Abstract
Although radiation effects have been extensively studied, the biological effects of low-dose radiation (LDR) are controversial. This study investigates LDR-induced alterations in locomotive behavior and gene expression profiles of Drosophila melanogaster. We measured locomotive behavior using larval pupation height and the rapid iterative negative geotaxis (RING) assay after exposure to 0.1 Gy γ-radiation (dose rate of 16.7 mGy/h). We also observed chronic LDR effects on development (pupation and eclosion rates) and longevity (life span). To identify chronic LDR effects on gene expression, we performed whole-genome expression analysis using gene-expression microarrays, and confirmed the results using quantitative real-time PCR. The pupation height of the LDR-treated group at the first larval instar was significantly higher (∼2-fold increase in PHI value, P < 0.05). The locomotive behavior of LDR-treated male flies (∼3 − 5 weeks of age) was significantly increased by 7.7%, 29% and 138%, respectively (P < 0.01), but pupation and eclosion rates and life spans were not significantly altered. Genome-wide expression analysis identified 344 genes that were differentially expressed in irradiated larvae compared with in control larvae. We identified several genes belonging to larval behavior functional groups such as locomotion (1.1%), oxidation reduction (8.0%), and genes involved in conventional functional groups modulated by irradiation such as defense response (4.9%), and sensory and perception (2.5%). Four candidate genes were confirmed as differentially expressed genes in irradiated larvae using qRT-PCR (>2-fold change). These data suggest that LDR stimulates locomotion-related genes, and these genes can be used as potential markers for LDR.
Keywords: locomotion, low-dose radiation, gene expression microarray, Drosophila behavior
INTRODUCTION
Individuals are frequently exposed to ionizing radiation (IR) from diagnostic, therapeutic, occupational and environmental sources. Health risks associated with exposure to low-dose radiation (LDR) have been estimated by extrapolating empirical linear fits for data on humans exposed to relatively high doses. LDR exposure from the nuclear bombing of Hiroshima and Nagasaki, and from the accidents at the Fukushima Dai-ichi nuclear power plant (NPP) and the Chernobyl NPP, is a global threat because LDR has negative effects on living beings [1]. LDR is reported to have various long-term biological effects such as adaptive responses [2] and low-dose hyper-radiosensitivity [3], in addition to reported beneficial effects [4, 5]. Therefore, it is difficult to evaluate and understand the biological effects of LDR.
Drosophila melanogaster is a powerful model system for genetic study due to its rapid development and relatively short life span, and it has been used to study the molecular mechanisms of a wide range of human diseases. Also, flies have advantages in experimental design due to easier scaling up and reproduction than many other organisms. Considerable progress in understanding life-span regulation has been achieved during the last two decades based on work in Drosophila; oxidative stress, food restriction, heat shock, and ionizing radiation can modulate life span [6]. Drosophila larvae have an intricate peripheral nervous system that detects odors, light, temperature, sound, and mechanical touch, enabling the study of sensory signaling [7]. Therefore, Drosophila melanogaster is an ideal model for LDR research.
So far, several studies have reported the biological effects of various kinds of irradiations. Our previous work showed that exposure to acute LDR enhanced Drosophila longevity and changed gene expression profiles [8]. Other reports indicate that LDR increases longevity by altering life span [9] and heat-shock protein (hsp) gene expression [10]. Several projects are measuring LDR effects using Drosophila. Most experiments with Drosophila suggest that LDR can induce hormesis and adaptive responses [11]. However, health risks associated with low-dose exposure (<0.1 Gy) are unknown. The biological mechanism of LDR effects on living organisms has not been resolved, and LDR effects induced by acute or chronic exposure can differ [12]. Chronic LDR induced butterfly abnormalities under field conditions in Japan [13]. Hiyama et al. [13] emphasize that their study investigates the consequences of chronic rather than acute exposure, and many laboratories conduct research on chronic LDR exposure. We explored the consequences of chronic LDR exposure to gamma radiation for Drosophila melanogaster. We examined behavioral responses to chronic LDR (0.1 Gy) and identified genes involved in LDR-mediated locomotive-behavior activation using microarray analysis and RT-PCR. This study of Drosophila melanogaster radiation biology can provide important clues to mechanisms mediating chronic-LDR responses.
MATERIALS AND METHODS
Fly strains and cultivation conditions
The wild-type Drosophila melanogaster Oregon-R strain was used for these studies. Flies were maintained at 25°C on medium containing dextrose anhydrous, dry yeast, cornmeal, agar, propionic acid, and tegosept in fly-food bottles with 60% relative humidity. Flies were maintained under a strict 12-h light/12-h dark photoperiod.
Larval collection and γ-irradiation
Eggs were collected from 5-day-old female flies and cultivated for 24 h on standard medium. Then, 20 larvae were manually selected and seeded on fresh standard medium in a new vial. After transfer, the experimental group of first-instar larvae was immediately irradiated with chronic γ-radiation at a dose rate of 16.7 mGy/h. After treatment, γ-irradiated flies and non-irradiated control flies were maintained in the same incubator at 25°C.
Larval pupation height assay
The larval pupation height assay was performed using a previously published method [7]. A total of 20 irradiated larvae were randomly collected and placed in a vial (2-cm diameter and 9-cm height) containing 2 ml of yeast–agar–sugar medium. After all larvae eclosed, pupation height was calculated as the height above the surface of the medium at which a larva pupates. The pupation-site height was recorded after completion of pupation. To calculate the pupation-height index (PHI), designated zones (1 − 6 cm) above the food zone were marked on the vials. Individual pupation-site heights were recorded to the nearest 1 cm, data for all pupae were combined, and the results were graphically displayed. Overall larval pupation height was also represented using PHI, which was calculated as follows: [(Number of pupae >3 cm height) − (Number of pupae <3 cm height)]/(Total number of pupae). The experiments were performed three times.
