Abstract
Introduction: The brain‐derived neurotrophic factor (BDNF) Val66Met variant and HMG‐COA reductase inhibitors (statins) have been implicated in insulin resistance with a possible increased risk of diabetes. We sought to determine the effect of the BDNF Met variant and statin medication use on insulin resistance in schizophrenia and bipolar disorder using the homeostasis model assessment of insulin resistance (HOMA‐IR).
Methods: A cross‐sectional design was used and patients with diabetes or on any medications affecting glucose regulation were ‐excluded. Associations between insulin resistance and genotype were then analyzed by ANOVA and regression analysis. Subjects were grouped by BDNF genotype as well as presence of statin.
Results: Two hundred fifty‐two subjects with a mean age of 44 years were included. The group was 53% male and 41% had a diagnosis of bipolar disorder; 78% and 19% were receiving atypical antipsychotics (AAPs) and statin medications, respectively. Analysis showed schizophrenia subjects with the BDNF met allele as well as schizophrenia subjects with both the BDNF met allele and were receiving a statin had significantly higher HOMA‐IR values compared to the other groups (p= 0.046 and p= 0.016, respectively).
Conclusions: Our results suggest that in the metabolically high‐risk population of schizophrenia the BDNF met allele alone and in combination with statin medications is associated with higher insulin resistance values. This was not seen in the bipolar population. Further validation of these associations remains necessary. Clin Trans Sci 2012; Volume 5: 486–490
Keywords: pharmacogenomic, diabetes, psychology
Introduction
Antipsychotic use is a common treatment for patients with schizophrenia and bipolar disorder. With the advent of the atypical antipsychotics (AAPs), the rates of undesirable metabolic complications like weight gain and type II diabetes mellitus has increased within these patient populations. 1 In addition, lifestyle habits and pharmacogenetics have been touted as having a role in developing type II diabetes within these populations. 2 , 3 Although several pharmacogenetic variants have been implicated in the metabolic dysregulations seen with AAP use, 4 , 5 one that is currently emerging within the general population is the brain‐derived neurotrophic factor (BDNF). 6 , 7 The BDNF Val66Met (196 G/A) variant has been implicated in both schizophrenia and bipolar disorder due to its role in neuronal health, development, and function. 8 , 9
Pharmacogenetically, a BDNF gene variant identified at codon 66 results in a methionine (Met) substitution for valine (Val) and results in a drastic impairment of BDNF packaging and distribution. 10 In animal and human models within the general population, the Met allele is associated with an increased risk of diabetes and insulin resistance through altered regulation of glucose metabolism and feeding habits. 11 Given the strong relationship between mental illness, AAP treatment, and glucose regulation, BDNF genetic variability may mediate development of type 2 diabetes (T2DM) in a large percentage of AAP treated patients. However, the relationship between this variant and glucose dysregulation seen in schizophrenia and bipolar disorder has not been studied. Although all metabolic complications carry a certain degree of risk, development of T2DM adds additional risks. Therefore, these patients require careful clinical monitoring.
It is standard of practice to treat cholesterol abnormalities with hydroxymethylglutaryl CoA reductase inhibitors also known as “statins.” These medications have recently been implicated in increasing the risk of diabetes and insulin resistance 12 and also have effects on circulating BDNF and the cleavage of pro‐BDNF in certain populations. 13 , 14 Their cautious use in populations at increased risk for diabetes like that of schizophrenia and bipolar disorder has been advised, however, work delineating the true extent of this risk in those with a serious mental illness is unknown.
This investigation sought to determine the relationship between the BDNF 66 Met allele and insulin resistance in schizophrenia and bipolar patients. In addition, we determined the relationship between statin medication use, BDNF 66Met allele carriers and insulin resistance in this same cohort.
