Abstract
Addiction is due to changes in the structure and function of the brain, including neuronal networks and the cells that comprise them. Within cells, gene expression changes can track and help explain their altered function. Transcriptional changes induced by addictive agents are dynamic and divergent and range from signal pathway-specific perturbations to widespread molecular and cellular dysregulation that can be measured by “omic” methods and that can be used to identify new pathways. The molecular effects of addiction depend on timing of exposure or withdrawal, the stage of adaptation, the brain region, and the behavioral model, there being many models of addiction. However, the molecular neural adaptations across different drug exposures, conditions, and regions are to some extent shared and can reflect common actions on pathways relevant to addiction. Epigenetic studies of DNA methylation and histone modifications and studies of regulatory RNA networks have been informative for elucidating the mechanisms of transcriptional change in the addicted brain.
1. INTRODUCTION
Substance abuse and addiction to drugs and other addictive agents lead to cellular and molecular changes and are also caused in part by adaptations in epigenetic regulation and gene expression that can be measured in cells. Addictive behaviors are the outcome of allostatic maladaptation of neural circuitries (Goldman, Oroszi, & Ducci, 2005; Koob & Le Moal, 2001). Although great efforts have been made to understand the molecular basis of addiction, the mechanisms are elusive, in part because they are likely to be multiple. However, the study of gene expression in the addicted brain has already yielded valuable insights to the molecular mechanisms of maladaption. In model organisms and cellular models, several important pathway-related changes induced by acute and chronic drug exposure have been discovered. Human studies enabled by the availability of postmortem brain tissues from addicted individuals (Albertson et al., 2004; Albertson, Schmidt, Kapatos, & Bannon, 2006; Bannon, Kapatos, & Albertson, 2005; Kristiansen, Bannon, & Meador-Woodruff, 2009; Lehrmann et al., 2003; Lewohl et al., 2011; Liu, Chen, Lerner, Brackett, & Matsumoto, 2005; Mash et al., 2007; Ponomarev, Wang, Zhang, Harris, & Mayfield, 2012; Tang, Fasulo, Mash, & Hemby, 2003; Zhou, Yuan, Mash, & Goldman, 2011) have also provided critical, although somewhat divergent results for the understanding of addiction. Overall, there appear to be many common neuronal changes in gene expression among individuals addicted to various agents (Lehrmann et al., 2006; Marie-Claire et al., 2007; Zhou et al., 2011) and some commonalities with observations from model organisms, reflecting impact on shared molecular pathways involved in neuronal adaptation as well as drug-specific changes (Albertson et al., 2006; Celentano et al., 2009; Zhou et al., 2011).
It is clear that several differences in type of exposure alter the pattern of altered gene expression. One such factor is course of the exposure. Specific changes in early response genes and signal transduction pathways are more visible in the early stages of drug-induced neural adaptive processes (Celentano et al., 2009; Marie-Claire et al., 2007; Zhou et al., 2011), whereas prolonged exposure leads to widespread transcriptional changes of genes involved in diverse cellular functions such as ion transport, chromosome remodeling, stress and immune response, cell adhesion, cell cycle, apoptosis, protein and lipid metabolism, and mitochondrial functions (Albertson et al., 2004; Bannon et al., 2005; Mash et al., 2007; Renthal et al., 2007; Zhou et al., 2011). The impact of drug exposure on transcription is also brain region specific. In two components of the mesolimbic system, the dorsal striatum and nucleus accumbens (NAc), the expression of genes involved in dopaminergic, glutamatergic, and GABAergic transmission (Ghasemzadeh, Mueller, & Vasudevan, 2009; Hyman & Malenka, 2001; McClung et al., 2005; Schumann & Yaka, 2009) and that play key roles in drug-reward and drug-seeking behavior is strongly altered. In the hippocampus, a brain region critical for associative learning and memory, addiction alters the expression of genes involved in long-term potentiation (LTP) (Zhou et al., 2011). Genetic studies, especially ones using genomic sequencing of animal models selectively bred for addiction phenotypes, have uncovered functional variants of genes involved in neural adaptation that are directly responsible for genetic differences in the propensity to use addictive agents and in response (Zhou et al., 2013). Using “omic” approaches, it has also become possible to analyze the whole transcriptome and epigenetic patterning of the genome, and new molecular adaptive processes that contribute to addiction have recently been revealed by applying these methods both in humans and in model organisms.
2. MOLECULAR ADAPTATIONS ACCOMPANYING EARLY RESPONSE AND LONG-TERM ADAPTATIONS IN THE ADDICTED BRAIN
Knowledge of early gene expression changes in response to drug exposure has largely derived from animal studies, many of which have focused on preselected candidate genes and pathways. The molecular targets are often drug specific, for example, the dopamine transporter for cocaine and amphetamine exposure (Calipari, Ferris, Salahpour, Caron, & Jones, 2013; Peraile et al., 2010), opioid receptors and propeptide genes for opioid exposure (Diaz, Barros, Antonelli, Rubio, & Balerio, 2006), and GABA and glutamate receptors for other drug and alcohol exposure (Enoch et al., 2012; Meinhardt et al., 2013; Nona, Li, & Nobrega, 2013; Schumann & Yaka, 2009; Swanson, Baker, Carson, Worley, & Kalivas, 2001; Zhang et al., 2009; Zhou et al., 2013). Certain aspects of cell signaling, early transcriptional response, and learning have been obvious, and fruitful, targets for study in the addictions. Acute exposure to cocaine induces expression of immediate-early genes such as Jun and Fos, which encode transcription factors. The transcripts of these gene return to control levels, and following repeated administration of the drugs, desentization is seen (Hope, Kosofsky, Hyman, & Nestler, 1992). The transcription factors FosB (Hope et al., 1992; Nestler, 2008) and CREB (Carlezon, Duman, & Nestler, 2005) have also been well documented as key components targeted by multiple signal transduction pathways and are involved in regulating expression of drug response genes. Binding of the Fos/Jun heterodimer to AP-1 sites and CREB to cAMP-response elements (CREs) in gene promoters activates transcription of the targeted genes. Another group of well-studied immediate-early gene products is the Nur transcription factors that bind to Nur-responsive elements. These are widely present in the hypothalamus–pituitary–adrenal axis and show rapid and transient increases in expression during acute exposure to addictive drugs (Campos-Melo, Galleguillos, Sanchez, Gysling, & Andres, 2013).
In the past decade, global analysis of gene expression using high-throughput microarrays and, more recently, the use of genomic sequencing, have been frequently applied to the problem of addiction (Hitzemann et al., 2013) and have shed new light on molecular pathways that are altered in the addicted brain. These studies have been conducted in diverse contexts including rodents, nonhuman primates, and postmortem human brain samples and have revealed some important divergences. The differences in what is observed appear to be mainly due to timing and exposure: each study is a snapshot of the addicted brain in dynamic processes. In rodents, some studies have profiled gene expression during drug-self administration, whereas others during withdrawal. An important distinction between the rodent models and human postmortem brain is that in rodents, “chronic exposure” usually refers to a few days or weeks, whereas in humans, it usually denotes many years of heavy use. This one fact appears to explain most of the differences observed in studies of rodents versus those on people. More widespread and divergent molecular and cellular changes have been observed in the chronically addicted human postmortem brain. In a study of postmortem prefrontal cortex from chronic cocaine abusers, Lehrmann and colleagues found expression alterations in multiple cellular functional domains, including energy metabolism, mitochondrial oxidative phosphorylation, oligodendrocyte function, cytoskeleton and related signaling, and neuronal plasticity (Lehrmann et al., 2003). Interestingly, they also noted two distinctive states of transcription regulation, an elevated gene expression profile in the recent active cocaine abusers and decreased expression state in the non-active abusers. Altered expression in cocaine addicts has also been shown in myelin-related genes. In a study by Albertson and colleagues (Albertson et al., 2004; Bannon et al., 2005) on human postmortem NAc, the most prominent changes were decreases of myelin basic protein (MBP), proteolipid protein, and myelin-associated oligodendrocyte basic protein. The expression changes were also consistent with a decrease in the number of MBP-immunoactive oligodendrocytes. A study by Mash and colleagues also found cocaine-induced expression changes in genes involved in regulating extracellular matrix integrity and angiogenesis (Mash et al., 2007). At the top of the list of affected genes was RECK, encoding a membrane-anchored glycoprotein serving as an inhibitor for matrix metalloproteinase-9. In addition, they also observed altered expression of genes involved in apoptosis and cell death, neurogenesis and axon guidance, signal transduction, transcriptional and translational regulation, and ion transport (Mash et al., 2007).
