such protein, CagA, into gastric epithelial cells has been linked to an increased risk for peptic ulcers and gastric carcinoma, and this has been shown to be a consequence of its ability to deregulate intercellular signal transduction pathways. Upon H. pylori infection, CagA becomes phosphorylated by the Src family kinases and then binds to and activates/inactivates multiple signaling proteins, including (but not limited to) other kinases and phosphatases. Numerous adhesion and cytoskeletal proteins such as vinculin are also affected; vinculin, in particular, undergoes dramatic dephosphorylation at tyrosine residues (Moese et al., 2007) which prevents it from binding to the Arp2/3 complex. Consequently, nascent adhesions do not link to the underlying actin cytoskeleton, and the number of focal complexes that anchor the gut epithelium to the underlying basement membrane is greatly reduced. Taken together with the findings from Shigella studies, these discoveries demonstrate that bacteria such as Shigella and H. pylori hijack the normal functions of vinculin in a cell to circumvent some of the constraints to bacterial entry. While these bacteria share a common target in vinculin, it is important to note that these effects occur via vastly different mechanisms. Moreover, these effects appear to be limited to a subset of bacteria, as vinculin accumulates only modestly at the entry site of the gram-negative bacterium Salmonella (Finlay et al., 1991), and its inhibition does not affect bacterial invasion. Likewise, vinculin is not present at the site of enteropathogenic Escherichia coli entry into cells (Finlay et al., 1992).
5. Modes of Vinculin Regulation
5.1. Regulation of conformational changes: Tyrosine phosphorylation
Vinculin was identified as the first substrate of the Src tyrosine kinase nearly 30 years ago (Sefton et al., 1981), yet the consequences of its phosphorylation remain poorly understood. Early studies correlated alterations in tyrosine phosphorylation on vinculin with a loss of cell–matrix and cell–cell adhesion (Ayalon and Geiger, 1997; Halegoua, 1987; Tidball and Spencer, 1993; Vostal and Shulman, 1993), but many of those were performed using tyrosine kinase inhibitors that alter the phosphorylation states of other proteins in addition to vinculin (Ayalon and Geiger, 1997). More recently, the Haimovich laboratory identified tyrosines Y100 and Y1065 as the major sites of tyrosine phosphorylation in platelets (Zhang et al., 2004). In the full-length crystal structure, Y100 is fully exposed, whereas Y1065 is occluded by the proline-rich linker domain (Fig. 5.1C) (Bakolitsa et al., 2004; Borgon et al., 2004). Tyrosine to phenylalanine mutations at Y100 and Y1065 inhibit cell spreading (Zhang et al., 2004), and a single substitution at Y1065 leads to decrease traction (Diez et al., 2009). The mechanism for the latter effect likely results from uncontrolled exchange of vinculin in and out of adhesion sites (Kupper et al., 2010; Mohl et al., 2009). Collectively, these findings suggest that Y1065 plays an important role in regulating adhesion dynamics.
How tyrosine phosphorylation regulates vinculin function remains to be fully explored. Phosphorylation at Y1065 reduces head–tail interactions, suggesting that vinculin activation might be affected by this modification (Zhang et al., 2004). Consistent with this notion, the Arp2/3 complex cannot bind the vinculin strap in cells expressing a mutant version of vinculin with a Y1065F substitution (Moese et al., 2007). A loss of vinculin binding to the Arp2/3 complex results in a decrease in the association of nascent focal adhesion complexes with the underlying actin cytoskeleton (DeMali et al., 2002), which might explain the decrease in cell spreading and traction observed in the presence of Y1065F. If tyrosine phosphorylation at Y1065 does regulate head–tail interactions, it seems unlikely that all the active vinculin in a cell is phosphorylated at Y1065; if it were, the amount of tyrosine-phosphory-lated vinculin should reflect the amount of active vinculin in a cell, yet in cells transformed with the rous sarcoma virus, only 2% of the total vinculin is phosphorylated (Sefton et al., 1981; Zhang et al., 2004), whereas the amount of active vinculin in a cell is much greater. Hence, it is unlikely that tyrosine phosphorylation of vinculin strictly mirrors vinculin activation. An alternative possibility is that only a subset of the active vinculin is phosphorylated. Consistent with this notion, the Craig lab elegantly demonstrated heterogeneity in the vinculin conformation that is present in focal adhesions, and that the ratio of the different conformers correlates with adhesion dynamics. It is also plausible that phosphorylation is an important step in the pathway to activation. For example, phosphorylation may be requisite for anchorage of vinculin to the lipid membrane (Diez et al., 2009). A scenario such as the latter would imply that the activation state of vinculin is tightly regulated by phosphorylation. A complete understanding of this phenomenon awaits the identification of the phosphatase that dephosphorylates vinculin.
5.2. Regulation of vinculin expression
While it has been appreciated for some time that vinculin function is regulated by conformational changes and posttranslational modification(s), recent work indicates that vinculin function may also be highly regulated at the expression level (Babic et al., 2009). Specifically, the 3′ untranslated region of vinculin binds polypyrimidine tract-binding protein (PTB), a widely expressed RNA-binding protein with roles in splicing, polyadenylation, mRNA stability, and translation initiation (Auweter and Allain, 2008; Sawicka et al., 2008). PTB function is regulated by a combination of events, including its subcellular distribution (it shuttles from the nucleus to the cytoplasm) and its interaction with additional proteins (Sawicka et al., 2008). In most cells in culture, PTB localizes to the nucleus. However, in cells undergoing adhesion and spreading, PTB transiently shifts to the cytoplasm where it concentrates in nascent adhesions and membrane protrusions (Babic et al., 2009). In these regions, it colocalizes with and binds to the vinculin mRNA. This interaction appears to be essential for vinculin expression at adhesion sites, as RNA interference-mediated PTB inhibition reduced the amounts of both the vinculin-encoding mRNA and vinculin protein localized to cell–matrix adhesions (Babic et al., 2009). In further support of the essential nature of the interaction between these proteins, cells lacking PTB exhibit many of the same phenotypes as cells devoid of vinculin: they are more rounded, have smaller focal adhesions, and reduced membrane protrusions. Hence, PTB is required for efficient expression of vinculin in nascent adhesions and for the adhesion assembly (Fig. 5.4).
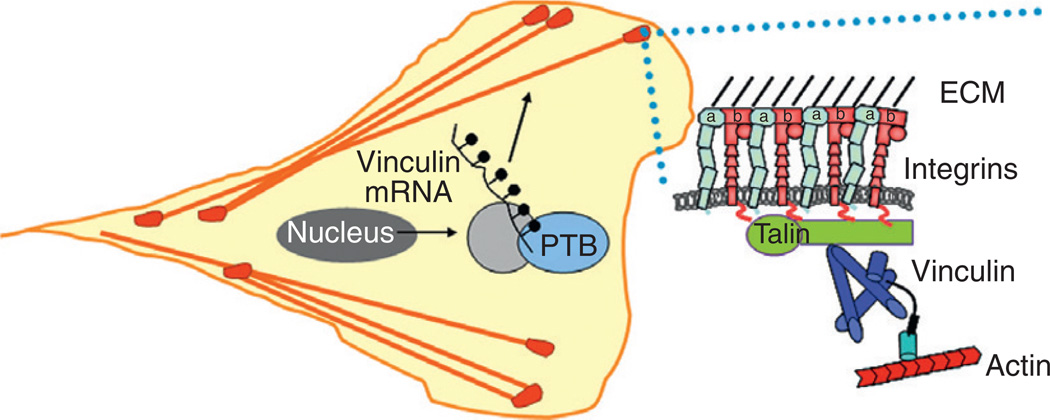
Figure 5.4. Vinculin mRNA translation is regulated spatiotemporally at sites of adhesion.
In response to migratory cues, vinculin mRNAs are transported to sites of membrane protrusion to allow for localized protein translation and the assembly of focal adhesions (dark circles). The delivery of mRNAs is likely regulated by a number of proteins including PTB and other unidentified proteins (gray circle).
Raver1 is another protein that likely plays a role in vinculin expression at cell adhesion sites. It is a heterogeneous nuclear ribonucleoprotein that was first identified as a vinculin tail binding protein in a yeast two-hybrid screen (Huttelmaier et al., 2001). It has three N-terminal RNA recognition motifs (RRM1–3), the first of which directly binds to the vinculin tail domain with micromolar affinity (Lee et al., 2009). In addition, it has two nuclear localization signals and one nuclear export sequence, which allow it to shuttle between the nucleus and the cytoplasm. When in the nucleus, raver1 interacts with PTB to regulate mRNA processing, but during cell differentiation it redistributes to the cytoplasm where it colocalizes with and binds to vinculin in focal adhesions, adherens junctions, and costameres (Huttelmaier et al., 2001). A recent crystal structure of RRM1–3 of raver1 bound to the isolated vinculin tail reveals that raver1 has the potential to bind RNA and vinculin simultaneously (Lee et al., 2009). Moreover, even in complex with vinculin, raver1 is able to interact with actin filaments. A possible role for the raver1–vinculin complex in cell adhesions has been proposed based on these findings (Lee et al., 2009). Specifically, active vinculin binds to the raver1 RRM1 domain, which may or may not already be bound to, or subsequently bind to, mRNA cargo, resulting in localization of the raver1–RNA complex to sites of cell adhesion where the vinculin–raver1 complex could engage the actin cytoskeleton. The raver1–vinculin interaction may thus play a dominant role in targeting bound vinculin mRNAs to adhesions to promote the localized translation of constituent proteins. Validation of this model awaits direct experimental evidence for simultaneous binding of the raver1 N-terminus to both vinculin and mRNAs.
