Abstract
Virtually structured detection (VSD) has been demonstrated to break the diffraction limit in scanning laser microscopy (SLM). VSD provides an easy, low-cost, and phase-artifact-free strategy to achieve super-resolution imaging. However, practical application of this method is challenging due to a limited image acquisition speed. We report here the combination of VSD and line-scanning microscopy (LSM) to improve the image acquisition speed. A motorized dove prism was used to achieve automatic control of four-angle (i.e., 0°, 45°, 90°, and 135°) scanning, thus ensuring isotropic resolution improvement. Both an optical resolution target and a living frog eyecup were used to verify resolution enhancement.
Advanced light microscopy plays an important role in biomedical research and clinical diagnoses due to its excellent ability to reveal fine structures invisible to the naked eye. Multiple imaging modalities such as phase contrast microscopy, differential interference contrast (DIC) microscopy, laser scanning confocal microscopy (LSCM), and fluorescence microscopy have been developed to extend the capabilities of light microscopy. However, the spatial resolution of conventional microscopy is restricted by light diffraction. In fluorescence imaging, multiple strategies have been developed to overcome the diffraction limit, such as stimulated emission-depletion (STED) microscopy, stochastic optical-reconstruction microscopy (STORM) [1], photoactivated localization microscopy (PALM) [2], fluorescence PALM (FPALM) [3], and structured-illumination microscopy (SIM) [4, 5]. Both STED and single-molecular imaging modalities require specific dyes or fluorescent proteins, and are not suitable for autofluorescence imaging or intrinsic signal (i.e., reflection or transmission) imaging.
The SIM is applicable to both fluorescence and intrinsic signal imaging. SIM requires a sinusoidal-patterned illumination to shift some high-frequency information beyond the diffraction limit to lower frequencies and, thus, expand the effective passband of the optical system. However, for super-resolution reconstruction, the fringe or grid projection [6] requires complicated mechanical manipulation to generate structured-illumination patterns with specific phases, which is particularly challenging for moving samples. In addition, SIM employs wide-field illumination, which has difficulty penetrating through thick (i.e., >100 μm) tissues. There are currently two strategies to modify wide-field illumination SIM for applications involving thick tissues. First, wide-field illumination is condensed to single-line illumination which consists of two superimposed lines of illumination [7, 8]. In the longitudinal direction, intensity is modulated with a sinusoidal function because there is interference between two illumination lines. Therefore, there is resolution enhancement along this direction, just as with wide-field illumination SIM. At the same time, the condensed illumination allows optical sectioning by reducing the out-of-focus light. However, in this scheme, axial resolution is compromised due to the reduced effective numerical aperture of the objective [7]. The second strategy is to combine the structured illumination with light sheet illumination [9]. Perpendicular to the direction of observation, a sheet of structured light pattern is used to illuminate the sample. Because only the volume close to the focal plane is illuminated, structured light sheet microscopy can provide unparalleled optical sectioning ability. However, it is not practical to employ a light sheet approach for in vivo applications such as imaging of the cortex or retina.
In theory, SIM can also be realized in a point-scanning system through spatiotemporal modulation, either by modulating the light source intensity in an illumination light path or by moving a physical mask in a detection light path [10]. However, spatiotemporal modulation of the illumination/detection light is technically complicated. We recently demonstrated virtually structured detection (VSD)-based super-resolution scanning laser microscopy [11, 12], which requires neither dynamic modulation of light source intensity in the illumination arm nor a physical mask in the light detection arm. However, the image acquisition speed was limited because of the point-by-point scanning method and use of a low-speed digital camera, which is required for capturing the light profile of individual sampling points. A high image acquisition speed is vital for imaging biological dynamics and moving samples. In this paper, we report an optical system that combines VSD and line scanning microscopy (LSM) to achieve super-resolution imaging with a frame speed up to 0.33 fps at frame resolution of 400 × 400 pixels.