Materials and Methods
Subjects and data collection
Participants were recruited from Southeastern Michigan mental health clinics using the following study inclusion criteria: (i) Diagnostic and Statistical Manual of Mental Disorders Fourth Edition (DSM‐IV) diagnosis of schizophrenia, schizophreniform disorder, schizoaffective disorder, or bipolar disorder, (ii) 18–90 years old, and (iii) no medication changes in the prior 6 months. Participants were excluded if they were unable to give informed consent, had diabetes before starting antipsychotic treatment, had any condition significantly affecting glucose and insulin measures or had an active substance abuse diagnosis. The protocol was approved by the University of Michigan Institutional Review Boards.
Study visits took place within 2 hours of the subject's usual wakening time, with most visits being completed by 11 am. Height, weight, and waist circumference were measured in all participants. Fasting plasma glucose, triglycerides, total cholesterol, high‐density lipoprotein and low‐density lipoprotein measures were obtained, and the homeostasis model assessment of insulin resistance (HOMA‐IR) 15 was calculated for each patient using the following formula:
Although various thresholds for insulin resistance have been identified, 16 a HOMA‐IR above 2.6 was used as a threshold for insulin resistance. 17
Subjects were excluded from this analysis if they had a documented diabetes diagnosis or were receiving diabetes treatment since treatment for diabetes would artificially lower insulin resistance values giving a false negative finding using the HOMA‐IR.
Each patient was assessed for metabolic syndrome using the National Cholesterol Education Program Adult Treatment Panel III (NCEP ATP‐III Criteria). 18 Structured Clinical Interview for DSM Diagnoses (SCID) conducted by a trained research assistant and medical record review was used to confirm diagnoses. Participant complete medication histories were collected by medical record and patient interview. Social histories were also completed. Subjects were segregated to account for metabolic side effects differences for a given antipsychotic as follows: (1) olanzapine or clozapine, (2) quetiapine, risperidone, paliperidone or iloperidone, and (3) all other antipsychotics or no antipsychotic.
Analytical methods
DNA was extracted from whole blood 19 and for genotyped for the BDNF Val66Met variant (DBSNP rs6265) using Pyrosequencing. 20 Pyrosequencing primers were designed using Pyrosequencing Nucleotide Polymorphism (SNP) primer design version 1.01 software (http://www.pyrosequencing.com). Conditions for the specific assay are available upon request.
Statistical analysis
Due to the anticipated low‐genotype frequency of the BDNF 66MetMet genotype, the statistical analysis was performed by forming two groups: ValVal genotype and those carrying a Met allele (ValMet or MetMet genotypes). For our second aim, the genotype groups were stratified by presence or absence of statin use. The primary group of interest within this second aim was those subjects taking statins and carrying a 66 Met Allele compared to all others. A power analysis showed that to detect a clinically meaningful difference of 1.0 in HOMA‐IR with a power of 80% a sample size approximately 56 was needed. Student t‐tests and chi‐square analyses were used for baseline demographics comparisons based on the two genotype groupings. ANOVA examined the relationship of genotype, statins, and HOMA‐IR. Regression analysis was performed to analyze the relationship between BDNF genotype status and HOMA‐IR, whereas controlling for variables we found to significantly effect HOMA‐IR. HOMA‐IR was used as the dependent variable while the genotype‐based groups and the variables affecting HOMA‐IR (detailed in results) were used as the independent variables for each aim stated above. We then analyzed based on each diagnosis given the differences in pathogenesis and medication use for these two diseases. All analyses utilized the JMP 9.0 statistical program. A p‐value < 0.05 with a 95% confidence interval was considered significant.
Results
After excluding based on the criteria above (diagnosis of diabetes or current diabetes treatment), our final sample size for data analysis was 252 (148 with a schizophrenia spectrum diagnosis and 104 with a bipolar diagnosis). See Table 1 for demographic characteristics. The BDNF Val66Met genotypes were in Hardy Weinberg Equilibrium for the entire cohort as well as for each diagnosis spectrum (all p > 0.26). Eight participants were unable to be genotyped.
Table 1.