While the studies carried out to date have provided significant insight into how vinculin expression is regulated, the connection, if any, between the PTB and raver1 pathways in this context has not been explored. Interestingly, raver1 colocalizes with and binds to PTB in the nucleus, and both proteins can translocate to cytoplasm (Huttelmaier et al., 2001; Sawicka et al., 2008). These findings raise the possibility that these proteins act in concert to regulate the expression of vinculin (and that of other focal adhesion proteins) (Huttelmaier et al., 2001). In support of this notion, the binding sites for vinculin and PTB on raver1 appear to be distinct, with vinculin binding to the raver1 N-terminus and PTB to the C-terminus (Babic et al., 2009; Huttelmaier et al., 2001; Lee et al., 2009). However, other evidence suggests that PTB can act independently of raver1 in some circumstances. For example, Babic et al. (2009) found that while PTB translocates to the cytoplasm during cell spreading and adhesion, raver1 remains in the nucleus. Whether this phenomenon is universal remains to be determined, as it is unknown if PTB remains in the nucleus under conditions that promote shuttling of raver1 to the cytoplasm.
6. Emerging Themes and Concepts
6.1. Vinculin in adherens junctions
6.1.1. A key member of the cadherin–adhesion complex
Since the identification and localization of vinculin to focal adhesions and cell–cell adhesions over 30 years ago, its presence in focal adhesions has commanded more attention. Thus, relatively little is known about its function in cell–cell junctions. Several specialized cell–cell attachment sites were originally defined as morphologically distinct structures by electron microscopy: tight junctions, adherens junctions, and desmosomes (Obrink, 1986). In the adherens junctions, the major cell-surface adhesion receptors are the cadherins, but the nectins are also present (Niessen and Gottardi, 2008; Nishimura and Takeichi, 2009; Ogita and Takai, 2008). The extracellular domain of cadherins mediate strong cell–cell adhesion by binding to cadherins on an adjacent cell; the cytoplasmic tail of cadherin interacts with several proteins, including β-catenin (via the distal portion of the cadherin cytoplasmic tail) and p120-catenin (via a more proximal region of the cadherin cytoplasmic tail; Perez-Moreno and Fuchs, 2006). β-Catenin in turn binds α-catenin, and both β- and α-catenin bind the vinculin head. The interaction between β-catenin and vinculin requires residues in the β-catenin N-terminus; key among these is methionine 8, which when mutated to a proline residue blocks vinculin binding, both in vitro and in cells (Peng et al., 2010). The α-catenin binding site for vinculin remains unclear, with several groups having shown that vinculin binds to α-catenin residues 273–510 (or smaller fragments, 326–510 amino acids) (Bakolitsa et al., 2004; Imamura et al., 1999; Watabe-Uchida et al., 1998; Yonemura et al., 2010), and others report that vinculin binds to α-catenin 697–906 (Weiss et al., 1998).
With regard to functional relevance, all the evidence to date suggests that vinculin is required to maintain the integrity of adherens junctions. The first hint of vinculin’s importance came from the observation that cell–cell adhesion is lost in numerous cancer cells during the initial stages of tumor formation, at the time when vinculin becomes mislocalized (Kawahara et al., 1999; Lifschitz-Mercer et al., 1997; Meyer and Brinck, 1997; Sadano et al., 1992; Somiari et al., 2003). Later it was found that defects in mice lacking vinculin were consistent with a role for vinculin in regulating adherens junction function (discussed in detail later). However, one limitation to interpreting vinculin’s importance using these approaches is that vinculin function at cell-matrix adhesions is also perturbed. Thus, it is impossible to draw definitive conclusions about the contribution of vinculin specifically to adherens junctions. Similarly, overexpression studies are uninformative as overexpressed vinculin neither integrates readily into preexisting cell–cell adhesions nor turns over in a manner consistent with cadherin–catenin dynamics (Yamada et al., 2005). Given that vinculin’s affinity for cell–matrix adhesions is higher than its affinity for cell–cell adhesions, and that only a small amount of vinculin is required to maintain cell–matrix adhesions (Bakolitsa et al., 2004; Xu et al., 1998a,b), it seemed likely that vinculin could be preferentially depleted from adherens junctions by RNA interference. Indeed, it is possible to disrupt vinculin colocalization with β-catenin and its binding to cadherin adhesion complexes through this approach, and the outcome is loss of epithelial morphology, adhesion to cadherin extracellular domains, and junctional integrity (Maddugoda et al., 2007; Peng et al., 2010). Moreover, these effects could be specifically attributed to the pool of vinculin in adherens junctions, as the residual vinculin localized to focal adhesions, and cell adhesion to the extracellular matrix was maintained at wild-type levels (Peng et al., 2010).
6.1.2. Roles for vinculin in adherens junctions
Although vinculin had been identified as an integral component of adherens junctions in various tissues throughout the years, it was only recently that attention was focused on understanding its role at this site (Bloch and Hall, 1983; Drenckhahn and Franz, 1986; Drenckhahn and Mannherz, 1983; Geiger et al., 1981; Koteliansky and Gneushev, 1983; Massa et al., 1995; Opas et al., 1985; Pardo et al., 1983a). These efforts have uncovered three roles for vinculin.
One line of evidence suggests that vinculin acts downstream of myosin VI, a minus end-directed motor necessary for the E-cadherin-dependent process of border-cell migration (Maddugoda et al., 2007). Depletion of myosin VI in epithelial cells resulted in a loss of vinculin binding to E-cadherin, and in a disruption of the actin cytoskeletal network. This observation, combined with the fact that cells devoid of vinculin (Watabe-Uchida et al., 1998) share phenotypes with cells lacking myosin VI, led to the idea that vinculin may be responsible for the effects of myosin VI. To test this possibility, it was determined if vinculin could rescue the phenotype of myosin VI knockdown cells using chimeras of the vinculin head or tail fused to the N-terminal region of α-catenin to overcome the potential complication of vinculin autoinhibition. Interestingly, both trans-genes rescue the junctional defects induced by a loss of myosin VI. These observations suggest that vinculin acts downstream of myosin VI; this idea is confirmed by the observation that exogenous myosin VI could not rescue the effects induced by a loss of myosin VI in the absence of vinculin expression (Maddugoda et al., 2007).
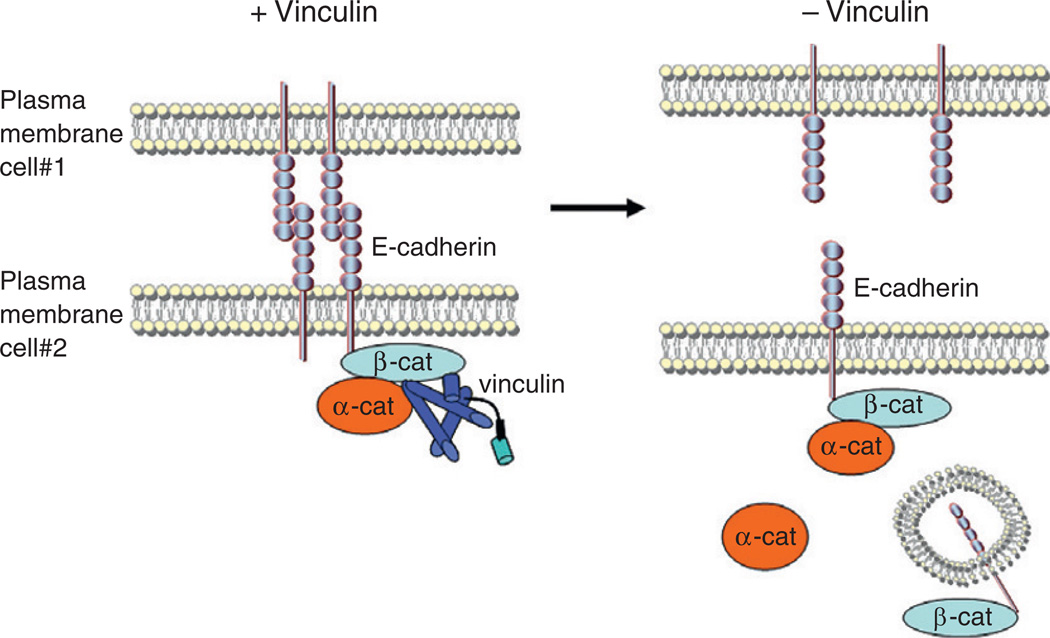
The second reported role for vinculin in cadherin-based adhesion is the regulation of E-cadherin surface expression. We found that in the absence of vinculin, the expression of E-cadherin at the cell surface was significantly reduced, whereas the total E-cadherin levels remained unchanged (Peng et al., 2010). This defect could be rescued by wild-type vinculin, but not by a mutant (A50I) form. Biochemical analysis showed that the A50I mutation blocked vinculin binding to β-catenin while maintaining the α-catenin– vinculin interaction, suggesting that the interaction between vinculin and β-catenin may account for the decreased expression of E-cadherin at the cell surface. Indeed, a β-catenin point mutant that specifically blocks vinculin binding recapitulated the loss of surface E-cadherin expression phenotype. Thus, vinculin regulates surface E-cadherin dynamics via its interaction with β-catenin (Fig. 5.5).
Figure 5.5. Vinculin is required for proper cell–cell adhesion.
In the presence of vinculin (left panel), E-cadherin is stably expressed at the cell surface and strong adhesions are formed between neighboring cells. Upon the loss of vinculin (right panel), cell–cell adhesions disassemble, and E-cadherin is lost from the cell surface and is internalized.
Third, vinculin has been reported to play a role in the mechanosensory response of E-cadherin (le Duc et al., 2010). In F9 cells, cadherin-mediated junctional stiffness increases in proportion to the applied stress, providing direct evidence that reinforcement of E-cadherin adhesion is dependent on force (le Duc et al., 2010). This response requires the contractile actin cytoskeleton, as treatment with latrunculin B, cytochalasin D, and blebbistatin largely ameliorated this effect. In the search for the protein(s) that transmit force between the cadherin complex and the actin cytoskeleton, vinculin was found to be recruited to cell–cell junctions in a myosin II-dependent manner (le Duc et al., 2010; Yonemura et al., 2010). More importantly, cells lacking vinculin had a ~50% reduction in stiffness and exhibited a dramatic decrease in the recruitment of phosphorylated myosin light chains (a downstream mediator of tension) to cell–cell junctions when stimulated with hepatocyte growth factor. Further, α-catenin binding to vinculin was recently reported to be dependent upon myosin II activity (Yonemura et al., 2010). This effect is similar to what has been observed when force is applied to talin (del Rio et al., 2009).