The detail principle of VSD has been described in a previous publication [11]. In conventional SIM, the acquired wide-field image can be represented as
| (1) |
where m is the modulation function, s is the reflectance ratio, and hil and hde are PSFs of the illumination path and detection path, respectively. ⊗ denotes convolution. In the VSD-based scanning laser microscopy (SLM), two dimensional light profiles of individual scanning lines are collected, modulated and then integrated. An image is generated as
| (2) |
Equation (1) is exactly equivalent to Eq. (2) for epi-illumination microscopy. In other words, the modulation of the illumination light pattern is equivalent to that of the detection light pattern. The structured illumination is to shift higher frequency information to the lower passing band. In Fourier domain, those resolved components containing super-resolution information are displaced and can be shifted back by multiplying the equivalent cosine wave in real space. The Wiener filter can be used to suppress the noise during combination of those components. The reassembled image is then retransformed to the real space to generate the final super-resolution image [4]. Only taking one dimension (i.e., along the y axis) in VSD-based LSM into consideration, Eq. (2) can be rewritten as
| (3) |
Along the y axis, the resolution can be doubled, while along the x axis, the resolution remains the same as in wide-field microscopy. Therefore, the image field needs to be rotated to achieve isotropic resolution doubling.
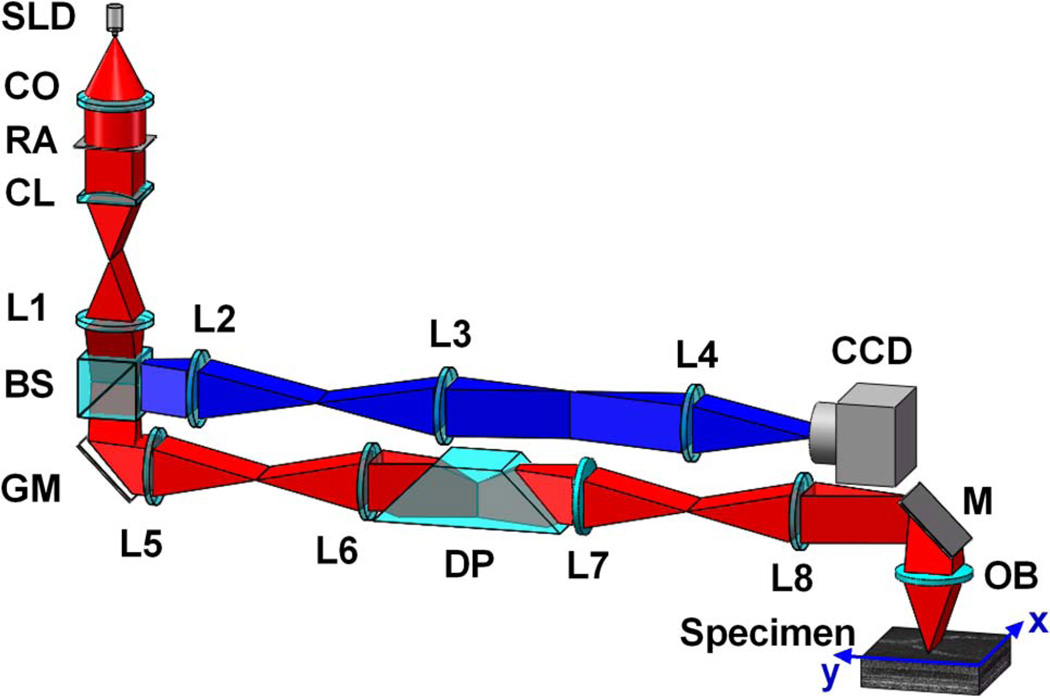
Figure 1 illustrates the schematic diagram of the experimental setup. A near-infrared light (NIR) source (SLD-35-HP, SUPERLUM), with a center wavelength at 830 nm (λ = 830 nm) and bandwidth at 60 nm (Δλ = 60 nm), was used. A cylindrical lens was used to focus the light into a line. The focused line was swept across the specimen using a 1D scanning galvo mirror (GVS001, THORLABS). The pivot point of the scanner was conjugate to the pupil plane of the objective to minimize the vignetting effect. The reflected light from the sample was descanned by the scanner and was relayed to the image plane with a 4-f system. A two-dimensional charge-coupled device (CCD) camera (Pike F-032B, Allied Vision) was used to record the light profile of individual sampling lines. To achieve isotropic resolution improvement, a dove prism was mounted to a motorized rotation stage to rotate the image field. The rotation of the prism with respect to the longitudinal axis rotated the image field at twice the rate of the prism’s rotation.