Analysis of demographic variables.
| Variable | Schizophrenia spectrum disorders (n= 148) | Bipolar disorder (n= 104) | Combined sample (n= 252) |
|---|---|---|---|
| Age (years ± s.d) | 44.8 ± 11.7 | 42.7 ± 12.0 | 43.9 ± 11.9 |
| % Male* | 66 | 35 | 53 |
| % Caucasian/% African American* | 61/30 | 84/11 | 71/22 |
| % On Atypical Antipsychotics* | 83 | 70 | 78 |
| % On Clozapine or Olanzapine/% On Risperidone, Quetiapine, Iloperidone or Paliperidone* | 28/37 | 15/56 | 23/43 |
| % On Statins | 19 | 18 | 19 |
| % Smoking* | 49 | 32 | 42 |
| % With ATP III/NCEP Metabolic Syndrome | 30 | 26 | 29 |
| BMI (kg/m2± s.d.) | 31.4 ± 7.15 | 32.1 ± 8.88 | 31.7 ± 7.9 |
| Blood pressure (mmHg ± s.d.) | 122/75 ± 16/12 | 124/73 ± 18/11 | 123/74 ± 17/11 |
| Total cholesterol (mg/dL ± s.d.)* | 176 ± 39.2 | 192 ± 44.7 | 182 ± 42.2 |
| Triglycerides (mg/dL ± s.d.) | 131 ± 89.3 | 138 ± 104 | 134 ± 95.7 |
| High‐density lipoprotein (mg/dL ± s.d)* | 52.7 ± 16.3 | 58 ± 14.9 | 54.9 ± 15.9 |
| Low‐density lipoprotein (mg/dL ± s.d.)* | 108 ± 30.6 | 118 ± 38.2 | 112 ± 34.3 |
| Insulin (mU/L ± s.d.) | 22.9 ± 15.2 | 24.2 ± 20.8 | 23.5 ± 17.7 |
| Glucose (mg/dL ± s.d.) | 96.1 ± 12.5 | 96.0 ± 12.0 | 96.0 ± 12.3 |
| HbA1C% | 5.63 | 5.55 | 5.60 |
| HOMA‐IR ± s.d. | 5.51 ± 3.92 | 5.91 ± 5.46 | 5.68 ± 4.62 |
| % BDNF Val66Met Genotype (%) | 73 ValVal/24 ValMet/3 MetMet | 77 ValVal/20 ValMet/3 MetMet | 74 ValVal/23 ValMet/3 MetMet |
| % With BDNF 66Met Allele | 28 | 23 | 26 |
*Indicates p < 0.05 when comparing the Schizophrenia and Bipolar samples.
Comparisons of all subject found a few significant differences. The schizophrenia cohort had a higher percentage of male and African American subjects compared to the bipolar cohort (p < 0.05 for both), there were more schizophrenia subjects receiving AAPs (p= 0.02), differences in the percentage of subjects who were current smokers (p= 0.01) and differences in lipid levels between the two cohorts (Total Cholesterol: p= 0.003; HDL: p= 0.01; LDL: p= 0.02). See Table 1 for percentage comparisons between the two groups.
Preliminary analysis found that in the combined sample, the Met allele carriers were older (46 vs. 43 years old, p= 0.043), less likely to be African Americans (5% vs. 26%, p= 0.005), and had higher AAP use (89% vs. 73%, p= 0.010). There were no significant differences for the schizophrenia or bipolar groups based on genotype
No significant relationship was found between the BDNF 66 Met allele and HOMA‐IR for the whole group (p= 0.3). However when segregated by diagnosis, we found a significant effect of the BDNF 66Met allele on HOMA‐IR in the schizophrenia sample where subjects carrying the Met allele had higher HOMA‐IRs (p= 0.046). No relationship was found for the bipolar group (analysis in Table 2 , Figure 1 ).
Table 2.
