Abstract
Background
This study was carried out to investigate whether sex-related differences exist in the adipocyte expression of clock genes from subcutaneous abdominal and visceral fat depots in severely obese patients.
Methods
We investigated 16 morbidly obese patients, eight men and eight women (mean age 45±20 years; mean BMI 46±6 kg/m2), undergoing laparoscopic gastric bypass surgery. Biopsies were taken as paired samples [subcutaneous and visceral adipose tissue (AT)] at the beginning of the surgical process at 11:00 h in the morning. Metabolic syndrome features such as waist circumference, plasma glucose, triglycerides, total cholesterol, high-density lipoprotein cholesterol (HDL-C), and low-density lipoprotein cholesterol (LDL-C) were also studied. The expression of clock genes (PER2, BMAL1, and CRY1) was measured by quantitative real-time PCR, Western blot, and immunohistochemical analysis.
Results
Gene expression was significantly higher in women than in men for the three genes studied in both ATs (P<0.05). In visceral fat, these differences were more marked. (P<0.001). Western blot analysis partially confirmed these results since statistical differences were observed for PER2 in both ATs and for CRY1 in subcutaneous adipose tissue. There were no differences in BMAL1 protein expression. Interestingly, clock gene expression level was correlated with LDL-C and HDL-C (P<0.05). Moreover, we found significant associations with body fat mass in women and with age in men.
Conclusions
Clock genes expression is sex dependent in human adipose tissue from morbidly obese subjects and correlates to a decreased in metabolic syndrome-related traits. These preliminary results make necessary to go deep into the knowledge of the molecular basis of the sexual dimorphism in chronobiology.
Keywords: Clock genes, Sex, Human adipose tissue, Obesity, Metabolic syndrome
Introduction
There is increasing evidence supporting a relation between chronobiology and obesity spanning a wide range of scientific approaches from experimental models to population studies [1]. Circadian and metabolic systems are intertwined to the point that alterations in either one of them influence the other [2]. Circadian rhythm of metabolic processes is driven by the products of clock genes, such as Clock, Bmal1, Per2, and Cry1, and these rhythms may be crucial to maintain whole-body homeostasis including energy balance [3].
The adipose tissue is key for energy storage and fat metabolism. This tissue synthesizes and releases a number of bioactive molecules known as adipokines [4]. In addition, we have reported the adipocyte expression of the clock genes aryl hydrocarbon receptor nuclear translocator-like (ARNTL, also called BMAL1), cryptochrome 1 (CRY1), and period homolog 2 (PER2), and have shown that their expression was correlated in men with some of the metabolic syndrome traits [5]. Moreover, others have shown that sex hormones modulate adipocyte gene expression [6] and fat distribution among different depots [7]. Sex-related differences in the lipolytic pathway of adipose tissue have also been demonstrated in severely obese patients [8].
Sexual dimorphism has been shown also in chronobiology. Morningness–eveningness preference is the individual difference that most clearly explains the variations in the rhythmic expression of biological and behavioural patterns [9]. Marked differences in circadian typology between men and women have been shown with eveningness preference being more common in men [9–11].
Along these lines, some reports suggest that shift workers have an increased risk of coronary heart disease (CHD) and metabolic syndrome (MetS), and that this risk differs between sexes [12]. More recently, a large clinical trial has revealed that sex can significantly affect the survival outcome of patients undergoing cancer chronotherapy [10]. Although many factors could be implicated in these results, differences in expression of genes that comprise the positive and the negative arms of circadian transcriptional feedback could be implicated in this reported sex differences.
Previous studies examining the expression of clock genes in different tissues [13–15] have either used only one sex or failed to do sex-specific analyses [14, 16, 17]. To our knowledge, no study has been reported demonstrating the sex-specific basal expression of different clock genes in brain or in peripheral tissues, including the adipose tissue.
The aim of the present study was to analyze the sex-specific expression of BMAL1, CRY1, and PER2 in two adipose tissue depots in morbidly obese men and women, and to relate this expression to MetS-related traits.
Subjects and Methods
Subjects
Visceral (VAT) and subcutaneous (SAT) abdominal adipose tissue biopsies were obtained from a total of 16 morbidly obese patients, eight men (mean age 43± 13 years; mean BMI 47±5 kg/m2) and eight women (mean age 47±7 years; mean BMI 45±6 kg/m2), undergoing obesity-related laparoscopic gastric bypass surgery after an overnight fast. Interventions were performed in the General Surgery Service of “Virgen de la Arrixaca” University Hospital. The day before surgery, all patients were synchronized having lunch at 14:30 and dinner at 21:00 h. AT biopsies were taken as paired samples from the two AT depots at the beginning of the surgical procedure (between 11:00 and 14:00 h). VAT was taken from the omental depot. The SAT was taken 5 cm lateral from the umbilicus. All biopsies were immediately frozen at −80°C.