Fig. 1.
Schematic diagram of experimental setup. CO, collimator; L1–L8, lenses; BS, beam splitter; RA, rectangular aperture; CL, cylindrical lens; GM, galvo mirror; DP, dove prism; M, mirror; OB, objective. Focal lengths of lenses L1–L8 are 100, 150, 40, 80, 100, 100, 75, and 100 mm, respectively. SLD, superluminescent laser diode.
A region of interest (ROI) for the camera using a virtual slit of 0.5× Airy disc diameter was selected for confocal LSM (CLSM) imaging [13]. A ROI for the camera using a virtual slit of 2× Airy disc diameter was selected for VSD reconstruction. In the x axis direction, a maximum length of 400 pixels was set. The ROI of the camera was set at 50 × 400 pixels. Along the y axis, 400 frames were sampled. Thus, the final reconstructed super-resolution image size was 400 × 400 pixels. The frame rate of the CCD was set at 1000 fps. Therefore, the line-scanning scheme required 0.4 s to finish one dimensional scanning. For isotropic resolution enhancement, a four-angle (i.e., 0°, 45°, 90°, and 135°) scanning pattern was used. The acquisition time was ~1.6 s for the camera, and the transition time was ~1.4 s for the rotation motor (NR360S, THROLABS) to control the dove prism at the desired angles in sequence (i.e., 0°, 22.5°, 45°, and 67.5°).
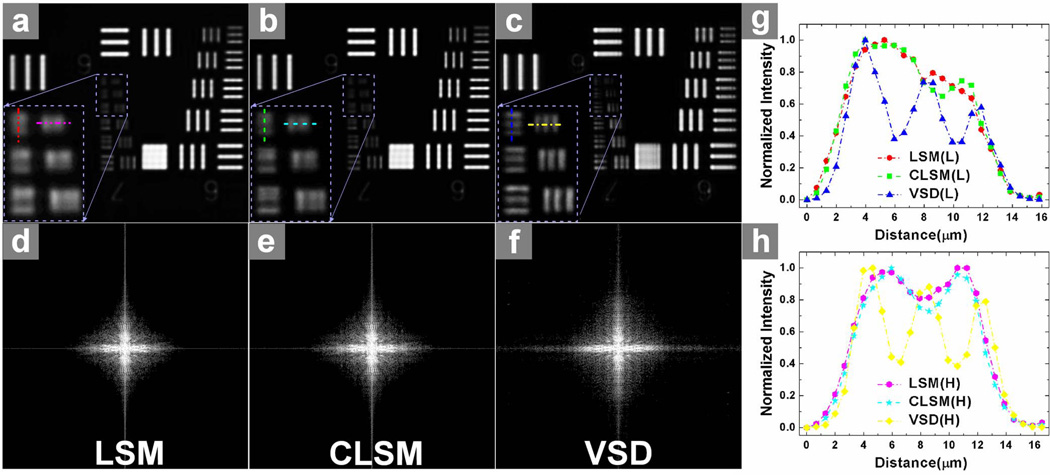
A standard resolution target (USAF 1951 1X, EDMOND) was employed to verify resolution enhancement of the VSD-based super-resolution LSM system. Figure 2 shows the comparison between conventional LSM [shown in Fig. 2(a)], confocal LSM [shown in Fig. 2(b)], and VSD-based super-resolution LSM [shown in Fig. 2(c)]. The smallest grid period was 4.4 μm. A 5× objective with 0.1 numerical aperture (NA) was used for this experiment. Conventional LSM, which has a theoretical resolution of ~5 μm, was not able to differentiate these 4.4-μm grids. Confocal LSM could barely differentiate them, as shown in Fig. 2(b). In contrast, the smallest grid could be resolved clearly in both longitudinal and horizontal directions after VSD reconstruction. The resolution enhancement was further confirmed by intensity profiles, as shown in Figs. 2(g) and 2(h). Three bumps were observed after VSD reconstruction, which were the intensity profiles along the horizontal and longitudinal directions, as shown in Figs. 2(a)–2(c). They were not differentiated by LSM and CLSM. The corresponding results in Fourier space [shown in Figs. 2(d)–2(f)] also demonstrated that more spectrum information was retained with VSD reconstruction, and there was higher super-resolution than with both conventional LSM and CLSM.
