Abstract
The mammalian alimentary tract harbors hundreds of species of commensal microorganisms (microbiota) that intimately interact with the host and provide it with genetic, metabolic, and immunological attributes. Recent reports have indicated that the microbiota composition and its collective genomes (microbiome) are major factors in predetermining the type and robustness of mucosal immune responses. In this review, we discuss the recent advances in our understanding of host-microbiota interactions and their effect on the health and disease susceptibility of the host.
Keywords: intestinal microbiota, Th17 cells, Treg cells, innate lymphoid cells, segmented filamentous bacteria, Clostridium
INTRODUCTION
The tissues of the gastrointestinal tract have the unique property of harboring an enormous number of microbes within the lumen. The concentration of microbes that reside in the small intestine is estimated to be from 103 to 109 cells/ml, whereas the large intestine contains abundant bacteria, which achieve a concentration of up to 1011 or 1012 cells/g of luminal contents (1). This concentration is similar to or even higher than that achieved in colonies growing under optimum conditions on laboratory plates, indicating that the colonic lumen provides a safe and nourishing environment and represents an extremely efficient natural bioreactor for bacteria. The estimated total number of bacteria carried by a healthy human in the gut is 1014, which, as a whole, constitutes the microbiota (also referred to as the microbial flora), an ecosystem in dynamic equilibrium. The members of the intestinal microbiota can be categorized as either allochthonous or autochthonous (1, 2). Allochthonous bacteria are only transiently present, whereas autochthonous bacteria are indigenous and preferentially colonize physical spaces or niches in particular animal species. In most cases, indigenous bacteria can attach to the epithelium or mucus layer and form a biofilm, and thereby significantly affect host development and physiology.
The microbiota allows for optimal breakdown of foods, uptake of nutrients, and enhancement of intestinal development, which have led to diet diversification and increased evolutionary fitness. Beyond digestion and metabolism, the microbiota also contributes to development and maintenance of the intestinal epithelial barrier, development of the immune system, and competition with pathogenic microorganisms, thus preventing their propagation. Indeed, studies of germfree (GF) animals indicate that intestinal microbes profoundly affect the development of the mucosal immune system in terms of the organization of Peyer's patches (PPs) [←AU: this abbreviation is introduced below. Is it for Peyer's patches?** Yes, it is for Peyer's patches.] and isolated lymphoid follicles (ILFs), secretion of antimicrobial peptides by epithelial cells, and accumulation of various immunocytes at mucosal sites (3--7). Collectively, the gut microbiota provides an indispensable internal ecosystem for numerous host physiological processes and can be considered to have coevolved with the host to form a superorganism (8).
GF: germfree
PP: Peyer's patch
ILF: isolated lymphoid follicle
[**AU: Please note that the terms temporarily placed at the end of various paragraphs will be typeset as “floating” elements in the margins of the typeset article. Also, note that colored words, phrases, acronyms, and numbers within the text will be hyperlinked in the online version, but will appear as normal text in the printed version.**]
The collective genome of intestinal microbes, termed the microbiome, is estimated to contain at least 100 times more genes than our own genome (9). Unlike the human genome, which is rarely altered by xenobiotic intervention, the gut microbiota composition is readily changeable by diet, antibiotic ingestion, infection by pathogens, and other life events. The plasticity of the microbiome has been implicated in numerous disease conditions, and an unfavorable alteration of the gut microbiota composition is called dysbiosis, which includes an outgrowth of potential pathogenic bacteria (pathobionts) and a decrease in the number of beneficial bacteria (10, 11). Multiple recent reports have shown a link between dysbiosis and immune disorders. Crohn's disease and ulcerative colitis are two chronic intestinal inflammatory conditions referred to as inflammatory bowel disease (IBD). IBD is a disease with an elusive etiology, and although many potential triggers have been invoked, one attractive hypothesis is that IBD may be a result of dysbiosis in the intestinal microbial community that promotes the overgrowth of bacteria that aberrantly stimulate the intestinal immune system. Indeed, many reports have shown that the microbial populations in the intestine of IBD patients are different from those of healthy individuals (12--14). Accumulating evidence suggests that a change in the gut microbiota composition has a key role not only in IBD, but also in the development of systemic immune diseases, such as rheumatoid arthritis (15, 16), encephalomyelitis (17, 18), type 1 diabetes (19, 20), and allergic diseases (21, 22).
Microbiome: a genetic catalog of the microbial species that inhabit a defined environment such as the human body
Dysbiosis: a condition with imbalance in the composition of the bacterial microbiota; this includes an outgrowth of potentially pathogenic bacteria and/or a decrease in bacterial diversity and bacteria beneficial to the host
Pathobiont: a symbiont or a commensal that is able to promote pathology only when genetic or environmental conditions are altered in the host
IBD: inflammatory bowel disease
The microbiota affects the host immune system through multiple factors, which include microbial components and their metabolites. The immune system recognizes these factors mostly through innate immune receptors. Constitutive signaling induced by the microbiota keeps the intestinal mucosa in a state of physiological inflammation, with continuous production of tissue repair factors, antimicrobial proteins, and immunoglobulin A (IgA) that, together, maintain intestinal barrier integrity and provide beneficial functions to the microbiota (23--25). Without constitutive innate signaling, intestinal barrier injury and bacterial translocation may occur. Innate immune recognition of the microbiota leads to the establishment of an arsenal of unique and diverse intestinal immune cell populations. IgA-producing plasma cells, intraepithelial lymphocytes (IELs), and T cell receptor (TCR) γδ-expressing T cells (γδ T cells) are the classically known lymphocytes unique to the mucosa. Recent studies have shown the presence of innate immune lymphocytes, such as CD4+CD3− lymphoid tissue–inducer cells (LTi cells) and interleukin (IL)-22-producing natural killer (NK)-like cells (26, 27). Furthermore, CD4+ T cells in the intestinal mucosa comprise significant numbers of IL-17-expressing cells (Th17 cells) and fork-head box P3 [**AU: lowercase “p” nearly everywhere else. OK to change here? → OK] (Foxp3)-expressing regulatory T cells (Treg cells) (28). All these cells are particularly abundant in the intestinal mucosa, even under steady-state conditions, and their accumulation and function are deeply affected by the presence of the microbiota.
LTi: lymphoid tissue inducer
Although it is not fully understood why and how the intestinal microbiota generates such a large variety of immune cell populations, recent studies using gnotobiotes (animals with a defined microbiological status) have suggested that specific components of the microbiota induce specific populations of immune cells. Below, we summarize the recent findings on how members of the microbiota provide an intestinal environment uniquely suited for the well-balanced development of the innate and adaptive immune system and discuss the role of the microbiota in infectious diseases and inflammation.
Gnotobiote: gnotobiotic comes from the Greek “known life” and refers to animals with defined microbiological status
COMPOSITION OF THE MICROBIOME
The establishment of the intestinal microbiota occurs progressively, beginning immediately after birth. The initial infant gut microbiota has a relatively simple composition, which is affected in large part by the maternal microbiota. Vaginally delivered infants acquire bacterial communities resembling their own mother's vaginal microbiota, dominated by Lactobacillus, Prevotella, or Sneathia spp., whereas infants delivered by Cesarean section harbor bacterial communities similar to those found on the skin surface, dominated by Staphylococcus, Corynebacterium, and Propionibacterium spp. (29). These pioneer bacteria may affect the composition of adult flora. After the weaning period, however, the microbiota markedly changes, and obligate anaerobes become prominent, with much lower numbers of facultative anaerobes.
Only four phyla dominate adult human intestinal habitats (30). Most (>90%) of them belong to Bacteroidetes (including Bacteroides) and Firmicutes (including Clostridium, Lactobacillus, and Bacillus). Firmicute bacteria in the gut include two major clostridial groups, namely the clostridial clusters IV and XIVa that comprise the Lachnospiraceae. Lower-abundance phyla are mainly composed of Proteobacteria (including Escherichia) and Actinobacteria (including Bifidobacterium). The mouse intestinal microbiota is similar to the human microbiota in broad terms. Such limited phylum predominance suggests the presence of strong selective forces over thousands, perhaps even millions, of years of coevolution. Notably, certain members of the Firmicutes, such as Clostridium and Bacillus genera, are found in a state of vegetative growth or as spores. The ability to make spores may be of ecological advantage to the organism as it enables it to survive under adverse conditions to efficiently colonize the intestine.
At lower taxonomic levels, there is considerable interindividual variation. Metagenomic approaches using massive parallel sequencing allow for the direct enumeration of the microbiota without having to isolate and cultivate bacteria. Using this technology, the international MetaHIT (Metagenomics of the Human Intestinal Tract) project has recently reported that each human individual carries on average 540,000 common genes in the intestine (9). This estimate suggests that only approximately 35% of bacterial genes are shared between individuals. Interestingly, the results from the MetaHIT consortium also suggested the existence of at least three enterotypes in the human population (31). Enterotypes, which can be compared to blood types, are defined by characteristic populations of bacterial species and the genes that they encode. It is not yet known how enterotypes affect metabolism or immune system homeostasis in the host.
MetaHIT (Metagenomics of the Human Intestinal Tract consortium): the MetaHIT project aims to understand the role of the human intestinal microbiota in health and disease; the consortium involves 13 research centers from eight countries
The microbiome is adaptable to environmental changes and host genotypes. Recent studies have shown that community membership and function of the microbiota can change owing to numerous variables including lifestyle, hygiene, diet, and use of antibiotics (32). Furthermore, it has recently become clear that the composition of the microbiota can influence onset and/or progression of several diseases. Indeed, the respective levels of the two main intestinal phyla, the Bacteroidetes and Firmicutes, are linked to obesity and metabolic disorders, both in humans and mice (33, 34). There has also been a substantial increase in the number of reports showing the relationship between the microbiota composition and the incidence of chronic inflammatory disease, including allergic conditions and autoimmune disorders (15--22). Furthermore, transplantation experiments in which the microbiota of diseased animals is grafted into healthy recipients have demonstrated the transfer of several disease phenotypes. These include obesity, metabolic disorders, and chronic colitis (35--37), all of which have complex etiologies affected by host genetic and environmental factors. Therefore, a better understanding of the functional properties of individual members of the microbiota is increasingly relevant to the treatment of complex chronic diseases.
Factors That Affect Community Membership of Microbiota
Diet
Diet is one of the most important factors shaping microbial diversity in the gut. Because members of the microbiota have their own substrate preference and there is intense competition for resources, alterations in the components of the diet, particularly the type and quantity of fat and polysaccharides, result in changes in community composition and function of the microbiota. Mouse studies revealed that feeding mice with a high-fat and high-carbohydrate diet (Western diet) resulted in an increase in the number of bacteria of the Firmicutes phylum and a decrease in that of bacteria of the Bacteroidetes phylum (38, 39). This increase in the number of Firmicutes was mainly due to the proliferation of the Erysipelotrichaceae family (38, 39). The abundance of this family of bacteria immediately diminished when the diet was changed to a diet low in fat and rich in plant polysaccharides. The decrease in the proportion of Firmicutes after a low-calorie diet was similarly observed in humans (40). Another human study of 19 obese volunteers showed that a decreased carbohydrate intake led to a decrease in the number of bacteria within a specific group of Firmicutes that included Roseburia spp. and Eubacterium rectale (41). Diet also influences fecal community enterotypes in human subjects (42). Individuals with long-term diets rich in protein and animal fat had an enterotype dominated by Bacteroides, whereas those on high carbohydrate diets had a prevalence of Prevotella. Although change in diet resulted in a rapid change in microbiota, this was not sufficient to shift the enterotype during a 10-day course. A similar distribution of fecal enterotypes was observed in a comparison of European and African children, who have diets rich in protein/animal fat and carbohydrates, respectively [← AU: should use “and” rather than “versus” for the “respective” relationship. OK as changed?** OK] (43). Whether enterotypes associated with long-term diets can be reversed by changes in the diet remains to be determined.
Changes in the diet and accompanied alterations in community membership of the microbiota, whether chronic or short-term, lead to changes in the gene expression profiles of the microbiota. For instance, alterations in availability of diet polysaccharides result in changes in the expression of genes for carbohydrate active enzymes (CAZymes), including glycoside hydrolases and polysaccharide lyases, in members of the microbiota, such as Bacteroides thetaiotaomicron (44). Seaweeds are major components of the Japanese diet; Bacteroides plebeius residing in the gut of Japanese individuals acquired the genes of enzymes that can metabolize the [**AU: “ph” is usually the British spelling. OK to change? → OK] sulfated polysaccharide porphyran of marine algae through the horizontal transfer from marine bacteria naturally colonizing dietary seaweeds (45). These changes in gene expression of constituents of the microbiota likely ensure that the microbial community can adapt to dietary changes and maximize energy harvest, while contributing to host fitness.
In some cases, changes in the diet and subsequent alterations in the microbiome may exert a detrimental effect on host physiology. Indeed, in individuals having a high-fat and high-carbohydrate diet, the microbiota is more heavily enriched with bacteria of the phylum Firmicutes and less with those of the phylum Bacteroidetes, and this condition may be more efficient at extracting energy from a given diet compared with the microbiota of lean individuals (33). This suggests a positive-feedback mechanism, in which an obesity-inducing diet can change the composition of the gut microbiota, which results in a shape of the microbiome more capable of extracting energy from the diet, thereby helping perpetuate obesity. It has been postulated that changes in the diet and associated changes in the gut microbiota may also lead to immune disorders (46). In fact, allergies and asthma are almost completely nonexistent in certain rural African communities, where people eat diets low in protein and animal fat and high in plant polysaccharides. Actinobacteria and Bacteroidetes are more abundant in African (Burkina Faso) than in EU children's microbiota, whereas Firmicutes and Proteobacteria are more abundant in EU than in African children (43). Importantly, the microbiome of African children exhibits a higher microbial richness and biodiversity than that of EU children. The African samples, but not the EU sample's, contain two bacterial species, Prevottela and Xylanibacter (43). These findings are consistent with the above-described diet/enterotype concept (42) and suggest that the African microbiomes are dominated by the Prevotella enterotype driven by the lowfat/high-fiber diet. Prevottela and Xylanibacter have enzymes necessary for the hydrolysis of cellulose and xylan and contribute to the production of high amounts of short-chain fatty acids (SCFAs) (43). As discussed below, SCFAs contribute to the maintenance of immune homeostasis in the intestine. Therefore, changes in the diet and gut microbial ecology are likely to affect the metabolic predisposition and immune status of the host.
SCFA: short-chain fatty acid
Inflammation
Alterations in community membership can also be induced by inflammation. In mice, enteric pathogens such as Citrobacter rodentium and Salmonella enterica subspecies 1 serovar Typhimurium actively induce intestinal inflammation, which then alters the composition of indigenous microbiota, reducing the number of strict anaerobes such as the Firmicutes and allowing proteobacteria to proliferate (47, 48). Intestinal inflammation caused by either a chemical inducer, such as dextran sulfate sodium (DSS), or a genetic deficiency, such as Il10 deficiency, can also alter the composition of the intestinal microbiota, resulting in a reduction in both the total number of resident intestinal bacteria and bacterial diversity (47). Studies have shown a change in composition of the microbiota in T-bet−/− Rag2−/− ulcerative colitis (TRUC) mice, which have a T-bet deficiency in the innate immune system and develop spontaneous colitis with high penetrance on a BALB/c background (36, 49). TRUC colitis is attributed to the hyperproduction of tumor necrosis factor (TNF)-α by dendritic cells (DCs), because T-bet is a negative regulator of TNF-α transcription (36). The colitis in TRUC mice is accompanied by a considerable alteration in microbial composition. TRUC colitis can be transmitted to wild-type mice when they are cross-fostered or cohoused with TRUC mice (36). Detailed analysis of altered microbiota has revealed a lower proportional representation of Firmicutes, including Clostridium spp., and the constitutive presence of colitogenic bacteria, such as Proteus mirabilis and Klebsiella pneumoniae (50), which are otherwise transient allochthonous bacteria and constitute very minor, if any, components of the microbiota. Interestingly, colitis in TRUC mice is improved by the consumption of a fermented milk product containing a probiotic strain, Bifidobacterium lactis (51). Consumption of B. lactis results in a change in the structure of the microbiota, namely, an increase in the number of lactate-consuming Desulfovibrio spp. and the butyrate-producing Anaerostipes caccae subgroup and Eubacteriumhallii, accompanied by a decrease in cecal pH and increases in acetic, propionic, and butyric acid levels. These conditions provide a nonpermissive environment for the colitogenic Proteus and Klebsiella in TRUC mice. These results suggest that inflammation mediated by the overproduction of inflammatory cytokines, such as TNF-α, leads to an increase in pH and a decrease in the amount of SCFAs in the intestinal lumen, resulting in an altered microbial community, including outgrowth of potentially pathogenic bacteria. Collectively, much like the positive-feedback mechanism in diet-mediated obesity described above, intestinal inflammation caused by infection or genetic predisposition can render the shape of the microbiome to be more prone to induce inflammation. This vicious cycle of dysbiosis and inflammation may be operative in IBD patients.
DC: dendritic cell
Probiotics: live microorganisms that, when administered in adequate amounts, confer a health benefit to the host
IBD patients exhibit substantial changes in the composition of the microbiota. A decrease in the numbers of bifidobacteria and lactobacilli in IBD patients has been known for many years (52). Overgrowth of Escherichia coli, the so-called adherent-invasive E. coli (AIEC) in particular, is frequently observed in human IBD patients and is considered to lead to the aggravation of disease symptoms. AIECs interact intimately with the inflamed ileal mucosa and even invade intestinal epithelial cells (IECs) (53, 54). Recent studies using high-throughput sequence analysis of 16S rRNA genes have shown that there is a decrease in Bacteroidetes and Firmicutes frequencies and an increase in Actinobacteria and Proteobacteria frequencies in IBD patients. In particular, it was shown that the family Lachnospiraceae, which comprises the Clostridium clusters IV and XIVa [also known as Clostridium leptum and coccoides groups (55)], was significantly less abundant in IBD patients than in healthy subjects (13). Other reports have provided data showing a reduction in the number of bacteria within the Clostridium cluster IV, particularly the species Faecalibacterium prausnitzii, in the gut of IBD patients (56, 57) and a correlation between low counts of F. prausnitzii and postsurgical recurrence (58).