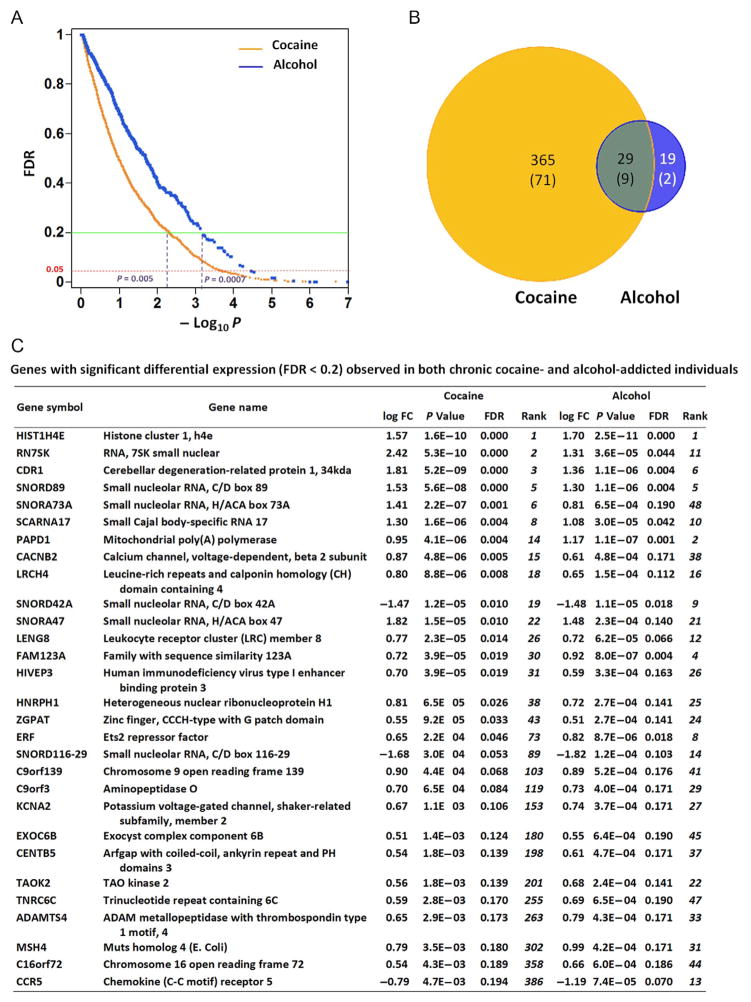
Our study (Zhou et al., 2011) with genomic sequencing directly examined mRNA-based transcriptome (RNA-Seq) in human postmortem hippocampal tissue from 24 men who were either cocaine addicts or alcoholics, or age-, ethnicity-, and postmortem interval-matched drug-free controls. Expression of 16,008 Refseq genes was detected. Among these, at an uncorrected P<0.05, we observed a total of 1994 differentially expressed genes in cocaine addicts, and 1275 differentially expressed genes in the alcoholics. After genome-wide multiple testing correction using a relatively stringent FDR cutoff of less than 0.2, there were 394 differentially expressed genes in the cocaine addicts and 48 in the alcoholics. At FDR<0.05, there were 80 differentially expressed genes in the cocaine addicts and 11 in the alcoholics (Fig. 10.1A and B).
Figure 10.1.
Differentially expressed genes in the hippocampus of cocaine- and alcohol-addicted individuals, as detected by RNA-Seq. (A) Scatter plot of –log10 P (uncorrected P value) versus FDR q value for all 16,008 expressed genes. FDR thresholds of 0.2 and 0.05 are marked, as well as corresponding uncorrected P values at an FDR of 0.2. (B) Genes differentially expressed (FDR <0.2) in cocaine only, alcohol only, and common to both. (C) Genes with significant differential expression (FDR <0.2) observed in both chronic cocaine- and alcohol-addicted individuals.
These genes differentially expressed in the human chronic cocaine brains we studied are involved in diverse cellular functions, but there were patterns that strongly implicated certain cellular functions. Cocaine depressed the transcript levels for all five members of the BEX gene family (BEX 1–5), which encodes brain expressed, X-linked proteins that are thought to mediate neurotrophin signaling and neuronal differentiation (Vilar et al., 2006). There were also significant expression changes for some histone protein genes. Genes involved in regulation of transcription, gene silencing, and chromatin modification were also affected. Several of these genes had been previously implicated in cocaine addiction, including DNMT3a, a DNA methyltransferase which was reported to play an important role in regulating cocaine response and spine plasticity in the NAc in the rat (LaPlant et al., 2010) and HDAC2, a histone deacetylase found to be involved in cocaine-induced transcription changes in rat NAc and cocaine seeking behavior (Chandrasekar & Dreyer, 2010). In addition, there was also convergent evidence that chronic cocaine exposure alters expression of genes involved in RNA processing, including significant alteration in the expression of genes encoding RNA-binding and processing proteins and enrichment of differentially expressed small nucleolar (sno) RNA genes, which are involved in both ribosomal RNA and mRNA processing (Kishore & Stamm, 2006).
A particularly salient effect of long-term cocaine exposure in postmortem brains we studied was alteration in the expression of genes involved in mitochondrial inner membrane functions and oxidative phosphorylation (Zhou et al., 2011). Interestingly, these genes integral to cellular energy production have also been implicated in neurodegenerative diseases (Cho, Nakamura, & Lipton, 2010). Among the 90 genes encoding components of oxidative phosphorylation whose expression could be reliably evaluated by RNA-Seq of hippocampal mRNA, 32 were differentially expressed (uncorrected P<0.05), and all were downregulated. Furthermore, 74 of the 90 genes (including the 32 genes that were significantly downregulated) displayed reduced expression levels in cocaine addicts. These findings were also highly consistent with previous brain imaging studies that have revealed negative effects of cocaine on brain glucose metabolism (London et al., 1990; Lyons, Friedman, Nader, & Porrino, 1996; Macey, Rice, Freedland, Whitlow, & Porrino, 2004; Thanos, Michaelides, Benveniste, Wang, & Volkow, 2008). Furthermore, alteration of certain genes encoding for mitochondrial components induced by cocaine (Lehrmann et al., 2003) and nicotine (Wang, Kim, Donovan, Becker, & Li, 2009) exposure had also been reported previously.
3. SUBSTANCE-SPECIFIC AND SHARED GENE EXPRESSION CHANGES IN ADDICTED BRAIN
Clearly, cellular and molecular changes of neural adaptation in addiction occur in a substance-specific fashion as well as through common transduction in neurotransmission pathways. Substance-specific effects are not only due to the direct action of certain drugs of abuse on specific receptors, but also by distinctive molecular and cellular changes related to the drugs that may be related to distinctive signaling mechanisms or differences in the modulation of specific neurons and circuits. Using microarray gene expression profiling, Bannon and colleagues (Albertson et al., 2006) observed decreased expression of many genes involved in presynaptic release of neurotransmitter in the NAc of chronic heroin abusers, but not in chronic cocaine abusers. Similarly, the prominent depressed expression of myelin-related genes found in cocaine abusers was not observed in heroin abusers. Their results suggested the divergent effects of cocaine and heroin on gene expression in the NAc, despite their common effects on dopaminergic transmission. Another study by Marie-Claire et al. (2007) also reported differential effects of cocaine and 3,4-methylenedioxymethamphetamine on expression of the Rnd gene family involved in actin cytoskeleton regulation in mouse striatum and noted that the two drugs might act through distinctive pathways to regulate these genes.