Analysis of HOMA‐IR based on BDNF Met carrier and Statin Status.
| Subject groups | Schizophrenia spectrum disorders | Bipolar disorder | Combined sample | |||
|---|---|---|---|---|---|---|
| HOMA‐IR ± s.d. | p‐value | HOMA‐IR ± s.d. | p‐value | HOMA‐IR ± s.d. | p‐value | |
| BDNF 66ValVal Genotype | 5.10 ± 0.400 | 0.046 | 6.04 ± 0.621 | 0.7 | 5.52 ± 0.352 | 0.3 |
| BDNF 66Met Allele Carriers | 6.63 ± 0.642 | 5.47 ± 1.12 | 6.18 ± 0.594 | |||
| BDNF Val66 with or without Statins and BDNF 66Met without Statins | 5.30 ± 0.350 | 0.016 | 5.99 ± 0.553 | 0.5 | 5.60 ± 0.312 | 0.2 |
| BDNF 66Met Carriers and Currently Taking a Statin | 8.45 ± 1.24 | 3.99 ± 2.74 | 7.15 ± 1.25 | |||
Figure 1.
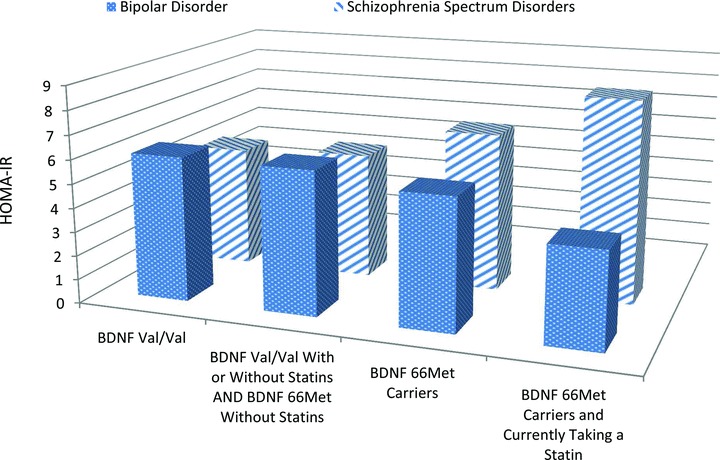
Graph of Analysis of HOMA‐IR based on BDNF Met carrier and Statin Status. This figure depicts the analysis detailed in Table 2 . The groups “BDNF Val/Val” and “BDNF 66Met Carriers” are from the primary analysis of this study looking at the pharmacogenetic effect of BDNF Val66Met on insulin resistance. The groups “BDNF Val/Val with or without statins and BDNF 66Met without Statins” and “BDNF 66Met Carriers and currently taking a Statin” are from the secondary analysis stratifying BDNF Val66Met status by current statin use. The figure depicts the differing effects seen in the bipolar and schizophrenia samples of adding these variables together on the insulin resistance measure of HOMA‐IR.
There were no effects of statin medications on BDNF 66Met allele carriers in the entire cohort (p= 0.2). However, the schizophrenia subjects currently taking statin medications and carrying the Met allele had significantly higher HOMA‐IRs compared to all other schizophrenia subjects (p= 0.016). No relationship was found in the bipolar cohort (analysis in Table 2 ).
In addition to the previous analysis, a linear regression was performed for each dependent variable (e.g., BDNF and statin status) controlling for age, race, AAP use, BMI and metabolic syndrome. In the combined sample, there was a significant interaction between BMI and BDNF 66Met carriers on HOMA‐IR (f[1,1]= 3.90, p= 0.049), where those with this allele, had higher levels of insulin resistance and similar BMIs compared to the Val/Val genotype groups. This analysis yielded no significant results for the bipolar sample. Within the schizophrenia subset, the effect of the BDNF 66Met allele on HOMA‐IR was significant by itself (f[11,135]= 4.80, p= 0.030) as well as 66Met's interaction with BMI on HOMA‐IR (f(1,1) = 9.04, p= 0.0032). Finally, only the interaction between BMI and genotype was seen in the schizophrenia sample when the analysis was stratified by statin use (f[1,1]= 6.26, p= 0.037), where those with the 66Met allele receiving a statin also had higher levels of insulin resistance at comparable BMIs.
