Abstract
Introduction
The success of obesity therapy is dependent on the genetic background of the patient. Circadian Locomotor Output Cycles Kaput (CLOCK), one of the transcription factors from the positive limb of the molecular clock, is involved in metabolic alterations.
Objective
To investigate whether five candidate polymorphisms from CLOCK were associated with anthropometric, metabolic measures and weight loss in response to a behavioural weight reduction programme based on the Mediterranean diet.
Methods
Five hundred overweight/obese subjects, aged 20–65 years, who attended outpatient clinics specializing in obesity, were studied. Anthropometric, biochemical and dietary intake variables were analysed. Effectiveness of the programme and weight loss progression during 28 weeks of treatment was assessed.
Results
Four of five CLOCK SNPs selected were significantly associated with obesity variables (P<0.05). The genetic variation in the rs1801260 CLOCK was associated with obesity at baseline and also affected weight loss. Patients with the variant allele (G) lost significantly less weight i(P = 0.008) compared with wild type. Repeated measures analysis showed that weight loss over time was significantly different between rs1801260 CLOCK variations (P = 0.038). Carriers of the G allele displayed greater difficulty in losing weight than non-carriers. In this particular polymorphism, the frequency of short-time sleepers (≤6 h per day) was greater in minor allele carriers than in non-carriers (59% vs 41%; P<0.05). CLOCK polymorphisms were also associated with significant differences in total plasma cholesterol at the completion of dietary treatment (P<0.05).
Conclusions
We have replicated previous studies showing a relationship between CLOCK gene polymorphisms and obesity. CLOCK rs1801260 SNP may predict the outcome of body weight reduction strategies based on low-energy diets.
Keywords: CLOCK, polymorphism, behavioural therapy
Introduction
Weight loss in response to obesity management strategies shows a wide range of interindividual variation that is largely influenced by nutritional, hormonal and psycho-behavioural factors. In addition, the success of obesity therapy may be modulated by the genetic background of the patient,1 as shown by studies carried out in monozygotic twins; weight loss seems to show substantial heritability. In addition, parental obesity has a role in the likelihood of obesity in the offspring.1 A number of potential candidate genes has been previously examined for associations with body weight loss and weight loss maintenance.2–5
The Human Obesity Gene Map lists about 250 loci possibly involved in the development of obesity.1 However, results from association studies for most of these loci are inconclusive for obesity and weight loss outcomes. Although some studies find no associations between weight loss and candidate genes,6 others have shown that changes in fat mass were predicted by polymorphisms at several obesity candidate genes, explaining up to 8.5% of the fat mass variance.1 Examples of genes related to changes in fat mass include leptin, G protein, ADRB3, PPARγ and perilipin.2–5
Recent clinical and epidemiological studies have shown significant relationships between chronobiology and obesity. For example, shift work, sleep deprivation and bright light exposure at night have been associated with increased adiposity.7 Although our understanding of the biological clock model continues to evolve, previous studies have established that Circadian Locomotor Output Cycles Kaput (CLOCK), one of the transcription factors from the positive limb of the molecular clock, is involved in altered metabolic function.8 Animal models have revealed that mice with Clock gene disruptions are prone to develop obesity.9 In 2004, Rudic et al. 10 showed that mutations in Clock gene were associated with impaired glucose tolerance. More recently, these research efforts have been extended to gain further understanding about the role of CLOCK gene variants in human obesity. Thus far, only two studies focused on obesity have shown interesting associations between different CLOCK gene polymorphisms and body mass index (BMI).11,12 However, no study has yet reported relationships between CLOCK gene polymorphisms and weight loss in response to a weight reduction programme.
On the basis of these previous studies and on a number of remaining unexplored questions, we investigated whether five candidate polymorphisms from CLOCK were associated with anthropometric, metabolic measures and weight loss in response to a behavioural weight reduction programme based on the Mediterranean diet in an obese population from southeastern Spain.
Subjects and methods
Subjects
We recruited overweight or obese subjects (BMI>25 kg m−2 and <40 kg m−2) within the age range of 20–65 years, (n = 500) who attended during 2008 five outpatient obesity clinics in the city of Murcia, located in southeastern Spain. Patients receiving thermogenic or lipogenic drugs, or those diagnosed with diabetes mellitus, chronic renal failure, hepatic diseases or cancer were excluded from the study (11%). The final sample consisted of 454 individuals.
All procedures were in accordance with good clinical practice. Written consent was obtained from each patient before participation and the study principles were approved by the Research Ethics Committee of the Virgen de la Arrixaca Hospital. Patient data were codified to guarantee anonymity.