Comparing cocaine exposure with alcohol exposure, our analysis (Zhou et al., 2011) of transcriptomes revealed a stronger shift in hippocampal mRNA expression in cocaine-addicted brains. This was manifested by both the larger number of differentially expressed genes (Fig. 10.1B) and changes in molecular and cellular functions defined by gene ontology. For example, the unidirectional depression of expression for the genes encoding for mitochondrial inner membrane and oxidative phosphorylation was only observed in the cocaine-addicted brain (Zhou et al., 2011). This strongly suggests that the inhibition is a specific effect of chronic cocaine exposure, with potential negative implications for brain energy metabolism and diverse brain functions that depend on it and with different metabolic consequences in alcoholism.
Drug-induced neuroplasticity involves some common molecular and cellular changes of the neurocircuitries (Hyman & Malenka, 2001; Kauer & Malenka, 2007; Koob & Volkow, 2010), such as dopaminergic transmission in mesolimbic system, and corticotropin-releasing factor and norepinephrine systems in the extended amygdale. We have also observed significant overlap of gene expression and pathway alteration in both cocaine- and alcohol-addicted brains. Among the 48 differentially expressed genes (FDR<0.2) in the alcoholics, 29 were common to cocaine addicts and in each case the change was in the same direction. More strikingly, for the 11 most significantly differentially expressed genes (FDR<0.05) in alcoholics, 9 were also altered to the same degree in the cocaine addicts, suggesting shared pathways impacted by both cocaine and alcohol in neuronal adaptation. It is apparent that these commonly affected protein-coding genes (Fig. 10.1C) play important roles in neuronal functions. They include CDR1, a cerebellar degeneration-related protein; LRCH4, a leucine-rich repeat-containing neuronal protein; CACNB2, a subunit of voltage-gated calcium channel and involved in neuronal functions; and FAM123A (AMER2), a member of the gene family involved in neurogenesis (Comai, Boutet, Neirijnck, & Schedl, 2010). Other commonly and most significantly affected genes also encode for proteins critical in cellular functions such as histone (HIST1H4E), transcription regulations (ZGPAT, ERF, and HIVEP3) (Li et al., 2007), and mitochrondrial poly(A) polymerase (PAPD1). A subsequent pathway-targeted analysis of GABAergic genes also revealed common expression changes in the cocaine addicts and alcoholics from our study, such as the downregulation of GABBR1, GABRG2, and GPHN, a gene encoding the associated scaffolding protein gephyrin (Enoch et al., 2012).
4. REGION-SPECIFIC GENE EXPRESSION CHANGES IN ADDICTED BRAIN
It is apparent that many neuronal gene expression changes in the drug-induced adaptive process are region specific and cell specific. The mesolimbic system is critically involved in drug-reward and drug-seeking behavior and has been a focus for studies of addiction. In the dorsal striatum and NAc, medium-sized spiny neurons mediate dopaminergic, glutamatergic, and GABAergic neurotransmission (Hyman & Malenka, 2001) and rodents exposed to cocaine or during withdrawal show significant changes in dopaminergic, glutamatergic, and GABAergic neurotransmission (Ghasemzadeh et al., 2009; Hyman & Malenka, 2001; Nestler, 2001). Expression changes of genes targeted by dopaminergic and glutamatergic transmissions or genes involved in mediating transmission were also initiated during adaptation to drug exposure. Some of these genes have been relatively well analyzed such as CART (Douglass, McKinzie, & Couceyro, 1995), the Fos family (Hope et al., 1992; Nestler, 2008), CREB (Carlezon et al., 2005), Arc (Fosnaugh, Bhat, Yamagata, Worley, & Baraban, 1995), EGR1 (O’Donovan, Tourtellotte, Millbrandt, & Baraban, 1999), Homer-1 (Swanson et al., 2001), MKP-1 (Ujike, Takaki, Kodama, & Kuroda, 2002), Narp (Hyman & Malenka, 2001), NFκB (Ang et al., 2001), and CdK5 (Bibb et al., 2001). During the adaptive process, changes in the striatum take place in synergy with changes in other brain regions, particularly with changes in dopaminergic neurons in the midbrain ventral tegmental area where cocaine-induced glutamate release activates calcium–calmodulin-dependent protein kinases such as CaMKII which are involved in the process of behavioral sensitization (Fernandez-Espejo, Ramiro-Fuentes, Portavella, & Moreno-Paublete, 2008).
In our study of the hippocampal transcriptome (Zhou et al., 2011) of cocaine addicts and alcoholics, we did not observe significant expression changes for some of the genes that have previously been shown to be altered in striatum, such as CART, FOSB, CdK5, NFκB, and HOMER. These differences may be a manifestation of brain region-specific changes or may also be the result of stage-specific alterations in response to drug exposure because the rodent studies were performed following relatively short-term drug exposure. However, in the cocaine addicts, we did observe expression changes in genes important for hippocampal functions, such as LTP. Hippocampal functions related to short- and long-term memory processes involve synaptic plasticity, and drug-associated learning and memories are important in craving. The hippocampus also directly projects excitatory efferents to the NAc and can also activate dopaminergic neurons of the ventral tegmental area, further implicating its involvement in drug-induced changes of neural plasticity. The genes involved in LTP include specific ionotropic and metabotropic glutamatergic receptors, calcium signaling-related proteins such as calmodulin, calcium/calmodulin-dependent protein kinase, protein phosphotase, adenylate cyclase, protein kinase A and C, mitogen-activated protein kinase, and cAMP-response element binding protein (CREB). Among these, the most significantly affected genes are the N-methyl D-aspartate (NMDA) receptor 2B (GRIN2B), a subunit of the ionotropic glutamate receptor; protein phosphatase 3 catalytic subunit α isoform (PPP3CA), a part of the calcium-dependent phosphatase calcineurin; and calcium/calmodulin-dependent protein kinase type II Δ chain subunit (CAMK2D). In addition, the list of genes relevant to LTP whose expression is altered by long-term cocaine exposure includes protein phosphatase 1 catalytic subunit β and γ isoforms (PPP1CB and PPP1CC), calmodulin 2 (CALM2), CREB (CREB1), adenylate cyclase 1 (ADCY1), protein kinase C β1 (PRKCB1), and an N-ras oncogene with intrinsic GTPase activity (NRAS). Although we did not observe significant changes of the LTP pathway in the alcoholics, the phosphatidylinositol signaling system, which is closely related to the LTP pathway, was significantly altered by chronic exposure to both cocaine and alcohol. These findings of gene expression changes, together with other studies that have shown the effects of cocaine on LTP (del Olmo et al., 2006; Dunwiddie, Proctor, & Tyma, 1988; Guan, Zhang, Xu, & Li, 2009; Huang, Lin, & Hsu, 2007; Smith, Browning, & Dunwiddie, 1993; Thompson, Gosnell, & Wagner, 2002; Thompson, Swant, & Wagner, 2005), provide evidence that chronic exposure to cocaine, and possibly alcohol, leads to long-term changes in the plasticity of the hippocampus and underlines the importance in addiction of molecular mechanisms for learning.
5. PERTURBATION OF THE GLUTAMATERGIC SYSTEM IN ADDICTED BRAIN
The glutamatergic system, the major excitatory system in the central nervous system, is of particular relevance to addiction through the network of interactions with dopaminergic and GABAergic transmission that underlie alcohol and drug craving and relapse. Glutamate receptors work in synergy with dopamine receptors in dendritic spines of medium-sized spiny neurons in the striatum (Cahill, Salery, Vanhoutte, & Caboche, 2014). Epi-static interactions of glutamatergic and dopaminergic genes have been claimed in alcoholics (Puls et al., 2008). Acute and chronic exposure to alcohol affects glutamate transmission (Ding, Engleman, Rodd, & McBride, 2012) and hyperfunctioning of glutamate transmission has been observed during ethanol or drug withdrawal (Hermann et al., 2012; Prior & Galduroz, 2011). Conditional knockout of the NMDA receptor GluN2B subunit in mice eliminates LTP in the bed nucleus of the stria terminalis (Wills et al., 2012) and makes the animals more sensitive to the locomotor effects of ethanol (Badanich et al., 2011). Pharmacological manipulations have demonstrated that activation of group II metabotropic glutamate receptors decreases alcohol (Rodd et al., 2006; Zhao et al., 2006) and cocaine (Jin et al., 2010) seeking and decreases alcohol-induced neurodegeneration (Cippitelli et al., 2010) in rats.