6.2. Vinculin recruitment to adhesion sites
6.2.1. Vinculin recruitment to focal adhesions
Talin has long been thought to be the protein that recruits vinculin to focal adhesions due to its early engagement with integrin (Horwitz et al., 1986). However, recent studies have shown that the above-mentioned vinculin mutant, A50I, which blocks talin binding is nevertheless able to localize to focal adhesions, leaving the question of how vinculin is recruited unanswered (Bakolitsa et al., 2004; Cohen et al., 2006; Humphries et al., 2007; Peng et al., 2010). Some evidence suggests that paxillin may be responsible for this recruitment, via its ability to bind both integrins and vinculin directly. Indeed, Pasapera et al. (2010) proposed a model for vinculin recruitment to focal adhesions. According to this model nascent adhesions are bound to talin and paxillin, an increase in myosin II-mediated FAK phosphorylation results in elevated paxillin phosphorylation, and phos-phorylated paxillin promotes vinculin recruitment to the adhesion site (Pasapera et al., 2010). The observation that paxillin and vinculin have different dynamics in mature focal adhesions (Hu et al., 2007; Humphries et al., 2007), together with the finding that vinculin localizes to large and stable adhesions at times when FAK expression is inhibited, suggests that vinculin recruitment is more complicated than what this model predicts (Pasapera et al., 2010). Consistent with this notion, vinculin fragments that lack an intact paxillin binding site still localizes to focal adhesions (Humphries et al., 2007).
6.2.2. Vinculin recruitment to adherens junctions
How vinculin is recruited to adherens junctions is controversial. Historically, α-catenin has been thought to recruit vinculin to intercellular junctions because it is lost from adherens junctions in cancer cells and hearts lacking α-catenin (Sheikh et al., 2006; Watabe-Uchida et al., 1998). However, in some cancer cell lines that lack α-catenin, vinculin can be coimmunoprecipitated with β-catenin–E-cadherin complexes (Hazan et al., 1997). We too have found that β-catenin is required for vinculin localization, based on two observations. The first is that a mutant version of vinculin (A50I) that retains the ability to bind α-catenin, but not β-catenin, is impaired in junctional localization. Second, shRNA-mediated inhibition of β-catenin expression results in a phenotype reminiscent of those obtained in vinculin knockdown cells; importantly, this phenotype could not be rescued by a mutant version of β-catenin that cannot bind vinculin (Peng et al., 2010). One possible explanation for these differences is that, in some contexts, the influence of α-catenin on vinculin is due to its effects on β-catenin. This notion is supported by the observation that the form of β-catenin that binds preferentially to cadherin also binds α-catenin, and that preassociation of recombinant α-catenin with β-catenin leads to enhanced binding of β-catenin to cadherin (Castano et al., 2002). α-Catenin expression is not disturbed in our vinculin knockdown cell lines, and the mutant version of β-catenin that fails to bind vinculin associates with α-catenin at wild-type levels (Peng et al., 2010). These observations indicate that if α-catenin is required for vinculin localization, it is secondary to β-catenin or is part of a molecular complex that contains β-catenin and is important for vinculin localization.
Myosin VI has also been implicated in vinculin recruitment to cell–cell junctions. Specifically, myosin VI is necessary for the incorporation of vinculin into stable cadherin-containing adhesions (Maddugoda et al., 2007). In the study that made this observation, it was noted that vinculin is recruited to cell–cell adhesions before myosin VI is detected at these sites (Maddugoda et al., 2007). This observation, in combination with our finding that vinculin recruitment requires β-catenin, suggests the intriguing possibility that the initial recruitment of vinculin to cell–cell adhesions is mediated by β-catenin, and that myosin VI is dominant at a later stage.
6.2.3. Different modes of recruitment
One question with respect to vinculin function that has remained unanswered for a number of years is how vinculin is differentially recruited to focal adhesions versus cell–cell adhesions. The vinculin mutant A50I which fails to localize to cell–cell adhesions is nevertheless able to localize to focal adhesions suggesting that ligand binding to this residue is dispensable for vinculin recruitment to cell–matrix adhesions (Bakolitsa et al., 2004; Cohen et al., 2006; Humphries et al., 2007; Peng et al., 2010). Hence, recruitment to different adhesions likely arises from differential binding to this amino acid residue. Future studies examining the mechanism of vinculin localization to focal adhesions and to adherens junctions will provide insight into how vinculin is directed to one site versus the other under distinct physiological conditions.
7. Interplay Between Cell–Cell and Cell–Matrix Adhesion: Vinculin in Development and Cardiomyopathy
Cell adhesions are pivotal in many morphogenic processes, including cell sorting, cell rearrangement, and cell movement (Gumbiner, 2005). Thus, it is not surprising that the disruption of cell adhesion components, such as vinculin, occurs in numerous diseased states. For example, vinculin expression is commonly lost in cancers (Kawahara et al., 1999; Lifschitz-Mercer et al., 1997; Meyer and Brinck, 1997; Sadano et al., 1992; Somiari et al., 2003), and mutations in vinculin are linked to a variety of cardio-myopathies (Maeda et al., 1997; Olson et al., 2002; Vasile et al., 2006a,b,c). Efforts to better understand the role of vinculin in these and other diseased states have included the development of animal models in which to study vinculin function. This work has confirmed and expanded our knowledge of vinculin’s role in cell–cell and cell–matrix adhesions and has underscored the importance of vinculin in vivo.
As described above, the loss of vinculin leads to the disruption of cell adhesion and cell migration, both of which are processes crucial to embryonic development. Therefore, it seemed likely that disrupting vinculin expression would lead to developmental abnormalities. To test this directly, homologous recombination was used to delete the vinculin gene in mouse (Xu et al., 1998a). Vinculin inactivation resulted in lethality at embryonic day E10, with abnormalities first observed as early as E8. The most prominent defect in the whole-mount animal was failure of the neural folds and head structures to fuse in the ventral cranial midline (Xu et al., 1998a). Another defect, and the most likely cause of lethality, was malformation of the heart (Xu et al., 1998a). At stage E10, a functional heart becomes very important because developing organs require the efficient vascular delivery of nutrients and the removal of waste. In the vinculin null embryo, the heart is only about half the size of that in normal littermates and is surrounded by a dilated pericardial cavity. The walls are thin, with too few cardiomyocytes in what should be the dense layer (Xu et al., 1998a). The endocardium is present, but the valves never form, and the heart never contracts (Xu et al., 1998a). The somites and limbs of the vinculin null embryos are also greatly reduced in size and retarded in development (Xu et al., 1998a). All these defects could potentially arise from improper adhesion and actin remodeling. These experiments suggest that these phenotypes may reflect a critical role for vinculin in mammalian embryogenesis, owing to its effects on cell– cell and cell–matrix adhesion.
The global vinculin knockout provides key insights into vinculin function and suggests that vinculin plays a crucial role in the regulation of heart function. In cardiac myocytes, vinculin is detected at the intercalated disks and costameres (Geiger et al., 1985; Koteliansky and Gneushev, 1983; Terracio et al., 1990; Volk and Geiger, 1984). The latter are heart-specific structures similar to cell–matrix adhesions and circumferentially align with the Z disk of the myofibrils (Samarel, 2005). They share many components of focal adhesions, including integrins, talin, vinculin, α-actinin, and focal adhesion kinase, among others. Like focal adhesions, costameres organize myofilaments, which consist primarily of actin, into a three-dimensional structure and link them to the extracellular matrix. Costameres are also sites that transduce mechanical information across the cell membrane. Intercalated disks, however, resemble intercellular junctions, such as the adherens junctions, desomosomes, and gap junctions (Noorman et al., 2009). These structures connect neighboring myocytes in a staggered fashion and are important for the mechanical and electrical coupling of cardiac myocytes. At both sites, vinculin is a major component of the mechanical transduction system, as its expression is upregulated in response to loading, and its localization in costameres is disrupted upon the unloading of mechanic force (Sharp et al., 1997).
As the vinculin homozygous knockout mouse fails to develop fully (Xu et al., 1998a), it has not been possible to use this model to study vinculin function in the adult heart. Fortunately, the vinculin hemizygous knockout mouse (vinculin+/− ) develops normally (Zemljic-Harpf et al., 2004), and this mouse model system has been used to study the effects of diminished vinculin expression on the structural and functional integrity of the heart (Zemljic-Harpf et al., 2004).In the basal state, the vinculin+/− hearts are histologically and functionally normal, with the exception of small differences in conduction. The fact that bigger differences were not observed is somewhat surprising, given that the intercalated disks of vinculin+/− hearts are disrupted and the myofibrils are not properly anchored in the intercalated disks and Z-lines (Zemljic-Harpf et al., 2004). Whether the dis-rupted ultrastructure of the vinculin+/− hearts reduces function was tested by measuring tolerance to pressure loading induced by transverse aortic constriction (a commonly used operation to experimentally induce cardiac hypertrophy and heart failure). Thirty-three percentage of the vinculin+/− mice died immediately following surgery, and many of the remaining mice later developed progressive left-ventricular dysfunction and subsequently died (Zemljic-Harpf et al., 2004).