Discussion
For this study, we found a significant relationship between BMI, the BDNF 66 Met allele and insulin resistance in a schizophrenia population primarily treated with AAPs. Our study showed that BDNF pharmacogenetics may not be the same across different populations prescribed AAPs or statins with the lack of a relationship seen in the combined and bipolar groups. This may suggest glucose regulation is more dependent on genetic factors like BDNF within schizophrenia. Of note, is that within our samples, the HOMA‐IR values are substantially higher than general population studies, which is not surprising given the rising incidence of diabetes and metabolic syndrome. Studies in other populations have HOMA‐IR averages of 1.8–2.9 whereas our average was much higher at 5.51 and 5.91 ( Table 1 ) for the schizophrenia and bipolar samples, respectively. 21 , 22 Contributing to the elevated HOMA‐IR may be high AAP use (78% overall) with the majority being AAPs with strong metabolic side effect profiles. Although not statistically significant, schizophrenia subjects on clozapine, olanzapine, risperidone, paliperidone or quetiapine (68%) had a mean HOMA‐IR of 5.8 versus those on ziprasidone, aripiprazole or 1st generation antipsychotics with a HOMA‐IR of 5.0 (p= 0.25) which is consistent with the literature. 23 Poor diet and a lack of activity could also be contributors to these high HOMA‐IR values. Furthermore, for the combined sample, 80% had a HOMA‐IR higher than a suggested threshold of 2.6 for insulin resistance..
Our study contradicts previous reports showing no association between the BDNF Val66Met polymorphism and HOMA‐IR in the diabetic population 6 as we found a significant association between BMI, the BDNF 66Met allele and insulin resistance within our schizophrenia sample. Thus, the BDNF Val66Met variant which has been studied for its psychiatric and cognitive effects 8 , 24 , 25 may also impact insulin resistance risk within schizophrenia, but perhaps not within other populations that uses AAPs. BMI and HOMA‐IR are easily measureable clinical variables that may represent risk factors in schizophrenia, along with the BDNF Val66Met genotype to help predict greatest insulin resistance risk. Subjects may then be targeted for specific interventions related to insulin resistance to prevent the development of diabetes and its detrimental comorbidities and cost. Although the bipolar sample did not show a correlation with the BDNF 66Met variant, their high insulin resistance values suggest that they should be studied further in order to determine personalized approaches to diabetes risk.
Statin medication use within the general population is becoming more controversial. We hypothesized that in subjects with the BDNF 66Met allele, statin use may result in greater insulin resistance. Based on our analysis, we found a significant relationship in schizophrenia patients taking statins and carrying the 66Met allele resulting in a HOMA‐IR measure about 1.5× higher than those patients either with the Val allele taking statins or those regardless of BDNF genotype not taking statins ( Table 2 ). Also, the combination of the 66Met allele and statin use resulted in a HOMA‐IR value more than 1.2 times higher than those patients carrying the 66Met allele alone. This was not seen in the bipolar population. After controlling for age, race, AAP use, BMI and metabolic syndrome, these relationships remained (p= 0.0032) suggesting that at any given BMI, schizophrenia BDNF 66Met allele carriers receiving a statin medication have significantly higher HOMA‐IR values. The combined effect of statin use and the BDNF 66Met allele on HOMA‐IR within this population could be a result of increased proteolytic cleavage (induced by statins) of the altered form of BDNF dictated by the Met variant whereas ValVal schizophrenia patients are unaffected by this increased cleavage since they are not producing the altered form of the BDNF protein. 10 , 13 Again, with further validation future personalized medicine approachs in schizophrenia could be developed when treating hyperlipidemia.