Alteration of gene expression has been linked to persistent behavioral changes in alcohol- or drug-dependent individuals in both animal and human studies (Edenberg et al., 2005; Heilig & Koob, 2007; Hwang, Stewart, Zhang, Lumeng, & Li, 2004; Liang et al., 2010; Zhou et al., 2011). In the “post-dependent” rats generated by intermittent alcohol vapor intoxication and withdrawal, Meinhardt et al. (2013) identified a pronounced deficit of the metabotropic glutamate receptor II (mGluR2) in the pyramidal neurons of the infralimbic cortex. Among a group of glutamatergic genes that showed enriched downregulation of expression, Grm2, which encodes for mGluR2, was one of the genes that were most significantly affected, although the expression of Grm3, which encodes mGluR3, the other member of the group II metabotropic glutamate receptors, was not altered in this region. Reduction of extracellular glutamate levels in the NAc, which was readily observed in control rats upon systemic injection of mGluR2/mGluR3 agonists, was also absent in the post-dependent rats, consistent with the lack of mGluR2 function as a presynaptic receptor to downregulate glutamate release upon activation. The role of mGluR2 was further demonstrated by restoring the receptor through bilateral injection of a lentiviral vector expressing mGluR2 into infralimbic cortex. Expression of the receptor significantly reduced alcohol seeking in the post-dependent rats during the cue-induced reinstatement tests.
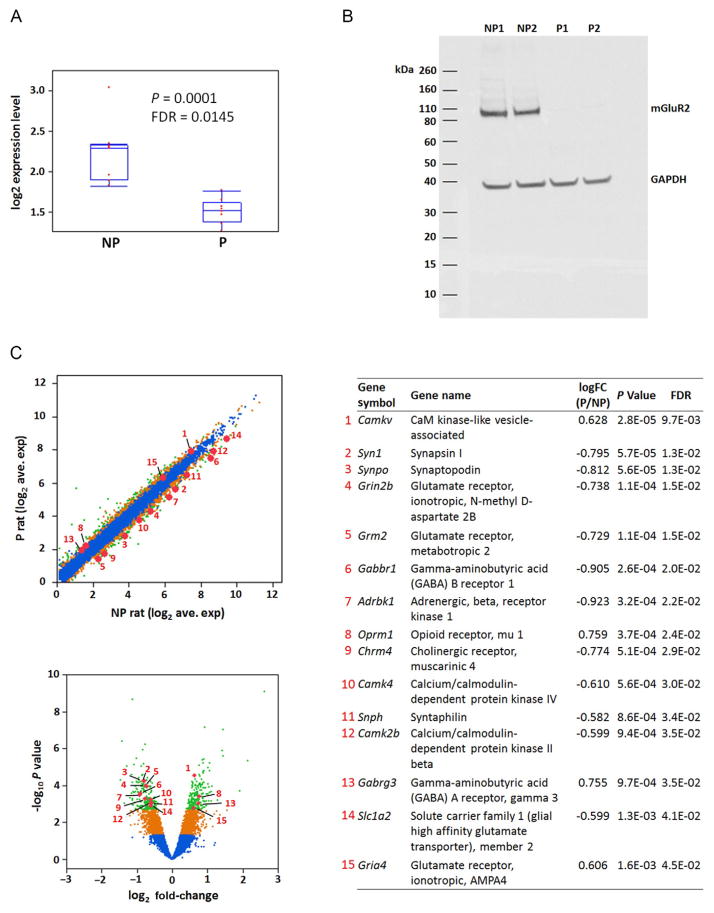
Using genomic sequencing, genetic linkage, functional validation, and transcriptome analysis, we found that a Grm2 stop codon functions as a genetic determinant for alcohol preference in selectively bred alcohol preferring (P) and nonpreferring (NP) rats (Zhou et al., 2013). In contrast, genetic studies in humans to identify genes and variants underlying complex disorders and addiction have achieved only limited success, largely due to genetic heterogeneity and the limited effect size of individual loci. Animals selectively bred for alcohol and drug dependence provide potentially powerful models for the identification of genetic variants influencing addiction behaviors both because the artificial selection may collect to high frequencies variants that are rare or uncommon in the ancestral population and because of the ability to control environmental exposures and test animals under the same conditions. By exome sequencing, we uncovered a Grm2 C407* variant from 25,715 SNPs that homozygously segregates between P and NP rats. All P rats were homozygous for this stop codon in the mGluR2 receptor ligand-binding domain, whereas none of the NP rats carried this allele. The levels of Grm2 transcript in both striatum and hippocampus were significantly lower, and expression of mGluR2 protein was undetectable in P rats (Fig. 10.2A and B). The loss of the mGluR2 receptor was also consistent with the observation of uncompensated impairment in synaptic depression in P rats, measured as field excitatory postsynaptic potential or population spike in dentate gyrus/hippocampal and striatal slices, upon activation of the receptor by the group II mGluR agonist LY379268.
Figure 10.2.
Differential gene expression in hippocampus of P and NP rats. (A) Hippocampal Grm2 mRNA levels in P and NP rats. (B) Western blot of hippocampal mGluR2 protein in NP and P rats (2 individuals in each group). (C) Average expression levels (normalized log2 read counts, upper left) of 11,406 genes measured by RNA-Seq are compared between P and NP rats. The fold change (P vs. NP) and nominal P values (–log10, t test) for each gene are also plotted (lower left). Significance of difference between P and NP for each gene is color coded: blue (black in the print version), not significant; orange (dark gray in the print version), P<0.05; green (light gray in the print version), false discovery rate <0.05. Fifteen differentially expressed genes among those in the over represented functional domains are highlighted and details of their expression differences are listed in the table (right).
The causal role of the mGluR2 stop codon in altered alcohol preference was supported by multiple layers of evidence (Zhou et al., 2013). Genetic linkage analysis in the F2 rats derived from intercrossed inbred P and NP rats showed that homozygous stop codon carriers had significantly increased alcohol consumption and preference. Pharmacological blockade of mGluR2 receptor by mGluR2/mGluR3 antagonist LY341495 also significantly escalated alcohol self-administration in Wistar rats trained in an operant self-reinforcement paradigm. To further validate whether the loss of mGluR2 causally contributes to excessive alcohol intake, we examined alcohol-drinking behavior in Grm2 knockout mice. Tested by a two-bottle free choice scheme and an escalation procedure, Grm2-null mice showed significantly higher levels of alcohol consumption and preference than the wild-type control.
To examine gene expression changes related to alcohol-drinking behavior, we also performed hippocampal transcriptome analysis in P and NP rats (Fig. 10.2C) using RNA-sequencing (Zhou et al., 2013). The results indicated an overall pattern of altered expression of genes involved in neural development and synaptic functions. A total of 485 genes were differentially expressed at FDR<0.05 following correction for genome-wide testing. Differentially expressed genes were significantly enriched with segregating SNPs located in the coding regions and UTRs, indicating the potential involvement of cis-regulatory elements in these genes. Using functional annotation analysis with twofold enrichment as a cutoff, we identified several functional domains among the 485 differentially expressed genes, including calmodulin binding, synapse, and neuronal projection. Of particular interest was overrepresentation of the genes that function in glutamate, GABA, opioid, cholinergic, and adrenergic transmission (Fig. 10.2C). This pattern was consistent with the loss of mGluR2 receptor in P rats, but also more readily points to overall neuronal differences between P and NP rats that influence alcohol-drinking behaviors. The gene expression differences between P and NP rats are thus consistent and convergent with their genetic and phenotypic differences and are likely to be influenced by their overall genetic differences or the interaction of Grm2 C407* with other loci.