IEC: intestinal epithelial cell
Importantly, individuals with chronic inflammation have a lower bacterial diversity of the microbiota than healthy individuals. The MetaHIT consortium reported that IBD patients harbored, on average, 25% fewer genes than individuals not suffering from IBD (9). Although it still remains unclear whether the decrease in microbial membership is a causative factor or a consequence of chronic inflammation in IBD patients, maintaining the diversity of the gut microbial community is likely a prerequisite for a stable and healthy gastrointestinal tract.
Host genotype
Host genotype can intrinsically affect the composition of the microbiota. For instance, genetically obese mice, such as ob/ob mice (leptin gene deficiency), have an altered microbiota with increased Firmicutes and decreased Bacteroidetes frequencies (33). Importantly, gut microbiota transferred from ob/ob mice to wild-type GF mice can induce obesity, presumably owing to the fact that obesity-associated microbes can extract more energy from the diet, which suggests that a change in the microbiota by [**AU: italics for gene name, as above? →] leptin gene deficiency may precede the obesity phenotype of ob/ob mice (33). Mice deficient in Toll-like receptor 5 (Tlr5−/−) similarly exhibit a change in the shape of the microbiota (35). Tlr5−/− mice display hyperphagia and signs of metabolic syndrome, including insulin resistance and increased adiposity. Wild-type mice inoculated with Tlr5−/− microbiota phenocopy the Tlr5−/− mice, displaying hyperphagia and obesity, suggesting that the Tlr5−/− phenotype is primarily due to the changes in the microbiota. NLRP6, a sensor of endogenous or exogenous stress, induces the formation of a multimolecular inflammasome complex upon binding of unknown ligand, leading to activation of caspase-1 and cleavage of pro-IL-1β and pro-IL-18 (37). In IECs, the inflammasome-mediated constitutive production of IL-18 is required for the maintenance of epithelial cell barrier function and regeneration. NLRP6−/− mice exhibit reduced IL-18 levels in IECs and are highly susceptible to DSS colitis (37). Importantly, NLRP6−/− mice show quantitative and qualitative changes in the microbiota, including increased representation of members of Prevotellaceae and TM7, and reductions in members of the genus Lactobacillus in the Firmicutes phylum. The transmission of this microbiota community confers similar susceptibility to DSS colitis onto wild-type mice. As described above, T-bet−/− Rag2−/− TRUC mice also have an abnormal microbial composition, and TRUC colitis is transmissible to wild-type mice (36). These reports indicate that obesitogenic or colitogenic microbiota can arise in the host depending on the genotype.
Nonobese diabetic [← AU: see note below re nucleotide oligomerization domain** OK for the deletion of ‘NOD’.] mice lacking Myd88, an adaptor molecule for TLRs and IL-1 receptor (IL-1R), did not develop type 1 diabetes when reared in specific pathogen-free (SPF) conditions, but littermate Myd88-sufficient mice exhibited diabetes under the same conditions (19). However, when rendered GF, Myd88−/− nonobese diabetic mice developed severe diabetes. Therefore, the attenuated phenotype of SPF Myd88−/− nonobese diabetic mice is not simply due to a defect in IL-1R or TLR signaling, but indicates a key role for the microbiota. Further analysis revealed that Myd88-deficient nonobese diabetic mice exhibit changes in the composition of the distal gut microbiota: a significantly lower Firmicutes/Bacteroidetes ratio compared with that of Myd88-sufficient control mice (19). Therefore, MyD88-dependent signaling affects the composition of the gut microbiota, and thereby affects the nonobese diabetic phenotype.
TLR: Toll-like receptor
The lack of activation-induced cytidine deaminase (AID), which results in defective class switch recombination and thereby lack of IgA-producing plasma cells in the intestine, leads to the excessive proliferation of anaerobic bacteria in the small intestine, particularly the segmented filamentous bacteria (SFB), accompanied by hyperplasia of ILFs (59). The addition of IgA prevented the aberrant SFB expansion and ILF hyperplasia in the mutant mice. Antimicrobial peptides produced by IECs also have important roles in controlling the growth of components of the microbiota. Thus, α-defensin deficiency was associated with decreased Bacteroidetes and increased Firmicutes frequencies, whereas the transgenic expression of human defensin 5 (HD5) in mice led to the reverse shift (60). Transgenic expression of HD5 also led to the loss of SFB, with a consequent decrease in the number of Th17 cells in the lamina propria (LP) of the small intestine (60) (discussed below).
LP: lamina propria
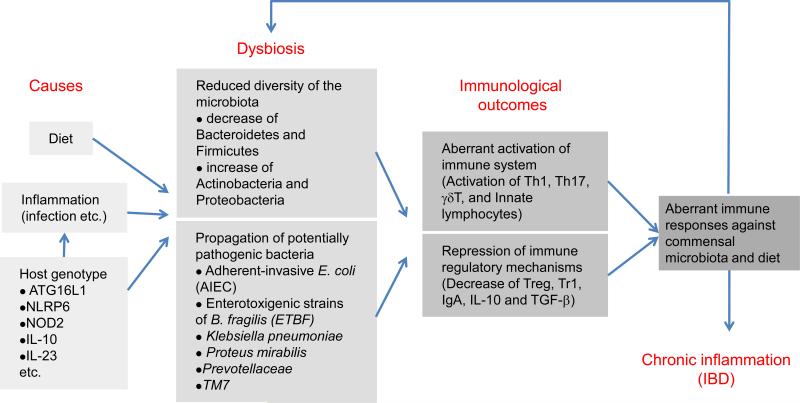
Collectively, many diseases, including metabolic diseases and chronic inflammatory conditions, may not be simply associated with host genetics, but may be mediated indirectly by the change in the microbiota (Figure 1). Recent genome-wide association studies (GWAS) have revealed more than 100 genetic loci that have significant association with IBD (61, 62). Several of these genes likely have a primary role in shaping the gut microbial community, which then affects IBD pathology.
Figure 1.
Causes and pathological outcomes of dysbiosis. The composition of the gut microbiota is readily affected by diet, inflammation (including infection by pathogens), and host genotypes. An unfavorable alteration of the community structure of the gut microbiota is termed dysbiosis, which includes an outgrowth of potential pathogenic bacteria (pathobionts) and a reduced diversity of the community structure of the microbiota. Dysbiosis is associated with the increased predisposition to immune system activation, which leads to aberrant immune responses against commensal microbiota and diet. The dysregulated activation of the immune system leads to the worsening of the dysfunction of the microbiota, which may result in sustained inflammation in the intestine (i.e., IBD) and other organs.
MICROBIAL SIGNALING THROUGH THE HOST INNATE IMMUNE SYSTEM
The gut mucosa has evolved multiple layers of protection to prevent the translocation of pathogenic as well as indigenous microbes. The intestinal epithelium is covered with layers of mucus, which is composed of mucin glycoproteins synthesized and secreted by goblet cells. The inner layer of the mucus gel in the colon is densely packed and firmly attached to the epithelium and creates a strict barrier that prevents large particles, including most bacteria, from directly contacting the epithelial cells (63). The intestinal epithelium is tightly sealed by [**AU: “TJ” only used a few times, and not consistently, and AJ not used again. OK if we just spell out instead? → OK] tight junctions and subjacent adherens junctions, which play critical roles in the prevention of microbe invasion (64). The small intestinal epithelium contains Paneth cells at the base of the crypts, which contribute to innate immunity by secreting a diverse repertoire of antimicrobial proteins. Beneath the epithelium, the LP contains DCs, which extend their dendrites between epithelial cells to continuously monitor the gut lumen content and activate LP lymphocytes. These in toto constitute the firewall that prevents the systemic penetration and spread of microbes (7, 65).
In the past, immune responses to commensal bacteria were considered to be completely prevented because of the sequestration of microbiota by physical barrier systems. Recent results have suggested, however, that constitutive and physiological inflammation induced by commensal microbes through pattern-recognition receptors (PRRs) is operative and is required for epithelial barrier functions and normal host-commensal homeostasis.
Pattern-recognition receptors (PRRs): receptors that recognize microbe- associated molecular patterns to initiate innate immune responses
Constitutive Activation of TLRs by Microbiota
TLRs expressed by hematopoietic and nonhematopoietic cells are involved in the recognition of the microbiota. When mice kept in SPF conditions are treated with broad-spectrum antibiotics, they become highly susceptible to DSS-induced intestinal inflammation (24). This is, at least in part, due to reduced constitutive TLR signaling in response to the microbiota, as oral administration of lipopolysaccharide (LPS, a TLR4 ligand) or lipoteichoic acid (a TLR2 ligand) restores resistance of antibiotic-treated mice to DSS-associated injury of the colonic epithelium (24). Mice lacking either Tlr2, Tlr4, Tlr9, or Myd88 are highly susceptible to DSS colitis (24, 66). Depending on the rearing condition, mice lacking Tlr5 develop spontaneous colitis, and bacterial translocation to the spleen and liver occurs. [← AU: which organ depends on environment? “or” OK then?**] OK (67). The absence of MyD88 signaling in IECs is associated with a decreased production of cytoprotective factors such as IL-6, KC-1, and heat shock proteins in IECs (24). TLR2 signaling plays critical roles in the maintenance of the functional tight junction barrier integrity of the intestinal epithelial layer (68). In addition, TLR2 signaling is required for the stimulation of goblet cells to produce trefoil factor 3 [← AU: acronym not used again, so deleted here** OK], which confers resistance to IECs against inflammation-induced damage (69). Accordingly, Tlr2−/− mice are susceptible to Citrobacter rodentium–induced colitis and exhibit severe mucosal ulcerations and crypt destruction (70). These findings all support the concept that TLRs play an important role in maintaining the functional integrity of mucosal barriers.
IECs produce antimicrobial proteins, including α-defensins, β-defensins, cathelicidins, C-type lectins, and lipocalin2. Most of these proteins can kill bacteria directly through enzymatic attack of the bacterial cell wall or by disrupting the bacterial inner membrane. The expression of the C-type lectin RegIIIγ is induced in the mouse small intestine, particularly in Paneth cells, by [**AU: per Webster's, we lowercase gram. “Gram's stain” would be cap, but not “gram” in this context. I have changed throughout, w/o tracking → OK] gram-negative commensal bacteria, such as B. thetaiotaomicron, but it is downregulated by the gram-positive probiotic bacterium Bifidobacterium longum (5, 71). RegIIIγ, whose induction depends on MyD88-dependent TLR activation, has a critical role in host defense against pathogenic gram-positive bacteria, including vancomycin-resistant Enterococcus (72). MyD88-dependent signaling is also required for the commensal-mediated expressions of RegIIIβ, CRP-ductin, and resistin-like molecule β (RELMβ) in small intestinal IECs (6). In addition, the production of antimicrobial proteins is indirectly regulated by TLRs via cytokines, such as IL-22 (73). Systemically injected flagellin stimulates TLR5 on hematopoietic cells, which leads to an increase in the concentration of IL-22 in the intestine. IL-22 then acts on IECs to produce RegIIIγ and lipocalin2, resulting in resistance to infection by vancomycin-resistant Enterococcus[← AU: abbreviation only used in these two places, so spelled out instead** OK] (73).
Microbiota-induced constitutive production of IgA is also dependent on TLR signaling. GF mice and mice deficient in TLR signaling have little luminal IgA. Microbial TLR ligands, such as LPS, can substitute for the CD40 ligand (CD40L) and act directly on B cells along with transforming growth factor-β (TGF-β) to initiate IgA class switching (74). TLR ligands stimulate DCs to express inducible nitric oxide synthase (iNOS) (75). The gaseous nitric oxide produced by iNOS then induces the expressions of B cell activating factor (BAFF; also known as BLyS) and a proliferation-inducing ligand (APRIL), which are functionally related to CD40L and can promote IgA class switching in a CD40L-independent manner. TLR5 signaling can stimulate CD11bhighCD11chigh DCs to express Raldh2, which catalyzes vitamin A to retinoic acid (76). Retinoic acid can act on B cells, causing them to differentiate into IgA-producing plasma cells. Thus, multiple TLR-dependent pathways are involved in the production of secretory IgA.
Autophagy is an evolutionarily conserved degradation system that orchestrates the engulfment of cytoplasmic proteins into double-membraned vesicles (autophagosomes) and coordinates the fusion of the autophagosomes with lysosomes. Autophagy plays a housekeeping role in removing misfolded or aggregated proteins and in clearing damaged organelles, such as mitochondria (77). Recent GWAS have revealed a strong association between the ATG16 autophagy-related 16-like 1 (ATG16L1) gene and Crohn's disease (78, 79). [**AU: otherwise “it” used the mice → OK] Using hypomorphic Atg16L1HM mice, in which the ATG16L1 expression level is low, Cadwell et al. (80) found that autophagy plays a key role in the granule exocytosis pathway in Paneth cells. Although there is no spontaneous inflammation in Atg16l1HM mice, these mice are more susceptible to DSS colitis when infected with a norovirus (81). Crohn's disease patients homozygous for the ATG16L1 risk allele have similar abnormalities in the secretion of granule contents from Paneth cells (80). TLRs can promote the induction of autophagy (82). TLR stimulation induces the interaction of Beclin-1, a key inducer of autophagosome formation, with MyD88 and TRIF (83). This interaction, in turn, dissociates Beclin-1 from antiapoptotic proteins Bcl-2 and Bcl-XL, thus leading to the induction of autophagy. Of interest, the autophagy pathway acts upstream of inflammasome activation. Atg16l1-deficient macrophages exhibit heightened inflammasome activation and IL-1β production when stimulated with LPS (84). Therefore, constitutive TLR signaling is likely required for facilitating the induction of autophagy, which regulates inflammasome activation as well as Paneth cell exocytosis pathways.
Regulation of TLR Signaling
We described above the importance of the constitutive activation of TLRs by commensal microbiota under normal steady-state conditions for the maintenance of IEC barrier integrity, production of antimicrobial proteins and IgA, and induction of autophagy. However, if the constitutive TLR signaling is not properly regulated, excessive and harmful inflammation may occur. In particular, inflammatory cytokines induced by TLRs, such as interferon (IFN)-γ and TNF-α, can act as precipitating factors for IBD by modifying tight junction function in IECs and increasing epithelial barrier leakage (64, 85). To avoid unnecessary inflammation and cytokine production, several molecules function as negative regulators of TLR signaling pathways. An example is A20, which is quickly induced following TLR stimulation and terminates TLR-dependent responses by removing ubiquitin moieties from the signaling molecule TNF-receptor-associated factor 6 [← AU: abbrev. not used again. OK to delete?** NO, most people recognize TRAF6 rather than the spelled out definition] (86). Tnfaip3 (the gene encoding A20)-deficient mice exhibit severe inflammation of multiple organs including the intestine. Spontaneous inflammation in Tnfaip3−/− mice is rescued in mice that are also deficient in Myd88. In addition, the depletion of intestinal bacteria by treatment with broad-spectrum antibiotics diminishes the inflammation in Tnfaip3−/− mice (87). Thus, the anti-inflammatory feedback loop becomes dispensable if microbiota-induced TLR signals are eliminated. A similar example is provided by the single immunoglobulin IL-1 receptor-related molecule (SIGIRR), which contains an intracellular Toll/interleukin-1 receptor (TIR) domain (88). SIGIRR is expressed on DCs and IECs and inhibits IL-1 and TLR signaling by acting as a decoy to sequester downstream signaling components. Mice lacking Sigirr are highly susceptible to intestinal inflammation induced by DSS, and IECs from Sigirr-deficient mice show increased cytokine production in response to commensal microbiota (89).
TLR-mediated responses are also counteracted by inhibitory cytokines, particularly IL-10. IL-10 is produced by Treg cells, DCs, and macrophages in the intestine in the steady state. Il10-deficient mice, or mice with myeloid cell–specific deficiency for STAT3, an essential signaling molecule acting downstream of the IL-10 receptor, spontaneously develop intestinal inflammation (90, 91). In these mutant mice, LP macrophages are in the state of constitutive and aberrant activation, and the blockade of TLR signaling by introducing a deficiency in TLR4 or MyD88 abrogates this intestinal inflammation (92, 93), indicating that IL-10 acts on myeloid cells and induces STAT3 activation to suppress excessive inflammation during TLR responses. In addition to A20, SIGIRR, and IL-10, TLR signals are regulated by many other molecules, including IRAK-M, suppressor of cytokine signaling-1 (SOCS1), and ST2 (94). These proteins can restrict the duration and/or intensity of TLR signals and modulate the cellular outcome of TLR signaling, thereby helping to determine whether TLR signals induce homeostatic or inflammatory responses.
TLR signaling in the mucosa exerts its appropriate function only when the integrity of the epithelial cell barrier is maintained. This was demonstrated by analyses of mice with IEC-specific ablation of NEMO [also called IκB kinase-γ (IKKγ)], or IKKα and IKKβ (95, 96). These conditional knockout mice spontaneously develop severe chronic intestinal inflammation. The defect in NF-κB activation leads to apoptosis of IECs, impaired expression of antimicrobial proteins, and translocation of bacteria into the mucosa. Intestinal inflammation is prevented by crossing these mice with Myd88-deficient mice. Similarly, mice with IEC-specific knockout of Fas-associated death domain (FADD) spontaneously develop IEC necrosis and severe erosive colitis; Tnfα-deficiency, Myd88-deficiency, or elimination of the microbiota prevent the colon inflammation (97). These findings support a model in which chronic activation of TLR by continuous bacterial translocation triggers the expression of proinflammatory cytokines, such as TNF-α and IL-1β, which causes further destruction of the epithelium and results in chronic intestinal inflammation. The breakdown of the epithelial cell barrier, along with persistent and aberrant TLR activation, may thus be one of the major etiologies of IBD.
Activation of NOD Receptors by Microbiota
Nucleotide oligomerization domain 1 (NOD1) and NOD2 [**AU: above “NOD” has been used for “nonobese mice.” I suggest spelling out the mice so that abbrev. can be used for this domain name.** OK] receptors constitute another family of PRRs that are constitutively activated by microbiota. NOD2 is a sensor of peptidoglycan (PGN) through the recognition of muramyl dipeptide (MDP), which is common to both gram-positive and gram-negative bacteria (98). Upon recognition of MDP, NOD2 forms oligomers and recruits receptor-interacting protein 2 (RIP2). RIP2 undergoes polyubiquitination and mediates the activation of NF-κB and induction of inflammatory cytokines. NOD2 is expressed in monocytes and Paneth cells and is required for the expression of antimicrobial proteins in Paneth cells (99). Indeed, Nod2−/− mice are susceptible to oral infection with Listeria monocytogenes and show poor expression of antimicrobial α-defensins, such as defensin-α (Defa) 4 and Defa-related sequence (Defa-rs) 10 [← AU: are these abbreviations meant to be related? YES, that is ok Is Defcr “ α defensin cryptdin”?** where is this mentioned??], in Paneth cells. Nod2−/− mice also have an increased amount of commensal bacteria as well as a reduced ability to remove newly colonizing bacteria, suggesting that NOD2 also plays an important role in maintaining the balance of commensal microbiota (100).