Characteristics of the treatment
The characteristics of the weight reduction programme have been described elsewhere.13,14 Briefly, during the initial 4 months, subjects attended a weekly 60-min therapy session in support groups (n = 10), followed by a 5-month maintenance period. Sessions were conducted by a nutritionist. Treatment was based on the following:
Dietary treatment
Dietary individual energy requirements were assessed by calculating (1) resting energy expenditure (REE) according to the Harris-Benedict formula and (2) total energy expenditure (TEE) according to the type and duration of physical activity. Next, about 2.6 MJ per day were subtracted from the TEE. The final dietary energy content ranged from 1200–1800 kcal per day for women and 1500– 2000 kcal per day for men to induce an approximate loss of 0.5–1 kg per week. The recommendations were consistent with the Mediterranean type of diet 14 and the macronutrient distribution followed the recommendations of the Spanish Society of Community Nutrition.15
Nutritional education was given during group therapy sessions to help subjects plan their own menus and to educate subjects to adopt appropriate lifetime eating habits.
Physical activity emphasised individual goals of 15–30 min or more of moderate intensity physical activity, at least 2 or 3 times a week. Patients were encouraged to use a pedometer to reach at least 10 000 steps per day.
Behavioural techniques included stimulus control, self-monitoring, positive reinforcement and cognitive behavioural therapy.
Anthropometric measurements
Subjects were weighed in barefoot wearing light clothes, with a digital scale to the nearest 0.1 kg, at the same time each day. Height was measured using a Harpenden digital stadiometer (rank 0.7–2.05). The subject was positioned upright, relaxed and with the head in the Frankfurt plane. BMI was calculated according to these measurements as weight(kg)/(height(m))2. Total body fat was measured by bioelectrical impedance using TANITA TBF-300 (TANITA Corporation of America, Arlington Heights, IL, USA) equipment. Body fat distribution was assessed by the measurement of different circumferences: waist circumference, at the level of the umbilicus, and hip circumference, as the widest circumference over the greater trochanters.16 All measurements were made with a flexible and inextensible tape measure. Waist–hip ratio (WHR) was then calculated.
Biochemical analysis
Plasma concentrations of glucose, total cholesterol, total haemoglobin and uric acid were determined from venous blood samples after overnight fast with commercial kits (Roche Diagnostics GmbH, Mannheim, Germany).
Habitual dietary intake
To evaluate food habits, initial nutrient intake was determined by a 24-h dietary recall. Interviews were conducted from monday to friday, including 24-h recalls of food intake from weekend and weekdays. Total energy intake and macronutrient composition from the initial 24-h recalls were analysed with the nutritional evaluation software program Grunumur17 on the basis of Spanish food composition tables.18
DNA isolation and clock genotyping
We selected tagSNPs as effective proxies for untyped SNPs in strong linkage disequilibrium (LD) by using the Tagger19 based on HapMap Caucasian European Utah data (www.hapmap.org) 20 with a minor allele frequency (MAF) ≥0.10 and a minimum r2 of 0.8. Tagger uses an algorithm that selects tagSNPs to construct single- and multi-marker tests to capture alleles of interest based on the computed correlation r2 between them.
DNA was isolated from blood samples using routine DNA isolation sets (Qiagen). We performed genotyping of CLOCK gene polymorphisms using a TaqMan assay with allele-specific probes on the ABI Prism 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA) according to the standardized laboratory protocols.21
Bioinformatics analysis
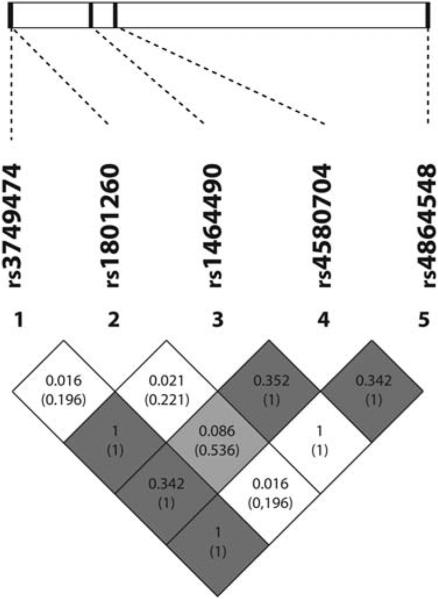
The LD plot between the studied SNPs is shown in Figure 1. SNPs were selected using three criteria: literature reports of genetic associations or biological function of interest, bioinformatics functional assessment and LD structure. In conjunction with the selection of tagSNPs, we also performed bioinformatics analysis of the genomic DNA sequence encompassing different SNPs to ascertain the putative biological consequences of different alleles. SNPs mapping to regions upstream of the transcription start site or within introns were studied with MAPPER22 to identify the potential allele-specific transcription factor binding sites. No intronic SNPs altered splice acceptor or donor sites or other signals recognised by the splicing machinery such as the poly-pyrimidine tract. Polymorphisms within the 3′-UTR of the mRNA can exert an effect on the folding of the mRNA with concomitant changes in mRNA stability. Potential effects of 3′-UTR SNPs were tested with RNAfold 23 within the Vienna RNA package.
