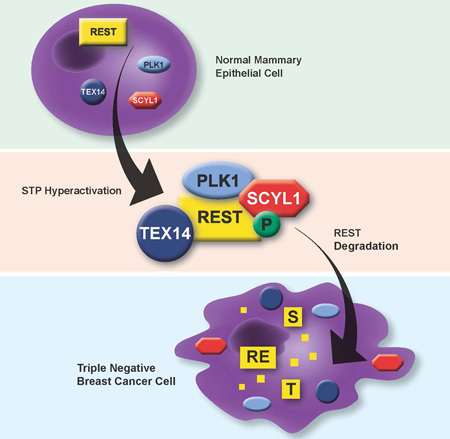
SUMMARY
Defining the molecular networks that drive breast cancer has led to therapeutic interventions and improved patient survival. However, the aggressive triple-negative breast cancer subtype (TNBC) remains recalcitrant to targeted therapies because its molecular etiology is poorly defined. In this study, we used a forward genetic screen to discover an oncogenic network driving human TNBC. SCYL1, TEX14, and PLK1 (“STP axis”) cooperatively trigger degradation of the REST tumor suppressor protein, a frequent event in human TNBC. The STP axis induces REST degradation by phosphorylating a conserved REST phospho-degron and bridging REST interaction with the ubiquitin-ligase βTRCP. Inhibition of the STP axis leads to increased REST protein levels and impairs TNBC transformation, tumor progression, and metastasis. Expression of the STP axis correlates with low REST protein levels in human TNBCs and poor clinical outcome for TNBC patients. Our findings demonstrate that the STP-REST axis is a new molecular driver of human TNBC.
Graphical Abstract
Introduction
Breast cancer is a heterogeneous disease comprised of three clinical subtypes: estrogen receptor positive (ER+), HER2 positive (HER2+), and triple negative breast cancer (TNBC). The identification of prominent molecular drivers of breast cancers (ER and HER2) has enabled the development of targeted therapies that have improved survival rates for patients with ER+ and HER2+ breast cancers (Arteaga et al., 2012; Osborne, 1998; Slamon et al., 1989). In contrast, there are currently no effective targeted therapies for patients with TNBC, which exhibit a poor prognosis due to high frequency of early relapse and metastasis (Di Cosimo and Baselga, 2010; Hurvitz and Finn, 2009). The molecular determinants driving TNBC are poorly understood, and this paucity in our understanding of pathways governing TNBC pathogenesis remains a major obstacle to developing therapies for TNBC patients.
The RE-1 silencing transcription factor (REST) was originally discovered as a transcriptional repressor of neural and neuronal genes (Chong et al., 1995; Schoenherr and Anderson, 1995). As a master regulator of neuronal gene programs, REST plays an important role in neural differentiation and has been suggested to restrain these programs in non-neuronal tissues (Ballas and Mandel, 2005). Surprisingly, REST also functions as a tumor suppressor in epithelial cancers (Westbrook et al., 2005), although the mechanisms by which REST restrains tumorigenesis remains undefined. Consistently, REST is frequently inactivated in epithelial malignancies such as colon and lung cancer via gene deletion, inactivating mutations (Westbrook et al., 2005), epigenetic silencing (Kreisler et al., 2010), and rarely through alternative splicing (Coulson et al., 2000; Wagoner et al., 2010). However, while REST potently suppresses transformation of cells derived from mammary epithelium (Westbrook et al., 2005), it has remained unclear whether REST is inactivated in breast cancer and what the molecular mechanisms of such misregulation might be.
The REST protein exhibits rapid turnover and is precisely regulated during neural development via the ubiquitin ligase SCFβTRCP and proteasomal degradation (Ballas et al., 2005). Like many SCFβTRCP substrates (Fuchs et al., 2004; Petroski and Deshaies, 2005; Winston et al., 1999), REST must be phosphorylated within a C-terminal phosphodegron for recognition, ubiquitylation and subsequent degradation (Guardavaccaro et al., 2008; Westbrook et al., 2008), suggesting REST stability is regulated by upstream kinase(s) that prime SCFβTRCP-mediated degradation. In this study, we report that REST protein is frequently lost through aberrant degradation in human breast cancer, most prevalently in TNBC. Using an unbiased genetic screen, we identified SCYL1, TEX14 and PLK1 as components of an oncogenic signaling axis (the STP axis) that prominently regulates REST abundance and stability. The STP axis supports cellular transformation, and inhibition of STP components leads to REST protein accumulation and impairment of TNBC at primary and metastatic sites. Collectively, this study uncovers the STP axis as a prominent regulator of REST stability and TNBC pathogenesis.
Results
REST is post-transcriptionally inactivated in human breast cancer
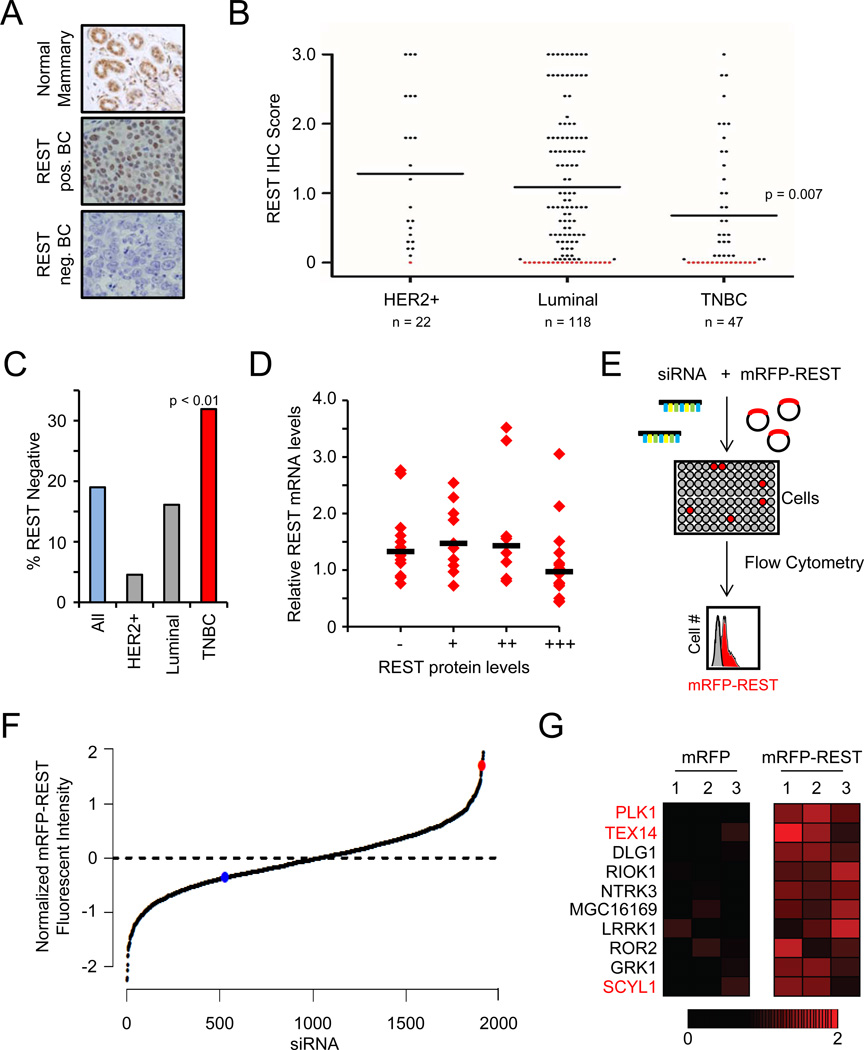
The REST tumor suppressor restrains cell transformation and is frequently inactivated by genetic and epigenetic mechanisms in epithelial cancers (Kreisler et al., 2010; Westbrook et al., 2005). In contrast, while REST potently suppresses transformation of mammary epithelium (Westbrook et al., 2005), analysis of public sequencing and copy-number data indicate that REST is not frequently deleted or mutated in human breast cancer (data not shown). To determine whether REST is compromised by alternative mechanisms in human breast cancer, we evaluated REST protein levels in primary human breast tumors (n=185) (Fig. 1A and Fig S1A). REST protein was readily detectable in all normal mammary tissue analyzed (n=8) and in normal mammary gland adjacent to tumor tissue (representative images in Fig 1A, S1B and S1C). In contrast, REST protein was undetectable in 19% of human breast cancers (Fig 1B, C). Strikingly, REST protein levels were lowest in TNBC (Fig 1B), with 32% of TNBCs exhibiting undetectable REST protein (Fig 1C), suggesting REST dysfunction is a common pathogenic event in this aggressive subtype of breast cancer.
Figure 1. Discovery of REST degradation regulators in human breast cancer.
- Analysis of REST protein expression. Representative images of normal mammary gland (top, n=8) and human tumors (n=185) exhibiting positive REST expression (middle panel) and lack of REST expression (bottom panel).
- REST protein expression is frequently lost in human breast cancer. The REST IHC scores from A are plotted by disease subtype. Each dot represents a tumor; the black line represents the median. Red dots are tumors with REST IHC score = 0 (19%).
- REST protein expression is frequently lost in TNBC. The percentage of tumors within each subtype with undetectable REST protein (REST IHC score = 0) is shown. TNBC = 32%
- REST mRNA and protein levels are not correlated in primary human breast tumors. REST mRNA levels were analyzed in 50 primary breast tumors (from A). Each dot represents a tumor; the black bar represents the median.
- RNAi-based screen for RDR genes. Cells were transfected with siRNAs targeting the kinome (3 siRNAs/gene) and the mRFP-REST or mRFP cDNA(n=4). Changes in fluorescence were assessed by flow cytometry.
- Primary genetic screen for RDR candidates. The normalized effect of each siRNA on mRFP-REST fluorescence from the primary screen is shown. The red and blue dots are the mean of the positive control (siβTRCP) and negative control siRNAs, respectively.