[**AU: italics needed for gene names? Please confirm throughout manuscript →]
NOD2 gene mutations are associated with an increased susceptibility to Crohn's disease in Western (but not Asian) populations (101, 102). Three polymorphisms are associated with Crohn's disease: R702W, G908R, and the frame-shift deletion mutation L1007fs (98). Although in vitro assays suggest that these NOD2 mutations result in a loss of function (103), whether these mutations predispose an individual to Crohn's disease in vivo through a loss or a gain of function remains an issue. To explain the discrepancy between the loss-of-function phenotype of patient monocytes and the reality of inflammation developing in the intestine, several hypotheses have been proposed (104, 105). The most favored explanation is that NOD2 variants cause a defect in mucosal barrier function, including a defect in the production of α-defensins by Paneth cells, which leads to an enhancement of bacterial translocation and subsequently induction of innate and adaptive immune responses. In any case, because Nod2-deficient mice have not been shown to develop spontaneous colitis (99), dysregulation of the NOD2 pathway alone is insufficient, and additional factors including environment and dysbiosis of the microbiota are required to induce Crohn's disease.
NOD1 detects diaminopimelate-containing PGN, which is found mostly in gram-negative bacteria (98). Similarly to NOD2, NOD1 regulates the RIP2-dependent NF-κB pathway. In contrast to the restricted cellular expression of NOD2, NOD1 is expressed more broadly, including in IECs. Therefore, NOD1 and NOD2 are postulated to play different roles in the intestinal immune system. In vitro, NOD1 is required for sensing Shigella flexneri and E. coli (106). In vivo, NOD1 plays a critical role in the host defense against infection by Helicobacter pylori (107). H. pylori–induced production of β-defensins is abolished in Nod1−/− mice, indicating that NOD1 plays a critical role in the production of β-defensins.
Constitutive NOD1 signaling by PGN from commensal gram-negative bacteria, such as the genus Bacteroides, is required for the induction and maintenance of ILFs (4). PGN recognition by NOD1 in IECs results in the production of CCL20 and recruitment of CCR6-expressing LTi cells that are required for the formation of ILFs. In the absence of NOD1 and/or ILFs, the total bacterial populations markedly expand and the composition of the intestinal bacterial community is profoundly altered: Clostridiales, Bacteroides, and Enterobacteriaceae [← AU: I removed the partial italics in this family name. OK?** OK] markedly expand, whereas Lactobacillaceae are reduced. Furthermore, NOD1 activation by intestinal microbiota under basal conditions is required for the systemic priming of the innate immune system, particularly for the activation of circulating neutrophils (108). Although the association of the [**AU: ital? → OK] NOD1 gene with IBD remains controversial, it is clear that NOD1 recognizes intestinal commensal and pathogenic bacteria and plays critical roles in the regulation, activation, and organization of both local and systemic innate and adaptive immune responses.
Signaling by Microbiota-Derived Free Fatty Acids
Emerging technologies, including metagenomics and metabolomics, are now being applied to study the role of microbiota in the biology of the host organism. Gene diversity in the microbiome provides various enzymes and metabolic pathways to break down polysaccharides, oligosaccharides, unabsorbed sugars, and alcohols, which are nondigestible to the host. One of the metabolic endpoints is the generation of SCFAs, particularly acetate (C2), propionate (C3), and butyrate (C4) (46). Bacteroidetes bacteria produce high levels of acetate and propionate, whereas Firmicutes bacteria produce high amounts of butyrate (109). Acetate and propionate are found in portal blood and are eventually metabolized by the liver or peripheral tissues, whereas butyrate is a major energy source of the colonocytes.
Metabolomics: the quantitative measurement of the dynamic multiparametric metabolic responses of multicellular systems to pathophysiological stimuli by nuclear magnetic resonance (NMR) spectroscopy or mass spectrometry (MS) [←AU: the stimuli are by NMR and MS? or the measurement is by NMR and MS?** the measurement is by NMR and MS]
Evidence suggests that butyrate metabolism is impaired in patients with ulcerative colitis and that topical treatment with sodium butyrate or butyrate enemas are effective for ulcerative colitis (110, 111). Butyrate treatment enhances the rate of mucin synthesis and restores the mucus lining by unknown mechanisms. Butyrate is transported into IECs via monocarboxylate transporters (such as MCT1 and SLC5A8) (112, 113) and also acts as an extracellular signaling molecule by activating G protein–coupled receptors (GPCRs), including GPR109A, which is expressed on the apical surface of IECs (114). GPR109A signaling in the colon suppresses NF-κB signaling and reduces production of TNF-α, IL-6, and IL-1β.
Other GPCRs, including GPR40, GPR41, GPR43, GPR84, and GPR120, can function as receptors for free fatty acids (FAs). GPR41 and GPR43 are responsive to SCFAs (2-6 carbons, C2–C6) [← AU: elsewhere abbreviated as SCFAs. Possible to apply abbrev. here? ** OK] (115), whereas GPR40, GPR84 and GPR120 recognize ligands with longer (>C6) acyl chains (116--118). GPR120 is highly expressed in the human and mouse intestinal tract and mouse enteroendocrine cells. Oral administration, or direct administration to the colon, of free FAs increases circulating glucagon-like peptide-1 and insulin levels in mice, and this effect of the free FAs is likely to be mediated by GPR120 (117). GPR120 is expressed not only in IECs, but also in numerous other cells, including adipocytes, DCs, and macrophages (119). In these cells, GPR120 can act as a functional receptor for ω-3 FAs, such as α-linolenic acid (C18), docosahexaenoic acid (C22, DHA), and eicosapentaenoic acid (C20, EPA) in fish oils and some plant oils (119). The signaling through GPR120 by DHA and EPA mediates anti-inflammatory effects to inhibit both TLR and TNF-α signaling pathways in macrophages. By preventing macrophage-induced chronic, low-grade tissue inflammation, GPR120 mediates anti-inflammatory and insulin-sensitizing effects in vivo (119).
GPR41 is expressed by a subset of enteroendocrine cells in the gut epithelium (120). GPR41 regulates the expression of peptide YY, an enteroendocrine cell–derived hormone, which inhibits gut motility, reduces the intestinal transit of food, and thereby enhances the harvest of energy from the diet (120). GPR43 is expressed predominantly in immune cells, particularly in neutrophils and eosinophils, and transmits microbial-derived immune modulatory signals (118, 121). GF mice are more susceptible to DSS colitis than mice colonized with normal microbiota. Treatment of GF mice with acetate in the drinking water activates GPR43 and markedly improves disease indices (121). Gpr43−/− neutrophils have an intrinsic hyperreactive phenotype, including hyperproduction of reactive oxygen species and high chemotactic activity. Acetate also has an antiapoptotic effect in colonic epithelium (122). Bifidobacterium longum and B. adrescentis possess glucose transporter(s) and can produce acetate from glucose in the proximal colon. In the distal colon, in contrast, only fructose is available, and thus, only B. longum, with its fructose transporters, can produce acetate. B. adrescentis lacks the fructose transporters and thus fails to produce sufficient acetate to prevent epithelial apoptosis induced by the pathogenic bacterium E. coli O157 (122). Collectively, FA-GPR signaling is one of the important molecular pathways activated by microbiota and diet and regulates immune and inflammatory responses that maintain the mucosal barrier.
INDUCTION AND FUNCTION OF INNATE AND ADAPTIVE INTESTINAL LYMPHOCYTES
The intestinal mucosa has a unique immune system composed of multiple innate and adaptive lymphocyte populations that are present in the steady state. Several studies have indicated that individual members of the commensal microbiota regulate the development and accumulation of these innate and adaptive lymphocytes through distinct mechanisms.
IgA-Producing Cells
Plasma cells that produce secretory IgA are, arguably, the best characterized among the numerous lymphocyte populations in the intestinal mucosa. IgA plays an important role in shaping gut microbial ecology, as exemplified by AID-deficient mice (59), as described above. In addition, IgA can affect gene expression by the microbiota. For instance, B. thetaiotaomicron is otherwise commensal to the host, but in the absence of IgA it elicits a robust innate immune response, including the induction of iNOS, and then adapts by inducing bacterial genes involved in nitric oxide metabolism (25). Therefore, mucosal IgA plays an essential role in determining the composition and nature of the microbiota and is required for maintaining the mutualistic benefit of bacterial-host interaction.
IgA-producing lymphocytes are generated by mechanisms that are both dependent and independent of T cells (74). IgA-positive B cells are generated in a T cell– and CD40L-dependent manner in the germinal centers of organized follicular structures of PPs [← AU: for Peyer's patches? OK to introduce abbrev. above?** OK], where antigens of commensal bacteria are taken up by M cells. IgA+ B cells then exit PPs and migrate into the draining mesenteric lymph nodes (MLNs), where they differentiate into IgA-secreting plasma cells, which then populate the intestinal LP. In contrast to this [**AU: we prefer T cell, w/o hyphen. I have changed throughout. → OK] T cell– and PP-dependent class switch mechanism, a large proportion of the intestinal IgA is induced locally in the intestinal LP in a T cell–independent manner (123). Furthermore, much attention has been focused on ILFs in the LP as the site where the T cell–independent induction of IgA class switching of B cells takes place (124). Indeed, Tcrb−/−Tcrd−/− mice cannot generate IgA+ B cells within PP follicles, but they have an almost normal level of luminal IgA. In these mice, a considerable number of IgA+ plasma cells can be observed in the LP of the small intestine, particularly in ILFs, but not in PPs (124).
The formation and maturation of ILFs are dependent on the presence of commensal bacteria, as is evident from the analysis of GF mice (4). In this context, GF mice also exhibit severe reductions in fecal IgA levels and in the numbers of LP IgA+ cells (3, 125). In gnotobiotic mice, individual bacterial species, such as SFB or clostridia, specifically stimulate the development of IgA-producing cells (126--128) (Figure 2). SFB adhere tightly to the follicle-associated epithelium of PPs and the absorptive epithelium of the villi in the ileum (129), and thereby induce IgA-producing cells in the ileum (126--128). In contrast, clostridia preferentially colonize in the cecum and proximal colon where their abundance correlates with an increase in the number of IgA-producing plasma cells (128) (Figure 3). Considering the fact that TLR and NOD1 signaling are indispensable for IgA production in the intestine, SFB and clostridia may provide TLR ligands that can induce the accumulation of IgA-producing cells in the ileum and colon, but this possibility has not yet been investigated. Further studies are required to elucidate the molecular basis for the induction of intestinal IgA-producing cells by the components of the microbiota.
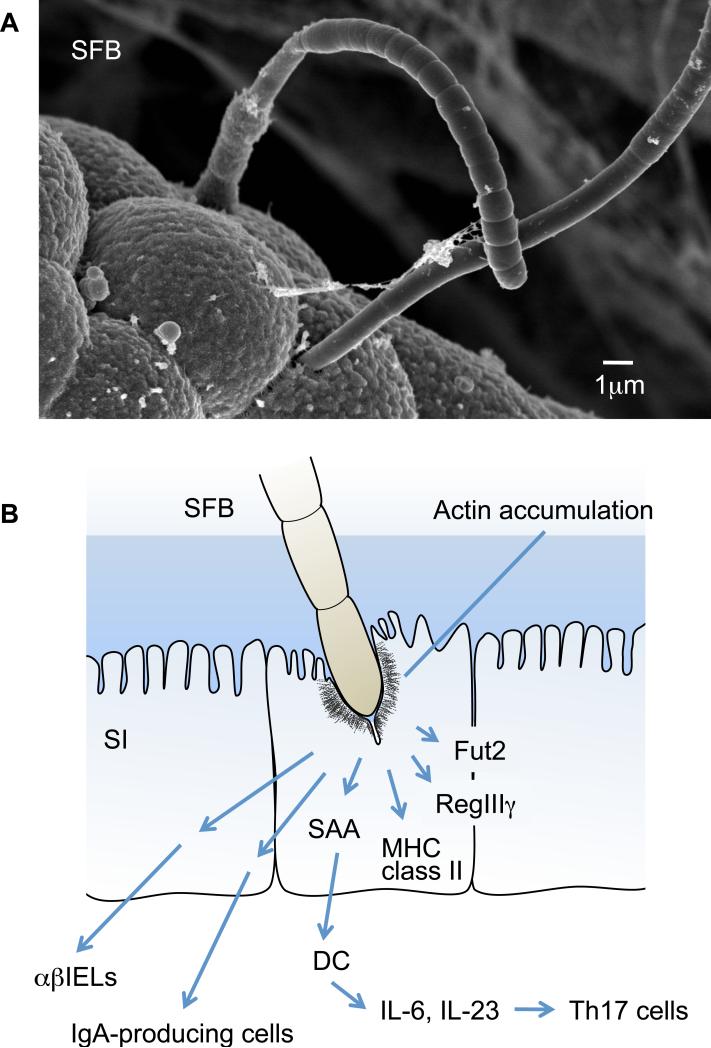
Figure 2.
Morphology (a) and immunological effects (b) of segmented filamentous bacteria (SFB). SFB are also known as Arthromitus. They are yet-to-be-cultured, gram-positive, spore-forming, and commonly considered nonpathogenic and host-specific members of the gut microbiota in numerous species including mammals, birds, fish, and insects (although it is currently unclear whether SFB are present in the human intestine). (a) Scanning electron microscopy shows that SFB colonize mainly the small intestine and exhibit a characteristic long filamentous morphology comprising multiple segments with well-defined septa. Each bacterium is likely to originate from a single cell that tightly adheres to and even embeds itself among the microvilli on the epithelial cell surface. (b) The attachment of SFB induces morphological changes in the epithelial cell, such as the accumulation of filamentous actin around the attachment site. SFB activate IECs to induce expression of MHC class II molecules and fucosyltransferase 2 (Fut2). Fut2 fucosylates host glycoproteins on the IECs, such as asialoGM1 glycolipids. SFB are potent stimuli of the IgA response and the recruitment of TCR αβ+ IELs in the small intestine. Furthermore, SFB induce the accumulation of Th17 cells. SFB colonization leads to an increase in local SAA production (perhaps in IECs), which can act on LP DCs to stimulate the induction of Th17 cells.
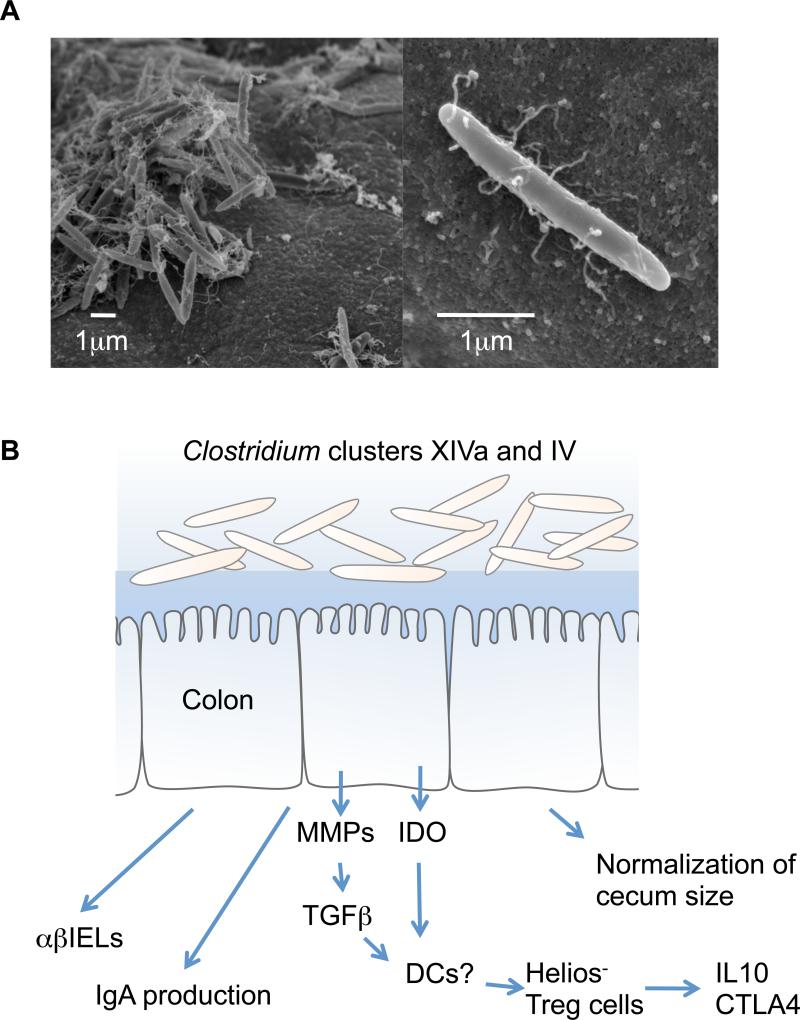
Figure 3.
Clostridium species from clusters XIVa and IV and their effect on the host immune system. (a) Clostridium spp. belonging to clusters XIVa and IV mainly colonize the cecum and proximal colon, have a fusiform shape, and form a thick layer on the mucosal epithelium as shown by scanning electron microscopy (left panel). The right panel shows a higher magnification of a representative bacterium. (b) Clostridium spp. belonging to clusters XIVa and IV can normalize the enlarged cecum of GF mice. The interaction between these Clostridium species and IECs results in the production of matrix metalloproteinases (MMPs) from colonic epithelial cells to convert TGF-β from the latent to the active form. Together with indoleamine 2,3-dioxygenase (IDO) produced by IECs, the active form of TGF-β induces the accumulation of Helios− Treg cells that express high levels of IL-10 and cytotoxic T lymphocyte antigen 4 ( CTLA-4). Clostridium spp. also act as potent stimuli for the IgA response and the recruitment of TCRαβ+ IELs in the colon.