6. EPIGENETIC REGULATION OF GENE EXPRESSION IN ADDICTED BRAIN
Epigenetics plays a key role in regulating gene expression. Studies have shown that drug exposure causes changes in DNA methylation that lead to alterations in transcription. Acute cocaine treatment was reported to increase the expression of DNA methyltransferase genes, Dnmt3a and Dnmt 3b, in mouse NAc, resulting in DNA hypermethylation and the increased binding of methyl CpG binding protein 2 (MeCP2) at the promoter of protein phosphatase-1 catalytic subunit gene (Pp1c). As a result, Pp1c expression was decreased (Anier, Malinovskaja, Aonurm-Helm, Zharkovsky, & Kalda, 2010). In contrast, chronic cocaine administration was found to decrease expression of Dnmt3a. The attenuation of DNA methylation led to potentiated cocaine reward (LaPlant et al., 2010). Acute and repeated cocaine administration was also shown to cause hypomethylation at the FosB promoter, leading to upregulation of FosB expression (Anier et al., 2010). It has also been shown that in heroin addicts, there was elevated methylation at several CpG sites in the promoter of a μ-opioid receptor gene, OPRM1, in lymphocytes, which might result in reduced expression of that gene (Nielsen et al., 2009). Differential DNA methylation at the promoter of the pro-opiomelanocortin gene (POMC) was found to be associated with alcoholism in a human study (Muschler et al., 2010). DNA hypomethylation was associated with activation of endogenous retroviruses in alcoholic brain (Ponomarev et al., 2012).
Chronic drug exposure also causes significant changes of histone modification. Histone acetylation is known to be associated with activated gene expression. Chronic cocaine exposure was shown to inhibit the function of Hdac5, a histone deacetylase, in mouse NAc (Renthal et al., 2007). Activation of dopamine D1 receptor induced upregulation of histone acetylation at the promoters of tyrosine hydroxylase (Th) and brain-derived neurotrophic factor (Bdnf) genes in mouse NAc and the expression of the two genes (Schroeder et al., 2008). There was a reported association in mice between histone H3 acetylation-activated transcription of addiction-related genes, such as CamkII-α and the motivation for cocaine (L. Wang et al., 2010). In mice chronically administered amphetamine, the Δ-FosB-mediated responses were also found to involve recruiting Hdac1 to its target gene promoters (Renthal et al., 2008). Inhibition of histone deacetylase reduced behavioral sensitization to morphine in mice (Jing et al., 2011). Alteration of histone methylation also plays important roles in neuronal adaptation of addicted brain. Repeated cocaine administration in mice was shown to repress the expression of lysine dimethyltransferase G9a, resulting in decrease of histone lysine 9 dimethylation (H3K9me2) in NAc (Maze et al., 2010).
Our chromatin immunoprecipitation and genomic sequencing (ChIP-Seq)-based analysis in the postmortem hippocampus of cocaine addicts and alcoholics revealed significant changes in histone H3 lysine 4 trimethylation (H3K4me3) (Zhou et al., 2011), a histone mark known to be associated with activation of gene expression. Similar to the changes observed in gene expression, there was a more widespread and greater impact in response to chronic cocaine exposure than to alcohol exposure. There were also concordant changes between H3K4me3 and gene expression at some loci. In cocaine addicts, these included components of the mitochondrial oxidative phosphorylation pathway or regulators of cellular energy metabolism such as NDUFS2, NDUFA12L, UQCRB, INSR, and IGF1R; genes involved in LTP and other neuronal functions such as calmodulin 2 (CALM2), Synaptophysin-like protein 2 (SYPL2), sodium/chloride-dependent neurotransmitter transporter (SLC6A15), and nociceptin (PNOC). In alcoholics, there were concordant changes between H3K4me3 and gene expression of Protocadherin alpha-7 (PCDHA7), Aquaporin-11 (AQP11), and potassium inwardly-rectifying channel, subfamily J, member 5 (CIR), all of which are involved in critical neuronal and cellular functions. Globally, among all 13,113 histone H3K4me3 peaks mapped to the promoters of hippocampal expressed genes, there was a trend of correlation between H3K4me3 and expression changes in cocaine addicts. However, this trend was not observed in alcoholics. Overall, there was no significant overlap between the genes with either significant H3K4me3 changes or expression changes in both cocaine addicts and alcoholics. This may reflect the fact that epigenetic regulation of gene expression through chromatin remodeling involves many different types of histone modifications at many different histone residues (Barski et al., 2007; Wang et al., 2008), and cocaine- and alcohol-induced expression changes are very likely the results of alterations of those many different histone modifications.
Gene expression changes caused by chronic drug exposure may also be mediated by regulatory RNAs, such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs). Regulatory RNAs modify gene expression through multiple means, such as altering mRNA stability, basal transcription machinery, translational efficiency, and chromosome modification. miRNA array analysis in human prefrontal cortex revealed upregulation of approximately 35 miRNAs in alcoholics relative to controls with predicted target genes implicated in apoptosis, cell cycle, cell adhesion, nervous system development, and cell–cell signaling (Lewohl et al., 2011). Exposure of zebrafish embryos to cocaine reduced the expression of miR-133b in the CNS, and this difference in miR signaling might in turn modulate expression of dopamine receptors, the dopamine transporter, and tyrosine hydroxylase (Barreto-Valer, Lopez-Bellido, Macho Sanchez-Simon, & Rodriguez, 2012). The let-7 miR family may also interact with the 3′-untranslated region of μ-opioid receptor mRNA to regulate opioid tolerance (He & Wang, 2012). miRNAs are both synaptically enriched and depleted by drug exposure. Cocaine modulates levels of the miR-8 family which is enriched at postsynaptic densities and regulates expression of cell adhesion molecules (Eipper-Mains, Eipper, & Mains, 2012). Differential expression of multiple lncRNAs was identified in the NAc of cocaine-conditioned mice and those lncRNAs were reported to regulate their target loci through both cis- and trans-actions (Bu et al., 2012).
7. CONCLUSION
Profiling gene expression in the addicted brain has revealed both agent-specific and common drug-induced neural adaptations, providing valuable insights for the understanding of the relevant molecular and cellular mechanisms. The development of transcriptome-based sequencing analysis has equipped us with potent tools that can be combined with neuroscience tools and approaches including the isolation of particular regions, circuits, and cells involved in addiction, genetic models including artificially selected strains and humans varying in vulnerability and response, and interventional models including pharmacological challenges and gene-based manipulations of pathway function and response. These approaches will further enable us to deconstruct the transcription machinery and epigenetic regulation in the addicted brain.