Rapid iterative negative geotaxis assay
The rapid iterative negative geotaxis (RING) assay was performed according to a previously published method [14]. Negative geotaxis of 20 flies in a tube was measured in RING assays. Flies were transferred to RING assay tubes, which were loaded into the RING apparatus. The flies were allowed 1 min rest, and then the apparatus was sharply struck on the table five times in rapid succession to initiate negative geotaxis responses. Images of fly positions were captured with a digital camera 6 s after initiating the test; the camera was located 30 cm from the RING apparatus for all experiments. Flies were assessed in five consecutive trials separated by 1 min of rest. After completion of the entire assay, flies were transferred to new tubes containing standard fly food and allowed to recover for at least 18 h. Each fly was subjected to the assay six times. Data from digital images were extracted, transformed with Image J, and analyzed for each tube. Prism 5.0 (GraphPad Software, CA) was used for statistical analysis of the data.
Longevity analysis
Flies were collected within 24 h of eclosion, separated under CO2 anesthesia according to gender, and allocated into groups of 20 to determine longevity (life-span analysis). A total of 100 flies were used for this study. Flies were cultivated at 25°C under controlled diurnal cycles and transferred to new food vials every 2 − 3 days. Dead flies were removed and counted. The experiments were performed three times.
Gene expression microarray analysis
Total RNA was extracted from irradiated and control larvae using TRIzol reagent (Invitrogen, Carlsbad, CA). Synthesis of target cRNA probes and hybridization were performed using Agilent's Low RNA Input Linear Amplification kit (Agilent Technology, Palo Alto, CA) according to the manufacturer's instructions. Labeled cRNA targets were quantified using an ND-1000 spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE). Labeled fragmented cRNA was resuspended with 2 × hybridization buffer and transferred onto assembled whole Drosophila genome oligo microarrays (44 K, Agilent Technology). Microarrays were hybridized at 65°C for 17 h, washed, and scanned according to the manufacturer's instructions.
Microarray data acquisition and analysis
Hybridized microarrays were scanned using a DNA microarray scanner and quantified with feature extraction software (Agilent Technologies). All data normalization and statistical/fold-change analyses were performed using GeneSpring GX 7.3 (Agilent Technologies). Data normalization was performed as follows: data transformation, set measurements <0.01 to 0.01; per chip, normalize to fiftieth percentile; per gene, normalize to median. To identify differentially expressed genes, >1.5 × fold-change analysis and the statistical t-test (P < 0.05, no multiple testing correction, no parametric test) were performed. Analyzed genes were functionally annotated according to the gene ontology (GO) consortium (http://www.geneontology.org). Analyzed genes were functionally classified based on DAVID bioinformatics (http://david.abcc.ncifcrf.gov) by uploading target gene lists and using the ‘Functional Annotation Clustering’ menu [15]. Raw and normalized DNA microarray data has been submitted to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) with the following accession number: GSE61759.
Quantitative RT-PCR analysis
Relative gene expression levels were analyzed by quantitative RT-PCR using the StepOnePlus instrument and power SYBR green master mix reagents (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. Primers for RT-PCR were designed with Primer Express V3.0 (Applied Biosystems, USA). PCR cycling conditions were: 95°C for 15 min for prewarming and 40 cycles of 95°C for 15 s and 59°C for 1 min. Relative mRNA expression levels were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The comparative cycle threshold (Ct) method was used for data analysis. Primers for target genes are listed in Table 1. Real-time PCR was repeated three times to verify results, and the mean mRNA expression level was used for subsequent analysis.
Table 1.
Oligonucleotide primer sequences for quantitative real-time PCR
| Primer name | Sequence (5′ → 3′) | Gene | GenBank acc. no. |
|---|---|---|---|
| gapdh1 forward | GAAGGGAATCCTGGGCTACA | gapdh | NM_080369 |
| gapdh1 reverse | CGTCGAACACAGACGAATGG | ||
| norpA forward | CGAAAACGCACCTGTATTCC | norpA | NM_001169190 |
| norpA reverse | GTAATGGGCCAGTGGCTGAT | ||
| Lsp2 forward | CTTTGTCTGCATCCACTTACC | Lsp2 | NM_080077 |
| Lsp2 reverse | ACAAGGTGGACTGACAAGCT | ||
| Est98B forward | ACCGGCGATGAACTATGATA | Est98B | NM_079811 |