Protocols were approved by the ethics committee of the “Virgen de la Arrixaca” University Hospital, and all participants signed a written informed consent before biopsies were obtained.
Clinical Characteristics
Anthropometric Measurements
The evaluation of obesity was carried out according to the criteria proposed by the Spanish Society for the Study of Obesity in 2007 [18].
Weight was determined in subjects wearing light clothes and bare-footed, using a digital electronic weighing scale. Height was determined using a Harpender digital stadiometer (range 0.70–2.05 m), with the subject upright and the head in the Frankfurt plane. From these data, the body mass index (BMI) was calculated.
Total body fat (%) was measured by bioimpedance with a TANITA Model TBF-300 (TANITA Corporation of America, Arlington Heights, IL, USA). The following skinfolds were measured: biceps, triceps, suprailiac, and subscapular. All measurements were obtained on the right side, with the subject upright and relaxed. A Harpenden caliper (Holtain Ltd, Bryberian, Crymmych, Pembroke-shire, UK) with a constant pressure of 10 g/mm2 was used. Body fat distribution was assessed using the waist circumference (WC) at the level of midway between the lower rib margin and the iliac crest and hip at the level of the widest circumference over the great trochanters. The waist-to-hip ratio (WHR) was calculated from both measurements. Anthropometric measurements were carried out three times by the same anthropometrist.
Plasma Determinations
Fasting blood samples were collected the day before surgery. Plasma and serum samples were obtained by centrifugation and stored at −80°C until analyzed.
Plasma concentrations of glucose, triacylglycerols, total cholesterol, high-density lipoprotein (HDL), and low-density lipoprotein (LDL) cholesterol were determined with commercial kits (Roche Diagnostics GmbH, Mannheim, Germany).
Analysis of Gene and Protein Expression
RNA Extraction from Adipose Tissue
Adipose tissue was collected in all patients around 11:00 h. Total RNA was extracted from adipose tissue using Trizol Reagent (Invitrogen, Paisley, UK) according to the instructions of the company. The fat cake was previously removed with a chloroform solution. RNA was quantified by measuring absorbance at 260 and 280 nm.
Real-Time PCR Measurement of Clock Genes mRNA
Reverse transcription was performed using random hexamers as primers and Thermoscript reverse transcriptase (Invitrogen, Cergy Pontoise, France) with 3 μg total RNA for each sample.
Quantitative real-time PCR was performed using an ABI PRISM 7900 HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA). PCR Master MIX (Perkin-Elmer, Norwalk, CT, USA) containing Hot Start Taq DNA polymerase was used. Taqman probes for human clock genes and 18S rRNA as internal control were also supplied by Applied Biosystems (Assay-by-Design®). The clock genes analyzed were CRY1, BMAL1, and PER2. All experimental samples and negative controls were measured in duplicate. In brief, clock genes mRNA and 18S RNA were amplified in separated wells at 95°C for 10 min and thereafter repeating cycles comprised of 95°C for 30 s and 60°C for 60 s for annealing and extension steps. During the extension step, increase in fluorescence was measured in real time.
Data were obtained as Ct values according to the manufacturer's guidelines and used to determine ΔCt values [ΔCt=Ct of the target gene−Ct of the housekeeping gene (18S)] of each sample. Fold changes of gene expression were calculated by the 2−ΔΔCt method [19].
Western Blot
The samples were homogenized in ice-cold buffer consisting of 0.1% sodium dodecylsulfate, 0.1% sodium deoxycholate, and 1% Triton X-100 in phosphate-buffered saline (PBS) with freshly added protease inhibitors (phenylmethylsulfonyl fluoride, aprotinin, pepstatin A, and 1,10-phenanthroline).
The protein content was measured by the bicinchoninic acid assay, using bovine serum albumin as standard. Samples were boiled for 4 min in Laemmli buffer, separated by 10% sodium dodecyl sulfate–polyacryl-amide gel electrophoresis (SDS–PAGE), electroblotted onto nitrocellulose or PVDF membranes, and blocked with 5% (w/v) nonfat dry milk in Tris-buffered saline. After incubation with PER2, BMAL1, and CRY1 or α-actin antibodies (Abcam, Madrid, Spain), the bands were detected by enhanced chemiluminescence (NEN, Zaventem, Belgium).
Statistical Analysis
Clinical and anthropometric data are presented as means ± SD. The results for clock genes expression, expressed in arbitrary units, are presented as means ± standard error of the mean. We used Student's t test to analyze possible differences between sexes. Pearson's correlations coefficients were used to analyze the relationships between clinical and anthropo-metric data and gene expression. All statistical analyses were carried out using SPSS for Windows (release 15.0; SPSS Inc., Chicago, IL, USA).
Results
Clinical Characteristics of the Population
Table 1 contains general characteristics of the 16 morbidly obese (BMI>40 kg/m2) subjects studied. Men had higher weight, height and waist-to-hip ratio (WHR) than women.