Figure 1.
Linkage disequilibrium (LD) plot across the CLOCK gene. The horizontal white bar depicts the 113 kb DNA segment of chromosome 4q12 analysed in the sample. The 5 tagSNP locations are indicated by hatch marks. An LD plot is depicted in the bottom part of the figure: each diamond represents the magnitude of LD for a single pair of markers. Black indicates strong LD (r2 = 1.0); white indicates no LD (r2 = 0) and the grey tones indicate intermediate LD. The numbers inside the diamonds indicate the r2 (D’) value.
Statistical analysis
We used Pearson's χ2 and Fisher's tests to test differences in frequencies. To assess the effectiveness of the program, changes in the characteristics of the population at the beginning and at the end of the treatment were assessed with the Student's paired t-test. We applied ANOVA and Student's t-test to compare crude means across genotype groups. We tested different genetic inheritance models and a dominant model was applied in the final analyses for all SNPs selected, except for rs4580704 that followed a recessive model. We performed multivariate adjustments of the associations by analysis of covariance and estimated adjusted means. We adjusted analyses for sex, age and centre. We also tested the statistical homogeneity of the effects by sex in the corresponding regression model with interaction terms. Statistical analyses were performed using SPSS 15.0 software (SPSS). A two-tailed P-value of <0.05 was considered statistically significant.
Results
Characteristics of the population
General characteristics of the population studied, including anthropometric measurements, biochemical variables, dietary intake and physical activity characteristics, are listed in Table 1. The population of 454 subjects (380 women and 67 men) had a mean age 39±12 years and BMI of 31±5.3 kg m−2 (mean±s.d.). Seventy-five percent of the population was considered sedentary (engaging in less than 1 h of physical activity per week). The mean number of hours of sleep per day was 7 h. Dietary habits obtained by the 24-h recall showed that the percentage of carbohydrate was lower than Spanish recommendations, whereas those of protein and fat were above the recommended levels.
Table 1.
Characteristics of the population studied
| Total population | |
|---|---|
| N | 447 |
| Anthropometric | Mean (s.d.) |
| Age (years) | 39 (12) |
| Weight (kg) | 84.61 (16.72) |
| BMI (kg m−2) | 31.3 (5.3) |
| Body fat (%) | 37.5 (7.5) |
| Hip (cm) | 114 (10) |
| Waist (cm) | 102 (14) |
| WHR | 0.89 (0.08) |
| Plasma values | |
| Glucose (mg per 100 ml) | 90.7 (22.4) |
| Cholesterol (mg per 100 ml) | 173.8 (40.6) |
| Uric acid (mg per 100 ml) | 4.4(1.5) |
| Haemoglobin (mg per 100 ml) | 14.6 (7.1) |
| Activity | |
| Sleep (hours per day) | 7.1 (1.5) |
| Exercise (METs) | 4140 (4426) |
| Dietary intake | |
| Total energy (kcal per day) | 1903 (709) |
| Carbohydrates (%) | 39.6 (10.5) |
| g per day | 148.6 (52.7) |
| Proteins (%) | 16.9 (3.6) |
| g per day | 65.6 (26.8) |
| Fats (%) | 43.5 (10.7) |
| g per day | 79.0 (38.3) |
| CLOCK polymorphism, n (%) | |
| rs4580704 | |
| GG | 65 (14.4) |
| CG | 202 (44.8) |
| CC | 166 (36.8) |
| rs1801260 | |
| GG | 33 (7.7) |
| AG | 170 (37.7) |
| AA | 220 (54.6) |
| rs3749474 | |
| TT | 46 (10.2) |
| TC | 178 (39.5) |
| CC | 204 (45.2) |
Abbreviation: WHR, weight-hip ratio.
Genotype frequency of clock variants in Garaulet subjects and associations with obesity parameters
SNP rs1464490 was selected because it was the tagSNP for a large LD block (LD1); rs3749474, also from LD1, was selected after bioinformatics functional assessment suggested important changes to mRNA structure; and rs4864584 because it was previously associated with overweight/obesity.11,12 From different SNPs comprising LD2, selection of rs4580704 was based on its previous relationship with BMI.12 From LD3, we selected rs1801260 SNP based on the previous reports showing associations between this SNP, sleep alterations24 and binge eating disorders.25 Their locations in the CLOCK gene, the Hardy-Weinberg equilibrium and minor allele frequency are listed in Table 2. CLOCK genotype frequencies did not deviate from the Hardy-Weinberg equilibrium. As SNPs, rs3749474, rs1464490 and rs4864548, were almost in complete LD and displayed a similar pattern of phenotypic associations, only the results for rs3749474 are presented.