Fig. 2.
Comparison of three different imaging results using the USAF standard resolution target: (a) by conventional LSM; (b) by confocal LSM (CLSM); and (c) by VSD reconstruction. The magnified views of the smallest bars are indicated by the rectangle boxes in (a)–(c). (d)–(f) show the corresponding results in Fourier domain, respectively. (g) Normalized intensity profiles along the longitudinal direction in (a)–(c). (h) Normalized intensity profiles along the horizontal direction in (a)–(c).
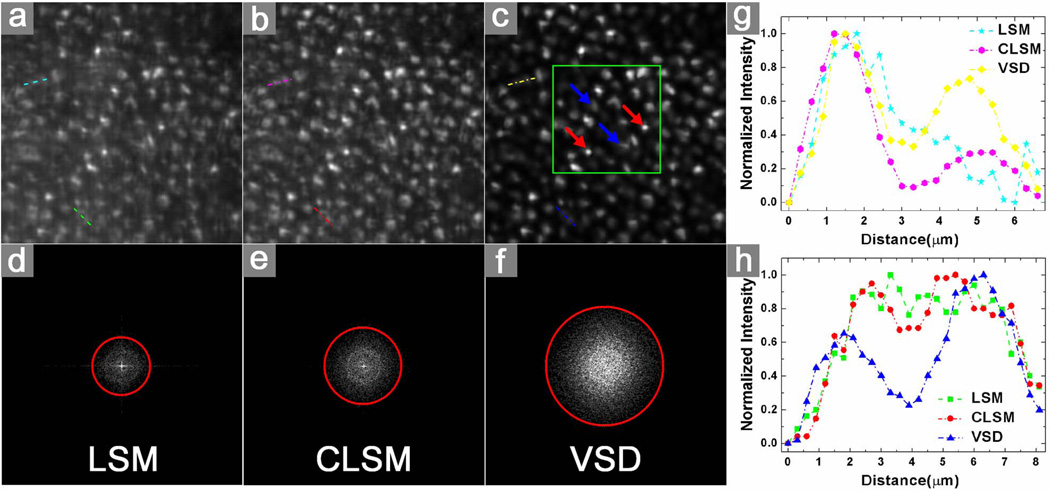
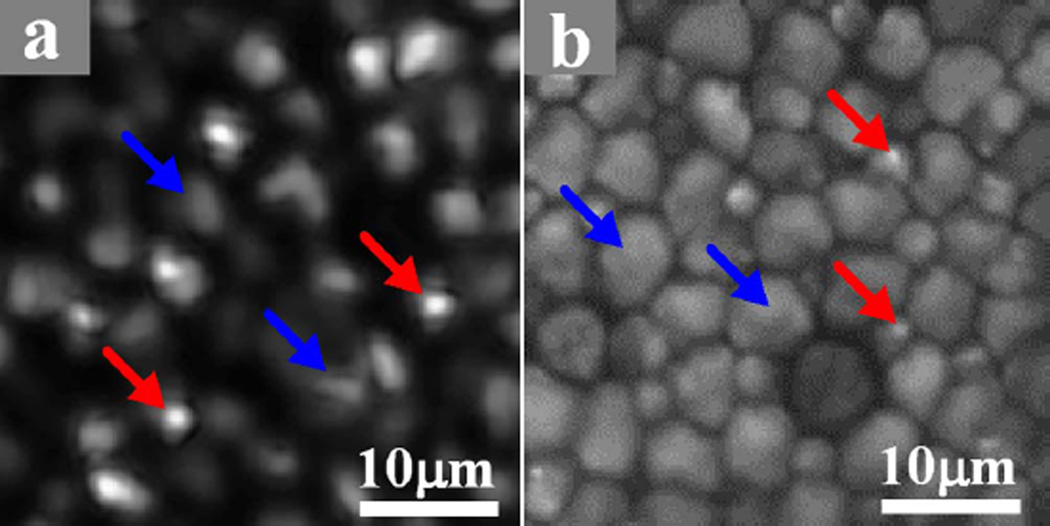
A freshly isolated frog (Rana pipiens) eyecup was also used for functional validation of the VSD-based super-resolution LSM system. The frog was euthanized by rapid decapitation and double pithing. After enucleating the eyes, the globe was hemisected below the equator with fine scissors. The lens and anterior structures were removed before the eyecup was moved into the chamber with Ringer’s solution for imaging. It has been established that the frog retina consists of photoreceptors with variable diameters (rods: ~5–8 μm; cones: ~1–3 μm) [14]. Therefore, the frog retina is a simple specimen that allows comparison of imaging performances of conventional LSM, confocal LSM, and super-resolution LSM. A 10× water immersion objective (NA = 0.25) was used in this experiment. Theoretical resolutions of conventional LSM, confocal LSM, and super-resolution LSM were estimated at 2.0, 1.5, and 1.0 μm, respectively. Figure 3(a) shows a retinal image acquired with conventional LSM. Figure 3(b) shows an image acquired with confocal LSM. Figure 3(c) shows a super-resolution result through VSD reconstruction. The two selected groups of normalized intensity profiles of the adjacent photoreceptor pairs in Figs. 3(a), 3(b), and 3(c) are plotted in Figs. 3(g) and 3(h), respectively. Both results showed that VSD reconstruction provides much improved lateral resolution over conventional LSM and confocal LSM. The corresponding results in Fourier domain are shown in Figs. 3(d)–3(f), respectively. Compared with conventional LSM [Fig. 3(d)], confocal LSM [Fig. 3(e)] can yield improved resolution by a factor of ~1.3, while