| Est98B reverse | TCCGTCTTCTTCATGATTCC | ||
| Fbp2 forward | CGCGATGGAGATGATGAAGA | Fbp2 | NM_078798 |
| Fbp2 reverse | ATCGTGGGCGTAACCTCCTT |
RESULTS
Low-dose γ-irradiation alters Drosophila larvae pupation height
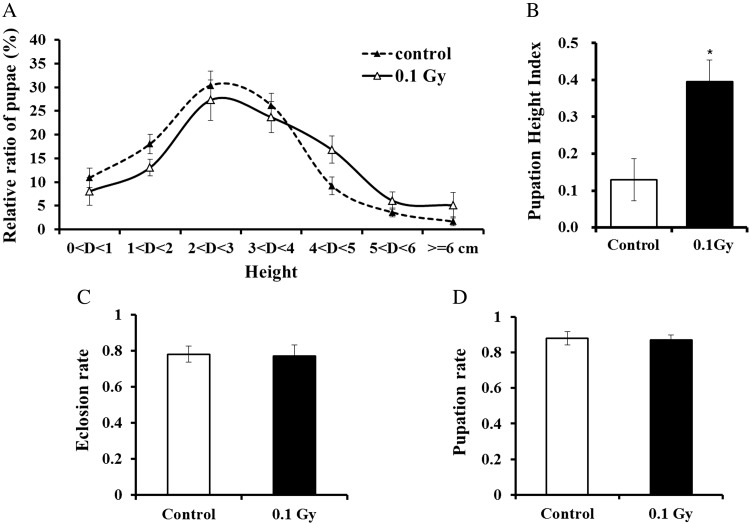
Drosophila larvae possess a complex peripheral nervous system that senses many environmental stimuli [16, 17]. Drosophila larvae are sensitive to dehydration and prefer moist food until the third larval instar exits the moist food to search for a dry pupation site [18]. To assess the potential effects of γ-radiation on larval pupation, we observed larval locomotive behavior after chronic LDR treatment with 0.1 Gy γ-radiation at the first larval instar stage. The pupation heights of irradiated flies were higher compared with those of control flies in the zone 4 − 6 cm above the medium surface (Fig. 1A). This behavior was expressed as the pupation-height index (PHI), which was calculated using the final pupation position above or below a designated height in the vial (Fig. 1B). The PHI of flies treated with chronic LDR was significantly higher compared with that of control flies by ∼2-fold in PHI value. The third larval instar searches for an appropriate pupation site. In general, the choice of pupation site can critically affect survival because the immobilized pupa is exposed to a variety of potential environmental insults during morphogenesis. Adult eclosion before completion of pupation may be lethal due to unfavorable environmental humidity [19]. Pupation within the food source under moist conditions may result in lethality due to asphyxiation or a physical block of adult eclosion. Conversely, premature movement to an area of extreme low humidity prior to pupation could result in lethality due to desiccation [18, 19]. To assess correlations between larval lethality, longevity, and pupation height, we examined pupation and eclosion rates in flies treated with 0.1-Gy γ-radiation and untreated control flies (Fig. 1C and D). Pupation and eclosion were not significantly different in chronic LDR and control flies. These results indicate that chronic LDR induces a slightly elevated shift in pupation-site height but does not increase larval lethality. The larval pupation height site by chronic LDR selected within the distance of movement away from a moister food source, not desiccation displaying a strong aversion to locomotion on dry surfaces severely limiting during morphogenesis.
Fig. 1.
Low-dose γ-radiation effects on pupation-height behavior and larval developmental processes in Drosophila. (A) Pupation sites of all tested larvae were recorded and the average population value was determined. Pupation height was higher in larvae exposed to 0.1 Gy of γ-radiation compared with that of control larvae. (B) The pupation-height index was greater in larvae exposed to 0.1 Gy of γ-radiation compared with that of control larvae. (C, D) To determine whether chronic LDR influenced fly development, we evaluated the pupation rate (C) and the eclosion rate (D) in irradiated and control larvae. Error bars represent SEM. Statistical significance was calculated using Student's t-test (*P < 0.05).
Adult flies exhibit enhanced climbing activity after low-dose γ-irradiation of larvae
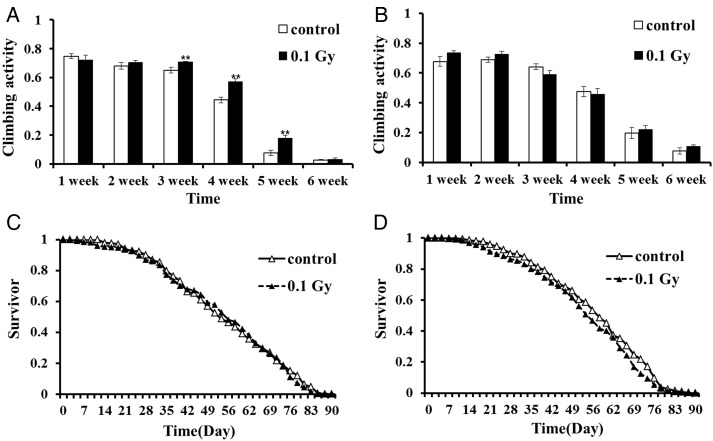
The behavioral effects of ionizing γ-radiation were analyzed by determining adult locomotion and the climbing ability of larvae that were irradiated at the first instar (Fig. 2). The RING assay is a suitable method for large-scale investigations of age-related locomotive behaviors [20]. The locomotive behavior of adult (1 − 2 weeks old) male flies was similar in chronic LDR-treated flies and sham-treated control flies. In male flies aged 3 − 5 weeks, chronic LDR-treated flies exhibited significantly greater locomotive behavior compared with control flies by 7.7%, 29% and 138%, respectively. In male flies aged ≥6 weeks, locomotive behavior did not differ significantly between irradiated and control flies. No significant differences in locomotive behaviors were observed in irradiated and control female flies irrespective of age. These results may reflect gender-specific differences in radiation responses. We assessed whether LDR-induced changes in locomotive behaviors influence longevity. The results showed that LDR did not affect male or female longevity, but significantly enhanced locomotive behavior in mid-life adult males. Locomotion capacity during aging is an important quality-of-life indicator. These results suggest that LDR may influence quality of life for adult male Drosophila.
Fig. 2.