- Identification of RDR candidate genes. Heatmaps represent the effect (relative change in fluorescence) of individual siRNAs on mRFP fluorescence (left) and mRFP-REST fluorescence (right). The red gradient represents the relative change in mRFP-REST fluorescence. The genes for which multiple siRNAs increased REST abundance are shown and those for which multiple siRNAs increased REST abundance upon retesting are labeled in red.
Because REST protein is frequently undetectable in TNBC and other breast cancers, we examined the mechanism by which REST protein is lost. Analysis of REST mRNA in primary human breast cancers revealed that REST mRNA was detectable in all samples analyzed (n=50) and did not correlate with REST protein levels (Fig 1D), suggesting REST is post-transcriptionally depleted in human breast cancer. Previous studies have detected a rare splice variant of REST, known as REST4 (Wagoner et al., 2010), in human breast cancer. However, while full-length REST was readily detectable in all breast cancers analyzed, the REST4 splice variant was rarely found to be expressed (Fig S1D). In tumors that express REST4, full-length REST mRNA was also expressed, and REST4 mRNA expression did not correlate with lower REST protein levels (Fig S1D), indicating that REST protein is not lost in human breast cancer via this alternative splicing event. Collectively, these data suggest that the REST tumor suppressor protein is post-transcriptionally depleted in human breast cancer.
Discovery of REST degradation regulators (RDRs)
Because REST protein was frequently lost in human TNBC without a concomitant loss of REST mRNA, we sought to identify post-transcriptional mechanisms by which REST may be compromised. REST is an unstable protein targeted for proteasomal-mediated degradation by the E3 ubiquitin ligase SCFβTRCP (Guardavaccaro et al., 2008; Westbrook et al., 2008). Like other SCFβTRCP substrates, phosphorylation of REST on a C-terminal phosphodegron is required for REST degradation (Guardavaccaro et al., 2008; Westbrook et al., 2008). Thus, we hypothesized that loss of REST protein in human TNBC may result from aberrant activation of one or more kinase(s), or REST degradation regulators (RDRs), leading to inappropriate REST degradation. To identify putative RDR kinases, we performed an RNAi-based screen that utilized the fluorescence intensity of an mRFP-REST fusion protein as a surrogate for relative REST abundance. Cells were transfected with a library of 1908 siRNAs targeting 636 kinase and kinase-like genes in the human genome (3 siRNAs/gene) and with a plasmid encoding mRFP-REST or mRFP (Fig 1E). Flow cytometry was used to identify siRNAs that increased mRFP-REST fluorescence without affecting control mRFP fluorescence. Depletion of SCFβTRCP, the REST-ubiquitin ligase, resulted in a 4-fold increase in mRFP-REST fluorescence without a significant change in the mRFP fluorescence (red dot, Fig 1F), confirming that mRFP-REST responds similarly to endogenous REST.
From the primary genetic screen, 74 siRNAs elicited a greater than 2-fold effect (p<0.01) on normalized mRFP-REST fluorescence. RDR genes were ranked using a modified two-way analysis of variance to summarize the effects of all siRNAs for each gene in the screen. We identified 10 genes for which multiple siRNAs increased REST abundance (Fig 1G). Subsequently, we retested siRNAs for each of these RDR candidates. These experiments revealed three candidates (PLK1, TEX14 and SCYL1) for which multiple siRNAs consistently increased mRFP-REST fluorescence (see Fig 2A and 3A). We focused subsequent studies on these RDR candidates.
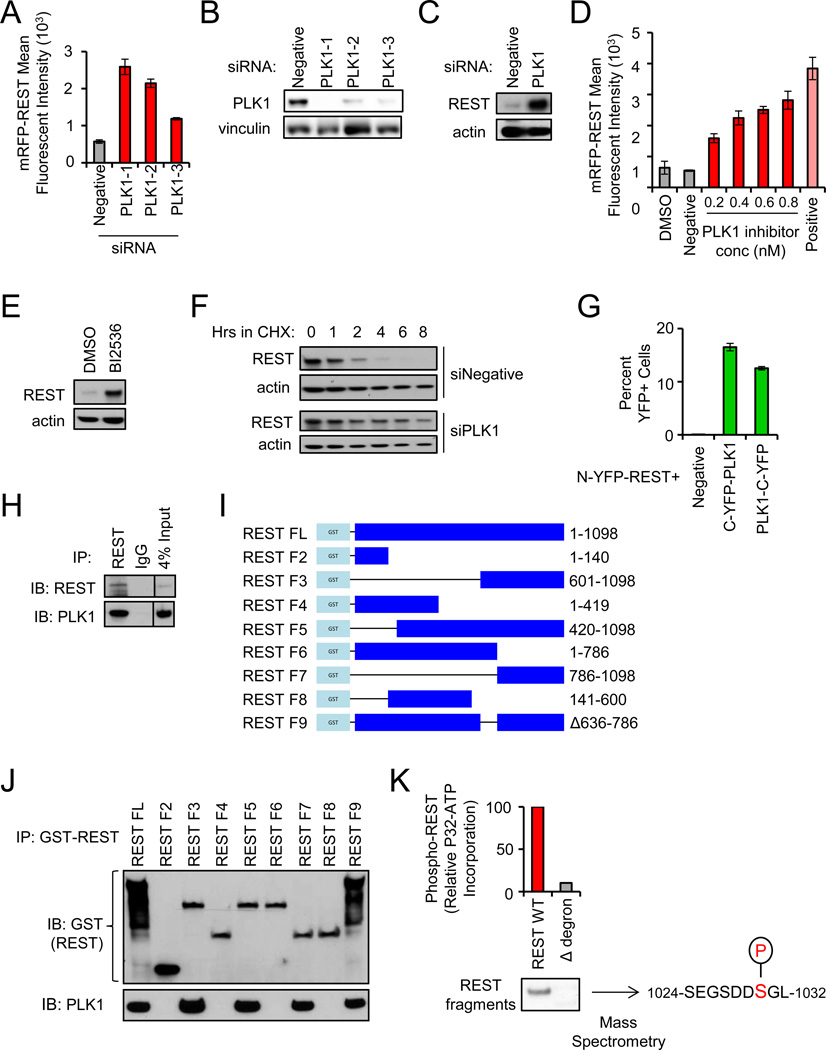
Figure 2. PLK1 phosphorylates the REST degron and regulates REST protein abundance.
- Depletion of PLK1 increases REST abundance. Cells were transfected with the indicated siRNAs and mRFP-REST (n=4). Cellular fluorescence was assessed by flow cytometry. Data presented as mean +/− SE.
- Western blot for PLK1 protein levels in cells transfected with the indicated siRNAs.
- Depletion of PLK1 increases endogenous REST abundance. 293T cells transfected with the indicated siRNAs were treated with monastrol for 8 hours (to enrich for cells with active PLK1). Endogenous REST protein levels were assessed via western blot.
- Pharmacologic inhibition of PLK1 increases REST abundance. 293T cells were treated with BI2536 (PLK1 inhibitor) as shown and transfected with mRFP-REST (n=4). Cellular fluorescence was assessed by flow cytometry. Data presented as mean +/− SE.
- Pharmacologic inhibition of PLK1 increases endogenous REST abundance. 293T cells were treated with BI2536 or DMSO. Endogenous REST protein levels were assessed via western blot.
- Depletion of PLK1 increases REST protein stability. 293T cells transfected with the indicated siRNAs were treated with cycloheximide and analyzed for REST protein levels.
- PLK1 interacts with REST. HMECs expressing N-YFP-REST were transduced with C-YFP-PLK1 or PLK1-C-YFP retroviruses. Cellular fluorescence was assessed by flow cytometry. Data presented as mean +/− SE.
- Endogenous REST was immunoprecipitated from monastrol-treated 293T cells and analyzed for REST and PLK1 protein by western blot.
- GST-REST fragments used for interaction studies. Numbers indicate amino acids included in each fragment.
- PLK1 interacts with the C-terminus of REST. 293T cells were transfected with the GST-REST fragments from I. After MG132 treatment (to inhibit proteasome function), interaction was assessed by GST pull down and western blot for GST (top) and PLK1 (bottom).
- REST is phosphorylated by PLK1 on serine-1030. Purified REST protein fragments (wild type or Δdegron) were incubated with active PLK1 and ATP [γ32P]. Audioradiography and mass spectrometry were performed. The serine within the REST phospho-degron phosphorylated by PLK1 is highlighted.
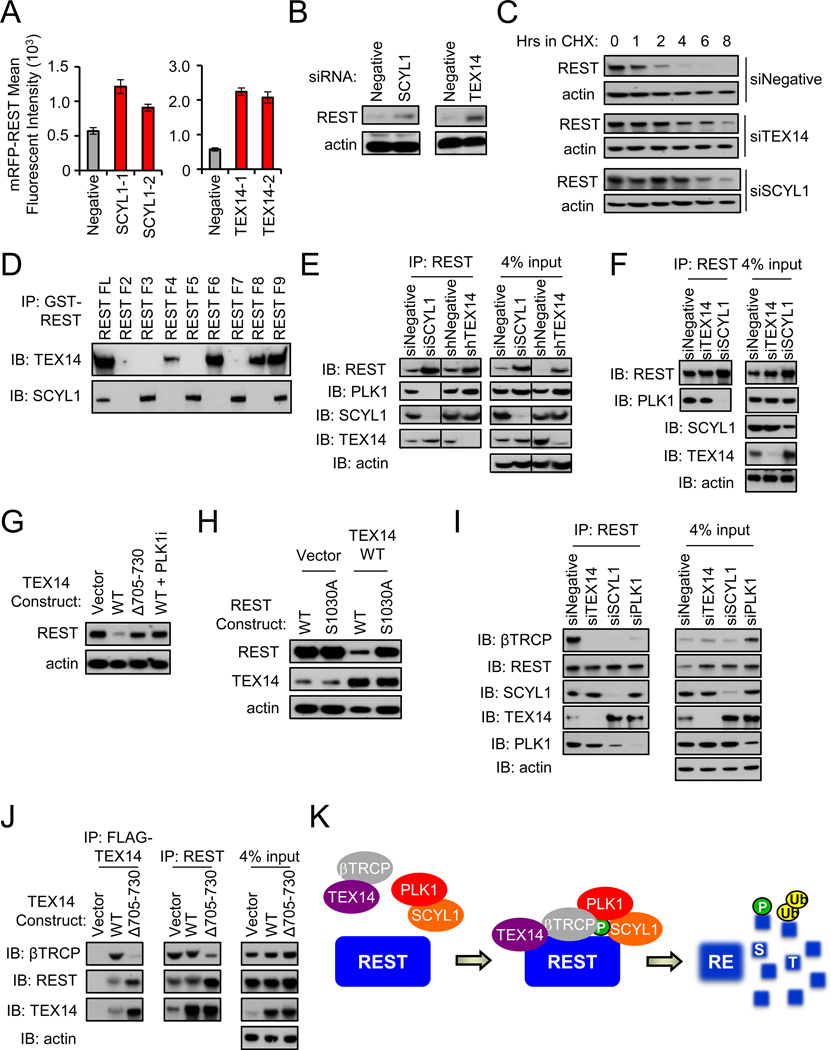
Figure 3. The STP axis (SCYL1, TEX14, PLK1) regulates REST abundance.