Table 1.
Clinical characteristics of the population studied
| Men (n=8) | Women (n=8) | P values | |
|---|---|---|---|
| Age (years) | 43±13 | 47±7 | 0.468 |
| Weight (kg) | 139±16 | 112±14 | 0.004 |
| Height (cm) | 172±7 | 158±7 | 0.001 |
| BMI (kg/m2) | 47±5 | 45±6 | 0.540 |
| Bicipital sinkfold (mm) | 21±6 | 27±3 | 0.014 |
| Tricipital sinkfold (mm) | 31±4 | 27±3 | 0.042 |
| Subscapular sinkfold (mm) | 37±2 | 33±3 | 0.008 |
| Suprailiac sinkfold (mm) | 30.02±3.06 | 30.57±3.44 | 0.741 |
| Body fat (TANITA) (%) | 47.67±6.16 | 42.25±6.37 | 0.105 |
| WC (cm) | 136.75±8.76 | 128.25±8.81 | 0.074 |
| HC (cm) | 137.25±12.49 | 142.11±8.40 | 0.376 |
| WHR | 1±0.03 | 0.9±0.03 | <0.001 |
| Cholesterol (mmol/l) | 4.7±0.8 | 5.1±1.0 | 0.578 |
| Triacylglycerols (mmol/l) | 1.8±0.6 | 1.9±1.2 | 0.876 |
| HDL cholesterol (mmol/l) | 1.1±0.2 | 1.2±0.3 | 0.480 |
| LDL cholesterol (mmol/l) | 2.8±0.8 | 3.2±0.6 | 0.352 |
| Glucose (mmol/l) | 5.9±1.5 | 6.3±1.1 | 0.550 |
| Systolic pressure (mmHg) | 154±36 | 144±19 | 0.498 |
| Diastolic pressure (mmHg) | 81±19 | 79±21 | 0.806 |
Data are presented as means ± SD. P values were calculated using Student's t test to compare men versus women. Significant differences (P<0.05) are shown in bold.
BMI body mass index, WC waist circumference, HC hip circumference, WHR waist-to-hip ratio, HDL high-density lipoprotein, LDL low-density lipoprotein
According to the International Diabetes Federation (IDF), subjects met the criteria to be defined as metabolic syndrome patients. Average values of waist, triglycerides, glucose, and systolic pressure fell outside the cutoff points proposed by the IDF [20].
Sex-Related Clock Genes and Proteins Expression
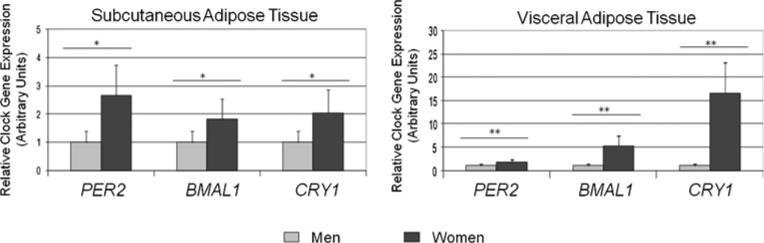
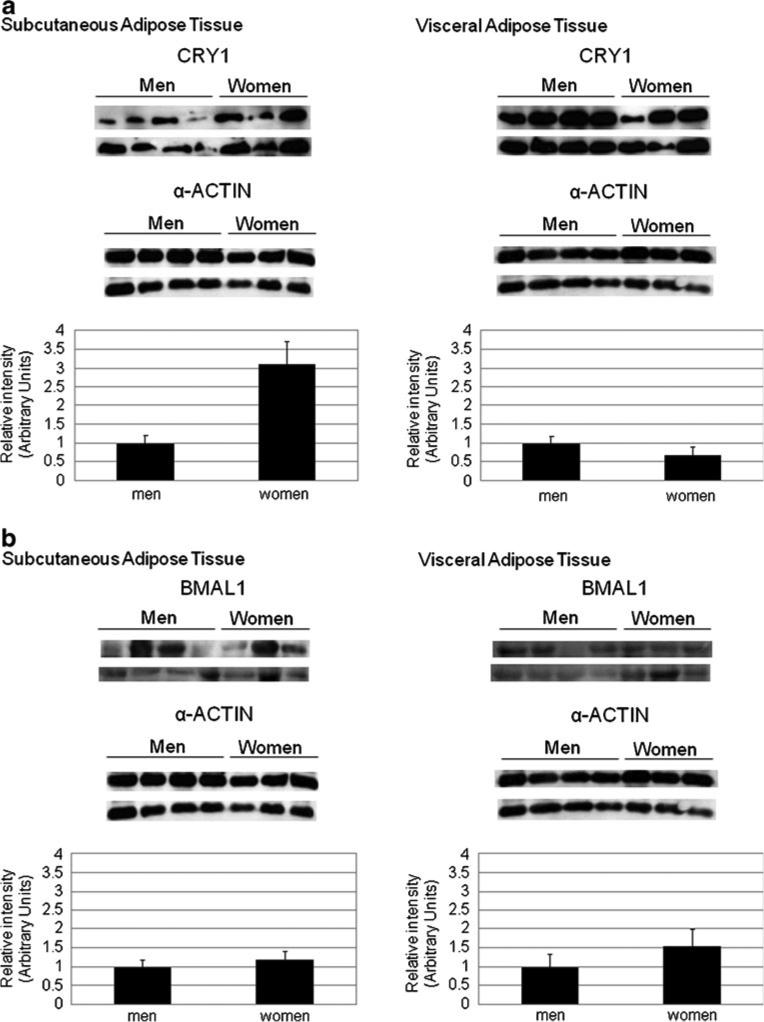
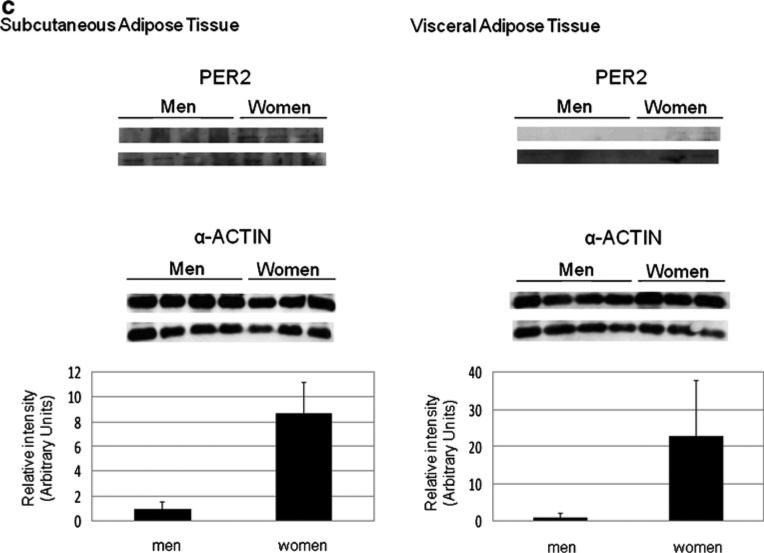