The Ross lab investigated whether the defects in vimculin+/− hearts are caused by a loss of cardiac myocyte function, using mice harboring a tissue-specific deletion of the vinculin gene (Zemljic-Harpf et al., 2007). Although the vinculin knockout mice appeared healthy initially, 50% died suddenly before reaching 14 weeks of age, due to ventricular tachycardia and disruptions in electrical conductance (Zemljic-Harpf et al., 2007). An analysis of tissues harvested prior to the loss of ventricular function showed highly serrated intercalated disks that were detached from the myofibrils (Zemljic-Harpf et al., 2007). The disruptions in intercalated disks arose from a loss of cadherin from this site, a phenomenon that is recapitulated in epithelial cells in which vinculin expression silenced (Peng et al., 2010). In addition, localization of connexin 43, a gap junction protein that is important for the electrical coupling of myocytes, was disrupted and is likely the cause of spontaneous arrhythmias that developed in these mice. Taken together, these findings demonstrate that vinculin is essential for proper heart function, owing to its effects on cell–cell adhesion in the intercalated disk.
8. Conclusions
Much progress has been made in our understanding of vinculin since its identification over 30 years ago. Recent advances in vinculin biology— specifically, elucidation of the full-length crystal structure and of the mechanisms, whereby vinculin regulates focal adhesion assembly and adherens junction function—will be critical in deciphering the function of vinculin at the organismal level and in disease. Accomplishing this task will require additional information about how these events are regulated in a cell. Specifically, the signal transduction pathways that control vinculin activation/inactivation, as well as those that trigger its incorporation into and dissociation from adhesions, will have to be elucidated. These feats will rely heavily upon technological advances. Mass spectrometry analyses will undoubtedly allow the full repertoire of vinculin binding partners to be uncovered, and high resolution imaging in combination with FRET should reveal how these interactions occur in a spatial and a temporal manner. A better understanding of these events will bring the scientific community one step closer to fully understanding how vinculin coordinates cell–matrix and cell–cell adhesion.
ACKNOWLEDGMENTS
This work is supported by National Institutes of Health Grant # 1K01CA111818 and American Cancer Society Research Scholar Grant # 115274 to K. A. D. and American Heart Association Predoctoral Fellowship # 0910127G to X. P. The UI Interdisciplinary Training Grant in Molecular Biology (NIH Grant #5T32GM073610) supported E. N.
REFERENCES
- Adey NB, Kay BK. Isolation of peptides from phage-displayed random peptide libraries that interact with the talin-binding domain of vinculin. Biochem. J. 1997;324(Pt. 2):523–528. doi: 10.1042/bj3240523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alenghat FJ, Fabry B, Tsai KY, Goldmann WH, Ingber DE. Analysis of cell mechanics in single vinculin-deficient cells using a magnetic tweezer. Biochem. Biophys. Res. Commun. 2000;277:93–99. doi: 10.1006/bbrc.2000.3636. [DOI] [PubMed] [Google Scholar]
- Auweter SD, Allain FH. Structure-function relationships of the polypyrimidine tract binding protein. Cell. Mol. Life Sci. 2008;65:516–527. doi: 10.1007/s00018-007-7378-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayalon O, Geiger B. Cyclic changes in the organization of cell adhesions and the associated cytoskeleton, induced by stimulation of tyrosine phosphorylation in bovine aortic endothelial cells. J. Cell Sci. 1997;110(Pt. 5):547–556. doi: 10.1242/jcs.110.5.547. [DOI] [PubMed] [Google Scholar]
- Babic I, Sharma S, Black DL. A role for polypyrimidine tract binding protein in the establishment of focal adhesions. Mol. Cell. Biol. 2009;29:5564–5577. doi: 10.1128/MCB.00590-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakolitsa C, de Pereda JM, Bagshaw CR, Critchley DR, Liddington RC. Crystal structure of the vinculin tail suggests a pathway for activation. Cell. 1999;99:603–613. doi: 10.1016/s0092-8674(00)81549-4. [DOI] [PubMed] [Google Scholar]
- Bakolitsa C, Cohen DM, Bankston LA, Bobkov AA, Cadwell GW, Jennings L, et al. Structural basis for vinculin activation at sites of cell adhesion. Nature. 2004;430:583–586. doi: 10.1038/nature02610. [DOI] [PubMed] [Google Scholar]
- Bass MD, Patel B, Barsukov IG, Fillingham IJ, Mason R, Smith BJ, et al. Further characterization of the interaction between the cytoskeletal proteins talin and vinculin. Biochem. J. 2002;362:761–768. doi: 10.1042/0264-6021:3620761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrier AL, Yamada KM. Cell-matrix adhesion. J. Cell. Physiol. 2007;213:565–573. doi: 10.1002/jcp.21237. [DOI] [PubMed] [Google Scholar]
- Bloch RJ, Hall ZW. Cytoskeletal components of the vertebrate neuromuscular junction: vinculin, alpha-actinin, and filamin. J. Cell Biol. 1983;97:217–223. doi: 10.1083/jcb.97.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bois PR, Borgon RA, Vonrhein C, Izard T. Structural dynamics of alpha-actinin-vinculin interactions. Mol. Cell. Biol. 2005;25:6112–6122. doi: 10.1128/MCB.25.14.6112-6122.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Bois PR, O’Hara BP, Nietlispach D, Kirkpatrick J, Izard T. The vinculin binding sites of talin and alpha-actinin are sufficient to activate vinculin. J. Biol. Chem. 2006;281:7228–7236. doi: 10.1074/jbc.M510397200. [DOI] [PubMed] [Google Scholar]
- Borgon RA, Vonrhein C, Bricogne G, Bois PR, Izard T. Crystal structure of human vinculin. Structure. 2004;12:1189–1197. doi: 10.1016/j.str.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Bourdet-Sicard R, Rudiger M, Jockusch BM, Gounon P, Sansonetti PJ, Nhieu GT. Binding of the Shigella protein IpaA to vinculin induces F-actin depolymerization. EMBO J. 1999;18:5853–5862. doi: 10.1093/emboj/18.21.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindle NP, Holt MR, Davies JE, Price CJ, Critchley DR. The focal-adhesion vasodilator-stimulated phosphoprotein (VASP) binds to the proline-rich domain in vinculin. Biochem. J. 1996;318(Pt. 3):753–757. doi: 10.1042/bj3180753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge K, Feramisco JR. Microinjection and localization of a 130K protein in living fibroblasts: a relationship to actin and fibronectin. Cell. 1980;19:587–595. doi: 10.1016/s0092-8674(80)80035-3. [DOI] [PubMed] [Google Scholar]
- Burridge K, Mangeat P. An interaction between vinculin and talin. Nature. 1984;308:744–746. doi: 10.1038/308744a0. [DOI] [PubMed] [Google Scholar]
- Calderwood DA, Zent R, Grant R, Rees DJ, Hynes RO, Ginsberg MH. The talin head domain binds to integrin beta subunit cytoplasmic tails and regulates integrin activation. J. Biol. Chem. 1999;274:28071–28074. doi: 10.1074/jbc.274.40.28071. [DOI] [PubMed] [Google Scholar]
- Carisey A, Ballestrem C. Vinculin, an adapter protein in control of cell adhesion signalling. Eur. J. Cell Biol. 2010;90:157–163. doi: 10.1016/j.ejcb.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castano J, Raurell I, Piedra JA, Miravet S, Dunach M, Garcia de Herreros A. Beta-catenin N- and C-terminal tails modulate the coordinated binding of adherens junction proteins to beta-catenin. J. Biol. Chem. 2002;277:31541–31550. doi: 10.1074/jbc.M204376200. [DOI] [PubMed] [Google Scholar]
- Ceccarelli DF, Song HK, Poy F, Schaller MD, Eck MJ. Crystal structure of the FERM domain of focal adhesion kinase. J. Biol. Chem. 2006;281:252–259. doi: 10.1074/jbc.M509188200. [DOI] [PubMed] [Google Scholar]
- Chandrasekar I, Stradal TE, Holt MR, Entschladen F, Jockusch BM, Ziegler WH. Vinculin acts as a sensor in lipid regulation of adhesion-site turnover. J. Cell Sci. 2005;118:1461–1472. doi: 10.1242/jcs.01734. [DOI] [PubMed] [Google Scholar]
- Chen H, Cohen DM, Choudhury DM, Kioka N, Craig SW. Spatial distribution and functional significance of activated vinculin in living cell. J. Cell Biol. 2005;169:459–470. doi: 10.1083/jcb.200410100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Choudhury DM, Craig SW. Coincidence of actin filaments and talin is required to activate vinculin. J. Biol. Chem. 2006;281:40389–40398. doi: 10.1074/jbc.M607324200. [DOI] [PubMed] [Google Scholar]
- Cohen DM, Chen H, Johnson RP, Choudhury B, Craig SW. Two distinct head-tail interfaces cooperate to suppress activation of vinculin by talin. J. Biol. Chem. 2005;280:17109–17117. doi: 10.1074/jbc.M414704200. [DOI] [PubMed] [Google Scholar]
- Cohen DM, Kutscher B, Chen H, Murphy DB, Craig SW. A conformational switch in vinculin drives formation and dynamics of a talin-vinculin complex at focal adhesions. J. Biol. Chem. 2006;281:16006–16015. doi: 10.1074/jbc.M600738200. [DOI] [PubMed] [Google Scholar]