- Depletion of SCYL1 and TEX14 increases REST abundance. Cells were transfected with the indicated siRNAs and mRFP-REST (n=4). Cellular fluorescence was assessed by flow cytometry. Data presented as mean +/− SE.
- Depletion of SCYL1 and TEX14 increases endogenous REST abundance. 293T cells transfected with the indicated siRNAs were treated with monastrol and endogenous REST protein was assessed via western blot.
- Depletion of SCYL1 and TEX14 increases REST protein stability. 293T cells transfected with the indicated siRNA were treated with cycloheximide for as shown. REST protein levels were assessed by western blot.
- SCYL1 interacts with the C-terminus of REST while TEX14 interacts with the N-terminus of REST. 293T cells were transfected with the GST-REST fragments (Fig 2I). After MG132 treatment, interaction was assessed by GST pull down and western blot.
- SCYL1 is required for the PLK1-REST interaction. Endogenous REST was immunoprecipitated from monastrol-treated 293T cells transfected with the indicated siRNAs and analyzed via western blot.
- SCYL1 is required for the PLK1-REST interaction in human TNBC cells. BT549 TNBC cells were treated with MLN4924 (to inhibit REST-ubiquitination) and transfected with the indicated siRNAs. Endogenous REST was immunoprecipitated and analyzed via western blot.
- TEX14-mediated regulation of REST abundance requires TEX14 amino acids 705–730. 293T cells were transfected with the indicated FLAG-TEX14 constructs and treated with the BI2536. Western blot analysis was performed for REST and actin (loading control).
- TEX14 regulates REST abundance through the PLK1-phosphorylated REST-degron. 293T cells were transfected with the indicated expression vectors and treated with monastrol. Western blot analysis was performed for REST, TEX14, and actin (loading control).
- TEX14 is required for the REST-βTRCP interaction. Endogenous REST was immunoprecipitated from MLN4924-treated 293T cells transfected with the indicated siRNAs and analyzed via western blot.
- TEX14 links REST and βTRCP. MLN4924-treated 293T cells were transfected with the indicated FLAG-TEX14 constructs. FLAG-TEX14 or endogenous REST was immunoprecipitated and analyzed via western blot analysis.
- Model of REST regulation by the STP axis. The STP axis (SCYL1-TEX14-PLK1) cooperates to regulate REST phosphorylation and βTRCP recruitment. REST is subsequently targeted for degradation by SCFβTRCP.
PLK1 phosphorylates REST and controls REST abundance
In the primary screen and validation experiments, depletion of Polo-like kinase 1 (PLK1) consistently yielded the strongest effects on mRFP-REST abundance. PLK1 is a serine/threonine kinase implicated in phosphorylation and priming of SCFβTRCP-substrates for ubiquitylation (Moshe et al., 2004; Watanabe et al., 2004). To confirm that PLK1 controls REST abundance, we tested whether genetic or pharmacologic inhibition of PLK1 affected REST levels. PLK1 siRNAs depleted PLK1 protein (Fig 2B) and led to increased mRFP-REST fluorescence (Fig 2A) and increased endogenous REST protein levels (Fig 2C). Additionally, the PLK1 inhibitor BI2536 (Steegmaier et al., 2007) caused a dose-dependent increase in REST abundance (Fig 2D and 2E), thus ruling out RNAi off-target effects. Notably, PLK1 depletion significantly increased REST protein half-life (Fig 2F, S2A and S2B), indicating PLK1 antagonizes REST abundance by restraining REST protein stability.
Because PLK1 directly phosphorylates and primes substrates for SCFβTRCP-mediated ubiquitylation and degradation (Moshe et al., 2004; Watanabe et al., 2004), we tested whether PLK1 interacts with and phosphorylates REST. To determine whether PLK1 interacts with REST in cells, we performed bimolecular fluorescence complementation (BiFC) (Kerppola, 2006). The C-terminal half of YFP was fused to either the N-terminal or C-terminal end of PLK1 (C-YFP-PLK1 and PLK1-C-YFP, respectively). These fusion cDNAs were transduced into human mammary epithelial cells (HMECs) engineered to express REST fused to the N-terminal half of YFP (N-YFP-REST). As indicated by cellular YFP fluorescence, PLK1 strongly interacts with REST (Fig 2G). Additionally, PLK1 was co-immunoprecipitated with a REST-specific antibody in 293T and human TNBC cells (Fig 2H and S3), suggesting endogenous PLK1 and REST interact in human breast cancer.
To determine which REST domains are required for interaction with PLK1, GST-tagged REST fragments were evaluated for interaction with endogenous PLK1 (Fig 2I). The C-terminus of REST (amino acids 786–1098), which includes the REST phospho-degron, was required for the PLK1 interaction (Fig 2J), supporting the hypothesis that PLK1 may phosphorylate REST. To directly test this hypothesis, we performed an in vitro kinase assay. Purified PLK1 was incubated with REST protein fragments that contain or lack the C-terminal phospho-degron (WT and Δdegron, respectively). PLK1 preferentially phosphorylated the REST WT fragment as compared to the REST Δdegron fragment (Fig 2K), suggesting PLK1 phosphorylates the REST C-terminus. Mass spectrometry revealed that PLK1 phosphorylates REST on serine-1030 (Fig 2K and Fig S4). While this analysis cannot definitively exclude phosphorylation by PLK1 on other phosphosites within the REST C-terminus (Guardavaccaro et al., 2008; Westbrook et al., 2008), it does indicate that PLK1 phosphorylates the critical serine within the REST phospho-degron (S1030) that is required for SCFβTRCP-binding, ubiquitylation, and degradation (Westbrook et al., 2008). Collectively, these data indicate that PLK1 directly phosphorylates the REST phospho-degron and is a potent regulator of REST stability.
The STP axis (SCYL1, TEX14, and PLK1) regulates REST abundance
In addition to PLK1, the primary genetic screen and subsequent validation revealed two RDR candidates, TEX14 and SCYL1, whose depletion consistently increased mRFP-REST fluorescence (Fig 1G). siRNA-mediated depletion of TEX14 or SCYL1 (Fig S5) led to increased levels of mRFP-REST fluorescence and endogenous REST protein in both 293T and human TNBC cells (Fig 3A, 3B and Fig 5C). Additionally, depletion of both SCYL1 and TEX14 resulted in a significant increase in REST protein half-life (Fig 3C, S2A and S2B), suggesting that SCYL1 and TEX14 regulate REST abundance and stability. Notably, while previous observations suggest that, in some contexts, REST is degraded as cells approach mitosis (Guardavaccaro et al., 2008), our data indicate that PLK1, SCYL1 and TEX14 regulate REST dosage and stability without synchronization in mitosis (Fig 2F & 3C), suggesting that the STP axis is capable of controlling REST protein abundance in interphase cells.
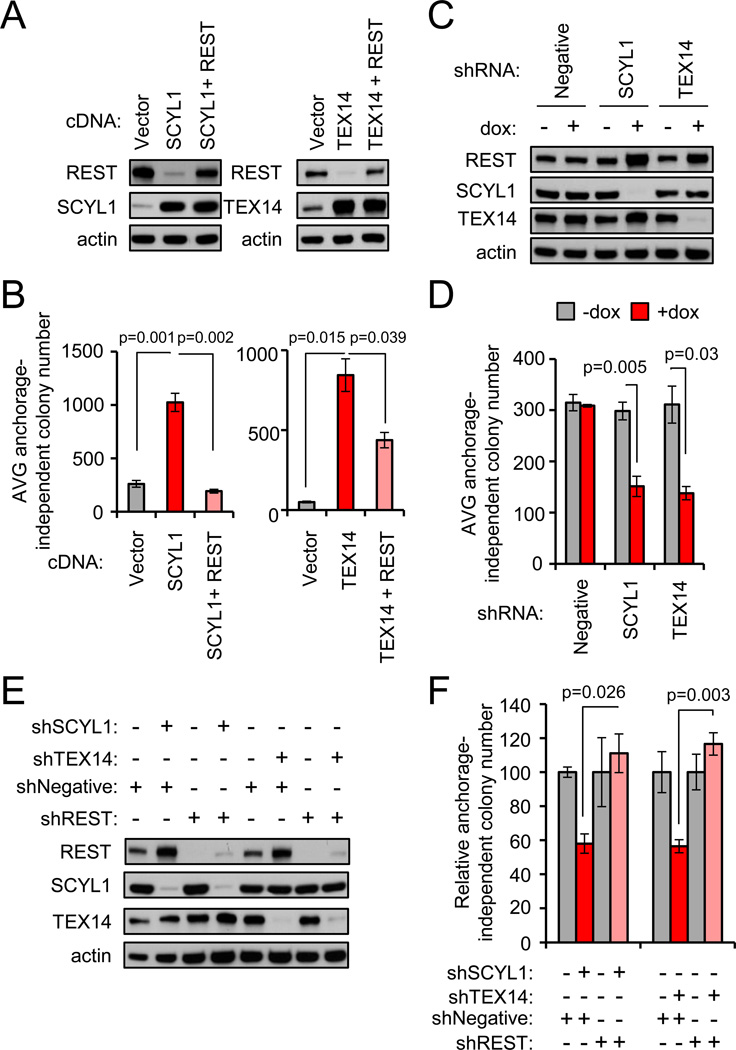
Figure 5. The STP axis supports the transformed state of breast cancer by restraining REST.