Table 2.
Characteristics of the population studied before and after behavioral treatment
| Measures | Total group | Total group | P |
|---|---|---|---|
| Before treatment (N = 447) | After treatment (N = 447) | ||
| Weight (kg) | 84.60 ± 0.79 | 75.27 ± 0.72 | 0.0001 |
| BMI (kg m−2) | 31.4 ± 0.3 | 28.1 ± 0.2 | 0.0001 |
| Hip (cm) | 115 ± 0.76 | 108 ± 0.86 | 0.0001 |
| Waist (cm) | 105 ± 1.17 | 97 ± 1.22 | 0.0001 |
| WHR | 0.91 ± 0.08 | 0.89 ± 0.08 | 0.0001 |
| Glucose (mg per 100 ml) | 89.4 ± 2.5 | 89.0 ± 3.0 | 0.897 |
| Cholesterol (mg per 100 ml) | 180.1 ± 3.78 | 163.8 ± 4.94 | 0.002 |
| Uric acid (mg per 100 ml) | 4.63 ± 0.15 | 4.01 ± 0.15 | 0.001 |
| Diastolic pressure (mmHg) | 70 ± 1.0 | 66 ± 1.1 | 0.010 |
| Systolic pressure (mmHg) | 112 ± 1.7 | 107 ± 1.6 | 0.005 |
Abbreviations: BMI, body mass index; WHR: waist-hip ratio. Data are presented as mean ± EEM. Student's t-test: P<0.05 considered significant.
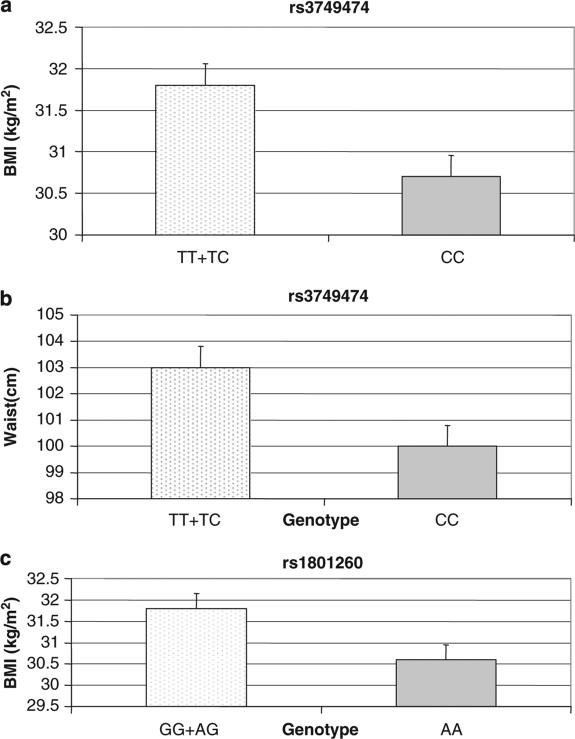
We first examined the association between the CLOCK SNPs and obesity parameters (Figure 2). We did not detect sex heterogeneity for any of the SNPs examined. Therefore, we present the results for men and women combined. We found significant associations with weight and BMI for SNPs rs3749474 and rs1801260 and with waist for rs3749474. A similar trend was found for rs4580704, although it did not reach statistical significance: weight (P = 0.077), waist (P = 0.077) and WHR (P = 0.097).
Figure 2.
Significant associations between CLOCK SNPs and obesity parameters. (a) CLOCK rs3749474 and BMI; (b) CLOCK rs3749474 and waist circumference; (c) CLOCK rs180126 (3111T>C) and BMI.
For rs3749474, carriers of the minor allele T had significantly higher weight, BMI and waist than CC subjects. Similar results were found for rs1801260 in which minor allele carriers also showed the highest BMI values. These differences remained statistically significant after additional adjustment for sex, age, treatment centre and initial BMI (Figure 2). For rs4580704, the trend was the opposite; subjects homozygous for the minor allele showed lower values for weight, waist and WHR than non-carriers.
Longitudinal data
Characteristics of the population before and after the dietary behavioural treatment are shown in Table 3. All of the anthropometric variables were significantly improved after treatment. Total cholesterol, uric acid and blood pressure were also significantly reduced after treatment.
Table 3.
Description of Clock SNPs
| Name | Location | HWPval | MAF | Alleles | Minor allele |
|---|---|---|---|---|---|
| rs3749474 | 3′-UTR | 0.444 | 0.32 | C/T | T |
| rs1801260 | 3′-UTR | 0.984 | 0.28 | A/G | G |
| rs1464490 | Intron 11 | 0.453 | 0.42 | C/T | T |
| rs4580704 | Intron 9 | 0.782 | 0.38 | C/G | G |
| rs4864548 | Promoter | 0.445 | 0.41 | G/A | A |
Abbreviations: HW Pval, Hardy-Weinberg equilibrium expectations P values; MAF, minor allele frequency.