Low-dose γ-radiation effects on adult climbing activity and longevity after irradiating Drosophila larvae. The rapid iterative negative geotaxis (RING) assay was used to measure fly locomotor activity. Climbing activity of (A) male and (B) female flies irradiated with 0.1 Gy γ-radiation was examined for flies aged 1 − 6 weeks and compared with that of control flies. Error bars represent SEM. Statistical significance was calculated using Student's t-test (*P < 0.05, **P < 0.01). Survival rates of control and irradiated (exposed to 0.1 Gy at the larval stage) flies were recorded. Age was measured in days after emergence. LDR had no effect on (C) male life span (P = 0.1592 with log rank test) or (D) female life span (P = 0.5750 with log rank test). Longevity analysis was performed three times, and data were analyzed with the Wilcoxon test.
Genome-wide expression analysis in Drosophila larvae
To identify LDR-modified gene expression, whole-genome expression of control and irradiated larvae was analyzed with gene-expression microarrays. A total of 32 222 probes were represented in the microarrays. We determined the normalized signal intensities of the control samples, and then filtered out probes that had normalized signal intensities equivalent to <15 for 50% of the control samples. The remaining 23 120 probes were statistically analyzed for significant differences between gene expression in control and irradiated larvae. The expression of 957 probes was statistically significant (t-test, P < 0.05), considering that sampling was performed in triplicate. Of these, 344 probes in irradiated larvae displayed a ≥1.5-fold difference in gene expression compared with those of the control (Table 2, ST1); 29 probes were downregulated and 315 probes were upregulated. Out of the 344 probes, 261 probes representing 229 genes were identified or had previous GenBank Accession numbers (http://www.ncbi.nlm.nih.gov/genbank). The remaining probes have not been annotated yet. *
Table 2.
Functional classification (biological process, molecular function) of 344 differentially expressed genes respond to LDR in Drosophila larvae (P < 0.05)
| Category | Term | Count | P-value | Genes |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Gene name | Fold ratio | GenBank Acc. No | Description | Genomic Coordinates | Cytoband | ||||
| Biological process | GO:0006952 defense response | 8 | 0.00199 | CG10345 | 0.45335618 | NM_142306 | – | chr3R:12422851-12422795 | dm|3R89D5 |
| SR-CIV | 1.8602393 | NM_134945 | Scavenger receptor class C, type IV | chr2L:3522653-3522594 | dm|2L23F6 | ||||
| DEF | 1.5728586 | NM_078948 | Defensin | chr2R:5941989-5941930 | dm|2R46D9 | ||||
| PROPO-A1 | 1.5037334 | NM_057464 | prophenol oxidase A1 | chr2R:13774816-13774757 | dm|2R54F6 | ||||
| CG1887 | 1.7244722 | NM_001104007 | chr3L:1893545-1893486 | dm|3L62B9 | |||||
| CG31741 | 1.7282056 | NM_165238 | chr2L:18086097-18086156 | dm|2L36E3 | |||||
| FON | 1.6542897 | NM_001169542 | fondue | chr2L:19385792-19385353 | dm|2L37D3 | ||||
| IM14 | 1.6513002 | 0 | Immune induced molecule 14 | chr2R:16758079-16758138 | dm|2R57B3 | ||||
| GO:0055114 oxidation reduction | 13 | 0.02467 | CG31809 | 1.5207597 | NM_001103698 | chr2L:16853737-16853678 | dm|2L36B2 | ||
| CG1434 | 1.500386 | NM_132722 | chrX:14134103-14134162 | dm|X12E5 | |||||
| CG17374 | 1.5812901 | NM_001015405 | chr3LHet:1140421-1140362 | ||||||
| CG3597 | 1.5520066 | NM_134830 | chr2L:2370297-2370238 | dm|2L22E1 | |||||
| CYP4E3 | 2.14801 | NM_078803 | Cytochrome P450-4e3 | chr2L:9747958-9747899 | dm|2L30C7 | ||||
| CG3699 | 1.540999 | NM_130519 | chrX:840783-840724 | dm|X1D2 | |||||
| FBP2 | 18.835476 | NM_078798 | Fat body protein 2 | chr2L:9427743-9427802 | dm|2L30B3 | ||||
| CG31810 | 1.987148 | NM_165198 | chr2L:16849144-16849085 | dm|2L36B2 | |||||
| EO | 1.5894746 | NM_132758 | Ecdysone oxidase | chrX:14810875-14810816 | dm|X13A1 | ||||
| PROPO-A1 | 1.5037334 | NM_057464 | prophenol oxidase A1 | chr2R:13774816-13774757 | dm|2R54F6 | ||||
| DHPR | 1.957855 | NM_001014580 | Dihydropteridine reductase | chr3L:9128701-9128760 | dm|3L67A1 | ||||
| CG34355 | 0.58311474 | NM_001260339 | chr3R:19646548-19646607 | dm|3R95C1 | |||||
| CG6910 | 1.9879138 | NM_140299 | chr3L:12180735-12180794 | dm|3L69A1 | |||||