- Ectopic SCYL1 and TEX14 reduce endogenous REST protein abundance. BT549 TNBC cells were transduced as indicated and analyzed by western blot.
- SCYL1 and TEX14 promote cellular transformation in a REST-dependent manner. Anchorage-independent growth was assessed in the cells from A. Data presented as mean +/− SE.
- SCYL1 and TEX14 loss of function increases REST protein abundance. MDA-MB231-LM2 TNBC cells were engineered with the indicated dox-inducible shRNAs. Protein levels were assessed by western blot analysis.
- SCYL1 and TEX14 loss of function suppresses cellular transformation. Anchorage-independent growth was assessed in the cells from C +/−dox. Data presented as mean +/− SE.
- SCYL1 and TEX14 support the transformed state by restraining REST. MDA-MB231-LM2 TNBC cells engineered with the indicated dox-inducible shRNAs (from C) were transduced with a constitutive control or REST shRNA retrovirus. Protein levels were assessed by western blot.
- SCYL1 and TEX14 support the transformed state by restraining REST. Anchorage-independent growth was assessed in the cells from E +/−dox. Data presented as mean +/− SE.
Notably, both TEX14 and SCYL1 encode proteins predicted to be kinase-deficient (based on amino acid sequence (Manning et al., 2002)), suggesting these proteins may play a non-catalytic role in suppressing REST levels. Because our data strongly suggest that PLK1 is a direct priming kinase for REST degradation, we hypothesized TEX14 and SCYL1 may govern REST levels by regulating the physical or functional interaction between PLK1 and REST. To elucidate the mechanism(s) by which TEX14 and SCYL1 regulate REST protein levels, we tested whether either candidate interacts with REST in cells. TEX14 and SCYL1 associated with both ectopic and endogenous REST by co-immunoprecipitation (first lane of Fig 3D and 3E, respectively), and this interaction was conserved in human TNBC cells (Fig S3). To determine which domains of REST mediate the interaction with these RDR candidates, we tested associations between GST-fused REST fragments (Fig 2I, 2J) and endogenous TEX14 and SCYL1. TEX14 associated with N-terminal zinc fingers within the DNA-binding domain of REST (amino acids 141–419) (Fig 3D). In contrast, SCYL1 interacted with the C-terminus of REST (amino acids 786–1098), which is the same region required for PLK1 interaction. These data indicate that SCYL1 and TEX14 associate with different regions of REST and further suggest that TEX14 and SCYL1 may regulate REST abundance through interaction with REST protein.
Based on their association with REST, we hypothesized TEX14 and/or SCYL1 may act as adaptors to bridge REST interaction with PLK1. To test this hypothesis, TEX14 or SCYL1 was depleted by RNAi, and the interaction between REST and PLK1 was examined via co-immunoprecipitation. Depletion of SCYL1 significantly reduced the REST-PLK1 association (Fig 3E). In contrast, depletion of TEX14 had no observable effect on the REST-PLK1 interaction (Fig 3E). Additionally, SCYL1, but not TEX14, is required for the REST-PLK1 interaction in human TNBC cells (Fig 3F and Fig S6). This suggests SCYL1 bridges or stabilizes the REST-PLK1 interaction, and that TEX14 may play other roles in facilitating REST degradation.
Since TEX14 does not mediate the REST-PLK1 interaction, we asked whether TEX14 functions with PLK1, or alternatively, in a PLK1-independent pathway to regulate REST abundance. While ectopic expression of TEX14 resulted in a significant decrease in REST protein abundance (Fig 3G, lane 2), inhibition of PLK1 completely reversed this effect (Fig 3G, lane 4). Furthermore, mutating the PLK1 phospho-site within the REST degron (S1030A) ameliorated the effects of TEX14 on REST protein levels (Fig 3H). Together, these data suggest that TEX14 reduces REST protein levels in a manner dependent on PLK1 activity and the REST phospho-degron.
REST is marked for degradation by the ubiquitin ligase βTRCP, and the REST phospho-degron serves as a requisite binding site for βTRCP-REST interaction (Guardavaccaro et al., 2008; Westbrook et al., 2008). However, additional evidence indicates that βTRCP interacts with some substrates through bipartite surfaces (Watanabe et al., 2004), suggesting other REST surfaces or interacting proteins may facilitate REST-βTRCP interaction. Thus, we hypothesized that TEX14, which associates with the N-terminus of REST, may act as an adaptor to bridge REST interaction with βTRCP. Consistent with this hypothesis, TEX14 associated with βTRCP by co-immunoprecipitation (Fig 3J, lane 2). We then determined whether TEX14 (and SCYL1 and PLK1) is required for the REST-βTRCP interaction. Consistent with their role in phosphorylating the REST phospho-degron (that facilitates βTRCP-binding), depletion of PLK1 or SCYL1 significantly impaired the REST-βTRCP interaction (Fig 3I). Interestingly, depletion of TEX14 also resulted in a significant reduction in the REST-βTRCP interaction (Fig 3I), suggesting TEX14 is also required for βTRCP interaction with REST.
Based on this, we hypothesized TEX14 may regulate REST levels by facilitating βTRCP-REST interaction. To test this hypothesis, we scanned a series of TEX14 deletion mutants (Mondal et al., 2012) for their ability to reduce REST protein levels. TEX14 Δ705–730 (lacking amino acids 705–730) was unable to negatively regulate REST protein abundance (Fig 3G, lane 3) as compared to full-length TEX14 (Fig 3G, lane 2). However, TEX14 Δ705–730 maintained its ability to associate with REST (Fig 3J, lane 3), suggesting REST-interaction is not sufficient for TEX14 to regulate REST abundance. Furthermore, in contrast to full-length TEX14, TEX14 Δ705–730 displayed significantly reduced interaction with βTRCP (Fig 3J, lane 3) and impeded endogenous REST-βTRCP interaction (Fig 3J, lane 6), suggesting TEX14-βTRCP interaction is required for βTRCP to interact with REST. Collectively, these data suggest TEX14 regulates REST abundance by facilitating the interaction between βTRCP and REST. More broadly, these data demonstrate that SCYL1, TEX14, and PLK1 function collectively in a signaling network, the STP axis, to promote βTRCP-interaction and REST degradation (see model in Fig 3K).
STP axis components are amplified and over-expressed in human breast cancer
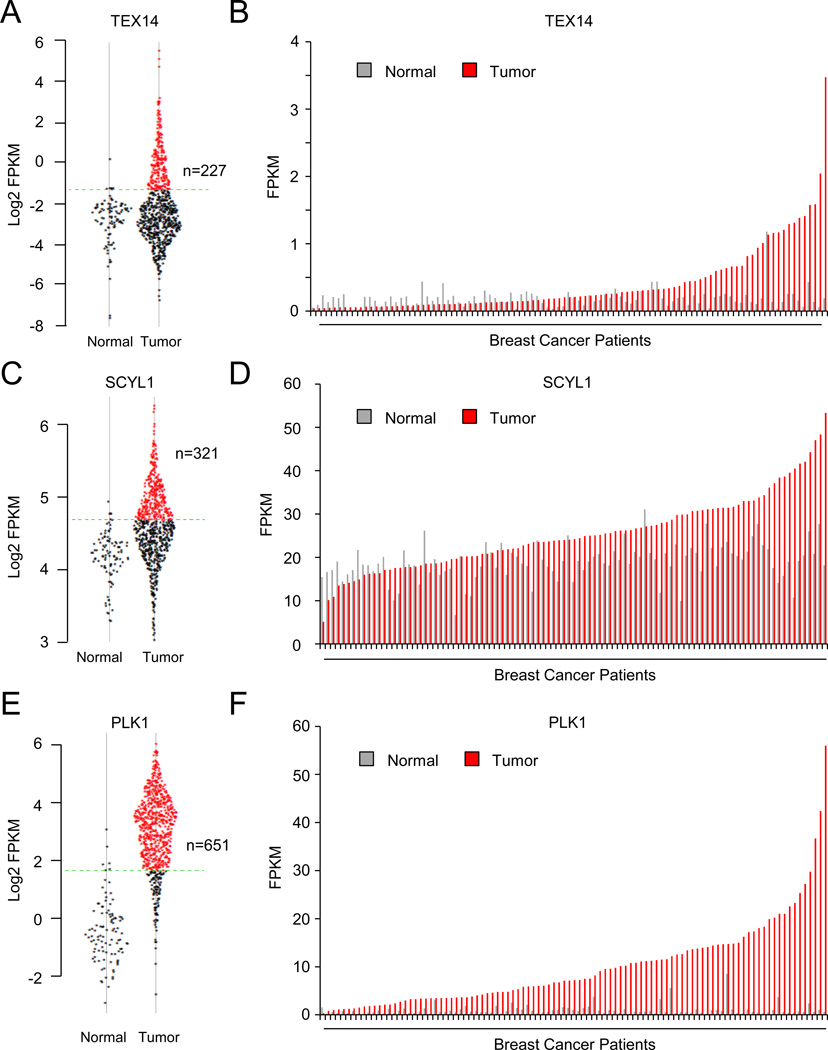
Our data indicate the STP axis negatively regulates the dosage of the tumor suppressor protein, REST, suggesting components of the STP axis may themselves function as oncogenes. Thus, we determined whether SCYL1, TEX14, and/or PLK1 have oncogenic characteristics and are aberrantly expressed in human breast cancers by analyzing a publicly available data set of 773 primary human breast tumors and 107 normal human breast tissue (Cancer Genome Atlas, 2012). Notably, TEX14, SCYL1, and PLK1 were over-expressed (candidate mRNA expression greater than the 95th percent confidence interval of that in normal mammary gland) in a substantial fraction of human breast cancers compared to normal human breast tissue (TEX14: 29.4%; SCYL1: 41.5%; PLK1: 84.2%) (Fig 4A, C, E). We also compared expression of TEX14, SCYL1, or PLK1 in the subset of breast tumors for which matched normal mammary tissue is available. Both TEX14 (up to 35-fold) and PLK1 (up to 93-fold) demonstrated remarkable over-expression in tumors as compared to normal (Fig 4B and 4F). SCYL1 exhibited modest over-expression at the mRNA level (maximum 3.8-fold) (Fig 4D). These data suggest a subset of breast cancers exhibit significantly elevated expression of STP components.