Gene expression for the three clock genes examined was significantly higher in morbidly obese women than in men in both AT (Fig. 1), but more markedly in VAT. The largest difference in SAT (2.7 times higher in women) was observed for PER2. Differences between sexes were maintained even after adjusting for age for every gene study (P<0.001). To further study the expression of PER2, BMAL1, and CRY1, a Western blot analysis was carried out (Fig. 2a). Our results showed that in women PER2 was increased in both AT, being the differences of expression of this protein (8.7-and 22.9-fold for SAT and VAT, respectively) the largest observed. CRY1 was increased only in SAT. No differences were found in BMAL1 between women and men in any of the tissues studied.
Fig. 1.
Clock genes expression in subcutaneous and visceral fat by sex. Relative gene expression of clock genes derived from subcutaneous and visceral adipose tissue in men (n=8) and women (n=8). Results are presented relative to the lowest basal relative expression for each gene (gene expression in men is set the value of 1 as reference). Data of relative expression are represented as arbitrary units (AU). Data are reported as means ± SEM. *P<0.05, **P<0.001 (Student's t test)
Fig. 2.
CRY1 (a), BMAL1 (b), and PER2 (c) protein expression in men (n=8) and women (n=6) studied. α-Actin was used as a loading control. Western blots and α-actin normalized quantification are shown
Relations Between Clinical Parameters and Clock Genes Expression
Correlations between gene expression anthropometric and clinical variables within each sex and AT depot are shown in Table 2. Gene expression level was negatively correlated with LDL-C in women and positively with HDL-C (P<0.05) in men studied in both AT depots. Moreover, gene expressions were inversely correlated with body fat mass in morbidly obese women in both AT. In men alone, we found a correlation between gene expression and age. From the three clock genes studied, only visceral PER2 and BMAL1 correlated significantly with body fat distribution in women, whereas in men, the trend did not reach statistical significance.
Table 2.