| Molecular function | GO:0005344 oxygen transporter activity | 5 | 0.00002 | LSP2 | 4.932235 | NM_080077 | Larval serum protein 2 | chr3L:12124737-12124796 | dm|3L68F5 |
| PROPO-A1 | 1.5037334 | NM_057464 | prophenol oxidase A1 | chr2R:13774816-13774757 | dm|2R54F6 | ||||
| LSP1ALPHA | 1.9465075 | NM_078583 | Larval serum protein 1 alpha | chrX:12386831-12386890 | dm|X11A12 | ||||
| CG8100 | 1.8050036 | NM_140436 | chr3L:14078775-14078834 | dm|3L70D2 | |||||
| GLOB3 | 2.3051763 | NM_141340 | globin 3 | chr3R:1657155-1657214 | dm|3R83C4 | ||||
| GO:0005044 scavenger receptor activity | 4 | 0.00280 | CG10345 | 0.45335618 | NM_142306 | chr3R:12422851-12422795 | dm|3R89D5 | ||
| SR-CIV | 1.8602393 | NM_134945 | Scavenger receptor class C, type IV | chr2L:3522653-3522594 | dm|2L23F6 | ||||
| CG1887 | 1.7244722 | NM_001104007 | – | chr3L:1893545-1893486 | dm|3L62B9 | ||||
| CG31741 | 1.7282056 | NM_165238 | – | chr2L:18086097-18086156 | dm|2L36E3 | ||||
| GO:0005096 GTPase activator activity | 5 | 0.01296 | DRONGO | 1.5596985 | NM_164392 | drongo | chr2L:833791-833732 | dm|2L21E2 | |
| RN-TRE | 2.21713 | NM_144124 | tre oncogene-related protein | chr2R:9874914-9874855 | dm|2R50C23 | ||||
| LOCO | 1.5090106 | NM_001260313 | locomotion defects | chr3R:18452089-18448623 | dm|3R94B7 | ||||
| NORPA | 1.636429 | NM_001169190 | no receptor potential A | chrX:4251717-4251776 | dm|X4C1 | ||||
| SPRI | 1.7273139 | NM_001103471 | Drosophila melanogaster sprint (spri), transcript variant H, mRNA | chrX:10390835-10390776 | dm|X9D2 | ||||
| Molecular Function | GO:0008047 enzyme activator activity | 5 | 0.02687 | DRONGO | 1.5596985 | NM_164392 | drongo | chr2L:833791-833732 | dm|2L21E2 |
| RN-TRE | 2.21713 | NM_144124 | tre oncogene-related protein | chr2R:9874914-9874855 | dm|2R50C23 | ||||
| LOCO | 1.5090106 | NM_001260313 | locomotion defects | chr3R:18452089-18448623 | dm|3R94B7 | ||||
| NORPA | 1.636429 | NM_001169190 | no receptor potential A | chrX:4251717-4251776 | dm|X4C1 | ||||
| SPRI | 1.7273139 | NM_001103471 | Drosophila melanogaster sprint (spri), transcript variant H, mRNA | chrX:10390835-10390776 | dm|X9D2 | ||||
| GO:0016298 lipase activity | 5 | 0.02687 | CG5966 | 1.5556251 | NM_132058 | – | chrX:5886453-5886512 | dm|X5D1 | |
| CG15533 | 1.5764413 | NM_143534 | Drosophila melanogaster CG15533 (CG15533), mRNA | chr3R:26257032-26257091 | dm|3R99F4 | ||||
| BMM | 1.597753 | NM_001169974 | brummer | chr3L:14770600-14770541 | dm|3L70F5 | ||||
| CG6283 | 1.7423133 | NM_143267 | – | chr3R:22846017-22845960 | dm|3R97D14 | ||||
| NORPA | 1.636429 | NM_001169190 | no receptor potential A | chrX:4251717-4251776 | dm|X4C1 | ||||
| GO:0015370 solute:sodium symporter activity | 4 | 0.04219 | CG13794 | 1.6694626 | NM_135293 | – | chr2L:7719922-7719863 | dm|2L28C2 | |
| CG13793 | 1.8739051 | NM_135292 | – | chr2L:7717111-7717052 | dm|2L28C1 | ||||
| CG9903 | 2.0286412 | NM_132904 | – | chrX:16434630-16434571 | dm|X14E1 | ||||
| CG7720 | 1.5052466 | NM_001170179 | – | chr3R:14585019-14584960 | dm|3R91C6 | ||||
Genes showing significant regulation from the microarray data were functionally classified, categorized, and placed in appropriate GO categories. A total of 14 terms were categorized with corrected P-values < 0.05. The functionally classified (biological process, molecular function) genes involved in each category are listed in Table 1. In the biological process subgroup, upregulated genes whose products are involved in a defense response (GO:0006952) and in oxidation–reduction (GO:0055114) were statistically significant (P < 0.05). In the cellular component subgroup, genes were grouped primarily as having an extracellular location (GO:0005576) or a plasma-membrane location (GO:0005886). In the molecular function subgroup, genes were categorized into six different subgroups. A total of 29 downregulated probes were not functionally categorized because of inadequate gene numbers and lack of gene-annotation information. Independently, seven genes that are involved in ‘Locomotion (GO:0040011)’ were identified out of 344 probes (Table 3). These seven genes included dally, norpA, spri, sli, Lsp2, RN-tre and Ets98B; all were upregulated at least 1.5-fold in response to LDR compared with those of controls.
Table 3.