Figure 4. STP axis components are aberrantly over-expressed in human breast cancer compared to normal mammary tissue.
A,C,E. TEX14, SCYL1, and PLK1 expression in normal mammary tissue and breast tumors. Expression level of the indicated gene (represented as log2 FPKM (Fragments Per Kilobase per Million fragments mapped)) in each normal and tumor sample in the TCGA data set is plotted. The 95% confidence interval for the normal samples is represented by the green dashed line.
B,D,F. TEX14, SCYL1, and PLK1 expression is higher in tumors than matched normal. The expression level of the indicated gene for each tumor normal pair in the TCGA data set (represented as FPKM).
Oncogenes are commonly over-expressed via genetic and epigenetic mechanisms. To determine whether gene amplification contributes to the aberrant expression of STP components in breast cancer, we compared genomic copy number and mRNA expression in the same TCGA breast cancer cohort. TEX14 is frequently amplified in human breast cancers (9.3%), with 6% of breast cancers harboring TEX14 focal amplifications of less than 10 Mb (median 8.2 Mb, Fig S7A and S7B). Amplifications of SCYL1 were also focal (median 2.6 Mb), but occurred less frequently (2.2% of breast cancers, Fig S7A and S7C). Notably, TEX14 and SCYL1 exhibited significant copy-number associated increases in mRNA expression (Fig S7E and S7F, respectively), suggesting amplifications drive aberrant expression of these candidate oncogenes in a subset of human breast cancers. While PLK1 is frequently amplified in human breast cancers (9.6%), these copy number gains exhibit a median size of 27.5 Mb, with only 0.9% of breast cancers harboring PLK1 amplifications less than 10 Mb (Fig S7A and S7D). The large size of the amplicon suggests that multiple oncogenic drivers may exist with PLK1 in this region of chromosome 16. Additionally, in accordance with previous reports of smaller breast cancer cohorts (Maire et al., 2013), copy number of the PLK1 locus was not correlated with PLK1 mRNA levels (Fig S7G). Thus, the frequent over-expression of PLK1 in human breast cancer is likely driven by other mechanisms. Collectively, these data suggest STP components have oncogenic characteristics and are frequently over-expressed in human breast cancer by both genetic and epigenetic mechanisms.
The STP axis supports the transformed state of TNBC cells by restraining REST
The observations described above implicate the STP axis as an oncogenic mechanism by which REST protein levels and function is suppressed in human TNBC. Thus, we hypothesized the STP axis may support the transformed state of TNBC cells by restraining REST. Because of the many described functions of PLK1 and potential pleiotropic effects of PLK1 inhibition (Lens et al., 2010; Petronczki et al., 2008), we tested this hypothesis by altering STP axis function via SCYL1 or TEX14 gain- or loss-of-function and measuring the effect on TNBC cell transformation. Ectopic expression of SCYL1 and TEX14 in TNBC cells (that exhibit low transformation potential) resulted in a decrease in REST abundance (Fig 5A, lane 2) and an increase in anchorage-independent proliferation (Fig 5B), indicating that SCYL1 and TEX14 promote TNBC cell transformation. Additionally, these data suggest that the aberrant over-expression of SCYL1 and TEX14 observed in human breast tumors may be a pathogenic mechanism driving transformation. Our model predicts SCYL1 and TEX14 promote cellular transformation through negative regulation of the REST tumor suppressor. Indeed, restoring REST levels/function (Fig 5A, lane 3) strongly suppressed TEX14- and SCYL1-mediated anchorage-independent proliferation in TNBC cells (Fig 5B), suggesting TEX14 and SCYL1 can promote TNBC cell transformation by reducing levels of the REST tumor suppressor protein.
To determine whether endogenous TEX14 or SCYL1 supports the transformed state of TNBC cells, we depleted TEX14 or SCYL1 by RNAi and measured the effects on anchorage independent proliferation in TNBC cells. Notably, inhibition of TEX14 or SCYL1 led to an increase in endogenous REST protein levels (Fig 5C) and a concomitant decrease in anchorage-independent proliferation (Fig 5D). Moreover, depletion of REST reversed the effects of SCYL1 and TEX14 depletion and restored TNBC cell transformation (Fig 5E, 5F). Taken together, these data suggest that the STP axis promotes transformation of TNBC cells by restraining REST tumor suppressor function.
Inhibition of the STP-REST axis impairs tumorigenicity and metastatic proclivity of TNBC
Our observations suggest TNBC cells are dependent on the STP axis to maintain low REST levels and the transformed state. To test whether the STP axis is required for the tumorigenic and metastatic potential of TNBC cells, we engineered the highly tumorigenic and metastatic TNBC cell line MDA-MB231-LM2 (Minn et al., 2005) to inducibly express dual shRNAs targeting TEX14 and SCYL1. We chose to disrupt both SCYL1 and TEX14 function to more significantly interfere with REST degradation. Indeed, combined depletion of TEX14 and SCYL1 resulted in a more significant increase in endogenous REST protein levels (Fig 6A) and a more substantial reduction in anchorage-independent proliferation (Fig 6B) compared to depletion of either TEX14 or SCYL1 alone (see Fig 5E and 5F).
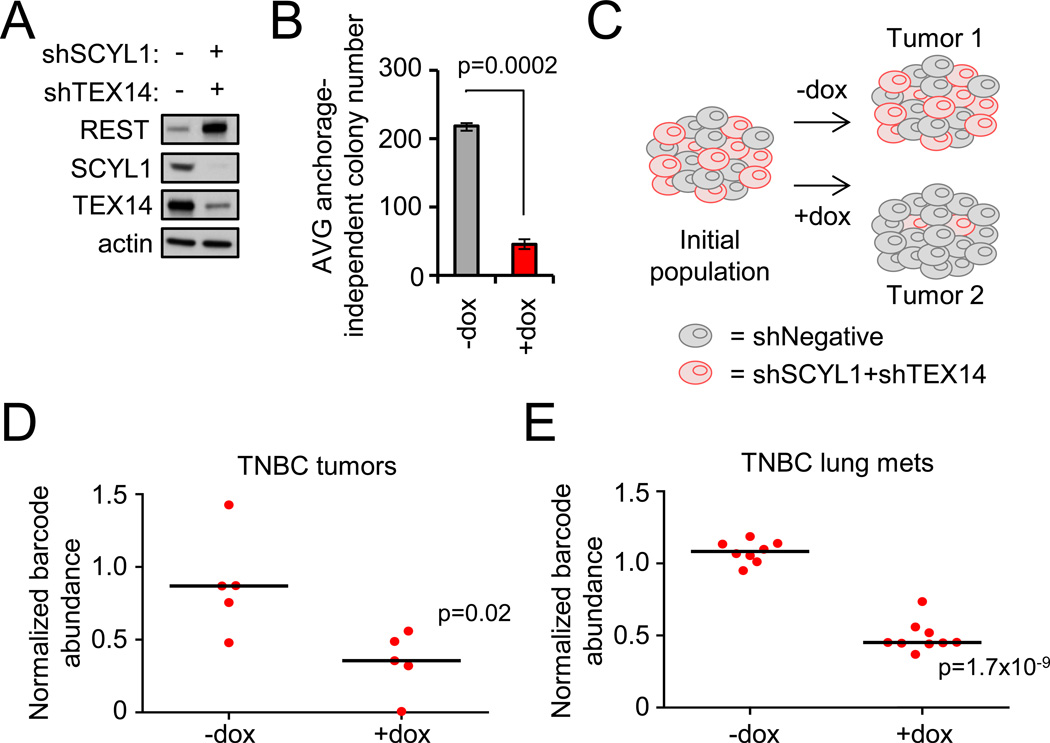
Figure 6. The STP-REST axis is required for primary tumor growth and metastatic expansion.
- MDA-MB231-LM2 TNBC cells were engineered to inducibly co-express SCYL1 and TEX14 shRNAs. Protein levels were analyzed by western blot.
- Depletion of SCYL1 and TEX14 impairs anchorage-independent growth. Cells from A were assessed for anchorage-independent growth. Data presented as mean +/− SE.
- In vivo barcode-based competition assay schematic. MDA-MB231-LM2 cells expressing inducible negative control shRNA or combination-shTEX14+shSCYL1 were mixed at an equal ratio. This population was transplanted into mice, and tumors were allowed to form +/−dox. At the experimental end point, genomic DNA was collected, and the relative abundance of each cell population was quantified via barcode-qPCR-analysis.
- Depletion of SCYL1 and TEX14 impairs primary tumor growth. The experiment was carried out as described in C. Barcode levels +/−dox are shown. Each dot represents an individual tumor. The black line represents the median.