Th17 Cells
Th17 cells are a subset of activated CD4+ T cells characterized by their production of IL-17A, IL-17F, IL-21, and IL-22 (28). IL-17 cytokines act on a broad range of immune and nonimmune cells and regulate granulopoiesis, neutrophil recruitment, and induction of antimicrobial peptides. IL-17 also plays a crucial role in class switching and germinal center formation by acting on B cells (130). IL-22 acts on IECs and triggers the production of antimicrobial peptides such as β-defensins; S100A7, A8, A9 and RegIIIγ; and the expression of genes involved in cellular differentiation and survival (131). By producing these cytokines, Th17 cells play a critical role in the homeostasis of epithelia and in the regulation of host defense against various extracellular pathogens, such as Candida albicans, Pseudomonas aeruginosa, Staphylococcus aureus, and Citrobacter rodentium (132--134). However, Th17 cells have been implicated in autoimmunity, and GWAS in humans have linked genes involved in Th17 cell differentiation and function with susceptibility to Crohn's disease, rheumatoid arthritis, and psoriasis (135--137).
In mice, both in vitro and in vivo, differentiation of Th17 cells from naive CD4+ T cells requires TGF-β and IL-6. TGF-β and IL-6 independently induce expression of the retinoic acid–related orphan receptor (ROR) γt, a ligand-regulated transcription factor pivotal for Th17 cell differentiation (28, 138). Despite its induction of RORγt, TGF-β alone is unable to initiate Th17 cell differentiation (139, 140). This is because TGF-β also induces Foxp3, which is an essential transcription factor for the differentiation and function of Treg cells. By binding to RORγt, Foxp3 inhibits RORγt-directed IL-17 expression. In the presence of IL-6, which activates STAT3 and suppresses the function and expression of Foxp3, the relative level and activity of RORγt is increased, favoring Th17 cell differentiation (140). In addition to IL-6 and TGF-β, several factors and cytokines affect the differentiation of Th17 cells. For example, Th17 cells express the aryl hydrocarbon receptor (AhR). Ligands for AhR, which may be derived from the activity of the microbiota (141), upregulate IL-22 and, to a lesser extent, IL-17 expression in Th17 cells (142). IL-21, which is produced by the differentiating Th17 cells, acts in an autocrine manner to upregulate the expression of IL-23R by activating STAT3 (139). Upon stimulation by IL-23, Th17 cells become fully enabled inflammatory cells. The production of granulocyte-macrophage colony-stimulating factor (GM-CSF) is one of the mechanisms by which Th17 cells mediate their effector functions (143). Th17 cells activated by IL-2 have a role in host defense against extracellular pathogens, such as C. rodentium (132), but IL-23-dependent innate lymphoid cells (ILCs) (discussed below) also have key functions (144). However, exposure to excess IL-23 may cause Th17 cells to become pathogenic, leading to autoinflammatory diseases. In fact, in a T cell transfer model of experimental autoimmune encephalomyelitis (EAE), Th17 cells expanded and induced disease only in the presence of IL-23 (145). Moreover, in humans, variants of IL23R are linked to IBD susceptibility (135). IL-1 also influences the differentiation and function of Th17 cells and is required for the effector function of these cells in vivo. IL-1R is specifically expressed on Th17 cells, and RORγt expression is enhanced by IL-1β (146). Interestingly, a mouse engineered to express an active form of NLRP3 (NLRP3-R258W), which is associated with Muckle-Wells syndrome, develops autoinflammatory disease (147). In this model, inflammation is mediated by Th17 cells induced by excess IL-1β produced by the hyperactivation of inflammasomes.
Although the cytokine requirements for human Th17 cell differentiation remain to be fully elucidated, similar mechanisms seem to be operative in both humans and mice (148, 149). A combination of TGF-β, IL-1β, and either IL-6, IL-21, or IL-23 can induce Th17 cell differentiation from naive CD4+ T cells isolated from adult or cord blood under serum-free conditions. RORγt expression is essential for human Th17 cell differentiation. Pharmacological inhibition of RORγt activity, e.g., by the cardiac glycoside digoxin and its derivatives, which bind specifically to RORγt, suppresses the differentiation of both mouse and human Th17 cells (150). These compounds are thought to prevent activation of RORγt by yet uncharacterized endogenous or diet/microbiota-derived ligand(s).
In the steady state, Th17 cells are present in the greatest numbers in the intestinal LP of mice, particularly in the small intestine (138, 151, 152). In extraintestinal sites, only a small percentage of CD4+TCRαβ+ T cells normally express IL-17 (152). In GF or antibiotic-treated mice, the percentage of LP Th17 cells is markedly reduced (151, 152). Therefore, the development of Th17 cells depends on stimulation by intestinal microbiota. TLR9- deficient mice have decreased numbers of LP Th17 cells (153), and in vitro differentiation of intestinal Th17 cells is enhanced by the addition of flagellin, a ligand for TLR5 (76) and NLRC4 inflammasomes (154, 155). Bacteria-infected apoptotic IECs also provide TLR ligands that trigger DCs to produce IL-6 and TGF-β, resulting in the promotion of Th17 differentiation (156). These results suggest a potential role for TLR and other PRRs in intestinal Th17 differentiation. In contrast, mice deficient in both Myd88 and Trif have normal numbers of LP Th17 cells in the small and large intestines (151, 152). Thus, individual TLRs may convey divergent signals that differentially affect Th17 cell differentiation. It is also possible that components of the intestinal microbiota are specifically altered in mice deficient for distinct TLRs or their adaptors, thus explaining the differences in the influence on Th17 cell number. Indeed, Myd88 deficiency was reported to cause a change in the composition of the gut microbiota (19). Further studies are needed to clarify the role of each TLR in the induction of intestinal Th17 cells. In addition to pathogen-associated molecules, extracellular adenosine 5′-triphosphate (ATP) derived from the microbiota can induce Th17 cells by activating purinergic receptors (P2X and P2Y receptors) expressed on CD70highCD11c+ LP DCs (151). Therefore, several signaling events likely contribute to the accumulation of Th17 cells in the intestine.
Because a small proportion of CD4+ T cells express IL-17 in the MLNs or PPs, it is likely that the differentiation of Th17 cells is induced within the LP in situ (138, 151, 152). In this context, the components of commensal bacteria are directly taken up via Fc receptor Rn (FcRn) by epithelial cells outside of the PPs (157). In addition, LP cells expressing CX3CR1 extend their cellular dendrites between the tight junctions of epithelial cells to take up luminal bacteria (158). CX3CR1+ cells are derived from monocytic precursors and include macrophages and DCs. CX3CR1+CD70highCD11c+ cells in the LP can induce Th17 differentiation in response to ATP stimulation via the production of IL-6, integrin-αV, and integrin-β8 (151). Integrin-αV and integrin-β8 contribute to the conversion of the latent form of TGF-β to the active form by triggering the degradation of the latency-associated protein (159).
Mice housed in different SPF facilities were found to have marked differences in the number of LP Th17 cells (152). This observation led to the identification of SFB as members of the indigenous microbiota responsible for gut Th17 cell accumulation (160) (Figure 2). Monocolonization of mice with SFB, but not other bacterial species, induced a marked accumulation of Th17 cells in the small intestine. In accordance with this observation, mice expressing transgenic human α-defensin 5 (DEFA5) in Paneth cells exhibited a loss of SFB, accompanied by a decrease in the number of Th17 cells in the small intestine (60). Although an effect of SFB on other T cell subsets was not observed in these studies, SFB colonization can also influence the accumulation of IFN-γ-producing cells in the LP (161). Fate-mapping studies have demonstrated that Th1 cells can be derived from IL-17+ LP cells, which may account for the apparent discrepancy in the reports (162).
Although SFB-mediated IgA production may be dependent on innate immune receptors, SFB-mediated Th17 cell differentiation is likely to occur through a mechanism independent of TLR, NOD receptors, or ATP signaling because the LP Th17 cells are normally present in Myd88-, Trif-, or Rip2-deficient mice and SFB colonization does not increase the luminal ATP levels (160). Notably, the attachment of SFB induces morphological changes in IECs such as the accumulation of actin around the attachment site (129, 163) (Figure 2). Moreover, colonization with SFB induces the expression of genes associated with inflammation, such as those encoding the serum amyloid A (SAA) proteins, MHC class II, fucosyltransferase 2 (Fut2), and defensins in IECs (160, 164, 165) (Figure 2). The addition of recombinant SAA to cocultures of naive CD4+ T cells and LP DCs induced differentiation of Th17 cells in vitro. Therefore, a plausible model is that SFB attachment to IECs induces an inflammatory cascade, which includes production of SAAs that act on LP DCs to stimulate Th17 cell differentiation (Figure 2). Further investigation is needed to establish the molecular basis underlying SFB-mediated Th17 cell differentiation.
Colonization with SFB, and consequent induction of Th17 cells, has a protective function in the gut mucosa against pathogenic bacteria, such as C. rodentium (160). Th17 cytokines, particularly IL-22, likely stimulate IECs to produce antimicrobial peptides, which limit the growth of C. rodentium. SFB also prevents colonization of enteropathogenic E. coli O103 in rabbits (166). Moreover, there is a strong correlation between the presence of SFB and a diabetes-free state in nonobese diabetic mice (20). These reports indicate a benefit of the constitutive presence of SFB and Th17 cells in the intestine and suggest that the manipulation of this commensal-regulated pathway may provide new opportunities for enhancing mucosal immunity.
Enterotoxigenic strains of Bacteroides fragilis (ETBF) colonize a proportion of the human population and can cause inflammatory diarrhea in both children and adults (167). ETBF also induce Th17 cells through STAT3 activation in the colon (168). Furthermore, ETBF colonization accelerates tumor formation in multiple intestinal neoplasia (Min) mice (heterozygous for the adenomatous polyposis coli, Apc, gene). Blocking of IL-23R inhibits ETBF-triggered pathologies. These results suggest that ETBF colonization induces IL-23 production in the intestine, resulting in the production of highly pathogenic Th17 cells that induce colitis and tumor formation in Min mice.
On the basis of these findings, one may envisage that SFB-mediated induction of Th17 cells is beneficial to mucosal immunity, whereas Th17 cells induced by ETBF are pathogenic. However, this is not always the case. In the K/BxN mouse model of autoimmune arthritis, colonization with SFB enhances the production of autoantibodies and accelerates disease progression through induction of Th17 cells (15). Furthermore, mice harboring SFB are highly susceptible to EAE symptoms compared with GF mice (17). These reports raise the possibility that the SFB-mediated Th17 induction may have harmful effects on the host, particularly in individuals genetically prone to autoimmune diseases. Conversely, Th17 cells induced by ETBF may be beneficial during an immune response against other pathogenic bacteria, such as C. rodentium. At present, the conditions that determine whether intestinal Th17 cells play a beneficial or harmful role in the host are not fully understood. IL-23 and IL-1β are possible candidate molecules that determine whether Th17 cells become pathogenic or protective. However, the conditions and cells in which IL-23 and IL-1β are produced are not fully understood. Interestingly, proinflammatory Th17 cells can be redirected to and controlled in the small intestine (169). During immune responses that preferentially induce the differentiation of Th17 cells, CCL20 expression is induced in the duodenum. CCL20 recruits CCR6-expressing Th17 cells to the duodenum, where they are eliminated or reprogrammed to acquire immunosuppressive and regulatory properties, including IL-10 expression. Further studies are required to address the cellular and molecular mechanisms that determine the inflammatory or regulatory character of Th17 cells.
The antigen specificity of intestinal Th17 cells also remains unknown. Considering that SFB-induced Th17 cells can contribute to autoimmune arthritis in K/BxN mice and host resistance to C. rodentium, Th17 cells present in the intestine may have a broad repertoire of TCRs and not be microbe-specific. Transgenic mice expressing a single transgenic TCR revealed that intestinal Th17 cells are generated in the absence of a cognate TCR antigen (170), although Foxp3+ Treg cells are not generated in such an environment and have been shown to be responsive to bacterial antigen (171) (discussed in more detail below). However, Th17 cells can develop in the thymus after selection in the medulla by high-affinity antigens, analogous to the development of natural Treg cells (172). These thymus-derived Th17 cells (referred to as natural Th17 or nTh17) express integrin α4β1, α4β7, and CCR6, preferentially migrate into mucosal sites, particularly to the intestine, have a biased TCR repertoire (preferential usage of TCR Vβ3), and are potentially self-reactive (172, 173). However, mice harboring a mutation in SLP-76 (SH2 domain--containing leukocyte protein of 76 kD) had a marked increase in the number of nTh17 cells but were defective in LP Th17 cell differentiation, suggesting that the LP Th17 cells may not derive from nTh17 cells (173). Further studies are required to assess the function and TCR repertoire of intestinal Th17 cells.
Intraepithelial Lymphocytes
IELs are T cells that are interspersed among the gut epithelial cells and whose character and function are different from those of conventional T cells (174). IELs are subdivided into two subpopulations: type a and type b IELs. Type a IELs are similar to conventional T cells and express TCRαβ together with CD4 or CD8αβ. However, unlike conventional T cells, type a IELs contain a higher ratio of CD8αβ+ cytotoxic–activated T cells to CD4+ T cells. These cells function as conventional MHC class I–restricted cytotoxic T cells and kill pathogen-infected epithelial cells. Type b IELs constitute a nonconventional T cell subset and express TCRαβ or TCRγδ with homodimers of CD8α (i.e., CD8αα). In the small intestine of mice, most IELs belong to this type b subset. Type b IELs remain a somewhat elusive cell type, with their function and development still poorly understood. TCRαβ+CD8αα+ IELs have an activated yet resting phenotype and have been suggested to serve a regulatory role in the gut (175). TCRαβ+ within this subset do not bind conventional peptide-MHC ligands, but instead bind several other ligands. CD8αα on this subset binds thymus leukemia antigen, which is expressed abundantly on IECs (176). They also express high levels of the inhibitory receptors cytotoxic T lymphocyte antigen 4 (CTLA-4), PD-1, and Lag3 and inhibitory NK receptors (177). Furthermore, TCRαβ+CD8αα+ IELs prevent colitis when transferred together with CD4+CD45RBhigh T cells into immune-compromised mice (178), and hence resemble Treg cells. TCRγδ+ type b IELs recognize nonmicrobial as well as microbial molecules through TCR or NKG2D (179). TCRγδ+ IELs play an essential role in promoting epithelial restitution following mucosal injury. This function has been linked to the upregulation of keratinocyte growth factor, which stimulates the proliferation of colonic epithelial progenitors (180). Furthermore, TCRγδ+ IELs express RegIIIγ, RegIIIβ, and chemokines, thereby limiting the bacterial penetration of mucosal surfaces during epithelial injury (181). Type b IELs in the small intestine of mice also include a unique cell population expressing both CD4 and CD8αα. CD4+CD8αα+ IELs increase in number with age and are considered to play a regulatory role in the intestine because they can suppress Th1-mediated intestinal inflammation in an IL-10-dependent manner (175).
The microbiota contributes to the proliferation and function of IELs. In fact, [**AU: correct that all these boxes should be + signs? I think there was a problem with the font ** Yes, these are +signs.] TCRαβ+ IELs are reduced in number, and the age-related increase in the CD4+CD8αα+ fraction does not occur in GF mice. In contrast, the localization of TCRγδ+ cells to the intestinal epithelium is independent of normal microbial colonization (182). Therefore, under GF conditions, the TCRγδ+ T subset remains as the only intestinal IEL population. Although the number of TCRγδ+ IELs is unaffected by GF conditions, those isolated from GF mice lack cytolytic activity. Furthermore, small intestinal TCRγδ+ IELs from GF mice show reduced RegIIIγ and β expression levels (181). The colonization of GF mice with whole intestinal microbiota (conventionalization) leads to an increase in the number of IELs and the enhancement of their cytotoxic activity. The decreased RegIIIγ and β expression levels are restored by the colonization of mice with a select subset of the microbiota, such as E. coli (181). The RegIII-expressing TCRγδ+ IELs prevent the penetration of pathogenic bacteria into the intestinal mucosa during the early phase of infection. Similar to the case of IgA-producing cells, SFB and Clostridium spp. induce the accumulation of TCRαβ+ IELs in the small and large intestines, respectively. The monocolonization of mice with SFB leads to an increase in the number of IELs and enhances their cytotoxic activity in the small intestine (128) (Figure 2). A defined mixture of 46 strains of Clostridium spp. belonging to clusters XIVa and IV can restore the number and function of CD8+TCRαβ+ IELs in the colon (128) (Figure 3). However, the molecular basis for IEL induction by these bacteria remains unknown. Of note, TCRαβ+CD8αα+ IELs and TCRγδ+ IELs express high levels of AhR, and its activation is required for their maintenance (141). Diet and bacterial metabolites are the major sources of AhR ligands; therefore, diet influences the number of IELs. AhR deficiency or the lack of AhR ligands compromise the maintenance of IELs and the control of the microbial burden and composition, resulting in heightened immune activation and increased vulnerability to epithelial damage (141). Therefore, communication between IELs and intestinal bacteria is bidirectional and required for intestinal homeostasis.
γδ T Cells
TCRγδ+ T cells (γδ T cells) [**AU: we have typically written this with a space: γδ T. OK to change throughout?** OK] are constituents not only of IELs, but also of peritoneal and LP lymphocytes. Although the antigenic targets of TCRγδ are still largely unknown, γδT cells recognize both self and nonself ligands, that is, a wide range of microbial products as well as stressed epithelial cells. γδT cells have many features of innate cells---for example, they express TLR2, TLR1, and dectin-1 (183). Within hours following infection, γδT cells produce elevated amounts of antibacterial compounds, chemokines, and cytokines that contribute to the recruitment of more specialized innate effectors and the functional polarization of TCRαβ+ conventional T cells to eradicate invading pathogens. Recent studies have shown that γδT cells are an important source of IL-17 during bacterial infection. During Mycobacterium tuberculosis infection, γδT cells are the main source of IL-17 in the lung (184). Another report showed that the antibody-mediated depletion of γδT cells during E. coli infection leads to decreased IL-17 production and neutrophil infiltration into the peritoneal cavity (185). IL-17-producing γδT cells represent a subset of γδT cells, with characteristics similar to those of Th17 cells, such as the expression of RORγt, IL-23R, and AhR (183). Similar to Th17 cells, IL-17-producing γδT cells express IL-22 upon activation of AhR and constitutively express IL-23R. By using IL-23R-GFP reporter mice, investigators showed that IL-23R GFP-positive cells [← AU: why is GFP in parens here? OK to delete?** because GFP is surrogate for IL-23R; could just use slash: IL-23R/GFP.] are prominent in the LP, and many of these are γδT cells (186). In the presence of IL-23, they produce IL-17 much more rapidly than do adaptive Th17 cells (187).