Seven differentially expressed genes involved in locomotion that are modulated by LDR
| Agilent probe ID | Normalized | Common | GenBank acc. no. | Description |
|---|---|---|---|---|
| A_09_P199635 | 1.57 | dally | NM_079259 | Division abnormally delayed (source, FlyBase; gene name, Acc:FBgn0011577) (FBtr0076583) |
| A_09_P145565 | 1.64 | norpA | NM_001169190 | No receptor potential A (source, FlyBase; gene name, Acc:FBgn0262738) (FBtr0301475) |
| A_09_P048936 | 1.73 | spri | NM_001103471 | Drosophila melanogaster sprint (spri), transcript variant H, mRNA (NM_001103471) |
| A_09_P044761 | 1.70 | sli | NM 057379 | Slit (CG43758; FBgn0264089) |
| A_09_P043376 | 4.93 | Lsp2 | NM_080077 | Larval serum protein 2 (source, FlyBase; gene name, Acc:FBgn0002565) (FBtr0089324) |
| A_09_P032511 | 2.22 | RN-tre | NM_144124 | Tre oncogene-related protein (source, FlyBase; gene name, Acc:FBgn0020620) (FBtr0087605) |
| A_09_P011736 | 1.78 | Ets98B | NM_079811 | Ets at 98B (source, FlyBase; gene name, Acc:FBgn0005659)(FBtr0085224) |
Quantitative real-time PCR of candidate LDR-marker genes
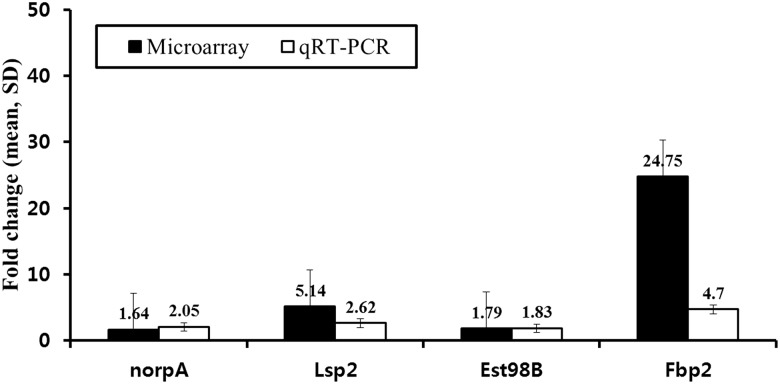
We randomly selected four target genes identified in locomotion or oxidation–reduction GO categories to verify and quantify the gene-expression microarray results using qRT-PCR. The four target genes include norpA, Lsp2, Ets98B and Fbp2. Primers and conditions are listed in Table 3; experiments were repeated three times. The results showed that Lsp2 and Fbp2 were strongly expressed in LDR-treated samples compared with controls, whereas Ets98B and norpA showed moderate gene-expression changes in response to LDR (Fig. 3). In microarray experiments, expression of Lsp2 and Fbp2 were upregulated 4.93- and 18.83-fold, respectively, in LDR-treated samples, whereas norpA and Ets98B expression showed 2-fold changes (Table 1). These results indicate that LDR-induced gene-expression changes are reproducibly detected using either microarray or qRT-PCR analysis.
Fig. 3.
Comparison of transcript levels in microarray and qRT-PCR analyses. Relative changes in transcript levels in microarray analysis (black bars) were compared with those of qRT-PCR analysis (white bars) (mean ± SD). Data from three individual experiments are presented.
DISCUSSION
Recent reports of the linear non-threshold (LNT) model for LDR (<100 mSv) have been subject to debate [21, 22]. A crucial point in the controversy is the lack of a biological mechanism [23]. Another crucial aspect for LDR ≤0.1 Gy is that the previous studies have shown variable results depending on the dose-rate. In particular, the impact of LDR on behavior is an emerging field, because the behavior changes on animal model systems are related to functional senescence. Functional senescence research on chronic LDR should identify the basic physiological declines that impinge on quality of life as well as length of life. Also, a wide range of behaviors in adult Drosophila can be quantitatively assessed in the laboratory. In this study, we demonstrated that chronic LDR (0.1 Gy) enhanced locomotion of Drosophila melanogaster larvae and adults, but did not affect longevity. Several studies recently investigated the effects of ionizing radiation on mouse locomotive activity [24] and rat motor-behavior [25]; the results indicated that acute-IR disrupted locomotive or social-exploratory behaviors. Other work reported that LDR (<0.1 Gy) treatments at both low- and high-dose rates affected fly longevity in Canton-S (low-dose, 0.04 Gy) and mutant flies, whereas chronic LDR induced adaptive responses in the Canton-S wild-type strain [10]. However, there are no reports investigating whether chronic LDR affects behavior in animal models. Our study explores chronic LDR effects on behavior in the Drosophila model system. Although we observed behavioral effects induced by chronic LDR, we did not observe LDR-mediated effects on longevity. Also, no significant differences in locomotive behaviors were observed in female flies. Although there are effects of gender on behavior senescence in some genetic backgrounds, not all genetic backgrounds showed consistent differences between males and females [20]. Also, a number of functions and its declines as age of flies can be assessed in the laboratory [20, 24]. It seems that there are complex interactions between the effects of gender and genetic background on Drosophila locomotor behavior. In our results, chronic LDR slightly induced climbing activity during a short period, and then recovered their movement within the arena. Thus, chronic LDR temporarily impinged on exploratory activity but not functional senescence in the fly.
Drosophila is a powerful model system for exploring correlations between genes and behavior. We performed whole-genome microarray expression analysis of control and chronic-LDR larvae. Only ∼90 probes (equivalent to ∼0.28% of all probes) were differentially expressed with > 2-fold changes (P < 0.05). Another study showed that LDR (0.1 Gy for 8 h) had little effect on gene expression [203 genes out of ∼24 000 (0.85%)] in human skin fibroblasts [26], so we reduced the fold-change stringency to >1.5-fold for further data analysis. LDR-induced modulation of gene expression in Drosophila larvae may correlate with a slight change in climbing activity of irradiated larvae compared with that of control larvae.
Functional grouping with 344 probes representing 313 genes revealed that conventional radiation-induced effects were reproduced in our study. We previously reported that LDR (20 cGy) treatment at the egg-stage potentially changed adult gene-expression profiles [8, 27]; LDR increased expression of genes that are functionally involved in oxidative stress, cell cycle, metabolism, ion transport, response to external stimuli, and immune responses. The results of the current study are consistent with those of the previous studies. Chronic LDR significantly modulated genes involved in the defense response (GO:0006952), oxidation–reduction (GO:0055114), oxygen-transporter activity (GO:0005344), scavenger-receptor activity (GO:0005044), and solute/sodium symporter activity (GO:0015370). The conventional effect of irradiation on gene-expression regulation seems to be largely dependent to model organisms. Different functional grouping results may be due to experimental differences in model life stages (larvae versus adult), radiation times, or radiation intensities. This suggests that larval responses to LDR may differ from those of adult flies.