Figure 1 depicts the differing HOMA‐IR trends between the groups as more insulin resistance risk factors (e.g., BDNF Val66Met polymorphism and statins) are added together. Originally, we hypothesized that BDNF's pharmacogenetic effects and statins would be the same on both populations. The different trends could be a result of differing baseline characteristics between the groups (gender, AAP use, lipid levels, etc.) that could not be fully controlled during analysis, pathophysiologic processes of the diseases or differences in baseline diabetes risk. Also, it may be possible that the BDNF protein may not be a major contributor to insulin resistance in the bipolar population and thus, the hypothesis of increased BDNF cleavage we proposed for the schizophrenia population would not have the same effect. Finally, although the mechanism for statin‐induced insulin resistance has not been fully elucidated, differences in transporter expression between schizophrenia and bipolar patients could be a future line of investigation into the differences seen within our study. 26 , 27
One major study limitation is that BDNF levels were not measured, however to date, the BDNF Val66Met has not been well correlated to BDNF levels. 28 Another limitation is use of a the HOMA‐IR to assess insulin resistance, however, this measurement correlates well with more invasive measures of insulin resistance such as the frequently sampled IV glucose tolerance test. 29 , 30 The cross‐sectional design of our study only gives us a snapshot of insulin resistance and needs replication in order to confirm our results. Our study population was also not drug naïve and polypharmacy was fairly common in regards to overall number of medications, making it difficult to confirm the interaction of the BDNF gene, AAPs and statins within the mental illness. Our candidate gene drive study only looked at the BDNF Val66Met polymorphism because other BDNF polymorphisms have not been studied in insulin resistance in the literature. However, possible associations with other variants cannot be ruled out. Lastly, we did not control for multiple comparisons, however, our most significant result in the schizophrenia population would sustain the more stringent multiple testing significance level of 0.01.
Conclusion
Overall our cross‐sectional study of schizophrenia and bipolar patients receiving antipsychotic treatment showed that in schizophrenia subjects, insulin resistance is associated with the BDNF 66Met allele, statin medications and BMI. No association was found for the bipolar disorder subjects. Although further validation is needed, the BDNF Val66Met variant, thought to be promising in predicting insulin resistance and glucose dysregulation, may play a role in predicting risk of developing insulin resistance and subsequently diabetes from AAP use in the schizophrenia population. This work helps to target genes of interest which can be followed up in future investigations, focusing on personalized medicine for those with schizophrenia.
Source of funding
The following funding sources were utilized for this publication: NIMH (R01 MH082784), NIH‐NCCR, GCRC Program (UL1RR024986), the Chemistry Core of the Michigan Diabetes Research and Training Center (NIH5P60 DK 20572), and the Washtenaw Community Health Organization (WCHO), The National Alliance for Research in Schizophrenia and Depression (NARSAD), and the Prechter Longitudinal Study of Bipolar Disorder.
References
- 1. Sernyak MJ, Leslie DL, Alarcon RD, Losonczy MF, Rosenheck R. Association of diabetes mellitus with use of atypical neuroleptics in the treatment of schizophrenia. Am J Psychiatry . 2002; 159(4): 561–566. [DOI] [PubMed] [Google Scholar]