References
- Albertson DN, Pruetz B, Schmidt CJ, Kuhn DM, Kapatos G, Bannon MJ. Gene expression profile of the nucleus accumbens of human cocaine abusers: Evidence for dysregulation of myelin. Journal of Neurochemistry. 2004;88(5):1211–1219. doi: 10.1046/j.1471-4159.2003.02247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson DN, Schmidt CJ, Kapatos G, Bannon MJ. Distinctive profiles of gene expression in the human nucleus accumbens associated with cocaine and heroin abuse. Neuropsychopharmacology. 2006;31(10):2304–2312. doi: 10.1038/sj.npp.1301089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang E, Chen J, Zagouras P, Magna H, Holland J, Schaeffer E, et al. Induction of nuclear factor-kappaB in nucleus accumbens by chronic cocaine administration. Journal of Neurochemistry. 2001;79(1):221–224. doi: 10.1046/j.1471-4159.2001.00563.x. [DOI] [PubMed] [Google Scholar]
- Anier K, Malinovskaja K, Aonurm-Helm A, Zharkovsky A, Kalda A. DNA methylation regulates cocaine-induced behavioral sensitization in mice. Neuropsychopharmacology. 2010;35(12):2450–2461. doi: 10.1038/npp.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badanich KA, Doremus-Fitzwater TL, Mulholland PJ, Randall PK, Delpire E, Becker HC. NR2B-deficient mice are more sensitive to the locomotor stimulant and depressant effects of ethanol. Genes, Brain, and Behavior. 2011;10(7):805–816. doi: 10.1111/j.1601-183X.2011.00720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannon M, Kapatos G, Albertson D. Gene expression profiling in the brains of human cocaine abusers. Addiction Biology. 2005;10(1):119–126. doi: 10.1080/13556210412331308921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto-Valer K, Lopez-Bellido R, Macho Sanchez-Simon F, Rodriguez RE. Modulation by cocaine of dopamine receptors through miRNA-133b in zebrafish embryos. PLoS One. 2012;7(12):e52701. doi: 10.1371/journal.pone.0052701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bibb JA, Chen J, Taylor JR, Svenningsson P, Nishi A, Snyder GL, et al. Effects of chronic exposure to cocaine are regulated by the neuronal protein Cdk5. Nature. 2001;410(6826):376–380. doi: 10.1038/35066591. [DOI] [PubMed] [Google Scholar]
- Bu Q, Hu Z, Chen F, Zhu R, Deng Y, Shao X, et al. Transcriptome analysis of long non-coding RNAs of the nucleus accumbens in cocaine-conditioned mice. Journal of Neurochemistry. 2012;123(5):790–799. doi: 10.1111/jnc.12006. [DOI] [PubMed] [Google Scholar]
- Cahill E, Salery M, Vanhoutte P, Caboche J. Convergence of dopamine and glutamate signaling onto striatal ERK activation in response to drugs of abuse. Frontiers in Pharmacology. 2014;4:172. doi: 10.3389/fphar.2013.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calipari ES, Ferris MJ, Salahpour A, Caron MG, Jones SR. Methylphenidate amplifies the potency and reinforcing effects of amphetamines by increasing dopamine transporter expression. Nature Communications. 2013;4:2720. doi: 10.1038/ncomms3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Melo D, Galleguillos D, Sanchez N, Gysling K, Andres ME. Nur transcription factors in stress and addiction. Frontiers in Molecular Neuroscience. 2013;6:44. doi: 10.3389/fnmol.2013.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlezon WA, Jr, Duman RS, Nestler EJ. The many faces of CREB. Trends in Neurosciences. 2005;28(8):436–445. doi: 10.1016/j.tins.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Celentano M, Caprioli D, Dipasquale P, Cardillo V, Nencini P, Gaetani S, et al. Drug context differently regulates cocaine versus heroin self-administration and cocaine- versus heroin-induced Fos mRNA expression in the rat. Psychopharmacology. 2009;204(2):349–360. doi: 10.1007/s00213-009-1467-x. [DOI] [PubMed] [Google Scholar]
- Chandrasekar V, Dreyer JL. The brain-specific neural zinc finger transcription factor 2b (NZF-2b/7ZFMyt1) suppresses cocaine self-administration in rats. Frontiers in Behavioral Neuroscience. 2010;4:14. doi: 10.3389/fnbeh.2010.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho DH, Nakamura T, Lipton SA. Mitochondrial dynamics in cell death and neurodegeneration. Cellular and Molecular Life Sciences. 2010;67(20):3435–3447. doi: 10.1007/s00018-010-0435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cippitelli A, Damadzic R, Frankola K, Goldstein A, Thorsell A, Singley E, et al. Alcohol-induced neurodegeneration, suppression of transforming growth factor-beta, and cognitive impairment in rats: Prevention by group II metabotropic glutamate receptor activation. Biological Psychiatry. 2010;67(9):823–830. doi: 10.1016/j.biopsych.2009.12.018. [DOI] [PubMed] [Google Scholar]
- Comai G, Boutet A, Neirijnck Y, Schedl A. Expression patterns of the Wtx/Amer gene family during mouse embryonic development. Developmental Dynamics. 2010;239(6):1867–1878. doi: 10.1002/dvdy.22313. [DOI] [PubMed] [Google Scholar]
- del Olmo N, Miguens M, Higuera-Matas A, Torres I, Garcia-Lecumberri C, Solis JM, et al. Enhancement of hippocampal long-term potentiation induced by cocaine self-administration is maintained during the extinction of this behavior. Brain Research. 2006;1116(1):120–126. doi: 10.1016/j.brainres.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Diaz SL, Barros VG, Antonelli MC, Rubio MC, Balerio GN. Morphine withdrawal syndrome and its prevention with baclofen: Autoradiographic study of mu-opioid receptors in prepubertal male and female mice. Synapse. 2006;60(2):132–140. doi: 10.1002/syn.20279. [DOI] [PubMed] [Google Scholar]
- Ding ZM, Engleman EA, Rodd ZA, McBride WJ. Ethanol increases glutamate neurotransmission in the posterior ventral tegmental area of female wistar rats. Alcoholism, Clinical and Experimental Research. 2012;36(4):633–640. doi: 10.1111/j.1530-0277.2011.01665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglass J, McKinzie AA, Couceyro P. PCR differential display identifies a rat brain mRNA that is transcriptionally regulated by cocaine and amphetamine. The Journal of Neuroscience. 1995;15(3 Pt 2):2471–2481. doi: 10.1523/JNEUROSCI.15-03-02471.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunwiddie TV, Proctor WR, Tyma J. Local anaesthetic actions of cocaine: Effects on excitatory and inhibitory synaptic responses in the hippocampus in vitro. British Journal of Pharmacology. 1988;95(4):1117–1124. doi: 10.1111/j.1476-5381.1988.tb11746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Strother WN, McClintick JN, Tian H, Stephens M, Jerome RE, et al. Gene expression in the hippocampus of inbred alcohol-preferring and -nonpreferring rats. Genes, Brain, and Behavior. 2005;4(1):20–30. doi: 10.1111/j.1601-183X.2004.00091.x. [DOI] [PubMed] [Google Scholar]
- Eipper-Mains JE, Eipper BA, Mains RE. Global approaches to the role of miRNAs in drug-induced changes in gene expression. Frontiers in Genetics. 2012;3:109. doi: 10.3389/fgene.2012.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch MA, Zhou Z, Kimura M, Mash DC, Yuan Q, Goldman D. GABAergic gene expression in postmortem hippocampus from alcoholics and cocaine addicts; corresponding findings in alcohol-naive P and NP rats. PLoS One. 2012;7(1):e29369. doi: 10.1371/journal.pone.0029369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Espejo E, Ramiro-Fuentes S, Portavella M, Moreno-Paublete R. Role for D-serine within the ventral tegmental area in the development of cocaine’s sensitization. Neuropsychopharmacology. 2008;33(5):995–1003. doi: 10.1038/sj.npp.1301495. [DOI] [PubMed] [Google Scholar]
- Fosnaugh JS, Bhat RV, Yamagata K, Worley PF, Baraban JM. Activation of arc, a putative “effector” immediate early gene, by cocaine in rat brain. Journal of Neurochemistry. 1995;64(5):2377–2380. doi: 10.1046/j.1471-4159.1995.64052377.x. [DOI] [PubMed] [Google Scholar]
- Ghasemzadeh MB, Mueller C, Vasudevan P. Behavioral sensitization to cocaine is associated with increased glutamate receptor trafficking to the postsynaptic density after extended withdrawal period. Neuroscience. 2009;159(1):414–426. doi: 10.1016/j.neuroscience.2008.10.027. [DOI] [PubMed] [Google Scholar]