The mean duration of the treatment was 26±17 weeks. The average weight loss was 8.85 kg. The percentage of weight loss compared with the initial body weight was 10.24 and the rate of weight loss was 450 g per week.
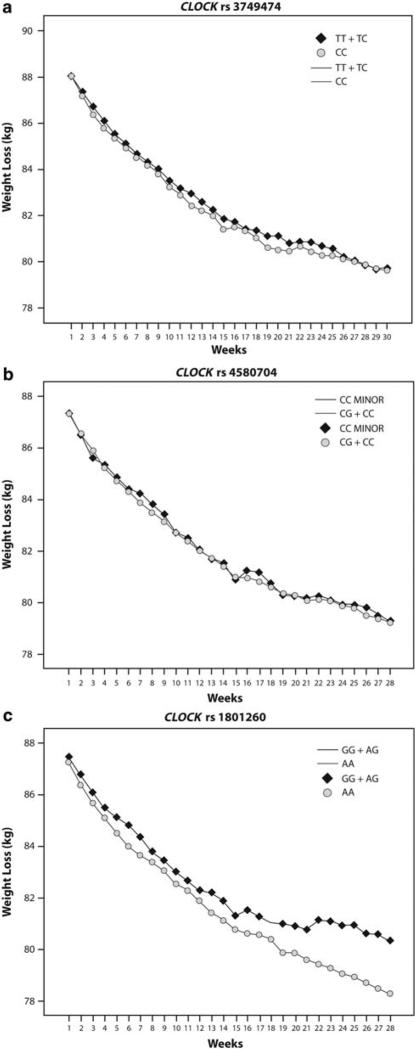
Genetic variation at the CLOCK rs1801260 SNP was associated with differences in body weight reduction. Patients with the minor allele were less successful losing weight after adjusting for baseline BMI (P = 0.008) (Table 4). The difficulty in losing weight was particularly evident after 12 weeks of treatment as evidenced by the analysis of repeated measurements after adjusting for baseline BMI (P = 0.038). No statistically significant differences were found for the remaining CLOCK SNPs analysed (P>0.05) (Figure 3 a–c). However, significant associations were found between CLOCK polymorphisms and changes in the total plasma cholesterol, even for those SNPS that showed no statistical associations with weight loss, such as rs374947 and rs4580704 (Table 4).
Table 4.
Associations of CLOCK polymorphisms with changes in weight and metabolic parameters
| CLOCK | Total weight loss (kg) | P-valueb | Serum cholesterol changes | P-value |
|---|---|---|---|---|
| rs4580704 | ||||
| GGa | 8.82 ± 5.13 | 0.877 | 6.80 ± 14.96 | 0.026 |
| CG+CC | 8.99 ± 5.54 | 44.61 ± 7.30 | ||
| rs1801260 | ||||
| GG+AGa | 7.96 ± 0.57 | 0.008 | 25.08 ± 9.65 | 0.064 |
| AA | 10.41 ± 0.53 | 52 ± 9.45 | ||
| rs3749474 | ||||
| TT+TCa | 9.37 ± 0.56 | 0.513 | 49.58 ± 9.54 | 0.047 |
| CC | 8.70 ± 0.62 | 22.42 ± 9.54 |
All data are given as mean ± s.e.m.
Recessive model.
Adjusted for initial BMI.
Figure 3.
CLOCK variants and weight loss evolution during 28 weeks of treatment: (a) CLOCK rs3749474; (b) CLOCK rs4580704; (c) CLOCK rs180126 (3111T>C).
When potential associations between sleep duration and CLOCK SNPs were examined, a trend was found for rs 1801260. Subjects carrying the minor allele were 2.10 times more likely to sleep ≤6 h per day than non-carriers (95% CI 0.95–4.45) (P = 0.060). Moreover, the frequency of short-time sleepers (≤6 h per day) was higher in minor allele carriers than in subjects homozygous for the major allele (59 vs 41%), whereas the opposite was true fore those who slept more than 6 h a day, 41% in minor allele carriers vs 69% in non-carriers (P<0.05).
Discussion
In this study, we have confirmed a role of the CLOCK gene in body weight regulation. Specifically, four out of five CLOCK SNPs genotyped, that is rs3749474, rs4864548, rs1464490 and rs1801260, were significantly associated with obesity parameters, and a trend was found for the fifth one (rs4580704). We have also shown that CLOCK rs1801260 was associated with response to a dietary and behavioural weight loss programme. Specifically, carriers of the G variant may be at higher risk for obesity, and they were least responsive to a weight loss intervention.