Intestinal microbiota affect the accumulation of IL-17-producing γδT cells, particularly by expanding IL-1R1-expressing IL-17-producing γδT cells (188). In the peritoneal cavity of SPF mice, about 60% of γδT cells are IL-1R1+, whereas the percentage of IL-1R1+ γδT cells drops to 30% or below under GF conditions (188). IL-1 acts synergistically with IL-23 to promote IL-17 production by IL-1R1+ γδT cells. Thus, constitutive induction of the IL-23R+ IL-1R1+ γδT cells by the intestinal microbiota provides an efficient mechanism for the rapid mucosal IL-17 response against infectious pathogens.
Although they are important in host defense, γδT cells have also been implicated in the induction of autoimmune disease. In a mouse model of collagen-induced arthritis, γδT cells serve as important sources of early IL-17 (189). In an EAE model, IL-23R+ γδT cells accumulate in the central nervous system and contribute substantially to the production of IL-17. In these autoimmune disease models, γδT cells promote the expansion of antigen-specific Th17 cells and limit Treg cell responses (187, 190). Therefore, similar to Th17 cells, innate IL-17-producing γδT cells, likely induced at least in part by the microbiota, can exacerbate inflammatory diseases under certain conditions.
Innate Lymphoid Cells
Numerous lines of evidence show the existence of several non-T ILCs in the intestine. These cells include, but are likely not limited to, NK-like NKp46+ cells, LTi-like cells, and Thy1highSCA1+Lin−Kit−CD3−CD4− cells (27, 191). These cells are abundant in the intestinal mucosa, express IL-23R and the transcription factor RORγt, and produce some combination of Th17 cell–associated cytokines, including IL-22 and, to a lesser extent, IL-17. Their cytokine production in response to IL-23 suggests that they represent innate versions of Th17 cells. ILC-derived IL-17 and IL-22 are likely to play critical roles in the maintenance of mucosal barrier functions, in maintaining homeostasis with the microbiota, and in the host response against pathogens, acting cooperatively with Th17 cells.
LTi-like cells
LTi cells were originally identified in the mouse embryo as critical for the organogenesis of LNs and PPs (192). LTi cells lack markers for specific hematopoietic lineage cells but typically express CD4, lymphotoxin-α (LT-α), LT-β, RANK ligand, CXCR5, CCR7, c-kit, and IL-7R (CD127). LTi cells require the helix-loop-helix inhibitor Id2 and RORγt for their development and IL-7 for their expression of LT-α and LT-β (193, 194). Human LTi cells have been detected in fetal MLNs, although they do not express CD4 (195). LTi cells accumulate in LN and PP anlagen and mediate LT-α/β-dependent upregulation of VCAM-1, ICAM-1, MADCAM-1, CXCL13, CCL19, and CCL21 in mesenchymal cells (referred to as organizer cells), which is an essential event for lymphoid organogenesis (196).
LTi-like cells are also present in the adult intestine and gut-associated lymphoid tissue (GALT) as RORγt+lin−c-kit+IL-7R+ cells (197). A fate mapping study showed that adult LTi-like cells are neither the direct progeny nor persistence of fetal LTi cells, but are perhaps derived from adult bone marrow progenitor cells (198). LTi-like cells within the intestinal LP of adult mice serve as components of cryptopatches and inducers of ILFs, which are required for T cell–independent IgA synthesis (124). LTi-like cells constitutively express high levels of OX40L and CD30L, which mediate the survival of memory CD4+ T cells (199). Moreover, upon stimulation with IL-23, LTi-like cells produce IL-22 and IL-17 and are involved in innate immune responses against pathogens, such as C. rodentium (144). Without CD4+ LTi-like cells, infection-induced expression of IL-22 and antimicrobial peptides is lost, resulting in exacerbated host mortality.
Considering the large number of LTi-like cells in the gut, their function and development are likely to be affected by the presence of gut microbiota. However, LTi-like cells are present in GF mice, and their function in cryptopatch formation does not appear to be impaired (198). Furthermore, the expression levels of IL-22 by LTi-like cells in GF mice was higher than in SPF mice (200). This was attributed to microbiota-dependent IEC-derived IL-25, which suppresses IL-22 expression in LTi-like cells (200). Therefore, the development of LTi-like cells may be genetically programmed, but their functions are regulated by the microbiota, presumably to maintain a necessary homeostasis between the microbiota and the host immune system. The effect of the microbiota on LTi-like cells still awaits further investigation.
NK-like cells
Several groups have described mouse NKp46 and human NKp44 expression in novel intestinal ILC subsets (26, 201--203). All intestinal NKp46+ subsets develop independently of the thymus and are found in Rag2-deficient mice. A minor subset of gut NKp46+ cells resembles mature splenic NK cells (NK1.1+CD127−) with the expression of typical NK cell markers (including NKG2D, DX5, CD122, CD11b, and Ly49 family members) (201). In contrast, a predominant NKp46+ subset (NK1.1−CD127+) in the gut expresses neither the above NK markers nor typical effectors of NK cells, such as perforin and IFN-γ. Furthermore, differently from conventional NK cells, NK1.1−CD127+ cells develop independently of IL-15 signaling (201, 202). Importantly, and similar to LTi-like cells, gut LP NKp46+CD127+NK1.1− cells express RORγt, which is required for their development (202, 203).
The LP CD3−NKp46+ cells may play a role in mucosal barrier function through the IL-22-mediated production of antimicrobial peptides from IECs. In fact, these cells are involved in the protection against various forms of experimental colitis by safeguarding epithelial homeostasis (204). In GF mice, the absolute number of intestinal NKp46+CD127+NK1.1− cells was reduced, whereas NKp46+NK1.1+ cells were unaffected (201, 202). Furthermore, IL-22 expression was severely suppressed in NKp46+CD127+NK1.1+ cells from GF mice (201). Therefore, indigenous intestinal microbiota may act to strengthen the mucosal barrier system by inducing the differentiation of these NKp46+ cells.
Thy1high SCA-1+ ILCs
In mice lacking all lymphocytes of the adaptive immune system (Rag2−/−), experimental colitis can be triggered by CD40-mediated activation of myeloid cells or by infection with Helicobacter hepaticus (205, 206). These colitis models are dependent on IL-23, IL-17, and IFN-γ and cannot be triggered in animals genetically lacking RORγt. Among RORγt+ cells, Thy1+SCA1+kit−CD4−NKp46− cells in the intestine are a critical source of IL-17 and IFN-γ (207). They express IL-17A and IFN-γ upon stimulation with IL-23. In addition to expressing RORγt, they express the Th1 cell– associated transcriptional regulator T-bet, which probably accounts for their ability to produce IFN-γ. Thy1highSCA-1+ ILCs also express LTi-related genes such as those encoding LT-α, LT-β, and CXCR5. Although this population of Thy1+SCA1+ cells play a pathological role in promoting intestinal inflammation induced by H. hepaticus or by antibody to CD40 (207), their function may be context dependent. Further investigation into the physiological functions of Thy1highSCA-1+ ILCs, their lineage relationship with other ILCs, and the reason they dominantly exist in the intestine will be required in mice and humans.
Treg Cells
Foxp3+ Treg cells play critical roles in maintaining immunological unresponsiveness to self-antigens and in suppressing excessive immune responses deleterious to the host (28, 208). They act to restrain activation, e.g., colitogenic activity, of both effector T cells (209, 210) and ILCs (211). Foxp3 is critical to the initiation and maintenance of a genetic program for the differentiation and function of Treg cells (212). Treg cells regulate immune responses through multiple mechanisms, including the expression of immune-suppressive molecules such as CTLA-4, IL-10, TGF-β, and IL-35, but also the depletion of ATP and IL-2 (28, 208). In addition, Treg cells are more mobile than naive T cells and can outcompete the latter by aggregating around DCs, thereby inhibiting the induction of effector T cells (213). Treg cells express OX40, which is required for the accumulation of Treg cells in the colon and the Treg-mediated suppression of colitis (214).
Treg cells have a plasticity that allows them to migrate to the inflamed tissue and selectively regulate a specific T cell lineage. For example, under conditions of Th1 inflammation, Treg cells upregulate T-bet, which promotes the expression of CXCR3 and allows Treg cells to accumulate at Th1 cell–inflamed sites (215). Likewise, the expression of interferon regulatory factor 4 (IRF4) in Treg cells is required for the expressions of ICOS and CCR8 and the suppression of Th2 responses (216). The expression of STAT3 in Treg cells is required for the suppression of Th17 responses (217). STAT3 activation confers upon Treg cells certain characteristics of Th17 cells, including the expression of CCR6, which allows Treg cells to migrate to the inflamed tissues in which a Th17 cell response is taking place. Interestingly, a population of Foxp3+ Treg cells in the intestine loses the ability to express Foxp3 and becomes follicular helper T cells (Tfh cells) that interact with B cells and facilitate class switch recombination to IgA (218).
Peripheral Foxp3+ Treg cells are derived from two sources: natural Treg cells (nTreg) arise and mature in the thymus, whereas induced Treg cells (iTreg) develop extrathymically (219). In the presence of IL-2 and TGF-β, naive CD4+ T cells differentiate into Foxp3+ iTreg cells in vitro (220). Retinoic acid can substitute for IL-2 and promote iTreg cell induction (221). In vivo, the peripheral conversion of Foxp3−CD4+ T cells to Foxp3+ Tregs can be observed experimentally in the intestinal LP and GALT after oral exposure to antigen, perhaps because the gut is an environment rich in TGF-β and retinoic acid (222--224). Although iTreg and nTreg cells suppress immune responses through similar cytokine- and contact-dependent mechanisms, iTreg cells can be distinguished from nTreg cells on the basis of gene expression differences. A lower level of Helios expression has been proposed to be characteristic of iTreg cells (225). Interestingly, Helios-negative Foxp3+ Treg cells are highly abundant in the intestinal LP (171, 226, 227), suggesting that the intestinal LP may be where peripheral conversion to Treg cells preferentially occurs. Alternatively, loss of Helios expression could represent a further activation or differentiation state of nTreg cells, and more detailed studies are required to fully characterize the Helios-negative Treg cells in the intestine. In any case, both Helios-positive and Helios-negative Treg cells are likely to cooperate to mediate peripheral tolerance or ignorance to antigens derived from microbial flora or diet.
IL-10 production by intestinal Treg cells
IL-10- or IL-10R-deficient mice develop colitis in the presence of gut microbiota, and recent GWAS have shown that genetic variation in the Il10 gene locus is associated with susceptibility to ulcerative colitis (61) and rare mutations in IL10R result in severe early onset IBD (227a). Using IL-10 reporter mice, many studies have unequivocally identified CD4+ T cells as the main source of IL-10 in the intestine (226, 228, 229). In particular, analysis of Treg cells has provided evidence that more than half of intestinal Foxp3+ Treg cells express high levels of IL-10 (226, 229). Because patients lacking functional IL10R develop colitis with an earlier onset and higher penetrance than patients with Foxp3+ Treg cell deficiency , it is highly likely that IL-10 secretion by both Treg cells and non-Treg cells plays a role in the control of intestinal immune homeostasis. Indeed, in the intestine there is a Foxp3-negative CD4+ T cell population that constitutively produces IL-10, called Tr1 cells (229). IL-10 produced by Treg and Tr1 cells is indispensable for the maintenance of intestinal homeostasis because CD4+ T cell– or Treg-specific disruption of IL-10 in mice (CD4-Cre or Foxp3-Cre x Il10flox/flox) results in severe colitis (230, 231). IL-10 acts on myeloid cells to suppress unnecessary inflammation via STAT3 activation, as exemplified by LysM-Cre x Stat3flox/flox mice, in which spontaneous intestinal inflammation develops (91). Furthermore, IL-10 from Treg cells is necessary for suppressing γδT cell proliferation (232). Deletion of phosphoinositide-dependent protein kinase 1 (Pdk1) in T cells (Cd4-Cre x Pdk1flox/flox) induces chronic inflammation of the intestine, with loss of IL-10 production by Treg cells and marked expansion of colonic CD8α+TCRγδ+ IELs expressing IL-17 (232). IL-10 production by Treg cells is also required for feed-forward autocrine activation of the Treg cells (231, 233). IL-10 binds to IL-10R on Treg cells and activates [**AU: sometimes Stat3 and other times STAT3. OK? Is one human and the other mouse? Or please advise. → YES, I have changed below since it refers to mouse studies] STAT3 to induce the regulatory function of Treg cells. Mice in which IL-10R has been selectively deleted in Treg cells spontaneously develop colitis. Deficiency of Stat3 or IL-10R in Treg cells selectively interferes with their capacity to suppress Th17 cell responses (233, 234).
Several molecules required for induction of IL-10 expression in Treg cells or the differentiation of Tr1 cells have been identified. TGF-β contributes to IL-10 production by Foxp3+ Treg cells and Tr1 cells in the gut (229). IL-27 induces the expressions of c-Maf, IL-21, and ICOS, which coordinately promote the differentiation of Tr1 cells (235). In addition, IL-27 upregulates the expression of AhR, which acts in synergy with c-Maf to promote the development of IL-10-producing cells (236). The transcription factors Blimp-1 and IRF4 are required for IL-10 production by mucosal Treg cells (237). IL-2 and proinflammatory cytokines such as IL-12, IL-6, and IL-4 induce Blimp1 expression and subsequent IL-10 expression in Treg cells.
Induction of Treg cells by LP DCs
Several DC subsets can induce Treg cells and Tr1 cells. In particular, a CD103 (also known as αE-integrin)-expressing DC population in the LP preferentially promotes the generation and homing of iTreg cells to the intestinal mucosa (222, 223, 238). CD103+ DCs express retinal dehydrogenase (RALDH) to produce retinoic acid. Retinoic acid induces expression of gut-homing receptors on T cells and enhances iTreg cell differentiation by cooperating with TGF-β (221). In addition to CD103+ DCs, CD11bhighCD11c− LP macrophages preferentially promote iTreg and Tr1 cell differentiation (239). LP macrophages express high levels of IL-10 and TGF-β and also preferentially induce the differentiation of naive CD4+ T cells into Treg cells. A more recent report showed that constitutive activation of β-catenin in DCs and macrophages in the intestinal LP is required to maintain intestinal homeostasis and tolerance (240). In mice selectively lacking β-catenin expression in DCs (β-catDC−/− mice), the LP DCs expressed reduced levels of RALDH, IL-10, and TGF-β, but high levels of IL-23 and IL-6. Accordingly, these mice had fewer Treg cells and an increase in Th1 and Th17 cells in the intestine, indicating that β-catenin expression by intestinal DCs is important for maintaining the balance between regulatory and effector T cell populations in the gut. Collectively, the preferential induction of iTreg cells and Tr1 cells in the intestinal mucosa may thus be attributed to the gut-specific presence of CD103+ DCs, CD11bhighCD11c− macrophages, and DCs with active β-catenin. Importantly, in the presence of IL-15, retinoic acid can activate LP DCs to release IL-12p70 and IL-23 (241). As a result, under these conditions, retinoic acid acts as an adjuvant that promotes Th1 and Th17 responses rather than induction of Treg cells. IL-15 is a cytokine markedly upregulated under inflammatory disease conditions, such as celiac disease. The functional conversion of retinoic acid is highly reminiscent of the action of TGF-β, which is otherwise a critical inducer of iTreg cell differentiation, but can lead to the generation of Th17 cells in the presence of IL-6. Therefore, the functional outcomes of the above-described factors and cells for Treg and Tr1 cell induction may all be context dependent.
Microbial effects on Treg cells
Foxp3+ Treg cells are present in essentially all organs, but their frequency is typically about 10% of total CD4+ T cells. In contrast, the frequency of Foxp3+ Treg cells in the gut LP is significantly higher (>30%) than those in other organs (226). In the colonic LP of antibiotic-treated or GF mice, the frequency and absolute number of Treg cells are considerably reduced (226). However, the numbers of Treg cells in the small intestine are unchanged or increased by the absence of the microbiota. Therefore, the intestinal microbiota has a profound effect on the number of colonic Treg cells, whereas different mechanisms are involved in the induction of small intestinal Treg cells.
As observed with Th17 cells, distinct constituents of the gut microbiota may also specifically induce Treg cells. Lactobacilli and bifidobacteria have been implicated in the induction of Treg cells. Treatment of mice with the probiotic mixture VSL#3 (a mixture of bifidobacteria, lactobacilli, and Streptococcus salivarius) or the probiotic strain Lactobacillus reuteri increases the percentage of Treg cells (242--244). Furthermore, daily ingestion of L. acidophilus, L. rhamnosus, L. reuteri, or B. infantis results in the modification of the inflammatory status or autoimmune responses in mice (245--248), presumably by inducing Treg and Tr1 cells. However, these probiotic strains are only transiently present (allochthonous) and do not exist at high levels in the intestines of adult mice or humans. Furthermore, it is yet to be demonstrated whether monocolonization with any of these probiotic strains induces Treg cells. Therefore, the effects of these probiotic strains on the induction of Treg cells are not well established and could be secondary to affecting the microbial ecology within the gut, rather than the direct induction of Treg cells.
The human commensal B. fragilis facilitates the functional maturation of Treg cells in mice (249). Monocolonization with B. fragilis boosted IL-10 production in Treg cells in the colon, although the effect on Foxp3+ Treg numbers was marginal. Furthermore, colonization with B. fragilis prevented Th1- and Th17-mediated inflammation caused by H. hepaticus or trinitrobenzene sulfonic acid in the intestine (250). The induction of IL-10 in Treg cells by B. fragilis is mediated by polysaccharide A (PSA), a unique surface polysaccharide of B. fragilis. Injecting or feeding mice with PSA was sufficient to replicate the B. fragilis immune effects. PSA was found to bind to TLR2 on CD4+ T cells to induce IL-10 production (249). This, in turn, led to suppression of Th17 cell responses, facilitating colonization of the intestinal epithelium with B. fragilis. Mice colonized with B. fragilis lacking PSA had higher levels of colonic Th17 cells and profoundly reduced numbers of IEC-associated bacteria than did animals colonized with wild-type B. fragilis (251). Therefore, PSA may be a critical factor that permits host-bacteria mutualism.