We observed enhanced climbing activity in larvae exposed to chronic LDR. We screened for genes that correlated with behavioral changes. We selected genes involved in locomotive behavior by performing gene-set enrichment analysis. Previous reports indicated that low-dose γ-radiation effects on locomotion activity differed according to the model organism, experimental methods, irradiation intensity, and irradiation duration. Jason et al. [24] reported that repeated exposures to LDR (≤200 cGy) in mice reduced spontaneous locomotor activity by ∼35%, but enhanced behavior 6 h after radiation [24]. In Caenorhabditis elegans, high-dose γ-radiation reduced body-bend locomotion [28].
Seven out of 344 differentially expressed genes in irradiated larvae are categorized in locomotion according to GSEA, and are reported to regulate locomotive behavior. Daily oscillations of norpA expression affect fly vision, which functions in circadian-clock regulation, nerve-cell differentiation, and behavioral activity [29, 30]. The Sprint gene is involved in receptor tyrosine kinase signaling in Drosophila; suppression of Sprint expression disrupts cell migration [31]. Slit affects Drosophila nervous system differentiation and modulates larval growth [32]. Lsp2 is a subunit of the Drosophila hexamerin protein; it functions during differentiation of the mosquito larval–pupa l stage [33]. The RN-tre gene product is thought to interact with Rab5 and Rab11 to regulate apical localization of the R-cell [34]. Ets family members are expressed in Drosophila oocytes and larvae, and are involved in cell migration [35]. These results suggest that genes involved in visual perception are sensitive to LDR. These genes may function in early responses to LDR.
Many factors have been identified that affect Drosophila locomotive behavior. Katherine et al. [36] reported that genes in functional groups such as lipid/steroid metabolism, response to external stimuli, defense response, and oxidoreductase activity often have altered expression in inbred Drosophila lines that have vigorous locomotive behavior [36]. Dry conditions reportedly inactivate sensory transduction pathways such as class IV multiple-dendritic nociceptors, degenerin/epithelial sodium channel subunit (pickpocket1), and transient receptor potential channel, which disrupts the larval aversion to dry environments [7]. Our results showed that defense-response and oxidation–reduction functional groups were strongly expressed in irradiated fruit-fly larvae (Table 2). We found that pickpocket gene expression was not activated by LDR (fold-change value of ∼0.99 − 1.10; data not shown).
The results of our studies are consistent with those of previous reports. Although the mechanism mediating LDR effects is unknown, we identified that LDR treatment of Drosophila larvae induces expression of genes in functional groups related to locomotive behavior. The interactions of these genes could enhance larval locomotion. Locomotion is a complex behavior, and locomotive phenotypes may result from interactions of complex gene pathways. A full understanding of mechanisms and regulatory pathways driving these phenotypes requires further research. The identification and annotation of additional genes involved in LDR responses will help to resolve the mechanisms involved in animal locomotion.
FUNDING
Funding to pay the Open Access publication charges for this article and to support this overall project was provided by the Ministry of Trade, Industry and Energy (MOTIE), Republic of Korea (20131610101840).
REFERENCES
- 1.Lubin JH, Boice JD., Jr Lung cancer risk from residential radon: meta-analysis of eight epidemiologic studies. J Natl Cancer Inst 1997;89:49–57. [DOI] [PubMed] [Google Scholar]
- 2.Ikushima T, Aritomi H, Morisita J. Radioadaptive response: efficient repair of radiation-induced DNA damage in adapted cells. Mutat Res 1996;358:193–8. [DOI] [PubMed] [Google Scholar]
- 3.Marples B, Wouters BG, Collis SJ, et al. Low-dose hyper-radiosensitivity: a consequence of ineffective cell cycle arrest of radiation-damaged G2-phase cells. Radiat Res 2004;161:247–55. [DOI] [PubMed] [Google Scholar]
- 4.Ina Y, Sakai K. Prolongation of life span associated with immunological modification by chronic low-dose-rate irradiation in MRL-lpr/lpr mice. Radiat Res 2004;161:168–73. [DOI] [PubMed] [Google Scholar]
- 5.Ina Y, Tanooka H, Yamada T, et al. Suppression of thymic lymphoma induction by life-long low-dose-rate irradiation accompanied by immune activation in C57BL/6 mice. Radiat Res 2005;163:153–8. [DOI] [PubMed] [Google Scholar]
- 6.Moskalev AA, Plyusnina EN, Shaposhnikov MV. Radiation hormesis and radioadaptive response in Drosophila melanogaster flies with different genetic backgrounds: the role of cellular stress-resistance mechanisms. Biogerontology 2011;12:253–63. [DOI] [PubMed] [Google Scholar]
- 7.Johnson WA, Carder JW. Drosophila nociceptors mediate larval aversion to dry surface environments utilizing both the painless TRP channel and the DEG/ENaC subunit, PPK1. PLoS One 2012;7:e32878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seong KM, Kim CS, Seo SW, et al. Genome-wide analysis of low-dose irradiated male Drosophila melanogaster with extended longevity. Biogerontology 2011;12:93–107. [DOI] [PubMed] [Google Scholar]