VSD reconstruction, as shown in Fig. 3(f), contains more information and can yield improved resolution by a factor of ~2. Given the improved resolution, bright sub-cellular spots (red arrowheads) were observed, as shown in Fig. 3(c). Figure 4(a) shows an enlarged illustration of the green color window in Fig. 3(c). We speculate that the bright sub-cellular spots may reflect hyper-reflectivity at a sub-cellular level, e.g., photoreceptor-connecting cilium (CC). Rods (blue arrowheads) and cones (red arrowheads) can be directly differentiated in Fig. 4(a) based on their diameters (rods: ~5–8 μm; cones: ~1–3 μm) [14]. It was observed that rod photoreceptors had relatively homogenous reflectivity at the cellular level. The distinct bright spots were typically observed in cone photoreceptors. Bright sub-cellular spots, which might be attributed to the CC, have been observed in two-photon-excited autofluorescence images [Fig. 4(b)] [15]. Since similar sub-cellular structures were observed in Figs. 4(a) and 4(b), we speculate that the bright hyper-reflectivity may be attributed to the same anatomic source as that of the bright autofluorescent spots seen in Fig. 4(b).
Fig. 3.
Comparison of three different imaging results using a living frog eyecup: (a) by conventional LSM, (b) by confocal LSM, and (c) by VSD reconstruction. The selected field of view was ~80 μm × 80 μm. The green color window in (c) marks the region of interest shown in greater detail in Fig. 4(a). The corresponding results in Fourier domain are shown, respectively, in (d)–(f). The measured frequency cutoff boundaries of the functions are delineated with red circles. (g) The normalized intensity profile of the selected adjacent photoreceptors (along upper lines) in (a), (b), and (c) are plotted, respectively. (h) The normalized intensity profile of the selected adjacent photoreceptors (along lower lines) in (a), (b), and (c) are plotted, respectively.
Fig. 4.
(a) Enlarged photoreceptor illustration of a sub-image marked by the green square in Fig. 3(c). (b) An example of two-photon-excited autofluorescence imaging of the flat-mounted retina. Blue arrowheads point to rods while the red arrowheads point to cones.