- 2. Suvisaari J, Perala J, Saarni SI, Härkänen T, Pirkola S, Joukamaa M, Koskinen S, Lönnqvist J, Reunanen A. Type 2 diabetes among persons with schizophrenia and other psychotic disorders in a general population survey. Eur Arch Psychiatry Clin Neurosci . 2008; 258(3): 129–136. [DOI] [PubMed] [Google Scholar]
- 3. Felker B, Yazel JJ, Short D. Mortality and medical comorbidity among psychiatric patients: A review. Psychiatr Serv . 1996; 47(12): 1356–1363. [DOI] [PubMed] [Google Scholar]
- 4. Ellingrod VL, Bishop JR, Moline J, Lin YC, Miller DD. Leptin and leptin receptor gene polymorphisms and increases in body mass index (BMI) from olanzapine treatment in persons with schizophrenia. Psychopharmacol Bull . 2007; 40(1): 57–62. [PubMed] [Google Scholar]
- 5. Ellingrod VL, Miller DD, Taylor SF, Moline J, Holman T, Kerr J. Metabolic syndrome and insulin resistance in schizophrenia patients receiving antipsychotics genotyped for the methylenetetrahydrofolate reductase (MTHFR) 677C/T and 1298A/C variants. Schizophr Res . 2008; 98(1–3): 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Krabbe KS, Nielsen AR, Krogh‐Madsen R, Plomgaard P, Rasmussen P, Erikstrup C, Fischer CP, Lindegaard B, Petersen AM, Taudorf S, et al Brain‐derived neurotrophic factor (BDNF) and type 2 diabetes. Diabetologia . 2007; 50(2): 431–438. [DOI] [PubMed] [Google Scholar]
- 7. Golden E, Emiliano A, Maudsley S, Windham BG, Carlson OD, Egan JM, Driscoll I, Ferrucci L, Martin B, Mattson MP. Circulating brain‐derived neurotrophic factor and indices of metabolic and cardiovascular health: Data from the baltimore longitudinal study of aging. PLoS One . 2010; 5(4): e10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buckley PF, Pillai A, Howell KR. Brain‐derived neurotrophic factor: Findings in schizophrenia. Curr Opin Psychiatry . 2011; 24(2): 122–127. [DOI] [PubMed] [Google Scholar]
- 9. Rybakowski JK. BDNF gene: Functional Val66Met polymorphism in mood disorders and schizophrenia. Pharmacogenomics . 2008; 9(11): 1589–1593. [DOI] [PubMed] [Google Scholar]
- 10. Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, Zaitsev E, Gold B, Goldman D, Dean M, et al The BDNF val66met polymorphism affects activity‐dependent secretion of BDNF and human memory and hippocampal function. Cell . 2003; 112(2): 257–269. [DOI] [PubMed] [Google Scholar]
- 11. Lyons WE, Mamounas LA, Ricaurte GA, Coppola V, Reid SW, Bora SH, Wihler C, Koliatsos VE, Tessarolio L. Brain‐derived neurotrophic factor‐deficient mice develop aggressiveness and hyperphagia in conjunction with brain serotonergic abnormalities. Proc Natl Acad Sci U S A . 1999; 96(26): 15239–15244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sattar N, Preiss D, Murray HM, Welsh P, Buckley BM, de Craen AJ, Seshasai SR, McMurray JJ, Freeman DJ, Jukema JW, et al Statins and risk of incident diabetes: A collaborative meta‐analysis of randomised statin trials. Lancet . 2010; 375(9716): 735–742. [DOI] [PubMed] [Google Scholar]
- 13. Tsai SJ. Statins may enhance the proteolytic cleavage of proBDNF: Implications for the treatment of depression. Med Hypotheses . 2007; 68(6): 1296–1299. [DOI] [PubMed] [Google Scholar]
- 14. Wu H, Lu D, Jiang H, Xiong Y, Qu C, Li B, Mahmood A, Zhou D. Chopp M. Simvastatin‐mediated upregulation of VEGF and BDNF, activation of the PI3K/Akt pathway, and increase of neurogenesis are associated with therapeutic improvement after traumatic brain injury. J Neurotrauma . 2008; 25(2): 130–139. [DOI] [PubMed] [Google Scholar]
- 15. Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: Insulin resistance and beta‐cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia . 1985; 28(7): 412–419. [DOI] [PubMed] [Google Scholar]
- 16. Radikova Z, Koska J, Huckova M, Ksinantova L, Imrich R, Vigas M, Trnovec T, Langer P, Sebokova E, Klimes I. Insulin sensitivity indices: A proposal of cut‐off points for simple identification of insulin‐resistant subjects. Exp Clin Endocrinol Diabetes . 2006; 114(5): 249–256. [DOI] [PubMed] [Google Scholar]