- Coll JL, Ben-Ze’ev A, Ezzell RM, Rodriguez Fernandez JL, Baribault H, Oshima RG, et al. Targeted disruption of vinculin genes in F9 and embryonic stem cells changes cell morphology, adhesion, and locomotion. Proc. Natl. Acad. Sci. USA. 1995;92:9161–9165. doi: 10.1073/pnas.92.20.9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutu MD, Craig SW. cDNA-derived sequence of chicken embryo vinculin. Proc. Natl. Acad. Sci. USA. 1988;85:8535–8539. doi: 10.1073/pnas.85.22.8535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMali KA, Burridge K. Coupling membrane protrusion and cell adhesion. J. Cell Sci. 2003;116:2389–2397. doi: 10.1242/jcs.00605. [DOI] [PubMed] [Google Scholar]
- DeMali KA, Barlow CA, Burridge K. Recruitment of the Arp2/3 complex to vinculin: coupling membrane protrusion to matrix adhesion. J. Cell Biol. 2002;159:881–891. doi: 10.1083/jcb.200206043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMali KA, Jue AL, Burridge K. IpaA targets beta1 integrins and rho to promote actin cytoskeleton rearrangements necessary for Shigella entry. J. Biol. Chem. 2006;281:39534–39541. doi: 10.1074/jbc.M605939200. [DOI] [PubMed] [Google Scholar]
- Diez G, Kollmannsberger P, Mierke CT, Koch TM, Vali H, Fabry B, et al. Anchorage of vinculin to lipid membranes influences cell mechanical properties. Biophys. J. 2009;97:3105–3112. doi: 10.1016/j.bpj.2009.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenckhahn D, Franz H. Identification of actin-, alpha-actinin-, and vinculin-containing plaques at the lateral membrane of epithelial cells. J. Cell Biol. 1986;102:1843–1852. doi: 10.1083/jcb.102.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenckhahn D, Mannherz HG. Distribution of actin and the actin-associated proteins myosin, tropomyosin, alpha-actinin, vinculin, and villin in rat and bovine exocrine glands. Eur. J. Cell Biol. 1983;30:167–176. [PubMed] [Google Scholar]
- Eimer W, Niermann M, Eppe MA, Jockusch BM. Molecular shape of vinculin in aqueous solution. J. Mol. Biol. 1993;229:146–152. doi: 10.1006/jmbi.1993.1014. [DOI] [PubMed] [Google Scholar]
- Fillingham I, Gingras AR, Papagrigoriou E, Patel B, Emsley J, Critchley DR, et al. A vinculin binding domain from the talin rod unfolds to form a complex with the vinculin head. Structure. 2005;13:65–74. doi: 10.1016/j.str.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Finlay BB, Ruschkowski S, Dedhar S. Cytoskeletal rearrangements accompanying salmonella entry into epithelial cells. J. Cell Sci. 1991;99(Pt. 2):283–296. doi: 10.1242/jcs.99.2.283. [DOI] [PubMed] [Google Scholar]
- Finlay BB, Rosenshine I, Donnenberg MS, Kaper JB. Cytoskeletal composition of attaching and effacing lesions associated with enteropathogenic Escherichia coli adherence to HeLa cells. Infect. Immun. 1992;60:2541–2543. doi: 10.1128/iai.60.6.2541-2543.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith CG, Yamada KM, Sheetz MP. The relationship between force and focal complex development. J. Cell Biol. 2002;159:695–705. doi: 10.1083/jcb.200204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant ND, Michael KE, Garcia AJ. Cell adhesion strengthening: contributions of adhesive area, integrin binding, and focal adhesion assembly. Mol. Biol. Cell. 2005;16:4329–4340. doi: 10.1091/mbc.E05-02-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Alvarez B, de Pereda JM, Calderwood DA, Ulmer TS, Critchley D, Campbell ID, et al. Structural determinants of integrin recognition by talin. Mol. Cell. 2003;11:49–58. doi: 10.1016/s1097-2765(02)00823-7. [DOI] [PubMed] [Google Scholar]
- Geiger B. A 130 K protein from chicken gizzard: its localization at the termini of microfilament bundles in cultured chicken cells. Cell. 1979;18:193–205. doi: 10.1016/0092-8674(79)90368-4. [DOI] [PubMed] [Google Scholar]
- Geiger B, Tokuyasu KT, Dutton AH, Singer SJ. Vinculin, an intracellular protein localized at specialized sites where microfilament bundles terminate at cell membranes. Proc. Natl. Acad. Sci. USA. 1980;77:4127–4131. doi: 10.1073/pnas.77.7.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger B, Dutton AH, Tokuyasu KT, Singer SJ. Immunoelectron microscope studies of membrane-microfilament interactions: distributions of alpha-actinin, tropomyosin, and vinculin in intestinal epithelial brush border and chicken gizzard smooth muscle cells. J. Cell Biol. 1981;91:614–628. doi: 10.1083/jcb.91.3.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger B, Volk T, Volberg T. Molecular heterogeneity of adherens junctions. J. Cell Biol. 1985;101:1523–1531. doi: 10.1083/jcb.101.4.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore AP, Burridge K. Regulation of vinculin binding to talin and actin by phosphatidyl-inositol-4-5-bisphosphate. Nature. 1996;381:531–535. doi: 10.1038/381531a0. [DOI] [PubMed] [Google Scholar]
- Gingras AR, Ziegler WH, Frank R, Barsukov IL, Roberts GC, Critchley DR, et al. Mapping and consensus sequence identification for multiple vinculin binding sites within the talin rod. J. Biol. Chem. 2005;280:37217–37224. doi: 10.1074/jbc.M508060200. [DOI] [PubMed] [Google Scholar]
- Gingras AR, Vogel KP, Steinhoff HJ, Ziegler WH, Patel B, Emsley J, et al. Structural and dynamic characterization of a vinculin binding site in the talin rod. Biochemistry. 2006;45:1805–1817. doi: 10.1021/bi052136l. [DOI] [PubMed] [Google Scholar]
- Goldmann WH, Guttenberg Z, Tang JX, Kroy K, Isenberg G, Ezzell RM. Analysis of the F-actin binding fragments of vinculin using stopped-flow and dynamic light-scattering measurements. Eur. J. Biochem. 1998;254:413–419. doi: 10.1046/j.1432-1327.1998.2540413.x. [DOI] [PubMed] [Google Scholar]
- Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang MT, et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groesch ME, Otto JJ. Purification and characterization of an 85 kDa talin-binding fragment of vinculin. Cell Motil. Cytoskeleton. 1990;15:41–50. doi: 10.1002/cm.970150107. [DOI] [PubMed] [Google Scholar]
- Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat. Rev. Mol. Cell Biol. 2005;6:622–634. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- Halegoua S. Changes in the phosphorylation and distribution of vinculin during nerve growth factor induced neurite outgrowth. Dev. Biol. 1987;121:97–104. doi: 10.1016/0012-1606(87)90142-4. [DOI] [PubMed] [Google Scholar]
- Hamiaux C, van Eerde A, Parsot C, Broos J, Dijkstra BW. Structural mimicry for vinculin activation by IpaA, a virulence factor of Shigella flexneri. EMBO Rep. 2006;7:794–799. doi: 10.1038/sj.embor.7400753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazan RB, Kang L, Roe S, Borgen PI, Rimm DL. Vinculin is associated with the E-cadherin adhesion complex. J. Biol. Chem. 1997;272:32448–32453. doi: 10.1074/jbc.272.51.32448. [DOI] [PubMed] [Google Scholar]
- Hemmings L, Rees DJ, Ohanian V, Bolton SJ, Gilmore AP, Patel B, et al. Talin contains three actin-binding sites each of which is adjacent to a vinculin-binding site. J. Cell Sci. 1996;109(Pt. 11):2715–2726. doi: 10.1242/jcs.109.11.2715. [DOI] [PubMed] [Google Scholar]
- Horwitz A, Duggan K, Buck C, Beckerle MC, Burridge K. Interaction of plasma membrane fibronectin receptor with talin—a transmembrane linkage. Nature. 1986;320:531–533. doi: 10.1038/320531a0. [DOI] [PubMed] [Google Scholar]
- Hu K, Ji L, Applegate KT, Danuser G, Waterman-Storer CM. Differential transmission of actin motion within focal adhesions. Science. 2007;315:111–115. doi: 10.1126/science.1135085. [DOI] [PubMed] [Google Scholar]
- Humphries JD, Wang P, Streuli C, Geiger B, Humphries MJ, Ballestrem C. Vinculin controls focal adhesion formation by direct interactions with talin and actin. J. Cell Biol. 2007;179:1043–1057. doi: 10.1083/jcb.200703036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttelmaier S, Bubeck P, Rudiger M, Jockusch BM. Characterization of two F-actin-binding and oligomerization sites in the cell-contact protein vinculin. Eur. J. Biochem. 1997;247:1136–1142. doi: 10.1111/j.1432-1033.1997.01136.x. [DOI] [PubMed] [Google Scholar]
- Huttelmaier S, Mayboroda O, Harbeck B, Jarchau T, Jockusch BM, Rudiger M. The interaction of the cell-contact proteins VASP and vinculin is regulated by phosphatidylinositol-4, 5-bisphosphate. Curr. Biol. 1998;8:479–488. doi: 10.1016/s0960-9822(98)70199-x. [DOI] [PubMed] [Google Scholar]
- Huttelmaier S, Illenberger S, Grosheva I, Rudiger M, Singer RH, Jockusch BM. Raver1, a dual compartment protein, is a ligand for PTB/hnRNPI and microfilament attachment proteins. J. Cell Biol. 2001;155:775–786. doi: 10.1083/jcb.200105044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huveneers S, Danen EH. Adhesion signaling—crosstalk between integrins, Src and Rho. J. Cell Sci. 2009;122:1059–1069. doi: 10.1242/jcs.039446. [DOI] [PubMed] [Google Scholar]