- Depletion of SCYL1 and TEX14 impairs metastatic progression. MDA-MB231-LM2 TNBC cells engineered to inducibly express SCYL1+TEX14 shRNAs or negative control shRNAs were mixed in equal ratio, intravenously injected, and monitored for lung metastatic growth +/− dox via bioluminescence imaging. Barcode-qPCR was performed on lung genoms the relative abundance of each cell population in the lung metastatic burden. Each dot represents lung metastases from an individual animal.
To assess the effect of TEX14 and SCYL1 on TNBC tumorigenicity, we used a barcode-based competition assay (Fig 6C) that enables one to compare the relative fitness of tumor cells harboring different shRNAs within a single tumor while controlling for inter-mouse and inter-tumor variability. MDA-MB231-LM2 cells harboring an inducible negative control shRNA or combination-shTEX14+shSCYL1 were mixed at equal cell ratios and transplanted into immunocompromised mice maintained in the presence or absence of dox. The relative abundance of each population was quantified from the genomic DNA of resultant tumors via barcode abundance. Notably, tumor cells harboring combination-shTEX14+shSCYL1 consistently dropped out of the tumor population selectively in the presence of dox (Fig 6D). This strongly suggests that loss of TEX14 and SCYL1 function impairs TNBC cell fitness and tumorigenic potential.
In addition to their aggressive tumorigenic potential, TNBC are known to metastasize to visceral organs such as the lungs (Di Cosimo and Baselga, 2010; Smid et al., 2008). To directly assess the effects of STP axis inhibition on TNBC metastatic colonization and expansion, we measured the effects of TEX14 and SCYL1 depletion in the context of an experimental metastasis model (tail vein injection) using the same barcode-based competition method described above. Once significant tumor burden was observed in the lung (using bioluminescence imaging), the relative abundance of tumor cells with negative control-shRNA or combination-shTEX14+shSCYL1 were quantified via barcode abundance. As shown in Figure 6E, dox-dependent depletion of TEX14 and SCYL1 significantly reduced fitness of these TNBC lung metastases. While these data do not address the role of the STP axis in the early stages of metastasis, these data support the model that TNBC cells are dependent on TEX14 and SCYL1 for their survival and expansion in the lung. Collectively, these data suggest that STP axis supports TNBC cell fitness in both primary and metastatic tumors.
The STP axis correlates with low REST protein levels and poor patient outcome in breast cancer
Taken together, our functional data suggest that some breast cancers are dependent on the STP axis for their tumorigenic and metastatic behavior. This hypothesis predicts that breast cancers with high STP axis activity may exhibit poor clinical outcome. As the STP axis may have broad downstream effects on gene expression, we tested this hypothesis by: (1) defining a transcriptional signature that correlates with SCYL1, TEX14, and PLK1 expression, and subsequently (2) testing the correlation of this STP signature on independent datasets with long-term clinical follow up. To define an STP signature, we used co-expression analyses to compute a gene set strongly correlated with STP expression in diverse human samples. Using the TCGA RNAseq dataset, we identified a gene signature whose expression significantly correlates with SCYL1, TEX14, and PLK1 across 773 primary human breast tumors. Expression of the STP signature was higher in TNBCs (Fig 7A) consistent with more frequent loss of REST protein in this subtype of breast cancers. Next, we compiled breast cancer data sets (n=1151 patients) for which gene expression data (Affymetrix U133 platform only) and metastasis free survival data were available. Notably, patients with tumors harboring high STP signature had significantly worse distant metastasis-free survival than those with low STP signature (Fig 7B, p<1.0×10−8). Furthermore, while STP expression correlates with the TNBC subtype, the STP signature also exhibited prognostic value independent of breast cancer subtype (Fig 7C, p=0.019).
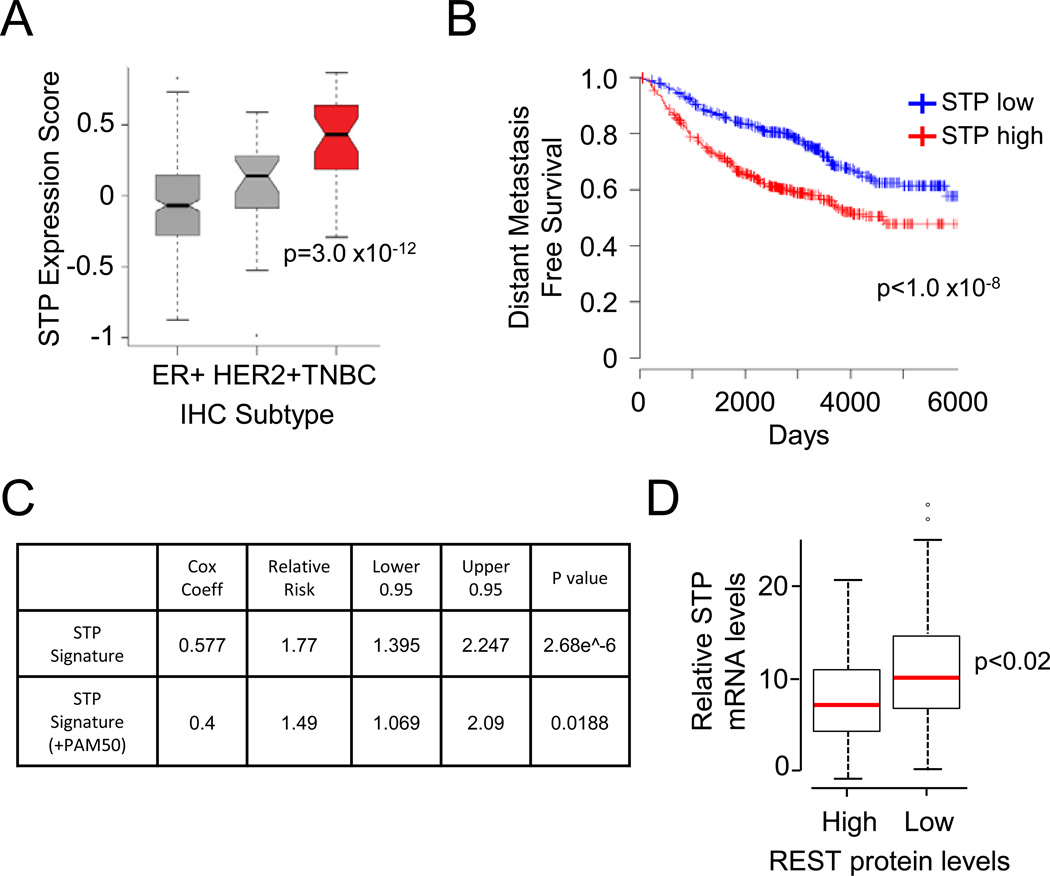
Figure 7. The STP axis correlates with low REST protein levels and poor patient outcome in human breast cancer.
- STP axis expression is correlated with TNBC subtype. The STP axis gene expression signature for each tumor was plotted by histologic subtype. The solid line represents the median. The boxes represent the 25th to 75th percentiles. Error bars represent 95% confidence interval. Outliers are represented as circles.
- STP axis expression correlates with poor patient outcome. Patients with the highest and lowest tertiles of STP axis gene expression signature are shown in red and blue, respectively (n=1151).
- STP axis expression correlates with increased metastatic recurrence in breast cancer patients. Hazard regression results for the STP expression signature are shown. Top: results for the STP signature alone. Bottom: results with an inference of PAM50 subtype.
- STP axis expression and REST protein levels are inversely correlated in human breast cancer. The normalized cumulative expression of SCYL1, TEX14, and PLK1 is shown for tumors exhibiting REST high and REST low protein (as determined by IHC). The red line represents the median. The boxes represent the 25th to 75th percentiles. Error bars represent 95% confidence interval. Outliers are represented as circles.
Our data indicate that the STP axis functions to inhibit REST protein accumulation. If the STP axis contributes to the frequent down-regulation of REST protein seen in breast cancer (Fig 1B and 1C), expression of STP components should inversely correlate with REST protein levels in human breast tumors. To test this hypothesis, we quantified the mRNA levels of each STP axis component in a subset of the primary human tumors (n=50) previously analyzed for REST protein levels (see Fig 1B and 1C). Since each member of the STP axis regulates REST abundance, we summed the normalized expression level of SCYL1, TEX14, and PLK1 into one cumulative STP expression value. Higher expression of the STP axis components strongly correlates with low REST protein levels (Fig 7D), suggesting this pathway negatively regulates REST protein levels in human breast cancer. Combined with our functional data, these data suggest that the STP axis supports the pathogenicity of TNBCs, at least in part, by reducing REST protein levels and function.
Discussion
The STP-REST axis as a driver of TNBC
TNBC is a collection of diseases with heterogeneous molecular features (Prat and Perou, 2011). Indeed, recent analyses of copy number variation and somatic mutations have revealed very few drivers that are common across TNBCs (Cancer Genome Atlas, 2012; Curtis et al., 2012). Such genetic heterogeneity has called into question whether there are common pathogenic mechanisms (and potential therapeutic targets) driving the TNBC subtype(s) of breast cancer (Brough et al., 2011; Collisson et al., 2012). Herein, we present evidence of a novel oncogene-tumor suppressor network that is frequently compromised in TNBC. Specifically, we demonstrate that the REST tumor suppressor is post-transcriptionally inactivated in human breast cancer, most commonly in TNBC. Using an unbiased genetic approach, we discovered the oncogenic STP axis (SCYL1, TEX14, and PLK1) that functions collectively to suppress REST protein levels via degradation. Components of this signaling axis are over-expressed in aggressive TNBCs and are required to support the transformed state of TNBC cells, at least in part, through negative regulation of the REST tumor suppressor protein. Notably, high STP axis expression correlates with low REST protein levels and poor clinical outcome in human breast cancers. Our data suggests that inactivation of REST (via protein degradation) may be a common driver of TNBC pathogenesis, and that the STP axis is a mediator of this REST dysfunction.