In summary, a line scanning strategy was combined with VSD to achieve rapid super-resolution imaging. Both standard resolution targets and living frog eyecups were used to verify resolution enhancement. High-speed imaging is important for practical applications of super-resolution LSM for imaging biological dynamics and moving samples. It takes, in total, ~3 s to scan four angles for isotropic reconstruction. This speed is significantly improved compared to our prototype VSD SLM with a single-point scanning scheme, which required ~160 s to cover the same field of view (400 × 400 pixels) [11, 12]. In this work, the imaging speed of the four-angle scanning that ensured isotropic resolution improvement, was determined by the camera and the rotation motor of the dove prism. If a faster camera (e.g., pco.dimax HS1, PCO-TECH) is used, it is possible to reduce the sampling time to millisecond level to finish one-angle scan while maintaining the same field of view. If a faster motorized rotation stage (e.g., RGV160BL, NEWPORT) is used, the time required to rotate the dove prism to specific angles in sequence (0°, 22.5°, 45°, and 67.5°) could be reduced to ~70 ms (23 ms × 3) Future integration of acousto-optic dove prism can provide fast angle rotation at microsecond speed [16]. Thus, in-frame movements (i.e., movements during a one-angle scan) can be minimized for in vivo applications. The displacement of the sample among different scanning angles can also be corrected by the tracking system or a global registration method. We anticipate that further development of VSD-based LSM could achieve super-resolution imaging in in vivo cortex or retinal imaging.
Acknowledgments
This research was supported in part by the National Institutes of Health (NIH) (R01 EY023522, R01 EY024628, and P30 EY001792) and the National Science Foundation (NSF) (CBET-1055889).
References
- 1.Rust MJ, Bates M, Zhuang X. Nat. Meth. 2006;3:793. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Science. 2006;313:1642. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 3.Hess ST, Girirajan TPK, Mason MD. Biophys. J. 2006;91:4258. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gustafsson MG. J. Microsc. 2000;198:82. doi: 10.1046/j.1365-2818.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- 5.Gustafsson MG. Proc. Natl. Acad. Sci. 2005;102:13081. doi: 10.1073/pnas.0406877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karadaglic D, Wilson T. Micron. 2008;39:808. doi: 10.1016/j.micron.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 7.Kim T, Gweon D, Lee JH. Meas. Sci. Technol. 2009;20:055501. [Google Scholar]
- 8.Mandula O, Kielhorn M, Wicker K, Krampert G, Kleppe I, Heintzmann R. Opt. Express. 2012;20:24167. doi: 10.1364/OE.20.024167. [DOI] [PubMed] [Google Scholar]
- 9.Planchon TA, Gao L, Milkie DE, Davidson MW, Galbraith JA, Galbraith CG, Betzig E. Nat. Methods. 2011;8:417. doi: 10.1038/nmeth.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu J, Min W, Conchello JA, Xie XS, Lichtman JW. Nano Lett. 2009;9:3883. doi: 10.1021/nl902087d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu RW, Wang BQ, Zhang QX, Yao XC. Biomed. Opt. Express. 2013;4:1673. doi: 10.1364/BOE.4.001673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang B, Lu R, Zhang Q, Yao X. Quant. Imaging Med. Surg. 2013;3:243. doi: 10.3978/j.issn.2223-4292.2013.10.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rector DM, Ranken DM, George JS. Methods. 2003;30:16. doi: 10.1016/s1046-2023(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 14.Nilsson SE. J. Ultrastruct. Res. 1964;10:390. doi: 10.1016/s0022-5320(64)80018-6. [DOI] [PubMed] [Google Scholar]
- 15.Lu RW, Li YC, Ye T, Strang C, Keyser K, Curcio CA, Yao XC. Biomed. Opt. Express. 2013;2:1494. doi: 10.1364/BOE.2.001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paek EG, Choe JY, Oh TK, Hong JH, Chang TY. Opt. Lett. 1997;22:1195. doi: 10.1364/ol.22.001195. [DOI] [PubMed] [Google Scholar]