In this study, we have replicated previous findings showing the association between different CLOCK SNPs and obesity, particularly for rs4864548 and rs4580704.11,12 In addition, we have shown that minor allele carriers carrying at least one copy of the T allele at the CLOCK rs3749474 displayed a significantly higher degree of obesity (weight and BMI) and abdominal obesity (waist circumference) than major allele carriers. This SNP was selected because of its location in the 3′-UTR. The mechanism through which this particular SNP is associated with obesity could be attributed to changes in mRNA structure. Indeed, our analysis suggests that this one-base change (T/C) in the CLOCK 3′-UTR transforms the structure of this particular region (Figure 2). The 3′-UTR is characterized by sequences such as SECIS or AU-rich elements (AREs), which are recognised by particular proteins that exert an effect on mRNA stability or location in the cell. Although specific binding sites are not elucidated in the analysis presented here, the difference in the predicted allele-specific structures is consistent with the hypothesis that this SNP strongly affects mRNA structure and stability. On the other hand, this SNP is in complete LD with rs4864548 (Figure 1), which is located in the promoter region. Variation in the promoter activity may also explain the results we observed.
These findings support the evolving view that the circadian oscillator has an important role in the development of obesity and metabolic syndrome.7 Recently, several studies have suggested that the disruption of the circadian system may contribute to obesity.26 Shift work, sleep deprivation and exposure to bright light at night are related to increased adiposity.27 Studies performed in animal models have revealed that mice with disruption of the Clock gene are prone to develop obesity and a phenotype resembling MetS.9 In a similar vein, we have recently provided evidence of CLOCK gene expression in human adipose tissue and shown its association with waist circumference and different components of the MetS.28 The precise mechanisms linking MetS to circadian system disruption are not well known. However, most hypotheses suggest that the internal desynchronization (ID) between different circadian rhythms involved in metabolism is a key factor in the development of MetS.29
Our dietary intervention study has revealed that minor allele carriers of rs1801260 at the CLOCK locus are more resistant to weight loss and to metabolic changes in response to an energy-restricted diet than AA homozygotes. Previous research supports the hypothesis that individual CLOCK genotypes may influence several variables linked with behaviour in both healthy individuals and in those with mental illness.30 Specifically, for rs1801260, it has been shown that this polymorphism is related to clinical features of mood disorders influencing diurnal preference in healthy humans and that it causes sleep phase delay and insomnia in people with depression and bipolar disorder.30–32 These alterations could be related not only to obesity but also to weight loss. Along the same lines, we have previously shown that most of the obstacles of weight loss are related to the subject's eating behaviours and psychological characteristics and that these obstacles are directly related to the difficulty in losing weight.14 Moreover, previous works have related rs1801260 to eating disorders and obesity.12,25
This idea is supported by animal findings suggesting a direct involvement of the Clock gene in the regulation of body weight, as homozygous Clock mutant mice developed obesity, hyperphagia and also suffered from changes in eating behaviour, sleep pattern and mood.9 Interestingly, we found that minor allele carriers of the rs1801260 variant had a tendency to sleep less than major allele subjects. Our data are consistent with earlier studies associating this SNP with insomnia 30 and also with epidemiological studies that indicate that sleep deprivation may increase the risk of obesity and weight gain.33 Although the molecular mechanisms underlying the observed results remain unknown, we can hypothesise from this study that carriers of the rs1801260 variant allele might experiment a disruption in the chronobiology system similar to that shown in the Clock knockout mouse. Loss of the Clock gene in the animal model affects obesity, sleep and eating behaviours that may, in human subjects, consequently influence weight loss as well.
In a previous study, investigators reported that polymorphisms in a panel of obesity-related candidate genes have a minor role, if any, in modulating weight changes induced by a moderately hypo-energetic diet.6 The investigators concluded that none of the genetic variants they examined had a significant association with weight loss in the intervention. Although the CLOCK gene was not included in that study, one important difference between the earlier study and this study was that in the earlier study the duration of interventions was limited to 10 weeks, whereas in this study the loss treatment was followed for 28 weeks. Sorenson et al. 6 have suggested that more prolonged interventions are necessary to effectively evaluate the rolls of particular polymorphisms in weight loss, and to detect differences among variants. Indeed, in this study, the most important differences between rs1801260 variants in weight loss were identified since the third month of treatment. Other studies that have identified associations between genetic variants and weight loss in obese populations have used intervention durations similar to those used in our study.1 Up to this point, most of the studies that have evaluated relationships between genetic variants and weight loss achieved through interventions have focused on genes related to energy expenditure, appetite control, adipogenesis or genes related to insulin resistance and lipid metabolism.1 Our study expands the realm of current knowledge by showing a potential role for circadian clock genes on weight loss.