Because B. fragilis is a human rather than a mouse commensal, its effects in the mouse may or may not reflect evolutionarily selected commensal functions. Among the microbiota indigenous to the murine colon, the spore-forming components, particularly the genus Clostridium belonging to clusters XIVa and IV, are outstanding inducers of colonic Treg cells (226) (Figure 3). The colonization of GF mice with a defined mixture of 46 strains of Clostridium, which were originally isolated from chloroform-treated fecal material (sporulated fraction) from conventionally reared mice, was sufficient to induce Treg cells (226). Those Clostridium spp. preferentially colonize and form a thick layer on the cecum and colon, where they promote the production of matrix metalloproteinases (MMPs) from IECs. MMPs have been implicated in the conversion of TGF-β from the latent to the active form (252). In addition, colonization with Clostridium enhances expression of indoleamine 2,3-dioxygenase (IDO), which has also been implicated in the induction of Treg cells (253) (Figure 3). As in mice with a normal microbiota, Treg cells in the colons of mice colonized with Clostridium consist of a large number of Helios-negative cells (226). It is possible that those Treg cells are derived from naive CD4+ T cells that have undergone conversion to express Foxp3 in the colon after receiving TGF-β and other Treg-inducing stimuli following colonization with Clostridium. Furthermore, a large subset of Treg cells induced by Clostridium expresses high levels of IL-10 and CTLA-4 (Figure 3). Therefore, Clostridium may affect the qualitative properties of colonic Treg cells in addition to their quantity. In addition to the induction of Treg cells, the 46 strains of Clostridium affect the accumulation of TCRαβ+ IELs and IgA-producing cells in the colon (128) (Figure 3). Therefore, Clostridium species modulate multiple facets of the intestinal immune system.
Increasing the frequency and abundance of Clostridium clusters XIVa and IV in mice with an otherwise normal immune system and microbiota enhances colonic Treg cell accumulation. Such mice are more resistant to experimental models of allergy and intestinal inflammation (226). Consistent with the importance of Clostridium spp., colonization with altered Shaedler flora, which includes Clostridium clostridioforme, results in Treg cell accumulation in the colonic LP (227). Furthermore, in humans, reduction in the number of F. prausnitzii, which belongs to the Clostridium cluster IV, is associated with a high risk of postoperative recurrence of Crohn's disease (58). Supernatants from F. prausnitzii cultures increased production of IL-10 by peripheral blood mononuclear cells in vitro (58). In addition, the proportion of Clostridium clusters XIVa and IV in the fecal community is smaller in IBD patients than in healthy controls (13). These reports raise the possibility that the Clostridium-dependent constitutive induction of Treg cells and their expression of IL-10 and CTLA-4 may contribute to the suppression of autoimmunity and deleterious inflammation in humans. Although it remains unknown whether defects in Treg cells actually contribute to human IBD, these findings suggest that prophylactic administration of human-gut-resident Clostridium species could reduce the susceptibility to chronic disease.
The antigen specificity of Treg cells, like that of other LP T cells, has remained largely unexplored. However, a recent study has described the ability of TCRs cloned from colonic Treg cells to mediate signals in response to individual commensal bacterial subsets (171). These TCRs endowed conventional T cells with colitogenic potential that was revealed upon their adoptive transfer into mice. Thus, specific commensal bacteria induce naive CD4+ T cells to differentiate into antigen-specific colonic Treg cells that, presumably, enforce immune system tolerance toward those bacteria. It is currently not known if effector T cells, e.g., Th17 cells, can similarly display TCR specificity for distinct bacterial species. If this is the case, it will be fascinating to learn how individual bacteria influence T cells with TCRs that target their antigens to become either Treg or Th17 cells. It also remains possible that many of the TCRs expressed by these cells are mostly reactive with self-peptide-MHC complexes and that environmental cytokines determine whether they become activated and polarized toward Treg or effector T cell profiles. Further studies will be needed to determine how the microenvironmental niches of individual commensal species influence T cell polarization.
IMPLICATIONS AND PERSPECTIVES
We emphasized in this review the interactions of the microbiota with the host mucosal immune system and the consequences of such interactions. Preserving the diversity of the microbiota and generating various mucosal immune cell populations are likely to be mutually beneficial for the maintenance of bacterial and host homeostasis in the intestine. Substantial progress has been made in understanding some of the relationships among diet, host genotypes, immune systems, and microbial ecology in the intestine. Recent metagenomic and metabolomic studies have been generating new insights and opportunities to reveal the components and functions of the gut microbiota in the host. In addition, gnotobiotic studies have implicated several specific components of gut flora in the induction of pro- or anti-inflammatory cells. However, we are still far from a comprehensive understanding of how this symbiotic ecosystem develops and functions. In particular, a key puzzle is how the host tolerates the enormous intestinal bacterial burden and what are the factors or conditions for keeping the luminal bacteria as symbionts. Recent reports have shown that specific components of the microbiota, such as B. fragilis and Clostridium spp., preferentially induce immune-suppressive cells; however, it is not known whether these immune cells contribute to tolerance toward commensal bacteria without reducing their numbers and functions. Moreover, even beneficial members of the microbiota can have detrimental effects, depending on the host's conditions. For instance, although B. fragilis can cause CD4+ cells to secrete IL-10 in mice, it opportunistically invades intestinal tissues, resulting in infectious diseases in immune-compromised patients. Furthermore, B. thetaiotaomicron, a well-characterized symbiotic species, can also induce colitis in mice with T cells deficient in Il10r2 and expressing a dominant-negative Tgfbr2 (254) or in mice deficient in IgA (25). Clearly, we have only just begun to understand the outline of the intestinal microbiota, and further investigation will likely provide new insights into the detailed interplay with the host during steady-state and disease processes.
Acronyms and Definitions.
DC: dendritic cell
Dysbiosis: a condition with imbalance in the composition of the bacterial microbiota; this includes an outgrowth of potentially pathogenic bacteria and/or a decrease in bacterial diversity and bacteria beneficial to the host
GF: germ-free
Gnotobiote: gnotobiotic comes from the Greek “known life” and refers to animals with defined microbiological status
IBD: inflammatory bowel disease
IEC: intestinal epithelial cell
ILF: isolated lymphoid follicle
LP: lamina propria
LTi: lymphoid tissue inducer
Metabolomics: the quantitative measurement of the dynamic multiparametric metabolic responses of multicellular systems to pathophysiological stimuli by nuclear magnetic resonance (NMR) spectroscopy or mass spectrometry (MS)
MetaHIT (Metagenomics of the Human Intestinal Tract consortium): the MetaHIT project aims to understand the role of the human intestinal microbiota in health and disease; the consortium involves 13 research centers from eight countries
Microbiome: a genetic catalogue of the microbial species that inhabit a defined environment such as the human body
Pathobiont: a symbiont or a commensal that is able to promote pathology only when genetic or environmental conditions are altered in the host
Pattern-recognition receptors: receptors that recognize microbe-associated molecular patterns to initiate innate immune responses
PP: Payer's patch
Probiotics: live microorganisms that, when administered in adequate amounts, confer a health benefit to the host
SCFA: short-chain fatty acid
TLR: Toll-like receptor
ACKNOWLEDGMENTS
The work was supported by Japan Science and Technology Agency PRESTO (Precursory Research for Embryonic Science and Technology) (K.H.), Japan Society for the Promotion Science NEXT program (K.H.), the Howard Hughes Medical Institute (D.R.L.), and National Institutes of Health grants RO1AI080885 and RC2 AR058986 (D.R.L.).
Footnotes
-
1)The authors suggest that Th17 differentiation takes place locally in the intestine. Not clear if they mean priming here-a recent Immunity ms (lewis et al., 2011) shows a reduction in Th17 cells in the intestine in the absence of migratory CD103+ DC suggesting a MLN priming and most likely further differentiation of the effector response in LP. May be good to at least mention this as a possibility.
-
2)Reference is made to the transfer of colitis from gentically susceptible mice with colitis (eg TRUC). However in these settings disease is transient suggesting in the absence of appropriate genetic predisposition it is in fact difficult to stably alter the microbiota
-
3)Discussion of the immune regulatory role of IL-10 in the intestine would benefit form inclusion of the Klien NEJM ms showing rare mutations in IL-10R induce severe early onset-IBD. **]
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Berg RD. The indigenous gastrointestinal microflora. Trends Microbiol. 1996;4:430–-35. doi: 10.1016/0966-842x(96)10057-3. [DOI] [PubMed] [Google Scholar]
- 2.Xu J, Gordon JI. Honor thy symbionts. Proc. Natl. Acad. Sci. USA. 2003;100:10452–-59. doi: 10.1073/pnas.1734063100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 2004;4:478–-85. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 4.Bouskra D, Brézillon C, Bérard M, Werts C, Varona R, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507–-10. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- 5.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–-30. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc. Natl. Acad. Sci. USA. 2008;105:20858–-63. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 2010;10:159–-69. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 8.Lederberg J. Infectious history. Science. 2000;288:287–-93. doi: 10.1126/science.288.5464.287. [DOI] [PubMed] [Google Scholar]
- 9.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–-65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009;9:313–-23. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu. Rev. Immunol. 2010;28:623–-67. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205–-11. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA. 2007;104:13780–-85. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peterson DA, Frank DN, Pace NR, Gordon JI. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe. 2008;3:417–-27. doi: 10.1016/j.chom.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–-27. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scher JU, Abramson SB. The microbiome and rheumatoid arthritis. Nat. Rev. Rheumatol. 2011;7:569–-78. doi: 10.1038/nrrheum.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. USA. 2011;108(Suppl. 1):4615–-22. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berer K, Mues M, Koutrolos M, Rasbi ZA, Boziki M, et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538–-41. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 19.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–-13. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc. Natl. Acad. Sci. USA. 2011;108:11548–-53. doi: 10.1073/pnas.1108924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong PY, Lee BW, Aw M, Shek LP, Yap GC, et al. Comparative analysis of fecal microbiota in infants with and without eczema. PLoS One. 2010;5:e9964. doi: 10.1371/journal.pone.0009964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakayama J, Kobayashi T, Tanaka S, Korenori Y, Tateyama A, et al. Aberrant structures of fecal bacterial community in allergic infants profiled by 16S rRNA gene pyrosequencing. FEMS Immunol. Med. Microbiol. 2011;63:397–-406. doi: 10.1111/j.1574-695X.2011.00872.x. [DOI] [PubMed] [Google Scholar]
- 23.Sansonetti PJ. War and peace at mucosal surfaces. Nat. Rev. Immunol. 2004;4:953–-64. doi: 10.1038/nri1499. [DOI] [PubMed] [Google Scholar]
- 24.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–-41. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–-39. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 26.Colonna M. Interleukin-22-producing natural killer cells and lymphoid tissue inducer-like cells in mucosal immunity. Immunity. 2009;31:15–-23. doi: 10.1016/j.immuni.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 27.Spits H, Di Santo JP. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat. Immunol. 2011;12:21–-27. doi: 10.1038/ni.1962. [DOI] [PubMed] [Google Scholar]
- 28.Littman DR, Rudensky AY. Th17 and regulatory T cells in mediating and restraining inflammation. Cell. 2010;140:845–-58. doi: 10.1016/j.cell.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 29.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA. 2010;107:11971–-75. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–-38. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–-80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 2011;9:279–-90. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 33.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–-31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 34.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–-84. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–-31. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–-45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–-57. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–-23. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang C, Zhang M, Wang S, Han R, Cao Y, et al. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 2010;4:232–-41. doi: 10.1038/ismej.2009.112. [DOI] [PubMed] [Google Scholar]
- 40.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–-23. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 41.Duncan SH, Belenguer A, Holtrop G, Johnstone AM, Flint HJ, Lobley GE. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl. Environ. Microbiol. 2007;73:1073–-78. doi: 10.1128/AEM.02340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–-8. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA. 2010;107:14691–-96. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–-59. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 45.Hehemann JH, Correc G, Barbeyron T, Helbert W, Czjzek M, Michel G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–-12. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- 46.Maslowski KM, Mackay CR. Diet, gut microbiota and immune responses. Nat. Immunol. 2011;12:5–-9. doi: 10.1038/ni0111-5. [DOI] [PubMed] [Google Scholar]
- 47.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:119–-29. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 48.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–-89. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ermann J, Garrett WS, Kuchroo J, Rourida K, Glickman JN, et al. Severity of innate immune-mediated colitis is controlled by the cytokine deficiency-induced colitis susceptibility-1 (Cdcs1) locus. Proc. Natl. Acad. Sci. USA. 2011;108:7137–-41. doi: 10.1073/pnas.1104234108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–-300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Veiga P, Gallini CA, Beal C, Michaud M, Delaney ML, et al. Bifidobacterium animalis subsp. lactis fermented milk product reduces inflammation by altering a niche for colitogenic microbes. Proc. Natl. Acad. Sci. USA. 2010;107:18132–-37. doi: 10.1073/pnas.1011737107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cummings JH, Macfarlane GT, Macfarlane S. Intestinal bacteria and ulcerative colitis. Curr. Issues Intest. Microbiol. 2003;4:9–-20. [PubMed] [Google Scholar]
- 53.Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology. 2004;127:412–-21. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 54.Barnich N, Carvalho FA, Glasser AL, Darcha C, Jantscheff P, et al. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J. Clin. Investig. 2007;117:1566–-74. doi: 10.1172/JCI30504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, et al. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 1994;44:812–-26. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 56.Willing B, Halfvarson J, Dicksved J, Rosenquist M, Jarnerot G, et al. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn's disease. Inflamm. Bowel Dis. 2009;15:653–-60. doi: 10.1002/ibd.20783. [DOI] [PubMed] [Google Scholar]
- 57.Sokol H, Seksik P, Rigottier-Gois L, Lay C, Lepage P, et al. Specificities of the fecal microbiota in inflammatory bowel disease. Inflamm. Bowel Dis. 2006;12:106–-11. doi: 10.1097/01.MIB.0000200323.38139.c6. [DOI] [PubMed] [Google Scholar]
- 58.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA. 2008;105:16731–-36. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fagarasan S, Muramatsu M, Suzuki K, Nagaoka H, Hiai H, Honjo T. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science. 2002;298:1424–-27. doi: 10.1126/science.1077336. [DOI] [PubMed] [Google Scholar]
- 60.Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjöberg J, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 2010;11:76–-83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Franke A, Balschun T, Karlsen TH, Sventoraityte J, Nikolaus S, et al. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat. Genet. 2008;40:1319–-23. doi: 10.1038/ng.221. [DOI] [PubMed] [Google Scholar]
- 62.McGovern DP, Gardet A, Torkvist L, Goyette P, Essers J, et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat. Genet. 2010;42:332–-37. doi: 10.1038/ng.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. USA. 2008;105:15064–-69. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Turner JR. Intestinal mucosal barrier function in health and disease. Nat. Rev. Immunol. 2009;9:799–-809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 65.Slack E, Hapfelmeier S, Stecher B, Velykoredko Y, Stoel M, et al. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science. 2009;325:617–-20. doi: 10.1126/science.1172747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Araki A, Kanai T, Ishikura T, Makita S, Uraushihara K, et al. MyD88-deficient mice develop severe intestinal inflammation in dextran sodium sulfate colitis. J. Gastroenterol. 2005;40:16–-23. doi: 10.1007/s00535-004-1492-9. [DOI] [PubMed] [Google Scholar]
- 67.Vijay-Kumar M, Sanders CJ, Taylor RT, Kumar A, Aitken JD, et al. Deletion of TLR5 results in spontaneous colitis in mice. J. Clin. Investig. 2007;117:3909–-21. doi: 10.1172/JCI33084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cario E, Gerken G, Podolsky DK. Toll-like receptor 2 controls mucosal inflammation by regulating epithelial barrier function. Gastroenterology. 2007;132:1359–-74. doi: 10.1053/j.gastro.2007.02.056. [DOI] [PubMed] [Google Scholar]
- 69.Podolsky DK, Gerken G, Eyking A, Cario E. Colitis-associated variant of TLR2 causes impaired mucosal repair because of TFF3 deficiency. Gastroenterology. 2009;137:209–-20. doi: 10.1053/j.gastro.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gibson DL, Ma C, Rosenberger CM, Bergstrom KS, Valdez Y, et al. Toll-like receptor 2 plays a critical role in maintaining mucosal integrity during Citrobacter rodentium-induced colitis. Cell Microbiol. 2008;10:388–-403. doi: 10.1111/j.1462-5822.2007.01052.x. [DOI] [PubMed] [Google Scholar]