- 17. Ascaso JF, Pardo S, Real JT, Lorente RI, Priego A, Carmena R. Diagnosing insulin resistance by simple quantitative methods in subjects with normal glucose metabolism. Diabetes Care . 2003; 26(12): 3320–3325. [DOI] [PubMed] [Google Scholar]
- 18. National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) . Third report of the national cholesterol education program (NCEP) expert panel on detection, evaluation, and treatment of high blood cholesterol in adults (adult treatment panel III) final report. Circulation . 2002; 106(25): 3143–3421. [PubMed] [Google Scholar]
- 19. Lahiri DK, Nurnberger JI, Jr. A rapid non‐enzymatic method for the preparation of HMW DNA from blood for RFLP studies. Nucleic Acids Res . 1991; 19(19): 5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marsh S, King CR, Garsa AA, McLeod HL. Pyrosequencing of clinically relevant polymorphisms. Methods Mol Biol . 2005; 311: 97–114. [DOI] [PubMed] [Google Scholar]
- 21. Britton KA, Mukamal KJ, Ix JH, Siscovick DS, Newman AB, de Boer IH, Thacker EL, Biggs ML, Gaziano JM, Djoussé L. Insulin resistance and incident peripheral artery disease in the cardiovascular health study. Vasc Med . 2012; 17: 85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Serfaty L, Forns X, Goeser T, Ferenci P, Nevens F, Carosi G, Drenth JP, Lonjon‐Domanec I, Demasi R, Picchio G, et al Insulin resistance and response to telaprevir plus peginterferon alpha and ribavirin in treatment‐naive patients infected with HCV genotype 1. Gut . 2012; 61: 1473–1480. [DOI] [PubMed] [Google Scholar]
- 23. Rummel‐Kluge C, Komossa K, Schwarz S, Hunger H, Schmid F, Lobos CA, Kissling W, Davis JM, Leucht S. Head‐to‐head comparisons of metabolic side effects of second generation antipsychotics in the treatment of schizophrenia: A systematic review and meta‐analysis. Schizophr Res . 2010; 123(2–3): 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tsai A, Liou YJ, Hong CJ, Wu CL, Tsai SJ, Bai YM. Association study of brain‐derived neurotrophic factor gene polymorphisms and body weight change in schizophrenic patients under long‐term atypical antipsychotic treatment. Neuromolecular Med . 2011; 13(4): 328–333. [DOI] [PubMed] [Google Scholar]
- 25. Zhang XY, Zhang WF, Zhou DF, Chen da C, Xiu MH, Wu HR, Haile DN, Kosten TA, Kosten TR. Cognitive and serum BDNF correlates of BDNF Val66Met gene polymorphism in patients with schizophrenia and normal controls. Biol Psychiatry . 2012; 72: 700–706. 22695185 [Google Scholar]
- 26. Sasaki J, Iwashita M, Kono S. Statins: Beneficial or adverse for glucose metabolism. J Atheroscler Thromb . 2006; 13(3): 123–129. [DOI] [PubMed] [Google Scholar]
- 27. Nakata M, Nagasaka S, Kusaka I, Matsuoka H, Ishibashi S, Yada T. Effects of statins on the adipocyte maturation and expression of glucose transporter 4 (SLC2A4): Implications in glycaemic control. Diabetologia . 2006; 49(8): 1881–1892. [DOI] [PubMed] [Google Scholar]
- 28. Terracciano A, Piras MG, Lobina M, Mulas A, Meirelles O, Sutin AR, Chan W, Sanna S, Uda M, Crisponi L, et al Genetics of serum BDNF: Meta‐analysis of the Val66Met and genome‐wide association study. World J Biol Psychiatry . 2011. Nov 2 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Muniyappa R, Lee S, Chen H, Quon MJ. Current approaches for assessing insulin sensitivity and resistance in vivo: Advantages, limitations, and appropriate usage. Am J Physiol Endocrinol Metab . 2008; 294(1): E15–E26. [DOI] [PubMed] [Google Scholar]
- 30. Rossner SM, Neovius M, Mattsson A, Marcus C, Norgren S. HOMA‐IR and QUICKI: Decide on a general standard instead of making further comparisons. Acta Paediatr . 2010; 99(11): 1735–1740. [DOI] [PubMed] [Google Scholar]