In conclusion, in this intervention trial performed in a Mediterranean population, we have replicated previous studies that reported relationships between CLOCK gene polymorphisms and obesity. Our results showed that rs1801260 may predict the outcome of weight loss strategies that are based on low-energy diets. Carriers of the G allele may exhibit a greater degree of obesity and experience greater difficulty in losing weight in response to a low-energy diet. The difficulty was consistently observed throughout the treatment period, although it was most evident since the third month of treatment. These data suggest that CLOCK gene polymorphisms may predict weight loss magnitude in response to a low-energy diet.
Acknowledgements
This work was supported by the Government of Education, Science and Research of Murcia (Project BIO/FFA 07/01– 0004) and by The Spanish Government of Science and Innovation (projects AGL2008–01655/ALI) and by NIDDK Grant DK075030, as well as by contracts 53-K06–5–10 and 58–1950–9–001 from the US Department of Agriculture Research Service.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Hainer V, Zamrazilová H, Spálová J, Hainerová I, Kunesová M, Aldhoon B, et al. Role of hereditary factors in weight loss and its maintenance. Physiol Res. 2008;57:S1–15. doi: 10.33549/physiolres.931485. [DOI] [PubMed] [Google Scholar]
- 2.Corella D, Qi L, Sorlí JV, Godoy D, Portolés O, Coltell O, et al. Obese subjects carrying the 11482G> polymorphism at the perilipin locus are resistant to weight loss after dietary energy restriction. J Clin Endocrinol Metab. 2005;90:5121–5126. doi: 10.1210/jc.2005-0576. [DOI] [PubMed] [Google Scholar]
- 3.Marti A, Moreno-Aliaga MJ, Zulet A, Martínez JA. Advances in molecular nutrition: nutrigenomics and/or nutrigenetics. Nutr Hosp. 2005;20:157–164. [PubMed] [Google Scholar]
- 4.Haupt A, Thamer C, Machann J, Kirchhoff K, Stefan N, Machicao F, et al. Impact of variation in the FTO gene on whole body fat distribution, ectopic fat and weight loss. Obesity. 2008;16:1969–1972. doi: 10.1038/oby.2008.283. [DOI] [PubMed] [Google Scholar]
- 5.Aberle J, Flitsch J, Beck NA, Mann O, Busch P, Peitsmeier P, et al. Genetic variation may influence obesity only under conditions of diet: analysis of three candidate genes. Mol Genet Metab. 2008;95:188–191. doi: 10.1016/j.ymgme.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 6.Sørensen TI, Boutin P, Taylor MA, Larsen LH, Verdich C, Petersen L, et al. NUGENOB Consortium. Genetic polymorphisms and weight loss in obesity: a randomised trial of hypo-energetic high- versus low-fat diets. PLoS Clin Trials. 2006;1:e12. doi: 10.1371/journal.pctr.0010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garaulet M, Madrid JA. Chronobiology, genetics and metabolic syndrome. Curr Opin Lipidol. 2009;20:127–134. doi: 10.1097/MOL.0b013e3283292399. [DOI] [PubMed] [Google Scholar]
- 8.Fuller PM, Lu J, Saper CB. Differential rescue of light- and food-entrainable circadian rhythms. Science. 2008;230:1074–1077. doi: 10.1126/science.1153277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turek FW, Joshu C, Kohsaka A. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rudic RD, McNamara P, Curtis AM. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLOS Biol. 2006;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott EM, Carter AM, Grant PJ. Association between polymorphisms in the Clock gene, obesity and the metabolic syndrome in man. Int J Obes. 2008;32:658–662. doi: 10.1038/sj.ijo.0803778. [DOI] [PubMed] [Google Scholar]
- 12.Sookoian S, Gemma C, Gianotti TF, Burgueño A, Castaño G, Pirola CJ. Genetic variants of Clock transcription factor are associated with individual susceptibility to obesity. Am J Clin Nutr. 2008;87:1606–1615. doi: 10.1093/ajcn/87.6.1606. [DOI] [PubMed] [Google Scholar]
- 13.Garaulet M, Pérez-Llamas F, Zamora S, Tébar FJ. Weight loss and possible reasons for dropping out of a dietary/behavioural programme in the treatment of overweight patients. J Hum Nutr Diet. 1999:219–227. [Google Scholar]
- 14.Corbalán MD, Morales EM, Canteras M, Espallardo A, Hernández T, Garaulet M. Effectiveness of cognitive-behavioral-therapy based on the Mediterranean diet for the treatment of obesity. Nutrition. 2009 doi: 10.1016/j.nut.2009.02.013. (in press)AQ. [DOI] [PubMed] [Google Scholar]
- 15.Serra-Majem L, Aranceta J. SENC working group on nutritional objectives for the Spanish population. Spanish Society of Community Nutrition. Nutritional objectives for the Spanish population. Consensus from the Spanish Society of Community Nutrition. Public Health Nutr. 2001;4:1409–1413. doi: 10.1079/phn2001229. [DOI] [PubMed] [Google Scholar]
- 16.Ferrario VF, Sforza C, Schmitz JH, Miani A, Taroni G. Fourier analysis of human soft tissue facial shape: sex differences in normal adults. J Anat. 1995;187:593–602. [PMC free article] [PubMed] [Google Scholar]
- 17.Perez-Llamas F, Garaulet M, Herrero F, Palma JT, Perez de Heredia F, Marin R, et al. Multivalent informatics application for studies of the nutricional status of the population. Assessment of food intake. Nutr Hosp. 2004;19:160–166. [PubMed] [Google Scholar]
- 18.Mataix J, Mañas M, Llopis J, Martinez E. Tabla de composición de alimentos españoles (Table of composition of Spanish foods) Instituto de Nutrición y tecnología. Universidad de Granada; Spain: 1995. [Google Scholar]
- 19.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Altshuler efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 20.Caucasian European Utah data. Internet www hapmap.org.