- Hytonen VP, Vogel V. How force might activate talin’s vinculin binding sites: SMD reveals a structural mechanism. PLoS Comput. Biol. 2008;4:e24. doi: 10.1371/journal.pcbi.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura Y, Itoh M, Maeno Y, Tsukita S, Nagafuchi A. Functional domains of alpha-catenin required for the strong state of cadherin-based cell adhesion. J. Cell Biol. 1999;144:1311–1322. doi: 10.1083/jcb.144.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenberg G, Goldmann WH. Peptide-specific antibodies localize the major lipid binding sites of talin dimers to oppositely arranged N-terminal 47 kDa subdomains. FEBS Lett. 1998;426:165–170. doi: 10.1016/s0014-5793(98)00336-6. [DOI] [PubMed] [Google Scholar]
- Isenberg G, Leonard K, Jockusch BM. Structural aspects of vinculin-actin interactions. J. Mol. Biol. 1982;158:231–249. doi: 10.1016/0022-2836(82)90431-4. [DOI] [PubMed] [Google Scholar]
- Izard T, Vonrhein C. Structural basis for amplifying vinculin activation by talin. J. Biol. Chem. 2004;279:27667–27678. doi: 10.1074/jbc.M403076200. [DOI] [PubMed] [Google Scholar]
- Izard T, Evans G, Borgon RA, Rush CL, Bricogne G, Bois PR. Vinculin activation by talin through helical bundle conversion. Nature. 2004;427:171–175. doi: 10.1038/nature02281. [DOI] [PubMed] [Google Scholar]
- Izard T, Tran Van Nhieu G, Bois PR. Shigella applies molecular mimicry to subvert vinculin and invade host cells. J. Cell Biol. 2006;175:465–475. doi: 10.1083/jcb.200605091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen ME, Kim E, Liu H, Fujimoto LM, Bobkov A, Volkmann N, et al. Three-dimensional structure of vinculin bound to actin filaments. Mol. Cell. 2006;21:271–281. doi: 10.1016/j.molcel.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Ji L, Lim J, Danuser G. Fluctuations of intracellular forces during cell protrusion. Nat. Cell Biol. 2008;10:1393–1400. doi: 10.1038/ncb1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jockusch BM, Isenberg G. Interaction of alpha-actinin and vinculin with actin: opposite effects on filament network formation. Proc. Natl. Acad. Sci. USA. 1981;78:3005–3009. doi: 10.1073/pnas.78.5.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RP, Craig SW. An intramolecular association between the head and tail domains of vinculin modulates talin binding. J. Biol. Chem. 1994;269:12611–12619. [PubMed] [Google Scholar]
- Johnson RP, Craig SW. The carboxy-terminal tail domain of vinculin contains a cryptic binding site for acidic phospholipids. Biochem. Biophys. Res. Commun. 1995a;210:159–164. doi: 10.1006/bbrc.1995.1641. [DOI] [PubMed] [Google Scholar]
- Johnson RP, Craig SW. F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature. 1995b;373:261–264. doi: 10.1038/373261a0. [DOI] [PubMed] [Google Scholar]
- Johnson RP, Craig SW. Actin activates a cryptic dimerization potential of the vinculin tail domain. J. Biol. Chem. 2000;275:95–105. doi: 10.1074/jbc.275.1.95. [DOI] [PubMed] [Google Scholar]
- Kawahara E, Tokuda R, Nakanishi I. Migratory phenotypes of HSC-3 squamous carcinoma cell line induced by EGF and PMA: relevance to migration of loosening of adhesion and vinculin-associated focal contacts with prominent filopodia. Cell Biol. Int. 1999;23:163–174. doi: 10.1006/cbir.1998.0331. [DOI] [PubMed] [Google Scholar]
- Kioka N, Sakata S, Kawauchi T, Amachi T, Akiyama SK, Okazaki K, et al. Vinexin: a novel vinculin-binding protein with multiple SH3 domains enhances actin cytoskeletal organization. J. Cell Biol. 1999;144:59–69. doi: 10.1083/jcb.144.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koteliansky VE, Gneushev GN. Vinculin localization in cardiac muscle. FEBS Lett. 1983;159:158–160. doi: 10.1016/0014-5793(83)80437-2. [DOI] [PubMed] [Google Scholar]
- Kroemker M, Rudiger AH, Jockusch BM, Rudiger M. Intramolecular interactions in vinculin control alpha-actinin binding to the vinculin head. FEBS Lett. 1994;355:259–262. doi: 10.1016/0014-5793(94)01216-4. [DOI] [PubMed] [Google Scholar]
- Kupper K, Lang N, Mohl C, Kirchgessner N, Born S, Goldmann WH, et al. Tyrosine phosphorylation of vinculin at position 1065 modifies focal adhesion dynamics and cell tractions. Biochem. Biophys. Res. Commun. 2010;399:560–564. doi: 10.1016/j.bbrc.2010.07.110. [DOI] [PubMed] [Google Scholar]
- le Duc Q, Shi Q, Blonk I, Sonnenberg A, Wang N, Leckband D, et al. Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II-dependent manner. J. Cell Biol. 2010;189:1107–1115. doi: 10.1083/jcb.201001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Rangarajan ES, Yogesha SD, Izard T. Raver1 interactions with vinculin and RNA suggest a feed-forward pathway in directing mRNA to focal adhesions. Structure. 2009;17:833–842. doi: 10.1016/j.str.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschitz-Mercer B, Czernobilsky B, Feldberg E, Geiger B. Expression of the adherens junction protein vinculin in human basal and squamous cell tumors: relationship to invasiveness and metastatic potential. Hum. Pathol. 1997;28:1230–1236. doi: 10.1016/s0046-8177(97)90195-7. [DOI] [PubMed] [Google Scholar]
- Lombardi ML, Knecht DA, Dembo M, Lee J. Traction force microscopy in Dictyostelium reveals distinct roles for myosin II motor and actin-crosslinking activity in polarized cell movement. J. Cell Sci. 2007;120:1624–1634. doi: 10.1242/jcs.002527. [DOI] [PubMed] [Google Scholar]
- Lotz MM, Burdsal CA, Erickson HP, McClay DR. Cell adhesion to fibronectin and tenascin: quantitative measurements of initial binding and subsequent strengthening response. J. Cell Biol. 1989;109:1795–1805. doi: 10.1083/jcb.109.4.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver SK. Myosin II function in non-muscle cells. Bioessays. 1996;18:179–182. doi: 10.1002/bies.950180304. [DOI] [PubMed] [Google Scholar]
- Maddugoda MP, Crampton MS, Shewan AM, Yap AS. Myosin VI and vinculin cooperate during the morphogenesis of cadherin cell cell contacts in mammalian epithelial cells. J. Cell Biol. 2007;178:529–540. doi: 10.1083/jcb.200612042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda M, Holder E, Lowes B, Valent S, Bies RD. Dilated cardiomyopathy associated with deficiency of the cytoskeletal protein metavinculin. Circulation. 1997;95:17–20. doi: 10.1161/01.cir.95.1.17. [DOI] [PubMed] [Google Scholar]
- Mandai K, Nakanishi H, Satoh A, Takahashi K, Satoh K, Nishioka H, et al. Ponsin/SH3P12: an l-afadin- and vinculin-binding protein localized at cell-cell and cell-matrix adherens junctions. J. Cell Biol. 1999;144:1001–1017. doi: 10.1083/jcb.144.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massa R, Silvestri G, Sancesario G, Bernardi G. Immunocytochemical localization of vinculin in muscle and nerve. Muscle Nerve. 1995;18:1277–1284. doi: 10.1002/mus.880181110. [DOI] [PubMed] [Google Scholar]
- McCann RO, Craig SW. The I/LWEQ module: a conserved sequence that signifies F-actin binding in functionally diverse proteins from yeast to mammals. Proc. Natl. Acad. Sci. USA. 1997;94:5679–5684. doi: 10.1073/pnas.94.11.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard R, Sansonetti PJ, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menkel AR, Kroemker M, Bubeck P, Ronsiek M, Nikolai G, Jockusch BM. Characterization of an F-actin-binding domain in the cytoskeletal protein vinculin. J. Cell Biol. 1994;126:1231–1240. doi: 10.1083/jcb.126.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer T, Brinck U. Immunohistochemical detection of vinculin in human rhab-domyosarcomas. Gen. Diagn. Pathol. 1997;142:191–198. [PubMed] [Google Scholar]
- Mierke CT, Kollmannsberger P, Zitterbart DP, Smith J, Fabry B, Goldmann WH. Mechano-coupling and regulation of contractility by the vinculin tail domain. Biophys. J. 2008;94:661–670. doi: 10.1529/biophysj.107.108472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierke CT, Kollmannsberger P, Zitterbart DP, Diez G, Koch TM, Marg S, et al. Vinculin facilitates cell invasion into three-dimensional collagen matrices. J. Biol. Chem. 2010;285:13121–13130. doi: 10.1074/jbc.M109.087171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milam LM. Electron microscopy of rotary shadowed vinculin and vinculin complexes. J. Mol. Biol. 1985;184:543–545. doi: 10.1016/0022-2836(85)90301-8. [DOI] [PubMed] [Google Scholar]
- Miller GJ, Dunn SD, Ball EH. Interaction of the N- and C-terminal domains of vinculin. Characterization and mapping studies. J. Biol. Chem. 2001;276:11729–11734. doi: 10.1074/jbc.M008646200. [DOI] [PubMed] [Google Scholar]
- Moese S, Selbach M, Brinkmann V, Karlas A, Haimovich B, Backert S, et al. The Helicobacter pylori CagA protein disrupts matrix adhesion of gastric epithelial cells by dephosphorylation of vinculin. Cell. Microbiol. 2007;9:1148–1161. doi: 10.1111/j.1462-5822.2006.00856.x. [DOI] [PubMed] [Google Scholar]
- Mohl C, Kirchgessner N, Schafer C, Kupper K, Born S, Diez G, et al. Becoming stable and strong: the interplay between vinculin exchange dynamics and adhesion strength during adhesion site maturation. Cell Motil. Cytoskeleton. 2009;66:350–364. doi: 10.1002/cm.20375. [DOI] [PubMed] [Google Scholar]