Associations between clock genes expression and clinical and anthropometric characteristics for both genders
| Men |
Women |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
PER2 expression |
BMAL1 expression |
CRY1 expression |
PER2 expression |
BMAL1 expression |
CRY1 expression |
|||||||
| P | r | P | r | P | r | P | r | P | r | P | r | |
| Visceral adipose tissue | ||||||||||||
| Age (years) | 0.011 | –0.914 | 0.012 | –0.914 | 0.187 | –0.622 | 0.824 | –0.118 | 0.824 | –0.118 | 0.941 | –0.039 |
| Body fat (%) | 0.105 | 0.723 | 0.107 | –0.752 | 0.956 | 0.029 | 0.005 | –0.939 | 0.003 | –0.941 | 0.186 | 0.624 |
| WHR | 0.892 | 0.072 | 0.831 | 0.080 | 0.988 | –0.008 | 0.012 | –0.910 | 0.012 | –0.910 | 0.244 | –0.564 |
| HDL cholesterol | 0.094 | –0.738 | 0.097 | –0.621 | 0.041 | 0.831 | 0.944 | 0.037 | 0.434 | –0.399 | 0.944 | 0.037 |
| LDL cholesterol | 0.543 | –0.315 | 0.517 | –0.302 | 0.527 | –0.327 | 0.047 | –0.866 | 0.036 | –0.898 | 0.398 | –0.428 |
| Subcutaneous adipose tissue | ||||||||||||
| Age (years) | 0.488 | 0.357 | 0.517 | 0.325 | 0.280 | 0.530 | 0.678 | –0.218 | 0.614 | –0.264 | 0.362 | 0.457 |
| Body fat (%) | 0.304 | –0.507 | 0.359 | –0.460 | 0.247 | –0.561 | 0.018 | –0.887 | 0.855 | –0.097 | 0.209 | –0.599 |
| WHR | 0.840 | 0.107 | 0.731 | 0.182 | 0.506 | 0.342 | 0.911 | 0.060 | 0.731 | 0.181 | 0.460 | 0.378 |
| HDL cholesterol | 0.369 | 0.452 | 0.003 | 0.954 | 0.523 | –0.330 | 0.236 | 0.571 | 0.619 | –0.260 | 0.906 | –0.063 |
| LDL cholesterol | 0.041 | –0.830 | 0.999 | –0.001 | 0.561 | –0.596 | 0.010 | –0.918 | 0.948 | 0.035 | 0.331 | 0.484 |
Data in bold represent statistical differences (P<0.05)
WHR waist-to-hip ratio, HDL high-density lipoprotein, LDL low-density lipoprotein
Discussion
The present study shows that in human adipose tissue from morbidly obese patients, quantitative expression of clock genes and proteins is sex dependent. Our data in humans are consistent with a previous report in mouse livers showing that the expression of clock genes differed between male and female, being higher in females [21].
The observed sex-related differential in expression might be expected in view of the effects that steroids exert on clock genes expression both in brain and in peripheral tissues [22, 23]. In addition, there is a loop regulation by which circulating levels of gonadal hormones influence circadian rhythms [24] and their levels are partially defined by the circadian system [25, 26]. Moreover, there is experimental evidence supporting that variations in ovarian hormones affect clock gene expression in the brain, introducing additional levels of complexity on the rhythmic modulation of behavior and metabolism [27].
The sexual dimorphism observed in this work could be due to differences in circadian phases between both sexes. Previous studies have shown that most hepatic clock genes had their peak mRNA levels 30 min earlier in female than in male mice [21]. In humans, it has been also reported that a phase advance exists in women's circadian rhythms compared to those of men [28, 29]. Relevant to this is our finding showing that both negative (PER2 and CRY1) and positive limb clock genes (BMAL1) were expressed more in women than in men in both fat depots. We could consider that if a phase advance were the cause of sex dimorphism in clock gene expression, the differences should be present only in the positive limb genes or the negative ones, but not in both groups of genes. In fact, the “two-loop” model of mouse clock indicates that CLOCK forms alternating, periodic associations with either BMAL1 or the negative regulators PER and CRY [30]. Our Western blot results, in agreement with this observation, showed an increase in protein expression of negative limb clock genes (PER2 in both AT and CRY1 in SAT), while no differences were observed in BMAL1 protein expression. It is well known that BMAL1 expression and degradation are tightly regulated [31, 32]. It is tempting to hypothesize that in morbidly obese women, even thought BMAL1 gene expression is increased in AT, the control of the translation or the stability of the protein results in a decrease in its abundance leading to a preponderance of negative limb clock gene products.
An alternative explanation for our results could be on the intrinsic characteristics of adipose tissue. It is known that adipose tissue fat accumulation [7] and mobilization [8] differs between sexes as well as in circadian rhythmicity of a number of adipokines such as adiponectin [33] and leptin [34]. Studies examining the circadian variability of these proteins in plasma have demonstrated a sexual dimorphism both in the 24-h profile as well as in the relative amplitude of its diurnal rhythm [34].
CRY1 showed a large sex difference (Fig. 1). Studies performed in mice have demonstrated that the circadian clock components, Cry1 and Cry2, are suggested as a prerequisite for sustaining sex dimorphism in other peripheral tissues [35]. PER2 also showed important and significant differences between sexes both in gene and protein expression (Figs. 1 and 2). In fact, studies performed in rats and mice have demonstrated a sexual dimorphism in Per2 function. Thus, Per2 appears to differentially regulate glucocorticoid secretion in females and males [36]. Moreover, the results obtained in the mPer2−/− mice and in humans [37] demonstrated the particular role of this gene in obesity and fat distribution [38].