Neural programs in breast cancer
Cancers often usurp developmental programs or signaling pathways during tumor inception and progression (ex. (Yang and Weinberg, 2008)). During normal embyrogenesis, REST serves as a master repressor of neural transcriptional programs and neural differentiation (Ballas et al., 2005). Our finding that REST is post-transcriptionally inactivated in TNBC and other breast cancers raises the important question of whether signaling networks promoting neural phenotypes are misregulated in and contribute to breast cancer pathogenesis. Indeed, many aspects of neural programs are misexpressed in human breast cancers. For example, neurotrophic receptors and their cognate ligands are commonly misexpressed in TNBC and other breast cancers and contribute to tumor growth and survival signaling (Davidson et al., 2004; Dolle et al., 2003; Hondermarck, 2012; Lagadec et al., 2009). Many of these neurotrophic receptors and ligands are targets of the REST repressor (Johnson et al., 2007). Our data is consistent with a model in which dysfunction of the STP-REST axis may contribute to such aberrant neural growth programs in breast cancer, although this hypothesis requires further testing.
During development, REST degradation is tightly regulated by SCFβTRCP-mediated ubiquitination and proteasomal degradation (Westbrook et al., 2008) and this process requires REST phosphorylation within its phospho-degron. In the current study, we have the identified the STP axis which phosphorylates the REST phospho-degron and controls REST protein degradation in human breast cancers. It will be important to determine whether the STP axis similarly controls REST protein levels in the context of normal development (early neural differentiation or other adult differentiation programs) or in the pathology of neurologic disorders as has been reported for casein kinase 1 (Kaneko et al., 2014). We note that while PLK1 has essential functions across cell types (Archambault and Glover, 2009), SCYL1 and TEX14 function primarily in the neural and germ cell compartments, respectively (Iwamori et al., 2010; Schmidt et al., 2007; Wu et al., 2003), suggesting developmental restrictions in their function. Importantly, changes in REST protein levels play causative roles in the response to neurologic insult (ex. ischemia) and neurodegenerative disorders such as Huntington’s disease and Alzheimer’s disease (Calderone et al., 2003; Lu et al., 2014; Zuccato et al., 2003). While STP components are clearly misregulated at the genetic level (i.e. amplification) in human breast cancer, it will be important to determine if they are similarly misregulated in neurologic pathologies.
New opportunities for TNBC therapy
Our discovery that the STP axis supports TNBC tumorigenic and metastatic proclivity provides new opportunities for therapeutic entry points for TNBC. It is noteworthy that pharmacologic inhibition of PLK1, one of the STP axis components, has recently been shown to impair breast cancer progression in pre-clinical (animal) models of TNBC (Maire et al., 2013). However, while several pharmacologic PLK1 inhibitors have been developed, these inhibitors have not been successfully employed in the clinic due, in large part, to toxicities in bone marrow-related tissues and other proliferative compartments (Degenhardt and Lampkin, 2010; Lens et al., 2010; Schoffski, 2009). PLK1 functions in many diverse signaling pathways and cellular processes, and the toxicities of PLK1 inhibitors suggest that many of the critical functions of PLK1 are shared between normal and malignant cells. Our data raise the important question of whether subset(s) of PLK1 functions may be specifically required in malignant cells (versus normal tissues) and thus may be exploited as more selective anti-cancer strategies.
Herein, we describe a new molecular pathway consisting of PLK1, SCYL1, and TEX14 that may be specifically required for breast cancer cell survival. Our data indicate PLK1 operates in conjunction with SCYL1 and TEX14 to lower protein levels of the REST tumor suppressor. PLK1 activity increases during mitosis which is consistent with previous studies suggesting that, in some contexts, REST may be degraded as cells transition through mitosis (Guardavaccaro et al., 2008). The cooperation between PLK1 and SCYL1 and TEX14 suggests there may be a relationship between the STP-REST axis and progression into or through mitosis, although future studies are required to explore the temporal and spatial regulation of REST degradation by the STP axis and its relation to mitotic progression. Importantly, inhibition of the STP axis strongly inhibits the fitness of TNBC cells in vitro and in vivo. While inhibition of PLK1 likely impairs normal and malignant cell viability via numerous mechanisms, inhibition of SCYL1 and TEX14 is deleterious to TNBC cells in a REST-dependent manner. Notably, increasing REST dosage significantly impairs the viability of TNBC and other cancer cell types (Gurrola-Diaz et al., 2003; Watanabe et al., 2006; Westbrook et al., 2005), but does not affect the fitness of normal adult cell types (Westbrook et al., 2008). Therefore, impairing the ability of PLK1 to reduce REST levels may be selectively deleterious to breast cancer cells (relative to normal tissues), while leaving the other cell essential functions of PLK1 intact. It will be important to test whether inhibition of TEX14 or SCYL1 function(s) has a wider therapeutic window than PLK1 inhibitors. Collectively, our studies identify the STP-REST axis as a new vulnerability in TNBC and warrant further exploration of whether this axis can be therapeutically exploited.
Experimental Procedures
siRNA Screen and Transfections
For the primary screen, Invitrogen’s Stealth Kinase siRNA library was used (3 siRNAs per gene, individually arrayed). 293T cells were transfected in 96 well format with 40nM siRNA using oligofectamine (Invitrogen) as recommended by the manufacturer. On the subsequent day, cells were transfected with CMV-mRFP-REST or CMV-mRFP plasmids (Westbrook et al., 2008) in quadruplicate using MIRUS Bio’s TransIT 293 under the manufacturer’s recommended conditions. Cells were analyzed for mRFP fluorescence after 48 hours using flow cytometry. The effects of each siRNA were normalized to mRFP and to the negative control.
Subsequent siRNA transfections were performed as described above. The Invitrogen Stealth siRNAs used were: GC Medium (negative control, Invitrogen); FBXW11 (positive control, HSS118532); PLK1-1 (VHS50337); PLK1–2 (VHS50338); PLK1–3 (VHS50340); TEX14-1 (HSS125434); TEX14-2 (HSS125435); SCYL1-1 (HSS126244); SCYL1–2 (HSS126245).
Anchorage-independent Proliferation Assays
For ectopic expression experiments, BT549 and MDA-MB231-LM2 breast cancer cells were transduced as described above and selected with the appropriate antibiotic. 1×104 BT549 breast cancer cells and 1.5×104 MDA-MB231-LM2 breast cancer cells were seeded per 6cm dish with a bottom layer of 0.6% agarose or noble agar in DMEM and a top layer of 0.3% agarose or noble agar in DMEM + 10% FBS, respectively. For shRNA experiments, MDA-MB231-LM2 breast cancer cells were transduced at equivalent M.O.I. (1.5) for each shRNA. Cells were treated with dox for 36 hours prior to seeding. All experiments were done in triplicate.
Supplementary Material
HIGHLIGHTS.
TNBCs commonly harbor post-transcriptional loss of the REST tumor suppressor protein
SCYL1, TEX14, and PLK1 constitute a new oncogenic signaling axis (STP axis)
The STP axis drives human TNBC transformation by reducing REST protein abundance
Inhibition of the STP axis impairs TNBC tumor progression and metastasis
Acknowledgements
We would like to thank X. Zhang, and members of the Westbrook, Couch, and Shaw laboratories for helpful comments. The authors also acknowledge the joint participation by Adrienne Helis Melvin Medical Research Foundation through its direct engagement in the continuous active conduct of medical research in conjunction with Baylor College of Medicine for cancer research. The Dan L. Duncan Cancer Center Shared Resources was supported by the NCI P30CA125123 Center Grant and provided technical assistance including Cell-Based Assay Screening Service (Dan Liu), Genomic and RNA Profiling Resource (Lisa White), Biostatistics & Informatics Shared Resource (Susan Hilsenbeck) and Cytometry and Cell Sorting (Joel Sederstrom). Authors would like to acknowledge Nexcelom for providing support for Celigo image data management system. Research at Mayo Clinic was supported by the Breast Cancer Research Foundation (BCRF), NIH Grant CA116167, an NCI specialized program of research excellence (SPORE) in Breast Cancer (P50 CA166201), the David and Margaret T. Grohne Family Foundation, and the Ting Tsung and Wei Fong Chao Foundation. K.L.K. was supported by DOD pre-doctoral fellowship (BC094077) and NIH training grant (5 T32 GM008231). T.F.W. was supported by CPRIT (RP120583), the Susan G. Komen for the Cure (KG090355), the NIH (1R01CA178039-01), and the DOD Breast Cancer Research Program (BC120604).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
K.K. and T.W. conceptualized the study and wrote the manuscript. K.K., G.M., J.H., S.T., S.K., C.B., T.H., J.F., I.M., C.C., A.N. and S.J. performed experiments. All authors contributed to the interpretation of the results.