- 71.Sonnenburg JL, Chen CT, Gordon JI. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 2006;4:e413. doi: 10.1371/journal.pbio.0040413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, et al. Vancomycin-resistant Enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–-7. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kinnebrew MA, Ubeda C, Zenewicz LA, Smith N, Flavell RA, Pamer EG. Bacterial flagellin stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. J. Infect. Dis. 2010;201:534–-43. doi: 10.1086/650203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cerutti A. The regulation of IgA class switching. Nat. Rev. Immunol. 2008;8:421–-34. doi: 10.1038/nri2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tezuka H, Abe Y, Iwata M, Takeuchi H, Ishikawa H, et al. Regulation of IgA production by naturally occurring TNF/iNOS-producing dendritic cells. Nature. 2007;448:929–-33. doi: 10.1038/nature06033. [DOI] [PubMed] [Google Scholar]
- 76.Uematsu S, Fujimoto K, Jang MH, Yang BG, Jung YJ, et al. Regulation of humoral and cellular gut immunity by lamina propria dendritic cells expressing Toll-like receptor 5. Nat. Immunol. 2008;9:769–-76. doi: 10.1038/ni.1622. [DOI] [PubMed] [Google Scholar]
- 77.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–-75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 2007;39:207–-11. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 79.Prescott NJ, Fisher SA, Franke A, Hampe J, Onnie CM, et al. A nonsynonymous SNP in ATG16L1 predisposes to ileal Crohn's disease and is independent of CARD15 and IBD5. Gastroenterology. 2007;132:1665–-71. doi: 10.1053/j.gastro.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 80.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–-63. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, et al. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–-45. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–-49. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shi CS, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J. Biol. Chem. 2008;283:33175–-82. doi: 10.1074/jbc.M804478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature. 2008;456:264–-68. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 85.Clayburgh DR, Musch MW, Leitges M, Fu YX, Turner JR. Coordinated epithelial NHE3 inhibition and barrier dysfunction are required for TNF-mediated diarrhea in vivo. J. Clin. Investig. 2006;116:2682–-94. doi: 10.1172/JCI29218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat. Immunol. 2004;5:1052–-60. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 87.Turer EE, Tavares RM, Mortier E, Hitotsumatsu O, Advincula R, et al. Homeostatic MyD88-dependent signals cause lethal inflammation in the absence of A20. J. Exp. Med. 2008;205:451–-64. doi: 10.1084/jem.20071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wald D, Qin J, Zhao Z, Qian Y, Naramura M, et al. SIGIRR, a negative regulator of Toll-like receptor-interleukin 1 receptor signaling. Nat. Immunol. 2003;4:920–-27. doi: 10.1038/ni968. [DOI] [PubMed] [Google Scholar]
- 89.Xiao H, Gulen MF, Qin J, Yao J, Bulek K, et al. The Toll-interleukin-1 receptor member SIGIRR regulates colonic epithelial homeostasis, inflammation, and tumorigenesis. Immunity. 2007;26:461–-75. doi: 10.1016/j.immuni.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 90.Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–-74. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 91.Takeda K, Clausen BE, Kaisho T, Tsujimura T, Terada N, et al. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity. 1999;10:39–-49. doi: 10.1016/s1074-7613(00)80005-9. [DOI] [PubMed] [Google Scholar]
- 92.Kobayashi M, Kweon MN, Kuwata H, Schreiber RD, Kiyono H, et al. Toll-like receptor-dependent production of IL-12p40 causes chronic enterocolitis in myeloid cell-specific Stat3-deficient mice. J. Clin. Investig. 2003;111:1297–-308. doi: 10.1172/JCI17085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rakoff-Nahoum S, Hao L, Medzhitov R. Role of toll-like receptors in spontaneous commensal-dependent colitis. Immunity. 2006;25:319–-29. doi: 10.1016/j.immuni.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 94.Liew FY, Xu D, Brint EK, O'Neill LA. Negative regulation of toll-like receptor-mediated immune responses. Nat. Rev. Immunol. 2005;5:446–-58. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- 95.Nenci A, Becker C, Wullaert A, Gareus R, van Loo G, et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. 2007;446:557–-61. doi: 10.1038/nature05698. [DOI] [PubMed] [Google Scholar]
- 96.Zaph C, Troy AE, Taylor BC, Berman-Booty LD, Guild KJ, et al. Epithelial-cell-intrinsic IKK-β expression regulates intestinal immune homeostasis. Nature. 2007;446:552–-56. doi: 10.1038/nature05590. [DOI] [PubMed] [Google Scholar]
- 97.Welz PS, Wullaert A, Vlantis K, Kondylis V, Fernández-Majada V, et al. FADD prevents RIP3-mediated epithelial cell necrosis and chronic intestinal inflammation. Nature. 2011;477:330–-34. doi: 10.1038/nature10273. [DOI] [PubMed] [Google Scholar]
- 98.Inohara N, Chamaillard M, McDonald C, Nuñez G. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu. Rev. Biochem. 2005;74:355–-83. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- 99.Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–-34. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 100.Petnicki-Ocwieja T, Hrncir T, Liu YJ, Biswas A, Hudcovic T, et al. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc. Natl. Acad. Sci. USA. 2009;106:15813–-18. doi: 10.1073/pnas.0907722106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–-6. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 102.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–-603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 103.van Heel DA, Ghosh S, Butler M, Hunt KA, Lundberg AM, et al. Muramyl dipeptide and toll-like receptor sensitivity in NOD2-associated Crohn's disease. Lancet. 2005;365:1794–-96. doi: 10.1016/S0140-6736(05)66582-8. [DOI] [PubMed] [Google Scholar]
- 104.Carneiro LA, Magalhaes JG, Tattoli I, Philpott DJ, Travassos LH. Nod-like proteins in inflammation and disease. J. Pathol. 2008;214:136–-48. doi: 10.1002/path.2271. [DOI] [PubMed] [Google Scholar]
- 105.Strober W, Kitani A, Fuss I, Asano N, Watanabe T. The molecular basis of NOD2 susceptibility mutations in Crohn's disease. Mucosal Immunol. 2008;1(Suppl. 1):S5–-9. doi: 10.1038/mi.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Girardin SE, Tournebize R, Mavris M, Page AL, Li X, et al. CARD4/Nod1 mediates NF-κB and JNK activation by invasive Shigella flexneri. EMBO Rep. 2001;2:736–-42. doi: 10.1093/embo-reports/kve155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, et al. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat. Immunol. 2004;5:1166–-74. doi: 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]
- 108.Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med. 2010;16:228–-31. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc. Nutr. Soc. 2003;62:67–-72. doi: 10.1079/PNS2002207. [DOI] [PubMed] [Google Scholar]
- 110.Scheppach W, Sommer H, Kirchner T, Paganelli GM, Bartram P, et al. Effect of butyrate enemas on the colonic mucosa in distal ulcerative colitis. Gastroenterology. 1992;103:51–-56. doi: 10.1016/0016-5085(92)91094-k. [DOI] [PubMed] [Google Scholar]
- 111.Vernia P, Annese V, Bresci G, d'Albasio G, D'Incà R, et al. Topical butyrate improves efficacy of 5-ASA in refractory distal ulcerative colitis: results of a multicentre trial. Eur. J. Clin. Investig. 2003;33:244–-48. doi: 10.1046/j.1365-2362.2003.01130.x. [DOI] [PubMed] [Google Scholar]
- 112.Miyauchi S, Gopal E, Fei YJ, Ganapathy V. Functional identification of SLC5A8, a tumor suppressor down-regulated in colon cancer, as a Na+-coupled transporter for short-chain fatty acids. J. Biol. Chem. 2004;279:13293–-96. doi: 10.1074/jbc.C400059200. [DOI] [PubMed] [Google Scholar]
- 113.Thibault R, Blachier F, Darcy-Vrillon B, de Coppet P, Bourreille A, Segain JP. Butyrate utilization by the colonic mucosa in inflammatory bowel diseases: a transport deficiency. Inflamm. Bowel Dis. 2010;16:684–-95. doi: 10.1002/ibd.21108. [DOI] [PubMed] [Google Scholar]
- 114.Thangaraju M, Cresci GA, Liu K, Ananth S, Gnanaprakasam JP, et al. GPR109A is a G-protein-coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res. 2009;69:2826–-32. doi: 10.1158/0008-5472.CAN-08-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, et al. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J. Biol. Chem. 2003;278:11312–-19. doi: 10.1074/jbc.M211609200. [DOI] [PubMed] [Google Scholar]
- 116.Briscoe CP, Tadayyon M, Andrews JL, Benson WG, Chambers JK, et al. The orphan G protein-coupled receptor GPR40 is activated by medium and long chain fatty acids. J. Biol. Chem. 2003;278:11303–-11. doi: 10.1074/jbc.M211495200. [DOI] [PubMed] [Google Scholar]
- 117.Hirasawa A, Tsumaya K, Awaji T, Katsuma S, Adachi T, et al. Free fatty acids regulate gut incretin glucagon-like peptide-1 secretion through GPR120. Nat. Med. 2005;11:90–-94. doi: 10.1038/nm1168. [DOI] [PubMed] [Google Scholar]
- 118.Ichimura A, Hirasawa A, Hara T, Tsujimoto G. Free fatty acid receptors act as nutrient sensors to regulate energy homeostasis. Prostaglandins Other Lipid Mediat. 2009;89:82–-88. doi: 10.1016/j.prostaglandins.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 119.Oh DY, Talukdar S, Bae EJ, Imamura T, Morinaga H, et al. GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell. 2010;142:687–-98. doi: 10.1016/j.cell.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc. Natl. Acad. Sci. USA. 2008;105:16767–-72. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–-86. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–-47. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 123.Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 2000;288:2222–-26. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- 124.Tsuji M, Suzuki K, Kitamura H, Maruya M, Kinoshita K, et al. Requirement for lymphoid tissue-inducer cells in isolated follicle formation and T cell-independent immunoglobulin A generation in the gut. Immunity. 2008;29:261–-71. doi: 10.1016/j.immuni.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 125.Cebra JJ. Influences of microbiota on intestinal immune system development. Am. J. Clin. Nutr. 1999;69:S1046–-51. doi: 10.1093/ajcn/69.5.1046s. [DOI] [PubMed] [Google Scholar]
- 126.Snel J, Bakker MH, Heidt PJ. Quantification of antigen-specific immunoglobulin A after oral booster immunization with ovalbumin in mice mono-associated with segmented filamentous bacteria or Clostridium innocuum. Immunol. Lett. 1997;58:25–-28. doi: 10.1016/s0165-2478(97)02715-6. [DOI] [PubMed] [Google Scholar]
- 127.Talham GL, Jiang HQ, Bos NA, Cebra JJ. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect. Immun. 1999;67:1992–-2000. doi: 10.1128/iai.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Umesaki Y, Setoyama H, Matsumoto S, Imaoka A, Itoh K. Differential roles of segmented filamentous bacteria and clostridia in development of the intestinal immune system. Infect. Immun. 1999;67:3504–-11. doi: 10.1128/iai.67.7.3504-3511.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Caselli M, Holton J, Boldrini P, Vaira D, Calo G. Morphology of segmented filamentous bacteria and their patterns of contact with the follicle-associated epithelium of the mouse terminal ileum: implications for the relationship with the immune system. Gut Microbes. 2010;1:367–-72. doi: 10.4161/gmic.1.6.14390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mitsdoerffer M, Lee Y, Jager A, Kim HJ, Korn T, et al. Proinflammatory T helper type 17 cells are effective B-cell helpers. Proc. Natl. Acad. Sci. USA. 2010;107:14292–-97. doi: 10.1073/pnas.1009234107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kolls JK, McCray PB, Jr, Chan YR. Cytokine-mediated regulation of antimicrobial proteins. Nat. Rev. Immunol. 2008;8:829–-35. doi: 10.1038/nri2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mangan PR, Harrington LE, O'Quinn DB, Helms WS, Bullard DC, et al. Transforming growth factor-β induces development of the T(H)17 lineage. Nature. 2006;441:231–-34. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 133.Conti HR, Shen F, Nayyar N, Stocum E, Sun JN, et al. Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J. Exp. Med. 2009;206:299–-311. doi: 10.1084/jem.20081463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ishigame H, Kakuta S, Nagai T, Kadoki M, Nambu A, et al. Differential roles of interleukin-17A and -17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity. 2009;30:108–-19. doi: 10.1016/j.immuni.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 135.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–-63. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nair RP, Duffin KC, Helms C, Ding J, Stuart PE, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-κB pathways. Nat. Genet. 2009;41:199–-204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet. 2010;42:508–-14. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, et al. The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–-33. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 139.Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, et al. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat. Immunol. 2007;8:967–-74. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 140.Zhou L, Lopes JE, Chong MM, Ivanov II, Min R, et al. TGF-β-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORγt function. Nature. 2008;453:236–-40. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li Y, Innocentin S, Withers DR, Roberts NA, Gallagher AR, et al. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell. 2011;147:629–-40. doi: 10.1016/j.cell.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 142.Veldhoen M, Hirota K, Christensen J, O'Garra A, Stockinger B. Natural agonists for aryl hydrocarbon receptor in culture medium are essential for optimal differentiation of Th17 T cells. J. Exp. Med. 2009;206:43–-49. doi: 10.1084/jem.20081438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.El-Behi M, Ciric B, Dai H, Yan Y, Cullimore M, et al. The encephalitogenicity of T(H)17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat. Immunol. 2011;12:568–-75. doi: 10.1038/ni.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sonnenberg GF, Monticelli LA, Elloso MM, Fouser LA, Artis D. CD4+ lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity. 2011;34:122–-34. doi: 10.1016/j.immuni.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.McGeachy MJ, Bak-Jensen KS, Chen Y, Tato CM, Blumenschein W, et al. TGF-β and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T(H)-17 cell-mediated pathology. Nat. Immunol. 2007;8:1390–-97. doi: 10.1038/ni1539. [DOI] [PubMed] [Google Scholar]
- 146.Chung Y, Chang SH, Martinez GJ, Yang XO, Nurieva R, et al. Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity. 2009;30:576–-87. doi: 10.1016/j.immuni.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Meng G, Zhang F, Fuss I, Kitani A, Strober W. A mutation in the Nlrp3 gene causing inflammasome hyperactivation potentiates Th17 cell-dominant immune responses. Immunity. 2009;30:860–-74. doi: 10.1016/j.immuni.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Manel N, Unutmaz D, Littman DR. The differentiation of human T(H)-17 cells requires transforming growth factor-β and induction of the nuclear receptor RORγt. Nat. Immunol. 2008;9:641–-49. doi: 10.1038/ni.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yang L, Anderson DE, Baecher-Allan C, Hastings WD, Bettelli E, et al. IL-21 and TGF-β are required for differentiation of human T(H)17 cells. Nature. 2008;454:350–-52. doi: 10.1038/nature07021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Huh JR, Leung MW, Huang P, Ryan DA, Krout MR, et al. Digoxin and its derivatives suppress TH17 cell differentiation by antagonizing RORγt activity. Nature. 2011;472:486–-90. doi: 10.1038/nature09978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, et al. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808–-12. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- 152.Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–-49. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hall JA, Bouladoux N, Sun CM, Wohlfert EA, Blank RB, et al. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity. 2008;29:637–-49. doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kofoed EM, Vance RE. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 2011;477:592–-95. doi: 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhao Y, Yang J, Shi J, Gong YN, Lu Q, et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477:596–-600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- 156.Torchinsky MB, Garaude J, Martin AP, Blander JM. Innate immune recognition of infected apoptotic cells directs T(H)17 cell differentiation. Nature. 2009;458:78–-82. doi: 10.1038/nature07781. [DOI] [PubMed] [Google Scholar]
- 157.Yoshida M, Claypool SM, Wagner JS, Mizoguchi E, Mizoguchi A, et al. Human neonatal Fc receptor mediates transport of IgG into luminal secretions for delivery of antigens to mucosal dendritic cells. Immunity. 2004;20:769–-83. doi: 10.1016/j.immuni.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 158.Niess JH, Brand S, Gu X, Landsman L, Jung S, et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254–-58. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- 159.Travis MA, Reizis B, Melton AC, Masteller E, Tang Q, et al. Loss of integrin α(v)β8 on dendritic cells causes autoimmunity and colitis in mice. Nature. 2007;449:361–-65. doi: 10.1038/nature06110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–-98. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–-89. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 162.Hirota K, Duarte JH, Veldhoen M, Hornsby E, Li Y, et al. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat. Immunol. 2011;12:255–-63. doi: 10.1038/ni.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yamauchi KE, Snel J. Transmission electron microscopic demonstration of phagocytosis and intracellular processing of segmented filamentous bacteria by intestinal epithelial cells of the chick ileum. Infect. Immun. 2000;68:6496–-504. doi: 10.1128/iai.68.11.6496-6504.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Umesaki Y, Okada Y, Matsumoto S, Imaoka A, Setoyama H. Segmented filamentous bacteria are indigenous intestinal bacteria that activate intraepithelial lymphocytes and induce MHC class II molecules and fucosyl asialo GM1 glycolipids on the small intestinal epithelial cells in the ex-germ-free mouse. Microbiol. Immunol. 1995;39:555–-62. doi: 10.1111/j.1348-0421.1995.tb02242.x. [DOI] [PubMed] [Google Scholar]
- 165.Shima T, Fukushima K, Setoyama H, Imaoka A, Matsumoto S, et al. Differential effects of two probiotic strains with different bacteriological properties on intestinal gene expression, with special reference to indigenous bacteria. FEMS Immunol. Med. Microbiol. 2008;52:69–-77. doi: 10.1111/j.1574-695X.2007.00344.x. [DOI] [PubMed] [Google Scholar]
- 166.Heczko U, Abe A, Finlay BB. Segmented filamentous bacteria prevent colonization of enteropathogenic Escherichia coli O103 in rabbits. J. Infect. Dis. 2000;181:1027–-33. doi: 10.1086/315348. [DOI] [PubMed] [Google Scholar]