- 9.Moskalev A. Radiation-induced life span alteration of Drosophila lines with genotype differences. Biogerontology 2007;8:499–504. [DOI] [PubMed] [Google Scholar]
- 10.Moskalev A, Shaposhnikov M, Turysheva E. Life span alteration after irradiation in Drosophila melanogaster strains with mutations of Hsf and Hsps. Biogerontology 2009;10:3–11. [DOI] [PubMed] [Google Scholar]
- 11.Shaposhnikov MV, Turysheva EV, Moskalev AA. Low-dose rate irradiation induced hormesis, hypersensitivity and adaptive response in Drosophila melanogaster of radiosensitive strains. Radiats Biol Radioecol 2009;49:46–54. [PubMed] [Google Scholar]
- 12.Tseng BP, Lan ML, Tran KK, et al. Characterizing low dose and dose rate effects in rodent and human neural stem cells exposed to proton and gamma irradiation. Redox Biol 2013;1:153–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiyama A, Nohara C, Taira W, et al. The Fukushima nuclear accident and the pale grass blue butterfly: evaluating biological effects of long-term low-dose exposures. BMC Evol Biol 2013;13:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nichols CD, Becnel J, Pandey UB. Methods to assay Drosophila behavior. J Vis Exp 2012;61:e3795:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 16.Gong Z. Behavioral dissection of Drosophila larval phototaxis. Biochem Biophys Res Commun 2009;382:395–9. [DOI] [PubMed] [Google Scholar]
- 17.Eberl DF, Hardy RW, Kernan MJ. Genetically similar transduction mechanisms for touch and hearing in Drosophila. J Neurosci 2000;20:5981–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bainbridge SP, Bownes M. Staging the metamorphosis of Drosophila melanogaster. J Embryol Exp Morphol 1981;66:57–80. [PubMed] [Google Scholar]
- 19.Rodriguez L, Sokolowski MB, Shore JS. Habitat selection by Drosophila melanogaster larvae. J Evol Biol 1992;5:61–70. [Google Scholar]
- 20.Gargano JW, Martin I, Bhandari P, et al. Rapid iterative negative geotaxis (RING): a new method for assessing age-related locomotor decline in Drosophila. Exp Gerontol 2005;40:386–95. [DOI] [PubMed] [Google Scholar]
- 21.Tubiana M, Arengo A, Averbeck D, et al. Low-dose risk assessment. Radiat Res 2007;167:742–4. [DOI] [PubMed] [Google Scholar]
- 22.Tubiana M, Aurengo A, Averbeck D, et al. Low-dose risk assessment: we still have much to learn. Radiat Res 2007;167:744. [DOI] [PubMed] [Google Scholar]
- 23.Tubiana M, Feinendegen LE, Yang C, et al. The linear no-threshold relationship is inconsistent with radiation biologic and experimental data. Radiology 2009;251:13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.York JM, Blevins NA, Meling DD, et al. The biobehavioral and neuroimmune impact of low-dose ionizing radiation. Brain Behav Immun 2012;26:218–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bogo V. Effects of bremsstrahlung and electron radiation on rat motor performance. Radiat Res 1984;100:313–20. [PubMed] [Google Scholar]
- 26.Ray M, Yunis R, Chen X, et al. Comparison of low and high dose ionizing radiation using topological analysis of gene coexpression networks. BMC Genomics 2012;13:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seong KM, Kim CS, Lee BS, et al. Low-dose radiation induces Drosophila innate immunity through Toll pathway activation. J Radiat Res 2012;53:242–9. [DOI] [PubMed] [Google Scholar]
- 28.Sakashita T, Hamada N, Ikeda DD, et al. Locomotion-learning behavior relationship in Caenorhabditis elegans following gamma-ray irradiation. J Radiat Res 2008;49:285–91. [DOI] [PubMed] [Google Scholar]
- 29.Rawson JM, Dimitroff B, Johnson KG, et al. The heparan sulfate proteoglycans Dally-like and Syndecan have distinct functions in axon guidance and visual-system assembly in Drosophila. Curr Biol 2005;15:833–8. [DOI] [PubMed] [Google Scholar]
- 30.Mealey-Ferrara ML, Montalvo AG, Hall JC. Effects of combining a cryptochrome mutation with other visual-system variants on entrainment of locomotor and adult-emergence rhythms in Drosophila. J Neurogenet 2003;17:171–221. [PubMed] [Google Scholar]
- 31.Jékely G, Sung HH, Luque CM, et al. Regulators of endocytosis maintain localized receptor tyrosine kinase signaling in guided migration. Dev Cell 2005;9:197–207. [DOI] [PubMed] [Google Scholar]
- 32.Dimitrova S, Reissaus A, Tavosanis G. Slit and Robo regulate dendrite branching and elongation of space-filling neurons in Drosophila. Dev Biol 2008;324:18–30. [DOI] [PubMed] [Google Scholar]
- 33.Korochkina SE, Gordadze AV, York JL, et al. Mosquito hexamerins: characterization during larval development. Insect Mol Biol 1997;6:11–21. [DOI] [PubMed] [Google Scholar]
- 34.Houalla T, Shi L, van Meyel DJ, et al. Rab-mediated vesicular transport is required for neuronal positioning in the developing Drosophila visual system. Mol Brain 2010;3:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsouna A, Watson DK, Hsu T. Developmental expression pattern of D-ets4, the Drosophila homologue of human Pdef. Gene Expr Patterns 2004;5:285–9. [DOI] [PubMed] [Google Scholar]
- 36.Jordan KW, Carbone MA, Yamamoto A, et al. Quantitative genomics of locomotor behavior in Drosophila melanogaster. Genome Biol 2007;8:R172. [DOI] [PMC free article] [PubMed] [Google Scholar]