References
- Archambault V, Glover DM. Polo-like kinases: conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol. 2009;10:265–275. doi: 10.1038/nrm2653. [DOI] [PubMed] [Google Scholar]
- Arteaga CL, Sliwkowski MX, Osborne CK, Perez EA, Puglisi F, Gianni L. Treatment of HER2-positive breast cancer: current status and future perspectives. Nat Rev Clin Oncol. 2012;9:16–32. doi: 10.1038/nrclinonc.2011.177. [DOI] [PubMed] [Google Scholar]
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and Its Corepressors Mediate Plasticity of Neuronal Gene Chromatin throughout Neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Ballas N, Mandel G. The many faces of REST oversee epigenetic programming of neuronal genes. Curr Opin Neurobiol. 2005;15:500–506. doi: 10.1016/j.conb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Brough R, Frankum JR, Sims D, Mackay A, Mendes-Pereira AM, Bajrami I, Costa-Cabral S, Rafiq R, Ahmad AS, Cerone MA, et al. Functional viability profiles of breast cancer. Cancer discovery. 2011;1:260–273. doi: 10.1158/2159-8290.CD-11-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderone A, Jover T, Noh KM, Tanaka H, Yokota H, Lin Y, Grooms SY, Regis R, Bennett MV, Zukin RS. Ischemic insults derepress the gene silencer REST in neurons destined to die. J Neurosci. 2003;23:2112–2121. doi: 10.1523/JNEUROSCI.23-06-02112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- Collisson EA, Cho RJ, Gray JW. What are we learning from the cancer genome? Nat Rev Clin Oncol. 2012;9:621–630. doi: 10.1038/nrclinonc.2012.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson JM, Edgson JL, Woll PJ, Quinn JP. A splice variant of the neuron-restrictive silencer factor repressor is expressed in small cell lung cancer: a potential role in derepression of neuroendocrine genes and a useful clinical marker. Cancer Res. 2000;60:1840–1844. [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson B, Reich R, Lazarovici P, Ann Florenes V, Nielsen S, Nesland JM. Altered expression and activation of the nerve growth factor receptors TrkA and p75 provide the first evidence of tumor progression to effusion in breast carcinoma. Breast Cancer Res Treat. 2004;83:119–128. doi: 10.1023/B:BREA.0000010704.17479.8a. [DOI] [PubMed] [Google Scholar]
- Degenhardt Y, Lampkin T. Targeting Polo-like kinase in cancer therapy. Clin Cancer Res. 2010;16:384–389. doi: 10.1158/1078-0432.CCR-09-1380. [DOI] [PubMed] [Google Scholar]
- Di Cosimo S, Baselga J. Management of breast cancer with targeted agents: importance of heterogenicity. Nat Rev Clin Oncol. 2010;7:139–147. doi: 10.1038/nrclinonc.2009.234. [DOI] [PubMed] [Google Scholar]
- Dolle L, El Yazidi-Belkoura I, Adriaenssens E, Nurcombe V, Hondermarck H. Nerve growth factor overexpression and autocrine loop in breast cancer cells. Oncogene. 2003;22:5592–5601. doi: 10.1038/sj.onc.1206805. [DOI] [PubMed] [Google Scholar]
- Fuchs SY, Spiegelman VS, Kumar KG. The many faces of beta-TrCP E3 ubiquitin ligases: reflections in the magic mirror of cancer. Oncogene. 2004;23:2028–2036. doi: 10.1038/sj.onc.1207389. [DOI] [PubMed] [Google Scholar]
- Guardavaccaro D, Frescas D, Dorrello NV, Peschiaroli A, Multani AS, Cardozo T, Lasorella A, Iavarone A, Chang S, Hernando E, et al. Control of chromosome stability by the beta-TrCP-REST-Mad2 axis. Nature. 2008;452:365–369. doi: 10.1038/nature06641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurrola-Diaz C, Lacroix J, Dihlmann S, Becker CM, von Knebel Doeberitz M. Reduced expression of the neuron restrictive silencer factor permits transcription of glycine receptor alpha1 subunit in small-cell lung cancer cells. Oncogene. 2003;22:5636–5645. doi: 10.1038/sj.onc.1206790. [DOI] [PubMed] [Google Scholar]
- Hondermarck H. Neurotrophins and their receptors in breast cancer. Cytokine Growth Factor Rev. 2012;23:357–365. doi: 10.1016/j.cytogfr.2012.06.004. [DOI] [PubMed] [Google Scholar]
- Hurvitz SA, Finn RS. What’s positive about ‘triple-negative’ breast cancer? Future Oncol. 2009;5:1015–1025. doi: 10.2217/fon.09.68. [DOI] [PubMed] [Google Scholar]
- Iwamori T, Iwamori N, Ma L, Edson MA, Greenbaum MP, Matzuk MM. TEX14 interacts with CEP55 to block cell abscission. Mol Cell Biol. 2010;30:2280–2292. doi: 10.1128/MCB.01392-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Kaneko N, Hwang JY, Gertner M, Pontarelli F, Zukin RS. Casein kinase 1 suppresses activation of REST in insulted hippocampal neurons and halts ischemia-induced neuronal death. J Neurosci. 2014;34:6030–6039. doi: 10.1523/JNEUROSCI.4045-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola TK. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat Protoc. 2006;1:1278–1286. doi: 10.1038/nprot.2006.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreisler A, Strissel PL, Strick R, Neumann SB, Schumacher U, Becker CM. Regulation of the NRSF/REST gene by methylation and CREB affects the cellular phenotype of small-cell lung cancer. Oncogene. 2010;29:5828–5838. doi: 10.1038/onc.2010.321. [DOI] [PubMed] [Google Scholar]
- Lagadec C, Meignan S, Adriaenssens E, Foveau B, Vanhecke E, Romon R, Toillon RA, Oxombre B, Hondermarck H, Le Bourhis X. TrkA overexpression enhances growth and metastasis of breast cancer cells. Oncogene. 2009;28:1960–1970. doi: 10.1038/onc.2009.61. [DOI] [PubMed] [Google Scholar]
- Lens SM, Voest EE, Medema RH. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat Rev Cancer. 2010;10:825–841. doi: 10.1038/nrc2964. [DOI] [PubMed] [Google Scholar]
- Lu T, Aron L, Zullo J, Pan Y, Kim H, Chen Y, Yang TH, Kim HM, Drake D, Liu XS, et al. REST and stress resistance in ageing and Alzheimer’s disease. Nature. 2014;507:448–454. doi: 10.1038/nature13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maire V, Nemati F, Richardson M, Vincent-Salomon A, Tesson B, Rigaill G, Gravier E, Marty-Prouvost B, De Koning L, Lang G, et al. Polo-like kinase 1: a potential therapeutic option in combination with conventional chemotherapy for the management of patients with triple-negative breast cancer. Cancer Res. 2013;73:813–823. doi: 10.1158/0008-5472.CAN-12-2633. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal G, Ohashi A, Yang L, Rowley M, Couch FJ. Tex14, a Plk1-regulated protein, is required for kinetochore-microtubule attachment and regulation of the spindle assembly checkpoint. Mol Cell. 2012;45:680–695. doi: 10.1016/j.molcel.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshe Y, Boulaire J, Pagano M, Hershko A. Role of Polo-like kinase in the degradation of early mitotic inhibitor 1, a regulator of the anaphase promoting complex/cyclosome. Proc Natl Acad Sci U S A. 2004;101:7937–7942. doi: 10.1073/pnas.0402442101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CK. Tamoxifen in the treatment of breast cancer. N Engl J Med. 1998;339:1609–1618. doi: 10.1056/NEJM199811263392207. [DOI] [PubMed] [Google Scholar]
- Petronczki M, Lenart P, Peters JM. Polo on the Rise-from Mitotic Entry to Cytokinesis with Plk1. Dev Cell. 2008;14:646–659. doi: 10.1016/j.devcel.2008.04.014. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Molecular oncology. 2011;5:5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt WM, Kraus C, Hoger H, Hochmeister S, Oberndorfer F, Branka M, Bingemann S, Lassmann H, Muller M, Macedo-Souza LI, et al. Mutation in the Scyl1 gene encoding amino-terminal kinase-like protein causes a recessive form of spinocerebellar neurodegeneration. EMBO Rep. 2007;8:691–697. doi: 10.1038/sj.embor.7401001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- Schoffski P. Polo-like kinase (PLK) inhibitors in preclinical and early clinical development in oncology. Oncologist. 2009;14:559–570. doi: 10.1634/theoncologist.2009-0010. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- Smid M, Wang Y, Zhang Y, Sieuwerts AM, Yu J, Klijn JG, Foekens JA, Martens JW. Subtypes of breast cancer show preferential site of relapse. Cancer Res. 2008;68:3108–3114. doi: 10.1158/0008-5472.CAN-07-5644. [DOI] [PubMed] [Google Scholar]
- Steegmaier M, Hoffmann M, Baum A, Lenart P, Petronczki M, Krssak M, Gurtler U, Garin-Chesa P, Lieb S, Quant J, et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr Biol. 2007;17:316–322. doi: 10.1016/j.cub.2006.12.037. [DOI] [PubMed] [Google Scholar]
- Wagoner MP, Gunsalus KT, Schoenike B, Richardson AL, Friedl A, Roopra A. The transcription factor REST is lost in aggressive breast cancer. PLoS Genet. 2010;6:e1000979. doi: 10.1371/journal.pgen.1000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H, Mizutani T, Haraguchi T, Yamamichi N, Minoguchi S, Yamamichi-Nishina M, Mori N, Kameda T, Sugiyama T, Iba H. SWI/SNF complex is essential for NRSF-mediated suppression of neuronal genes in human nonsmall cell lung carcinoma cell lines. Oncogene. 2006;25:470–479. doi: 10.1038/sj.onc.1209068. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Arai H, Nishihara Y, Taniguchi M, Watanabe N, Hunter T, Osada H. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFbeta-TrCP. Proc Natl Acad Sci U S A. 2004;101:4419–4424. doi: 10.1073/pnas.0307700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook TF, Hu G, Ang XL, Mulligan P, Pavlova NN, Liang A, Leng Y, Maehr R, Shi Y, Harper JW, et al. SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature. 2008;452:370–374. doi: 10.1038/nature06780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook TF, Martin ES, Schlabach MR, Leng Y, Liang AC, Feng B, Zhao JJ, Roberts TM, Mandel G, Hannon GJ, et al. A genetic screen for candidate tumor suppressors identifies REST. Cell. 2005;121:837–848. doi: 10.1016/j.cell.2005.03.033. [DOI] [PubMed] [Google Scholar]
- Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW. The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev. 1999;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MH, Rajkovic A, Burns KH, Yan W, Lin YN, Matzuk MM. Sequence and expression of testis-expressed gene 14 (Tex14): a gene encoding a protein kinase preferentially expressed during spermatogenesis. Gene expression patterns : GEP. 2003;3:231–236. doi: 10.1016/s1567-133x(03)00036-x. [DOI] [PubMed] [Google Scholar]
- Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Tartari M, Crotti A, Goffredo D, Valenza M, Conti L, Cataudella T, Leavitt BR, Hayden MR, Timmusk T, et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.