- Molony L, Burridge K. Molecular shape and self-association of vinculin and metavinculin. J. Cell. Biochem. 1985;29:31–36. doi: 10.1002/jcb.240290104. [DOI] [PubMed] [Google Scholar]
- Niessen CM, Gottardi CJ. Molecular components of the adherens junction. Biochim. Biophys. Acta. 2008;1778:562–571. doi: 10.1016/j.bbamem.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Takeichi M. Remodeling of the adherens junctions during morphogenesis. Curr. Top. Dev. Biol. 2009;89:33–54. doi: 10.1016/S0070-2153(09)89002-9. [DOI] [PubMed] [Google Scholar]
- Nolz JC, Medeiros RB, Mitchell JS, Zhu P, Freedman BD, Shimizu Y, et al. WAVE2 regulates high-affinity integrin binding by recruiting vinculin and talin to the immunological synapse. Mol. Cell. Biol. 2007;27:5986–6000. doi: 10.1128/MCB.00136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noorman M, van der Heyden MA, van Veen TA, Cox MG, Hauer RN, de Bakker JM, et al. Cardiac cell-cell junctions in health and disease: electrical versus mechanical coupling. J. Mol. Cell. Cardiol. 2009;47:23–31. doi: 10.1016/j.yjmcc.2009.03.016. [DOI] [PubMed] [Google Scholar]
- Obrink B. Epithelial cell adhesion molecules. Exp. Cell Res. 1986;163:1–21. doi: 10.1016/0014-4827(86)90554-9. [DOI] [PubMed] [Google Scholar]
- Ogita H, Takai Y. Cross-talk among integrin, cadherin, and growth factor receptor: roles of nectin and nectin-like molecule. Int. Rev. Cytol. 2008;265:1–54. doi: 10.1016/S0074-7696(07)65001-3. [DOI] [PubMed] [Google Scholar]
- Ohmori T, Kashiwakura Y, Ishiwata A, Madoiwa S, Mimuro J, Honda S, et al. Vinculin activates inside-out signaling of integrin alphaIIbbeta3 in Chinese hamster ovary cells. Biochem. Biophys. Res. Commun. 2010;400:323–328. doi: 10.1016/j.bbrc.2010.08.056. [DOI] [PubMed] [Google Scholar]
- Olson TM, Illenberger S, Kishimoto NY, Huttelmaier S, Keating MT, Jockusch BM. Metavinculin mutations alter actin interaction in dilated cardiomyopathy. Circulation. 2002;105:431–437. doi: 10.1161/hc0402.102930. [DOI] [PubMed] [Google Scholar]
- Opas M, Turksen K, Kalnins VI. Adhesiveness and distribution of vinculin and spectrin in retinal pigmented epithelial cells during growth and differentiation in vitro. Dev. Biol. 1985;107:269–280. doi: 10.1016/0012-1606(85)90310-0. [DOI] [PubMed] [Google Scholar]
- Otto JJ. The lack of interaction between vinculin actin. Cell Motil. Cytoskeleton. 1986;6:48–55. doi: 10.1002/cm.970060107. [DOI] [PubMed] [Google Scholar]
- Pantaloni D, Le Clainche C, Carlier MF. Mechanism of actin-based motility. Science. 2001;292:1502–1506. doi: 10.1126/science.1059975. [DOI] [PubMed] [Google Scholar]
- Papagrigoriou E, Gingras AR, Barsukov IL, Bate N, Fillingham IJ, Patel B, et al. Activation of a vinculin-binding site in the talin rod involves rearrangement of a five-helix bundle. EMBO J. 2004;23:2942–2951. doi: 10.1038/sj.emboj.7600285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo JV, Siliciano JD, Craig SW. A vinculin-containing cortical lattice in skeletal muscle: transverse lattice elements (“costameres”) mark sites of attachment between myofibrils and sarcolemma. Proc. Natl. Acad. Sci. USA. 1983a;80:1008–1012. doi: 10.1073/pnas.80.4.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo JV, Siliciano JD, Craig SW. Vinculin is a component of an extensive network of myofibril-sarcolemma attachment regions in cardiac muscle fibers. J. Cell Biol. 1983b;97:1081–1088. doi: 10.1083/jcb.97.4.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasapera AM, Schneider IC, Rericha E, Schlaepfer DD, Waterman CM. Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J. Cell Biol. 2010;188:877–890. doi: 10.1083/jcb.200906012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel B, Gingras AR, Bobkov AA, Fujimoto LM, Zhang M, Liddington RC, et al. The activity of the vinculin binding sites in talin is influenced by the stability of the helical bundles that make up the talin rod. J. Biol. Chem. 2006;281:7458–7467. doi: 10.1074/jbc.M508058200. [DOI] [PubMed] [Google Scholar]
- Peng X, Cuff LE, Lawton CD, DeMali KA. Vinculin regulates cell-surface E-cadherin expression by binding to beta-catenin. J. Cell Sci. 2010;123:567–577. doi: 10.1242/jcs.056432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Moreno M, Fuchs E. Catenins: keeping cells from getting their signals crossed. Dev. Cell. 2006;11:601–612. doi: 10.1016/j.devcel.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokutta S, Drees F, Takai Y, Nelson WJ, Weis WI. Biochemical and structural definition of the l-afadin- and actin-binding sites of alpha-catenin. J. Biol. Chem. 2002;277:18868–18874. doi: 10.1074/jbc.M201463200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Prutzman KC, Gao G, King ML, Iyer VV, Mueller GA, Schaller MD, et al. The focal adhesion targeting domain of focal adhesion kinase contains a hinge region that modulates tyrosine 926 phosphorylation. Structure. 2004;12:881–891. doi: 10.1016/j.str.2004.02.028. [DOI] [PubMed] [Google Scholar]
- Ramarao N, Le Clainche C, Izard T, Bourdet-Sicard R, Ageron E, Sansonetti PJ, et al. Capping of actin filaments by vinculin activated by the Shigella IpaA carboxyl-terminal domain. FEBS Lett. 2007;581:853–857. doi: 10.1016/j.febslet.2007.01.057. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, et al. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Rodriguez Fernandez JL, Geiger B, Salomon D, Sabanay I, Zoller M, Ben-Ze’ev A. Suppression of tumorigenicity in transformed cells after transfection with vinculin cDNA. J. Cell Biol. 1992;119:427–438. doi: 10.1083/jcb.119.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld GC, Hou DC, Dingus J, Meza I, Bryan J. Isolation and partial characterization of human platelet vinculin. J. Cell Biol. 1985;100:669–676. doi: 10.1083/jcb.100.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadano H, Inoue M, Taniguchi S. Differential expression of vinculin between weakly and highly metastatic B16-melanoma cell lines. Jpn. J. Cancer Res. 1992;83:625–630. doi: 10.1111/j.1349-7006.1992.tb00135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarel AM. Costameres, focal adhesions, and cardiomyocyte mechanotransduction. Am. J. Physiol. Heart Circ. Physiol. 2005;289:H2291–H2301. doi: 10.1152/ajpheart.00749.2005. [DOI] [PubMed] [Google Scholar]
- Saunders RM, Holt MR, Jennings L, Sutton DH, Barsukov IL, Bobkov A, et al. Role of vinculin in regulating focal adhesion turnover. Eur. J. Cell Biol. 2006;85:487–500. doi: 10.1016/j.ejcb.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Sawicka K, Bushell M, Spriggs KA, Willis AE. Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein. Biochem. Soc. Trans. 2008;36:641–647. doi: 10.1042/BST0360641. [DOI] [PubMed] [Google Scholar]
- Schindeler A, Lavulo L, Harvey RP. Muscle costameric protein, Chisel/Smpx, associates with focal adhesion complexes and modulates cell spreading in vitro via a Rac1/p38 pathway. Exp. Cell Res. 2005;307:367–380. doi: 10.1016/j.yexcr.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Schwartz MA, DeSimone DW. Cell adhesion receptors in mechanotransduction. Curr. Opin. Cell Biol. 2008;20:551–556. doi: 10.1016/j.ceb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwienbacher C, Jockusch BM, Rudiger M. Intramolecular interactions regulate serine/threonine phosphorylation of vinculin. FEBS Lett. 1996;384:71–74. doi: 10.1016/0014-5793(96)00286-4. [DOI] [PubMed] [Google Scholar]
- Sefton BM, Hunter T, Ball EH, Singer SJ. Vinculin: a cytoskeletal target of the transforming protein of Rous sarcoma virus. Cell. 1981;24:165–174. doi: 10.1016/0092-8674(81)90512-2. [DOI] [PubMed] [Google Scholar]
- Sharp WW, Simpson DG, Borg TK, Samarel AM, Terracio L. Mechanical forces regulate focal adhesion and costamere assembly in cardiac myocytes. Am. J. Physiol. 1997;273:H546–H556. doi: 10.1152/ajpheart.1997.273.2.H546. [DOI] [PubMed] [Google Scholar]
- Sheikh F, Chen Y, Liang X, Hirschy A, Stenbit AE, Gu Y, et al. Alpha-E-catenin inactivation disrupts the cardiomyocyte adherens junction, resulting in cardiomyopathy and susceptibility to wall rupture. Circulation. 2006;114:1046–1055. doi: 10.1161/CIRCULATIONAHA.106.634469. [DOI] [PubMed] [Google Scholar]
- Somiari RI, Sullivan A, Russell S, Somiari S, Hu H, Jordan R, et al. High-throughput proteomic analysis of human infiltrating ductal carcinoma of the breast. Proteomics. 2003;3:1863–1873. doi: 10.1002/pmic.200300560. [DOI] [PubMed] [Google Scholar]
- Steimle PA, Hoffert JD, Adey NB, Craig SW. Polyphosphoinositides inhibit the interaction of vinculin with actin filaments. J. Biol. Chem. 1999;274:18414–18420. doi: 10.1074/jbc.274.26.18414. [DOI] [PubMed] [Google Scholar]