Other studies performed in Drosophila have demonstrated a sexual dimorphism in Bmal1 towards a higher prominence in females. These results have been related with the differences in recovery sleep between women and men after deprivation [39] and with the sex-specific differences in sleep pattern [40]. BMAL expression has been also related to longevity [39]. Similarly to our data, Ando et al. have previously described that BMAL1 mRNA levels were correlated with age, suggesting that increased age independently affects the intracellular clock in humans [41].
In general, our data show an inverse correlation between expression of clock genes and MetS traits, i.e., decreases in body fat percentage and waist-to-hip ratio and increases in HDL cholesterol concentrations, and this relationship was sex dependent. Previously, we have reported results in men [5], and our current data show similar findings in women. Several groups have evaluated clock genes in the adipose tissue of obese mice and indicated that obesity attenuated and/or disrupted clock gene expression [16, 42]. Moreover, it has been reported that low expression of clock genes due to a disruption of the circadian system could lead to an increase risk of MetS [3, 43]. Indeed, epidemiological studies have shown that chronodisruption (induced by shift work, sleep deprivation, or by shifting the normal feeding time to night hours) is associated with high risk of developing abdominal obesity, hypertriglyceridemia, low HDL, diabetes, and many characteristics of MetS [3]. One limitation of the current study is that data have been obtained in morbidly obese patients. Further studies should be performed in lean subjects in order to be able to extrapolate these results to the general population.
The findings of this study provide some leads about the molecular basis of the sexual dimorphisms in chronobiology already demonstrated in clinical and epidemiology studies. Moreover, they supply the basis for further studies investigating sex specificities in the interaction of the circadian clock and adipose tissue metabolism, and their contribution on body fat distribution and metabolic risk. These results could lead to a practical application in the treatment of obesity and MetS, with different chronobiological approaches depending on sexes.
Acknowledgments
This work was supported by the Government of Education, Science and Research of Murcia (Project BIO/FFA 07/01-0004) and by The Spanish Government of Science and Innovation (projects AGL2008-01655/ALI and AGL2008-04332/ALI and SAF2008-01432). CIBERehd is funded by the Instituto de Salud Carlos III, National Heart, Lung, and Blood Institute grants HL-54776, National Institute of Diabetes and Digestive and Kidney Diseases, grant number DK075030, and by contracts 53-K06-5-10 and 58-1950-9-001 from the US Department of Agriculture Research.
Footnotes
Disclosure statement There is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Contributor Information
P. Gómez-Abellán, Department of Physiology, Faculty of Biology, University of Murcia, Campus de Espinardo, 30100 Murcia, Spain
J. A. Madrid, Department of Physiology, Faculty of Biology, University of Murcia, Campus de Espinardo, 30100 Murcia, Spain
J. A. Luján, General Surgery Service, University Hospital “Virgen de la Arrixaca”, Murcia, Spain
M. D. Frutos, General Surgery Service, University Hospital “Virgen de la Arrixaca”, Murcia, Spain
R. González, CIBERehd—Department of Pharmacology, University of Granada, Granada, Spain
O. Martínez-Augustín, CIBERehd—Department of Biochemistry and Molecular Biology II, University of Granada, Granada, Spain
F. Sánchez de Medina, CIBERehd—Department of Pharmacology, University of Granada, Granada, Spain.
J. M. Ordovás, Jean Mayer US Department of Agriculture Human Nutrition Research Center on Aging, Tufts University School of Medicine, Boston, MA, USA Department of Epidemiology and Population Genetics, “Centro Nacional Investigación Cardiovascular” (CNIC), Madrid, Spain; IMDEA Alimentación, Madrid, Spain.