- 167.Sears CL, Islam S, Saha A, Arjumand M, Alam NH, et al. Association of enterotoxigenic Bacteroides fragilis infection with inflammatory diarrhea. Clin. Infect. Dis. 2008;47:797–-803. doi: 10.1086/591130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 2009;15:1016–-22. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Esplugues E, Huber S, Gagliani N, Hauser AE, Town T, et al. Control of TH17 cells occurs in the small intestine. Nature. 2011;475:514–-18. doi: 10.1038/nature10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lochner M, Berard M, Sawa S, Hauer S, Gaboriau-Routhiau V, et al. Restricted microbiota and absence of cognate TCR antigen leads to an unbalanced generation of Th17 cells. J. Immunol. 2011;186:1531–-37. doi: 10.4049/jimmunol.1001723. [DOI] [PubMed] [Google Scholar]
- 171.Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–-54. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Marks BR, Nowyhed HN, Choi JY, Poholek AC, Odegard JM, et al. Thymic self-reactivity selects natural interleukin 17-producing T cells that can regulate peripheral inflammation. Nat. Immunol. 2009;10:1125–-32. doi: 10.1038/ni.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kim JS, Smith-Garvin JE, Koretzky GA, Jordan MS. The requirements for natural Th17 cell development are distinct from those of conventional Th17 cells. J. Exp. Med. 2011;208:2201–-7. doi: 10.1084/jem.20110680. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 174.van Wijk F, Cheroutre H. Intestinal T cells: facing the mucosal immune dilemma with synergy and diversity. Semin. Immunol. 2009;21:130–-38. doi: 10.1016/j.smim.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Das G, Augustine MM, Das J, Bottomly K, Ray P, Ray A. An important regulatory role for CD4+CD8αα T cells in the intestinal epithelial layer in the prevention of inflammatory bowel disease. Proc. Natl. Acad. Sci. USA. 2003;100:5324–-29. doi: 10.1073/pnas.0831037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Leishman AJ, Naidenko OV, Attinger A, Koning F, Lena CJ, et al. T cell responses modulated through interaction between CD8αα and the nonclassical MHC class I molecule, TL. Science. 2001;294:1936–-39. doi: 10.1126/science.1063564. [DOI] [PubMed] [Google Scholar]
- 177.Denning TL, Granger SW, Mucida D, Graddy R, Leclercq G, et al. Mouse TCRαβ+CD8αα intraepithelial lymphocytes express genes that down-regulate their antigen reactivity and suppress immune responses. J. Immunol. 2007;178:4230–-39. doi: 10.4049/jimmunol.178.7.4230. [DOI] [PubMed] [Google Scholar]
- 178.Poussier P, Ning T, Banerjee D, Julius M. A unique subset of self-specific intraintestinal T cells maintains gut integrity. J. Exp. Med. 2002;195:1491–-97. doi: 10.1084/jem.20011793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bauer S, Groh V, Wu J, Steinle A, Phillips JH, et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285:727–-29. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- 180.Boismenu R, Havran WL. Modulation of epithelial cell growth by intraepithelial γδ T cells. Science. 1994;266:1253–-55. doi: 10.1126/science.7973709. [DOI] [PubMed] [Google Scholar]
- 181.Ismail AS, Severson KM, Vaishnava S, Behrendt CL, Yu X, et al. γδ intraepithelial lymphocytes are essential mediators of host-microbial homeostasis at the intestinal mucosal surface. Proc. Natl. Acad. Sci. USA. 2011;108:8743–-48. doi: 10.1073/pnas.1019574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bandeira A, Mota-Santos T, Itohara S, Degermann S, Heusser C, et al. Localization of γ/δ T cells to the intestinal epithelium is independent of normal microbial colonization. J. Exp. Med. 1990;172:239–-44. doi: 10.1084/jem.172.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Martin B, Hirota K, Cua DJ, Stockinger B, Veldhoen M. Interleukin-17-producing γδ T cells selectively expand in response to pathogen products and environmental signals. Immunity. 2009;31:321–-30. doi: 10.1016/j.immuni.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 184.Lockhart E, Green AM, Flynn JL. IL-17 production is dominated by γδ T cells rather than CD4 T cells during Mycobacterium tuberculosis infection. J. Immunol. 2006;177:4662–-69. doi: 10.4049/jimmunol.177.7.4662. [DOI] [PubMed] [Google Scholar]
- 185.Shibata K, Yamada H, Hara H, Kishihara K, Yoshikai Y. Resident Vδ1+ γδ T cells control early infiltration of neutrophils after Escherichia coli infection via IL-17 production. J. Immunol. 2007;178:4466–-72. doi: 10.4049/jimmunol.178.7.4466. [DOI] [PubMed] [Google Scholar]
- 186.Awasthi A, Riol-Blanco L, Jager A, Korn T, Pot C, et al. Cutting edge: IL-23 receptor GFP reporter mice reveal distinct populations of IL-17-producing cells. J. Immunol. 2009;182:5904–-8. doi: 10.4049/jimmunol.0900732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sutton CE, Lalor SJ, Sweeney CM, Brereton CF, Lavelle EC, Mills KH. Interleukin-1 and IL-23 induce innate IL-17 production from γδ T cells, amplifying Th17 responses and autoimmunity. Immunity. 2009;31:331–-41. doi: 10.1016/j.immuni.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 188.Duan J, Chung H, Troy E, Kasper DL. Microbial colonization drives expansion of IL-1 receptor 1-expressing and IL-17-producing γ/δ T cells. Cell Host Microbe. 2010;7:140–-50. doi: 10.1016/j.chom.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ito Y, Usui T, Kobayashi S, Iguchi-Hashimoto M, Ito H, et al. γ/δ T cells are the predominant source of interleukin-17 in affected joints in collagen-induced arthritis, but not in rheumatoid arthritis. Arthritis Rheum. 2009;60:2294–-303. doi: 10.1002/art.24687. [DOI] [PubMed] [Google Scholar]
- 190.Petermann F, Rothhammer V, Claussen MC, Haas JD, Blanco LR, et al. γδ T cells enhance autoimmunity by restraining regulatory T cell responses via an interleukin-23-dependent mechanism. Immunity. 2010;33:351–-63. doi: 10.1016/j.immuni.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Cua DJ, Tato CM. Innate IL-17-producing cells: the sentinels of the immune system. Nat. Rev. Immunol. 2010;10:479–-89. doi: 10.1038/nri2800. [DOI] [PubMed] [Google Scholar]
- 192.Mebius RE, Rennert P, Weissman IL. Developing lymph nodes collect CD4+CD3-LTβ+ cells that can differentiate to APC, NK cells, and follicular cells but not T or B cells. Immunity. 1997;7:493–-504. doi: 10.1016/s1074-7613(00)80371-4. [DOI] [PubMed] [Google Scholar]
- 193.Eberl G, Marmon S, Sunshine MJ, Rennert PD, Choi Y, Littman DR. An essential function for the nuclear receptor RORγ(t) in the generation of fetal lymphoid tissue inducer cells. Nat. Immunol. 2004;5:64–-73. doi: 10.1038/ni1022. [DOI] [PubMed] [Google Scholar]
- 194.Ivanov II, Diehl GE, Littman DR. Lymphoid tissue inducer cells in intestinal immunity. Curr. Top. Microbiol. Immunol. 2006;308:59–-82. doi: 10.1007/3-540-30657-9_3. [DOI] [PubMed] [Google Scholar]
- 195.Cupedo T, Crellin NK, Papazian N, Rombouts EJ, Weijer K, et al. Human fetal lymphoid tissue-inducer cells are interleukin 17-producing precursors to RORC+ CD127+ natural killer-like cells. Nat. Immunol. 2009;10:66–-74. doi: 10.1038/ni.1668. [DOI] [PubMed] [Google Scholar]
- 196.Honda K, Nakano H, Yoshida H, Nishikawa S, Rennert P, et al. Molecular basis for hematopoietic/mesenchymal interaction during initiation of Peyer's patch organogenesis. J. Exp. Med. 2001;193:621–-30. doi: 10.1084/jem.193.5.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Eberl G, Littman DR. Thymic origin of intestinal αβ T cells revealed by fate mapping of RORγt+ cells. Science. 2004;305:248–-51. doi: 10.1126/science.1096472. [DOI] [PubMed] [Google Scholar]
- 198.Sawa S, Cherrier M, Lochner M, Satoh-Takayama N, Fehling HJ, et al. Lineage relationship analysis of RORγt+ innate lymphoid cells. Science. 2010;330:665–-69. doi: 10.1126/science.1194597. [DOI] [PubMed] [Google Scholar]
- 199.Kim MY, Gaspal FM, Wiggett HE, McConnell FM, Gulbranson-Judge A, et al. CD4+CD3− accessory cells costimulate primed CD4 T cells through OX40 and CD30 at sites where T cells collaborate with B cells. Immunity. 2003;18:643–-54. doi: 10.1016/s1074-7613(03)00110-9. [DOI] [PubMed] [Google Scholar]
- 200.Sawa S, Lochner M, Satoh-Takayama N, Dulauroy S, Berard M, et al. RORγt+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat. Immunol. 2011;12:320–-26. doi: 10.1038/ni.2002. [DOI] [PubMed] [Google Scholar]
- 201.Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, Sawa S, Lochner M, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–-70. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 202.Sanos SL, Bui VL, Mortha A, Oberle K, Heners C, et al. RORγt and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat. Immunol. 2009;10:83–-91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Luci C, Reynders A, Ivanov II, Cognet C, Chiche L, et al. Influence of the transcription factor RORγt on the development of NKp46+ cell populations in gut and skin. Nat. Immunol. 2009;10:75–-82. doi: 10.1038/ni.1681. [DOI] [PubMed] [Google Scholar]
- 204.Zenewicz LA, Yancopoulos GD, Valenzuela DM, Murphy AJ, Stevens S, Flavell RA. Innate and adaptive interleukin-22 protects mice from inflammatory bowel disease. Immunity. 2008;29:947–-57. doi: 10.1016/j.immuni.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Uhlig HH, McKenzie BS, Hue S, Thompson C, Joyce-Shaikh B, et al. Differential activity of IL-12 and IL-23 in mucosal and systemic innate immune pathology. Immunity. 2006;25:309–-18. doi: 10.1016/j.immuni.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 206.Hue S, Ahern P, Buonocore S, Kullberg MC, Cua DJ, et al. Interleukin-23 drives innate and T cell-mediated intestinal inflammation. J. Exp. Med. 2006;203:2473–-83. doi: 10.1084/jem.20061099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Buonocore S, Ahern PP, Uhlig HH, Ivanov II, Littman DR, et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464:1371–-75. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–-87. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 209.Izcue A, Coombes JL, Powrie F. Regulatory T cells suppress systemic and mucosal immune activation to control intestinal inflammation. Immunol. Rev. 2006;212:256–-71. doi: 10.1111/j.0105-2896.2006.00423.x. [DOI] [PubMed] [Google Scholar]
- 210.Barnes MJ, Powrie F. Regulatory T cells reinforce intestinal homeostasis. Immunity. 2009;31:401–-11. doi: 10.1016/j.immuni.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 211.Maloy KJ, Salaun L, Cahill R, Dougan G, Saunders NJ, Powrie F. CD4+CD25+ T(R) cells suppress innate immune pathology through cytokine-dependent mechanisms. J. Exp. Med. 2003;197:111–-19. doi: 10.1084/jem.20021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Zheng Y, Rudensky AY. Foxp3 in control of the regulatory T cell lineage. Nat. Immunol. 2007;8:457–-62. doi: 10.1038/ni1455. [DOI] [PubMed] [Google Scholar]
- 213.Onishi Y, Fehervari Z, Yamaguchi T, Sakaguchi S. Foxp3+ natural regulatory T cells preferentially form aggregates on dendritic cells in vitro and actively inhibit their maturation. Proc. Natl. Acad. Sci. USA. 2008;105:10113–-18. doi: 10.1073/pnas.0711106105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Griseri T, Asquith M, Thompson C, Powrie F. OX40 is required for regulatory T cell-mediated control of colitis. J. Exp. Med. 2010;207:699–-709. doi: 10.1084/jem.20091618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat. Immunol. 2009;10:595–-602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Zheng Y, Chaudhry A, Kas A, deRoos P, Kim JM, et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–-56. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, et al. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–-91. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Tsuji M, Komatsu N, Kawamoto S, Suzuki K, Kanagawa O, et al. Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer's patches. Science. 2009;323:1488–-92. doi: 10.1126/science.1169152. [DOI] [PubMed] [Google Scholar]
- 219.Curotto de Lafaille MA, Lino AC, Kutchukhidze N, Lafaille JJ. CD25- T cells generate CD25+Foxp3+ regulatory T cells by peripheral expansion. J. Immunol. 2004;173:7259–-68. doi: 10.4049/jimmunol.173.12.7259. [DOI] [PubMed] [Google Scholar]
- 220.Chen W, Jin W, Hardegen N, Lei KJ, Li L, et al. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-β induction of transcription factor Foxp3. J. Exp. Med. 2003;198:1875–-86. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Mucida D, Park Y, Kim G, Turovskaya O, Scott I, et al. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007;317:256–-60. doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- 222.Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-β and retinoic acid-dependent mechanism. J. Exp. Med. 2007;204:1757–-64. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, et al. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J. Exp. Med. 2007;204:1775–-85. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Curotto de Lafaille MA, Kutchukhidze N, Shen S, Ding Y, Yee H, Lafaille JJ. Adaptive Foxp3+ regulatory T cell-dependent and -independent control of allergic inflammation. Immunity. 2008;29:114–-26. doi: 10.1016/j.immuni.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 225.Thornton AM, Korty PE, Tran DQ, Wohlfert EA, Murray PE, et al. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J. Immunol. 2010;184:3433–-41. doi: 10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–-41. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–-806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 228.Kamanaka M, Kim ST, Wan YY, Sutterwala FS, Lara-Tejero M, et al. Expression of interleukin-10 in intestinal lymphocytes detected by an interleukin-10 reporter knockin tiger mouse. Immunity. 2006;25:941–-52. doi: 10.1016/j.immuni.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 229.Maynard CL, Harrington LE, Janowski KM, Oliver JR, Zindl CL, et al. Regulatory T cells expressing interleukin 10 develop from Foxp3+ and Foxp3– precursor cells in the absence of interleukin 10. Nat. Immunol. 2007;8:931–-41. doi: 10.1038/ni1504. [DOI] [PubMed] [Google Scholar]
- 230.Roers A, Siewe L, Strittmatter E, Deckert M, Schluter D, et al. T cell-specific inactivation of the interleukin 10 gene in mice results in enhanced T cell responses but normal innate responses to lipopolysaccharide or skin irritation. J. Exp. Med. 2004;200:1289–-97. doi: 10.1084/jem.20041789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Rubtsov YP, Rasmussen JP, Chi EY, Fontenot J, Castelli L, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–-58. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 232.Park SG, Mathur R, Long M, Hosh N, Hao L, et al. T regulatory cells maintain intestinal homeostasis by suppressing γδ T cells. Immunity. 2010;33:791–-803. doi: 10.1016/j.immuni.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Huber S, Gagliani N, Esplugues E, O'Connor W, Jr, Huber FJ, et al. Th17 cells express interleukin-10 receptor and are controlled by Foxp3 and Foxp3+ regulatory CD4+ T cells in an interleukin-10-dependent manner. Immunity. 2011;34:554–-65. doi: 10.1016/j.immuni.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Chaudhry A, Samstein RM, Treuting P, Liang Y, Pils MC, et al. Interleukin-10 signaling in regulatory T cells is required for suppression of Th17 cell-mediated inflammation. Immunity. 2011;34:566–-78. doi: 10.1016/j.immuni.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Pot C, Jin H, Awasthi A, Liu SM, Lai CY, et al. Cutting edge: IL-27 induces the transcription factor c-Maf, cytokine IL-21, and the costimulatory receptor ICOS that coordinately act together to promote differentiation of IL-10-producing Tr1 cells. J. Immunol. 2009;183:797–-801. doi: 10.4049/jimmunol.0901233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Apetoh L, Quintana FJ, Pot C, Joller N, Xiao S, et al. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat. Immunol. 2010;11:854–-61. doi: 10.1038/ni.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Cretney E, Xin A, Shi W, Minnich M, Masson F, et al. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat. Immunol. 2011;12:304–-11. doi: 10.1038/ni.2006. [DOI] [PubMed] [Google Scholar]
- 238.Johansson-Lindbom B, Svensson M, Pabst O, Palmqvist C, Marquez G, et al. Functional specialization of gut CD103+ dendritic cells in the regulation of tissue-selective T cell homing. J. Exp. Med. 2005;202:1063–-73. doi: 10.1084/jem.20051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Denning TL, Wang YC, Patel SR, Williams IR, Pulendran B. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T cell responses. Nat. Immunol. 2007;8:1086–-94. doi: 10.1038/ni1511. [DOI] [PubMed] [Google Scholar]
- 240.Manicassamy S, Reizis B, Ravindran R, Nakaya H, Salazar-Gonzalez RM, et al. Activation of β-catenin in dendritic cells regulates immunity versus tolerance in the intestine. Science. 2010;329:849–-53. doi: 10.1126/science.1188510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.DePaolo RW, Abadie V, Tang F, Fehlner-Peach H, Hall JA, et al. Co-adjuvant effects of retinoic acid and IL-15 induce inflammatory immunity to dietary antigens. Nature. 2011;471:220–-24. doi: 10.1038/nature09849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Di Giacinto C, Marinaro M, Sanchez M, Strober W, Boirivant M. Probiotics ameliorate recurrent Th1-mediated murine colitis by inducing IL-10 and IL-10-dependent TGF-β-bearing regulatory cells. J. Immunol. 2005;174:3237–-46. doi: 10.4049/jimmunol.174.6.3237. [DOI] [PubMed] [Google Scholar]
- 243.Karimi K, Inman MD, Bienenstock J, Forsythe P. Lactobacillus reuteri-induced regulatory T cells protect against an allergic airway response in mice. Am. J. Respir. Crit. Care Med. 2009;179:186–-93. doi: 10.1164/rccm.200806-951OC. [DOI] [PubMed] [Google Scholar]
- 244.Livingston M, Loach D, Wilson M, Tannock GW, Baird M. Gut commensal Lactobacillus reuteri 100--23 stimulates an immunoregulatory response. Immunol. Cell Biol. 2009;88:99–-102. doi: 10.1038/icb.2009.71. [DOI] [PubMed] [Google Scholar]
- 245.Torii A, Torii S, Fujiwara S, Tanaka H, Inagaki N, Nagai H. Lactobacillus acidophilus strain L-92 regulates the production of Th1 cytokine as well as Th2 cytokines. Allergol. Int. 2007;56:293–-301. doi: 10.2332/allergolint.O-06-459. [DOI] [PubMed] [Google Scholar]
- 246.Foligne B, Zoumpopoulou G, Dewulf J, Ben Younes A, Chareyre F, et al. A key role of dendritic cells in probiotic functionality. PLoS One. 2007;2:e313. doi: 10.1371/journal.pone.0000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Forsythe P, Inman MD, Bienenstock J. Oral treatment with live Lactobacillus reuteri inhibits the allergic airway response in mice. Am. J. Respir. Crit. Care Med. 2007;175:561–-69. doi: 10.1164/rccm.200606-821OC. [DOI] [PubMed] [Google Scholar]
- 248.O'Mahony C, Scully P, O'Mahony D, Murphy S, O'Brien F, et al. Commensal-induced regulatory T cells mediate protection against pathogen-stimulated NF-κB activation. PLoS Pathog. 2008;4:e1000112. doi: 10.1371/journal.ppat.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. USA. 2010;107:12204–-9. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–-25. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 251.Round JL, Lee SM, Li J, Tran G, Jabri B, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–-77. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.D'Angelo M, Billings PC, Pacifici M, Leboy PS, Kirsch T. Authentic matrix vesicles contain active metalloproteases (MMP). a role for matrix vesicle-associated MMP-13 in activation of transforming growth factor-β. J. Biol. Chem. 2001;276:11347–-53. doi: 10.1074/jbc.M009725200. [DOI] [PubMed] [Google Scholar]
- 253.Matteoli G, Mazzini E, Iliev ID, Mileti E, Fallarino F, et al. Gut CD103+ dendritic cells express indoleamine 2,3-dioxygenase which influences T regulatory/T effector cell balance and oral tolerance induction. Gut. 2010;59:595–-604. doi: 10.1136/gut.2009.185108. [DOI] [PubMed] [Google Scholar]
- 254.Bloom SM, Bijanki VN, Nava GM, Sun L, Malvin NP, et al. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe. 2011;9:390–-403. doi: 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]