- Stricker J, Falzone T, Gardel ML. Mechanics of the F-actin cytoskeleton. J. Biomech. 2010;43:9–14. doi: 10.1016/j.jbiomech.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subauste MC, Pertz O, Adamson ED, Turner CE, Junger S, Hahn KM. Vinculin modulation of paxillin-FAK interactions regulates ERK to control survival and motility. J. Cell Biol. 2004;165:371–381. doi: 10.1083/jcb.200308011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Mitsushima M, Okada N, Ito T, Aizawa S, Akahane R, et al. Role of interaction with vinculin in recruitment of vinexins to focal adhesions. Biochem. Biophys. Res. Commun. 2005;336:239–246. doi: 10.1016/j.bbrc.2005.08.064. [DOI] [PubMed] [Google Scholar]
- Terracio L, Simpson DG, Hilenski L, Carver W, Decker RS, Vinson N, et al. Distribution of vinculin in the Z-disk of striated muscle: analysis by laser scanning confocal microscopy. J. Cell. Physiol. 1990;145:78–87. doi: 10.1002/jcp.1041450112. [DOI] [PubMed] [Google Scholar]
- Tidball JG, Spencer MJ. PDGF stimulation induces phosphorylation of talin and cytoskeletal reorganization in skeletal muscle. J. Cell Biol. 1993;123:627–635. doi: 10.1083/jcb.123.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran Van Nhieu G, Ben-Ze’ev A, Sansonetti PJ. Modulation of bacterial entry into epithelial cells by association between vinculin and the Shigella IpaA invasin. EMBO J. 1997;16:2717–2729. doi: 10.1093/emboj/16.10.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremuth L, Kreis S, Melchior C, Hoebeke J, Ronde P, Plancon S, et al. A fluorescence cell biology approach to map the second integrin-binding site of talin to a 130-amino acid sequence within the rod domain. J. Biol. Chem. 2004;279:22258–22266. doi: 10.1074/jbc.M400947200. [DOI] [PubMed] [Google Scholar]
- Turner CE, Glenney JR, Jr, Burridge K. Paxillin: a new vinculin-binding protein present in focal adhesions. J. Cell Biol. 1990;111:1059–1068. doi: 10.1083/jcb.111.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasile VC, Edwards WD, Ommen SR, Ackerman MJ. Obstructive hyper-trophic cardiomyopathy is associated with reduced expression of vinculin in the intercalated disc. Biochem. Biophys. Res. Commun. 2006a;349:709–715. doi: 10.1016/j.bbrc.2006.08.106. [DOI] [PubMed] [Google Scholar]
- Vasile VC, Ommen SR, Edwards WD, Ackerman MJ. A missense mutation in a ubiquitously expressed protein, vinculin, confers susceptibility to hypertrophic cardiomyopathy. Biochem. Biophys. Res. Commun. 2006b;345:998–1003. doi: 10.1016/j.bbrc.2006.04.151. [DOI] [PubMed] [Google Scholar]
- Vasile VC, Will ML, Ommen SR, Edwards WD, Olson TM, Ackerman MJ. Identification of a metavinculin missense mutation, R975W, associated with both hypertrophic and dilated cardiomyopathy. Mol. Genet. Metab. 2006c;87:169–174. doi: 10.1016/j.ymgme.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Verkhovsky AB, Svitkina TM, Borisy GG. Network contraction model for cell translocation and retrograde flow. Biochem. Soc. Symp. 1999;65:207–222. [PubMed] [Google Scholar]
- Volberg T, Geiger B, Kam Z, Pankov R, Simcha I, Sabanay H, et al. Focal adhesion formation by F9 embryonal carcinoma cells after vinculin gene disruption. J. Cell Sci. 1995;108(Pt. 6):2253–2260. doi: 10.1242/jcs.108.6.2253. [DOI] [PubMed] [Google Scholar]
- Volk T, Geiger B. A 135-kd membrane protein of intercellular adherens junctions. EMBO J. 1984;3:2249–2260. doi: 10.1002/j.1460-2075.1984.tb02123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vostal JG, Shulman NR. Vinculin is a major platelet protein that undergoes calcium ion-dependent tyrosine phosphorylation. Biochem. J. 1993;294:675–680. doi: 10.1042/bj2940675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe-Uchida M, Uchida N, Imamura Y, Nagafuchi A, Fujimoto K, Uemura T, et al. alpha-Catenin-vinculin interaction functions to organize the apical junctional complex in epithelial cells. J. Cell Biol. 1998;142:847–857. doi: 10.1083/jcb.142.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weekes J, Barry ST, Critchley DR. Acidic phospholipids inhibit the intramolecular association between the N- and C-terminal regions of vinculin, exposing actin-binding and protein kinase C phosphorylation sites. Biochem. J. 1996;314(Pt. 3):827–832. doi: 10.1042/bj3140827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss EE, Kroemker M, Rudiger AH, Jockusch BM, Rudiger M. Vinculin is part of the cadherin-catenin junctional complex: complex formation between alpha-catenin and vinculin. J. Cell Biol. 1998;141:755–764. doi: 10.1083/jcb.141.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen KK, Rubenstein PA, DeMali KA. Vinculin nucleates actin polymerization and modifies actin filament structure. J. Biol. Chem. 2009;284:30463–30473. doi: 10.1074/jbc.M109.021295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins JA, Lin S. High-affinity interaction of vinculin with actin filaments in vitro. Cell. 1982;28:83–90. doi: 10.1016/0092-8674(82)90377-4. [DOI] [PubMed] [Google Scholar]
- Wilkins JA, Lin S. A re-examination of the interaction of vinculin with actin. J. Cell Biol. 1986;102:1085–1092. doi: 10.1083/jcb.102.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler J, Jockusch BM. 3-Dimensional organization of the N-terminal vinculin head fragment. Eur. J. Cell Biol. 2001;80:201–206. doi: 10.1078/0171-9335-00152. [DOI] [PubMed] [Google Scholar]
- Winkler J, Lunsdorf H, Jockusch BM. The ultrastructure of chicken gizzard vinculin as visualized by high-resolution electron microscopy. J. Struct. Biol. 1996;116:270–277. doi: 10.1006/jsbi.1996.0042. [DOI] [PubMed] [Google Scholar]
- Winkler J, Lunsdorf H, Jockusch BM. Energy-filtered electron microscopy reveals that talin is a highly flexible protein composed of a series of globular domains. Eur. J. Biochem. 1997;243:430–436. doi: 10.1111/j.1432-1033.1997.0430a.x. [DOI] [PubMed] [Google Scholar]
- Wood CK, Turner CE, Jackson P, Critchley DR. Characterisation of the paxillin-binding site and the C-terminal focal adhesion targeting sequence in vinculin. J. Cell Sci. 1994;107(Pt. 2):709–717. [PubMed] [Google Scholar]
- Xing B, Jedsadayanmata A, Lam SC. Localization of an integrin binding site to the C terminus of talin. J. Biol. Chem. 2001;276:44373–44378. doi: 10.1074/jbc.M108587200. [DOI] [PubMed] [Google Scholar]
- Xu W, Baribault H, Adamson ED. Vinculin knockout results in heart and brain defects during embryonic development. Development. 1998a;125:327–337. doi: 10.1242/dev.125.2.327. [DOI] [PubMed] [Google Scholar]
- Xu W, Coll JL, Adamson ED. Rescue of the mutant phenotype by reexpression of full-length vinculin in null F9 cells; effects on cell locomotion by domain deleted vinculin. J. Cell Sci. 1998b;111(Pt. 11):1535–1544. doi: 10.1242/jcs.111.11.1535. [DOI] [PubMed] [Google Scholar]
- Yamada S, Pokutta S, Drees F, Weis WI, Nelson WJ. Deconstructing the cadherin-catenin-actin complex. Cell. 2005;123:889–901. doi: 10.1016/j.cell.2005.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Dokurno P, Tonks NK, Barford D. Crystal structure of the M-fragment of alpha-catenin: implications for modulation of cell adhesion. EMBO J. 2001;20:3645–3656. doi: 10.1093/emboj/20.14.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylanne J, Scheffzek K, Young P, Saraste M. Crystal structure of the alpha-actinin rod reveals an extensive torsional twist. Structure. 2001;9:597–604. doi: 10.1016/s0969-2126(01)00619-0. [DOI] [PubMed] [Google Scholar]
- Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M. alpha-Catenin as a tension transducer that induces adherens junction development. Nat. Cell Biol. 2010;12:533–542. doi: 10.1038/ncb2055. [DOI] [PubMed] [Google Scholar]
- Zemljic-Harpf AE, Ponrartana S, Avalos RT, Jordan MC, Roos KP, Dalton ND. Heterozygous inactivation of the vinculin gene predisposes to stress-induced cardiomyopathy. Am. J. Pathol. 2004;165:1033–1044. doi: 10.1016/S0002-9440(10)63364-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemljic-Harpf AE, Miller JC, Henderson SA, Wright AT, Manso AM, Elsherif L, et al. Cardiac-myocyte-specific excision of the vinculin gene disrupts cellular junctions, causing sudden death or dilated cardiomyopathy. Mol. Cell. Biol. 2007;27:7522–7537. doi: 10.1128/MCB.00728-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Izaguirre G, Lin SY, Lee HY, Schaefer E, Haimovich B. The phosphorylation of vinculin on tyrosine residues 100 and 1065, mediated by SRC kinases, affects cell spreading. Mol. Biol. Cell. 2004;15:4234–4247. doi: 10.1091/mbc.E04-03-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler WH, Tigges U, Zieseniss A, Jockusch BM. A lipid-regulated docking site on vinculin for protein kinase C. J. Biol. Chem. 2002;277:7396–7404. doi: 10.1074/jbc.M110008200. [DOI] [PubMed] [Google Scholar]
- Ziegler WH, Liddington RC, Critchley DR. The structure and regulation of vinculin. Trends Cell Biol. 2006;16:453–460. doi: 10.1016/j.tcb.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Zimerman B, Volberg T, Geiger B. Early molecular events in the assembly of the focal adhesion-stress fiber complex during fibroblast spreading. Cell Motil. Cytoskeleton. 2004;58:143–159. doi: 10.1002/cm.20005. [DOI] [PubMed] [Google Scholar]