M. Garaulet, Department of Physiology, Faculty of Biology, University of Murcia, Campus de Espinardo, 30100 Murcia, Spain
References
- 1.Garaulet M, Gómez-Abellán P, Madrid JA. Chronobiology and obesity: the orchestra out of tune. Clin Lipidol. 2010;5(2):181–8. Review. [Google Scholar]
- 2.Marcheva B, Ramsey KM, Affinati A, et al. Clock genes and metabolic disease. J Appl Physiol. 2009;107(5):1638–46. doi: 10.1152/japplphysiol.00698.2009. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garaulet M, Madrid JA. Chronobiology, genetics and metabolic syndrome. Curr Opin Lipidol. 2009;20(2):127–34. doi: 10.1097/MOL.0b013e3283292399. [DOI] [PubMed] [Google Scholar]
- 4.Whitehead JP, Richards AA, Hickman IJ, et al. Adiponectin—a key adipokine in the metabolic syndrome. Diabetes Obes Metab. 2006;8:264–80. doi: 10.1111/j.1463-1326.2005.00510.x. [DOI] [PubMed] [Google Scholar]
- 5.Gómez-Abellán P, Hernández-Morante JJ, Luján JA, et al. Clock genes are implicated in the human metabolic syndrome. Int J Obes. 2008;32(1):121–8. doi: 10.1038/sj.ijo.0803689. [DOI] [PubMed] [Google Scholar]
- 6.Klaus S, Keijer J. Gene expression profiling of adipose tissue: individual, depot-dependent, and sex-dependent variabilities. Nutrition. 2004;20(1):115–20. doi: 10.1016/j.nut.2003.09.020. [DOI] [PubMed] [Google Scholar]
- 7.Garaulet M, Pérez-Llamas F, Fuente T, et al. Anthropometric, computed tomography and fat cell data in an obese population: relationship with insulin, leptin, tumor necrosis factor-alpha, sex hormone-binding globulin and sex hormones. Eur J Endocrinol. 2000;143(5):657–66. doi: 10.1530/eje.0.1430657. [DOI] [PubMed] [Google Scholar]
- 8.Ramis JM, Salinas R, García-Sanz JM, et al. Depot- and gender-related differences in the lipolytic pathway of adipose tissue from severely obese patients. Cell Physiol Biochem. 2006;17(3–4):173–80. doi: 10.1159/000092079. [DOI] [PubMed] [Google Scholar]
- 9.Adan A, Natale V. Gender differences in morningness–eveningness preference. Chronobiol Int. 2002;19(4):709–20. doi: 10.1081/cbi-120005390. [DOI] [PubMed] [Google Scholar]
- 10.Lévi F, Filipski E, Iurisci I, et al. Cross-talks between circadian timing system and cell division cycle determine cancer biology and therapeutics. Cold Spring Harb Symp Quant Biol. 2007;72:465–75. doi: 10.1101/sqb.2007.72.030. [DOI] [PubMed] [Google Scholar]
- 11.Mongrain V, Lavoie S, Selmaoui B, et al. Phase relationships between sleep–wake cycle and underlying circadian rhythms in morningness–eveningness. J Biol Rhythms. 2004;19(3):248–57. doi: 10.1177/0748730404264365. [DOI] [PubMed] [Google Scholar]
- 12.Sopowski MJ, Hampton SM, Ribeiro DC, et al. Postprandial triacylglycerol responses in simulated night and day shift: gender differences. J Biol Rhythms. 2001;16(3):272–6. doi: 10.1177/074873040101600310. [DOI] [PubMed] [Google Scholar]
- 13.Panda S, Antoch MP, Miller BH, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–20. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 14.Storch KF, Lipan O, Leykin I, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 15.Yoo SH, Yamazaki S, Lowrey PL, et al. PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101:5339–46. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ando H, Yanagihara H, Hayashi Y, et al. Rhythmic mRNA expression of clock genes and adipocytokines in mouse visceral adipose tissue. Endocrinology. 2005;146(12):5631–6. doi: 10.1210/en.2005-0771. [DOI] [PubMed] [Google Scholar]
- 17.Teboul M, Barrat-Petit MA, Li XM, et al. Atypical patterns of circadian clock gene expression in human peripheral blood mononuclear cells. J Mol Med. 2005;83:693–9. doi: 10.1007/s00109-005-0697-6. [DOI] [PubMed] [Google Scholar]
- 18.Sociedad Española para el Estudio de la Obesidad (SEEDO) Consenso SEEDO 2007 para la evaluación del sobrepeso y la obesidad y el establecimiento de criterios de intervención terapéutica [article in Spanish]. Med Clin (Barc) 2007;128:184–96. doi: 10.1016/s0025-7753(07)72531-9. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C (T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Alberti KGMM, Zimmet P, Shaw J. Metabolic syndrome a new world-wide definition. A consensus statement from the International Diabetes Federation. Diabet Med. 2006;23:469–80. doi: 10.1111/j.1464-5491.2006.01858.x. [DOI] [PubMed] [Google Scholar]
- 21.Yang X, Zhang YK, Esterly N, et al. Gender disparity of hepatic lipid homoeostasis regulated by the circadian clock. J Biochem. 2009;145(5):609–23. doi: 10.1093/jb/mvp018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Metz RP, Qu X, Laffin B, et al. Circadian clock and cell cycle gene expression in mouse mammary epithelial cells and in the developing mouse mammary gland. Dev Dyn. 2006;235:263–71. doi: 10.1002/dvdy.20605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller BH, Olson SL, Turek FW, et al. Circadian clock mutation disrupts estrous cyclicity and maintenance of pregnancy. Curr Biol. 2004;14:1367–73. doi: 10.1016/j.cub.2004.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albers HE. Gonadal hormones organize and modulate the circadian system of the rat. Am J Physiol. 1981;241:R62–6. doi: 10.1152/ajpregu.1981.241.1.R62. [DOI] [PubMed] [Google Scholar]