- 21.HelixTree Manual 5 3 0. Internet http://goldenhelix.com/HelixTreeManual/compositehaplotypemethodchm.htmlx133-75400023.7)
- 22.Marinescu VD, Kohane IS, Riva A. MAPPER: a search engine for the computational identification of putative transcription factor binding sites in multiple genomes. BMC Bioinform. 2005;6:79. doi: 10.1186/1471-2105-6-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The Vienna RNA Websuite. Nucleic Acids Res. 2008;36(Web Server issue):W70–W74. doi: 10.1093/nar/gkn188. Internet http://rna.tbi.univie.ac.at/cgibin/ RNAfold.cgi) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwase T, Kajimura N, Uchiyama M. Mutation screening of the human Clock gene in circadian rhythm sleep disorders. Psychiatry Res. 2002;109:121–128. doi: 10.1016/s0165-1781(02)00006-9. [DOI] [PubMed] [Google Scholar]
- 25.Monteleone P, Tortorella A, Docimo L. Investigation of 3111T/C polymorphism of the CLOCK gene in obese individuals with or without binge eating disorder: association with higher body mass index. Neurosci Lett. 2008;435:30–33. doi: 10.1016/j.neulet.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 26.Hamet P, Tremblay J. Genetics of the sleep–wake cycle and its disorders. Metabolism. 2006;55(Suppl 2):S7–S12. doi: 10.1016/j.metabol.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 27.Kohler HP, Grant PJ. Plasminogen-activator inhibitor type 1 and coronary artery disease. N Engl J Med. 2000;342:1792–1801. doi: 10.1056/NEJM200006153422406. [DOI] [PubMed] [Google Scholar]
- 28.Gomez-Abellan P, Hernandez-Morante JJ, Lujan JA. Clock genes are implicated in the human metabolic syndrome. Int J Obesity. 2008;32:121–128. doi: 10.1038/sj.ijo.0803689. [DOI] [PubMed] [Google Scholar]
- 29.Hernandez-Morante JJ, Gomez-Santos C, Milagro F. Expression of cortisol metabolism-related genes show circadian rhythmic patterns in human adipose tissue. Int J Obes. 2009 doi: 10.1038/ijo.2009.4. [DOI] [PubMed] [Google Scholar]
- 30.Benedetti F, Radaelli D, Bernasconi A, Dallaspezia S, Falini A, Scotti G, Lorenzi C, et al. Clock genes beyond the clock: CLOCK genotype biases neural correlates of moral valence decision in depressed patients. Genes Brain Behav. 2008;7:20–25. doi: 10.1111/j.1601-183X.2007.00312.x. [DOI] [PubMed] [Google Scholar]
- 31.Benedetti F, Dallaspezia S, Fulgosi MC, Lorenzi C, Serretti A, Barbini B, et al. Actimetric evidence that CLOCK 3111 T/C SNP influences sleep and activity patterns in patients affected by bipolar depression. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:631–635. doi: 10.1002/ajmg.b.30475. [DOI] [PubMed] [Google Scholar]
- 32.Lee HJ, Paik JW, Kang SG, Lim SW, Kim L. Allelic variants interaction of CLOCK gene and G-protein beta3 subunit gene with diurnal preference. Chronobiol Int. 2007;24:589–597. doi: 10.1080/07420520701534632. [DOI] [PubMed] [Google Scholar]
- 33.Van Cauter E, Knutson KL. Sleep and the epidemic of obesity in children and adults. Eur J Endocrinol. 2008;159(Suppl 1):S59–S66. doi: 10.1530/EJE-08-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]