- Goldman D, Oroszi G, Ducci F. The genetics of addictions: Uncovering the genes. Nature Reviews. Genetics. 2005;6(7):521–532. doi: 10.1038/nrg1635. [DOI] [PubMed] [Google Scholar]
- Guan X, Zhang R, Xu Y, Li S. Cocaine withdrawal enhances long-term potentiation in rat hippocampus via changing the activity of corticotropin-releasing factor receptor subtype 2. Neuroscience. 2009;161(3):665–670. doi: 10.1016/j.neuroscience.2009.04.035. [DOI] [PubMed] [Google Scholar]
- He Y, Wang ZJ. Let-7 microRNAs and opioid tolerance. Frontiers in Genetics. 2012;3:110. doi: 10.3389/fgene.2012.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilig M, Koob GF. A key role for corticotropin-releasing factor in alcohol dependence. Trends in Neurosciences. 2007;30(8):399–406. doi: 10.1016/j.tins.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann D, Weber-Fahr W, Sartorius A, Hoerst M, Frischknecht U, Tunc-Skarka N, et al. Translational magnetic resonance spectroscopy reveals excessive central glutamate levels during alcohol withdrawal in humans and rats. Biological Psychiatry. 2012;71(11):1015–1021. doi: 10.1016/j.biopsych.2011.07.034. [DOI] [PubMed] [Google Scholar]
- Hitzemann R, Bottomly D, Darakjian P, Walter N, Iancu O, Searles R, et al. Genes, behavior and next-generation RNA sequencing. Genes, Brain, and Behavior. 2013;12(1):1–12. doi: 10.1111/gbb.12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope B, Kosofsky B, Hyman SE, Nestler EJ. Regulation of immediate early gene expression and AP-1 binding in the rat nucleus accumbens by chronic cocaine. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(13):5764–5768. doi: 10.1073/pnas.89.13.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Lin HJ, Hsu KS. Repeated cocaine administration promotes long-term potentiation induction in rat medial prefrontal cortex. Cerebral Cortex. 2007;17(8):1877–1888. doi: 10.1093/cercor/bhl096. [DOI] [PubMed] [Google Scholar]
- Hwang BH, Stewart R, Zhang JK, Lumeng L, Li TK. Corticotropin-releasing factor gene expression is down-regulated in the central nucleus of the amygdala of alcohol-preferring rats which exhibit high anxiety: A comparison between rat lines selectively bred for high and low alcohol preference. Brain Research. 2004;1026(1):143–150. doi: 10.1016/j.brainres.2004.08.042. [DOI] [PubMed] [Google Scholar]
- Hyman SE, Malenka RC. Addiction and the brain: The neurobiology of compulsion and its persistence. Nature Reviews. Neuroscience. 2001;2(10):695–703. doi: 10.1038/35094560. [DOI] [PubMed] [Google Scholar]
- Jin X, Semenova S, Yang L, Ardecky R, Sheffler DJ, Dahl R, et al. The mGluR2 positive allosteric modulator BINA decreases cocaine self-administration and cue-induced cocaine-seeking and counteracts cocaine-induced enhancement of brain reward function in rats. Neuropsychopharmacology. 2010;35(10):2021–2036. doi: 10.1038/npp.2010.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing L, Luo J, Zhang M, Qin WJ, Li YL, Liu Q, et al. Effect of the histone deacetylase inhibitors on behavioural sensitization to a single morphine exposure in mice. Neuroscience Letters. 2011;494:169–173. doi: 10.1016/j.neulet.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Kauer JA, Malenka RC. Synaptic plasticity and addiction. Nature Reviews. Neuroscience. 2007;8(11):844–858. doi: 10.1038/nrn2234. [DOI] [PubMed] [Google Scholar]
- Kishore S, Stamm S. Regulation of alternative splicing by snoRNAs. Cold Spring Harbor Symposia on Quantitative Biology. 2006;71:329–334. doi: 10.1101/sqb.2006.71.024. [DOI] [PubMed] [Google Scholar]
- Koob GF, Le Moal M. Drug addiction, dysregulation of reward, and allostasis. Neuropsychopharmacology. 2001;24(2):97–129. doi: 10.1016/S0893-133X(00)00195-0. [DOI] [PubMed] [Google Scholar]
- Koob GF, Volkow ND. Neurocircuitry of addiction. Neuropsychopharmacology. 2010;35(1):217–238. doi: 10.1038/npp.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansen LV, Bannon MJ, Meador-Woodruff JH. Expression of transcripts for myelin related genes in postmortem brain from cocaine abusers. Neurochemical Research. 2009;34(1):46–54. doi: 10.1007/s11064-008-9655-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPlant Q, Vialou V, Covington HE, 3rd, Dumitriu D, Feng J, Warren BL, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nature Neuroscience. 2010;13(9):1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrmann E, Colantuoni C, Deep-Soboslay A, Becker KG, Lowe R, Huestis MA, et al. Transcriptional changes common to human cocaine, cannabis and phencyclidine abuse. PLoS One. 2006;1:e114. doi: 10.1371/journal.pone.0000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrmann E, Oyler J, Vawter MP, Hyde TM, Kolachana B, Kleinman JE, et al. Transcriptional profiling in the human prefrontal cortex: Evidence for two activational states associated with cocaine abuse. The Pharmacogenomics Journal. 2003;3(1):27–40. doi: 10.1038/sj.tpj.6500146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewohl JM, Nunez YO, Dodd PR, Tiwari GR, Harris RA, Mayfield RD. Up-regulation of microRNAs in brain of human alcoholics. Alcoholism, Clinical and Experimental Research. 2011;35(11):1928–1937. doi: 10.1111/j.1530-0277.2011.01544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YJ, Deng J, Mayhew GM, Grimsley JW, Huo X, Vance JM. Investigation of the PARK10 gene in Parkinson disease. Annals of Human Genetics. 2007;71(Pt 5):639–647. doi: 10.1111/j.1469-1809.2007.00353.x. [DOI] [PubMed] [Google Scholar]
- Liang T, Kimpel MW, McClintick JN, Skillman AR, McCall K, Edenberg HJ, et al. Candidate genes for alcohol preference identified by expression profiling in alcohol-preferring and -nonpreferring reciprocal congenic rats. Genome Biology. 2010;11(2):R11. doi: 10.1186/gb-2010-11-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Chen GD, Lerner MR, Brackett DJ, Matsumoto RR. Cocaine up-regulates Fra-2 and sigma-1 receptor gene and protein expression in brain regions involved in addiction and reward. The Journal of Pharmacology and Experimental Therapeutics. 2005;314(2):770–779. doi: 10.1124/jpet.105.084525. [DOI] [PubMed] [Google Scholar]
- London ED, Cascella NG, Wong DF, Phillips RL, Dannals RF, Links JM, et al. Cocaine-induced reduction of glucose utilization in human brain. A study using positron emission tomography and [fluorine 18]-fluorodeoxyglucose. Archives of General Psychiatry. 1990;47(6):567–574. doi: 10.1001/archpsyc.1990.01810180067010. [DOI] [PubMed] [Google Scholar]
- Lyons D, Friedman DP, Nader MA, Porrino LJ. Cocaine alters cerebral metabolism within the ventral striatum and limbic cortex of monkeys. The Journal of Neuroscience. 1996;16(3):1230–1238. doi: 10.1523/JNEUROSCI.16-03-01230.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macey DJ, Rice WN, Freedland CS, Whitlow CT, Porrino LJ. Patterns of functional activity associated with cocaine self-administration in the rat change over time. Psychopharmacology. 2004;172(4):384–392. doi: 10.1007/s00213-003-1676-7. [DOI] [PubMed] [Google Scholar]
- Marie-Claire C, Salzmann J, David A, Courtin C, Canestrelli C, Noble F. Rnd family genes are differentially regulated by 3,4-methylenedioxymethamphetamine and cocaine acute treatment in mice brain. Brain Research. 2007;1134(1):12–17. doi: 10.1016/j.brainres.2006.11.065. [DOI] [PubMed] [Google Scholar]