- 25.Wiegand SJ, Terasawa E. Discrete lesions reveal functional heterogeneity of suprachiasmatic structures in regulation of gonadotropin secretion in the female rat. Neuroendocrinology. 1982;34:395–404. doi: 10.1159/000123335. [DOI] [PubMed] [Google Scholar]
- 26.Wiegand SJ, Terasawa E, Bridson WE. Persistent estrus and blockade of progesterone-induced LH release follows lesions which do not damage the suprachiasmatic nucleus. Endocrinology. 1978;102:1645–8. doi: 10.1210/endo-102-5-1645. [DOI] [PubMed] [Google Scholar]
- 27.Perrin JS, Segall LA, Harbour VL, et al. The expression of the clock protein PER2 in the limbic forebrain is modulated by the estrous cycle. Proc Natl Acad Sci USA. 2006;103(14):5591–6. doi: 10.1073/pnas.0601310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Dongen HPA. Inter- and intra-individual differences in circadian phase. Leiden University Press; The Netherlands: 1998. [Google Scholar]
- 29.Wever RA. Characteristics of circadian rhythms in human functions. In: Wurtman RJ, Waldhause F, editors. Journal of Neural Transmission (Suppl.) Melatonin in Humans. Springer; New York: 1986. pp. 323–335. [PubMed] [Google Scholar]
- 30.Maywood ES, O'Brien JA, Hastings MH. Expression of mCLOCK and other circadian clock-relevant proteins in the mouse suprachiasmatic nuclei. J Neuroendocrinol. 2003;15(4):329–34. doi: 10.1046/j.1365-2826.2003.00971.x. [DOI] [PubMed] [Google Scholar]
- 31.Shende VR, Goldrick MM, Ramani S, et al. Expression and rhythmic modulation of circulating microRNAs targeting the clock gene Bmal1 in mice. PLoS One. 2011;6(7):e22586. doi: 10.1371/journal.pone.0022586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee J, Lee Y, Lee MJ, et al. Dual modification of BMAL1 by SUMO2/3 and ubiquitin promotes circadian activation of the CLOCK/BMAL1 complex. Mol Cell Biol. 2008;28(19):6056–65. doi: 10.1128/MCB.00583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cnop M, Havel PJ, Utzschneider KM, et al. Relationship of adiponectin to body fat distribution, insulin sensitivity and plasma lipoproteins: evidence for independent roles of age and sex. Diabetologia. 2003;46(4):459–69. doi: 10.1007/s00125-003-1074-z. [DOI] [PubMed] [Google Scholar]
- 34.Saad MF, Riad-Gabriel MG, Khan A, et al. Diurnal and ultradian rhythmicity of plasma leptin: effects of gender and adiposity. J Clin Endocrinol Metab. 1998;83(2):453–9. doi: 10.1210/jcem.83.2.4532. [DOI] [PubMed] [Google Scholar]
- 35.Bur IM, Cohen-Solal AM, Carmignac D, et al. The circadian clock components CRY1 and CRY2 are necessary to sustain sex dimorphism in mouse liver metabolism. J Biol Chem. 2009;284(14):9066–73. doi: 10.1074/jbc.M808360200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pilorz V, Steinlechner S, Oster H. Age and oestrus cycle-related changes in glucocorticoid excretion and wheel-running activity in female mice carrying mutations in the circadian clock genes Per1 and Per2. Physiol Behav. 2009;96(1):57–63. doi: 10.1016/j.physbeh.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 37.Garaulet M, Corbalán MD, Madrid JA, et al. PER2 variants are associated with abdominal obesity, psycho-behavioral factors and attrition in the dietary treatment of obesity. J Am Diet Assoc. 2010;110(6):917–21. doi: 10.1016/j.jada.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang S, Liu A, Weidenhammer A, et al. The role of mPer2 clock gene in glucocorticoid and feeding rhythm. Endocrinology. 2009;150(5):2153–60. doi: 10.1210/en.2008-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Armitage R, Smith C, Thompson S, et al. Sex differences in slow-wave activity in response to sleep deprivation. Sleep Res Online. 2001;4:33–41. [Google Scholar]
- 40.Manber R, Armitage R. Sex, steroids, and sleep: a review. Sleep. 1999;22:540–55. [PubMed] [Google Scholar]
- 41.Ando H, Ushijima K, Kumazaki M, et al. Influence of age on clock gene expression in peripheral blood cells of healthy women. J Gerontol A Biol Sci Med Sci. 2010;65(1):9–13. doi: 10.1093/gerona/glp160. [DOI] [PubMed] [Google Scholar]
- 42.Kohsaka A, Laposky AD, Ramsey KM, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6(5):414–21. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 43.Karlsson B, Knutsson A, Lindahl B. Is there an association between shift work and having a metabolic syndrome? Results from a population based study of 27,485 people. Occup Environ Med. 2001;58:747–52. doi: 10.1136/oem.58.11.747. [DOI] [PMC free article] [PubMed] [Google Scholar]