- Mash DC, ffrench-Mullen J, Adi N, Qin Y, Buck A, Pablo J. Gene expression in human hippocampus from cocaine abusers identifies genes which regulate extra-cellular matrix remodeling. PLoS One. 2007;2(11):e1187. doi: 10.1371/journal.pone.0001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Covington HE, 3rd, Dietz DM, LaPlant Q, Renthal W, Russo SJ, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327(5962):213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CA, Sidiropoulou K, Vitaterna M, Takahashi JS, White FJ, Cooper DC, et al. Regulation of dopaminergic transmission and cocaine reward by the Clock gene. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(26):9377–9381. doi: 10.1073/pnas.0503584102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt MW, Hansson AC, Perreau-Lenz S, Bauder-Wenz C, Stahlin O, Heilig M, et al. Rescue of infralimbic mGluR2 deficit restores control over drug-seeking behavior in alcohol dependence. The Journal of Neuroscience. 2013;33(7):2794–2806. doi: 10.1523/JNEUROSCI.4062-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschler MA, Hillemacher T, Kraus C, Kornhuber J, Bleich S, Frieling H. DNA methylation of the POMC gene promoter is associated with craving in alcohol dependence. Journal of Neural Transmission. 2010;117(4):513–519. doi: 10.1007/s00702-010-0378-7. [DOI] [PubMed] [Google Scholar]
- Nestler EJ. Molecular basis of long-term plasticity underlying addiction. Nature Reviews. Neuroscience. 2001;2(2):119–128. doi: 10.1038/35053570. [DOI] [PubMed] [Google Scholar]
- Nestler EJ. Review. Transcriptional mechanisms of addiction: Role of DeltaFosB. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 2008;363(1507):3245–3255. doi: 10.1098/rstb.2008.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen DA, Yuferov V, Hamon S, Jackson C, Ho A, Ott J, et al. Increased OPRM1 DNA methylation in lymphocytes of methadone-maintained former heroin addicts. Neuropsychopharmacology. 2009;34(4):867–873. doi: 10.1038/npp.2008.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nona CN, Li R, Nobrega JN. Altered NMDA receptor subunit gene expression in brains of mice showing high vs. low sensitization to ethanol. Behavioural Brain Research. 2013;260C:58–66. doi: 10.1016/j.bbr.2013.11.037. [DOI] [PubMed] [Google Scholar]
- O’Donovan KJ, Tourtellotte WG, Millbrandt J, Baraban JM. The EGR family of transcription-regulatory factors: Progress at the interface of molecular and systems neuroscience. Trends in Neurosciences. 1999;22(4):167–173. doi: 10.1016/s0166-2236(98)01343-5. [DOI] [PubMed] [Google Scholar]
- Peraile I, Torres E, Mayado A, Izco M, Lopez-Jimenez A, Lopez-Moreno JA, et al. Dopamine transporter down-regulation following repeated cocaine: Implications for 3,4-methylenedioxymethamphetamine-induced acute effects and long-term neurotoxicity in mice. British Journal of Pharmacology. 2010;159(1):201–211. doi: 10.1111/j.1476-5381.2009.00522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. The Journal of Neuroscience. 2012;32(5):1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior PL, Galduroz JC. Glutamatergic hyperfunctioning during alcohol withdrawal syndrome: Therapeutic perspective with zinc and magnesium. Medical Hypotheses. 2011;77(3):368–370. doi: 10.1016/j.mehy.2011.05.017. [DOI] [PubMed] [Google Scholar]
- Puls I, Mohr J, Wrase J, Priller J, Behr J, Kitzrow W, et al. Synergistic effects of the dopaminergic and glutamatergic system on hippocampal volume in alcohol-dependent patients. Biological Psychology. 2008;79(1):126–136. doi: 10.1016/j.biopsycho.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Renthal W, Carle TL, Maze I, Covington HE, 3rd, Truong HT, Alibhai I, et al. Delta FosB mediates epigenetic desensitization of the c-fos gene after chronic amphetamine exposure. The Journal of Neuroscience. 2008;28(29):7344–7349. doi: 10.1523/JNEUROSCI.1043-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Maze I, Krishnan V, Covington HE, 3rd, Xiao G, Kumar A, et al. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56(3):517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Rodd ZA, McKinzie DL, Bell RL, McQueen VK, Murphy JM, Schoepp DD, et al. The metabotropic glutamate 2/3 receptor agonist LY404039 reduces alcohol-seeking but not alcohol self-administration in alcohol-preferring (P) rats. Behavioural Brain Research. 2006;171(2):207–215. doi: 10.1016/j.bbr.2006.03.032. [DOI] [PubMed] [Google Scholar]
- Schroeder FA, Penta KL, Matevossian A, Jones SR, Konradi C, Tapper AR, et al. Drug-induced activation of dopamine D(1) receptor signaling and inhibition of class I/II histone deacetylase induce chromatin remodeling in reward circuitry and modulate cocaine-related behaviors. Neuropsychopharmacology. 2008;33(12):2981–2992. doi: 10.1038/npp.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann J, Yaka R. Prolonged withdrawal from repeated noncontingent cocaine exposure increases NMDA receptor expression and ERK activity in the nucleus accumbens. The Journal of Neuroscience. 2009;29(21):6955–6963. doi: 10.1523/JNEUROSCI.1329-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DA, Browning M, Dunwiddie TV. Cocaine inhibits hippocampal long-term potentiation. Brain Research. 1993;608(2):259–265. doi: 10.1016/0006-8993(93)91466-6. [DOI] [PubMed] [Google Scholar]
- Swanson CJ, Baker DA, Carson D, Worley PF, Kalivas PW. Repeated cocaine administration attenuates group I metabotropic glutamate receptor-mediated glutamate release and behavioral activation: A potential role for Homer. The Journal of Neuroscience. 2001;21(22):9043–9052. doi: 10.1523/JNEUROSCI.21-22-09043.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WX, Fasulo WH, Mash DC, Hemby SE. Molecular profiling of midbrain dopamine regions in cocaine overdose victims. Journal of Neurochemistry. 2003;85(4):911–924. doi: 10.1046/j.1471-4159.2003.01740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanos PK, Michaelides M, Benveniste H, Wang GJ, Volkow ND. The effects of cocaine on regional brain glucose metabolism is attenuated in dopamine transporter knockout mice. Synapse. 2008;62(5):319–324. doi: 10.1002/syn.20503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AM, Gosnell BA, Wagner JJ. Enhancement of long-term potentiation in the rat hippocampus following cocaine exposure. Neuropharmacology. 2002;42(8):1039–1042. doi: 10.1016/s0028-3908(02)00059-x. [DOI] [PubMed] [Google Scholar]
- Thompson AM, Swant J, Wagner JJ. Cocaine-induced modulation of long-term potentiation in the CA1 region of rat hippocampus. Neuropharmacology. 2005;49(2):185–194. doi: 10.1016/j.neuropharm.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Ujike H, Takaki M, Kodama M, Kuroda S. Gene expression related to synaptogenesis, neuritogenesis, and MAP kinase in behavioral sensitization to psychostimulants. The Annals of the New York Academy of Sciences. 2002;965:55–67. doi: 10.1111/j.1749-6632.2002.tb04151.x. [DOI] [PubMed] [Google Scholar]
- Vilar M, Murillo-Carretero M, Mira H, Magnusson K, Besset V, Ibanez CF. Bex1, a novel interactor of the p75 neurotrophin receptor, links neurotrophin signaling to the cell cycle. The EMBO Journal. 2006;25(6):1219–1230. doi: 10.1038/sj.emboj.7601017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Kim JM, Donovan DM, Becker KG, Li MD. Significant modulation of mitochondrial electron transport system by nicotine in various rat brain regions. Mitochondrion. 2009;9(3):186–195. doi: 10.1016/j.mito.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lv Z, Hu Z, Sheng J, Hui B, Sun J, et al. Chronic cocaine-induced H3 acetylation and transcriptional activation of CaMKIIalpha in the nucleus accumbens is critical for motivation for drug reinforcement. Neuropsychopharmacology. 2010;35(4):913–928. doi: 10.1038/npp.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nature Genetics. 2008;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills TA, Klug JR, Silberman Y, Baucum AJ, Weitlauf C, Colbran RJ, et al. GluN2B subunit deletion reveals key role in acute and chronic ethanol sensitivity of glutamate synapses in bed nucleus of the stria terminalis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(5):E278–E287. doi: 10.1073/pnas.1113820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GC, Vu K, Parelkar NK, Mao LM, Stanford IM, Fibuch EE, et al. Acute administration of cocaine reduces metabotropic glutamate receptor 8 protein expression in the rat striatum in vivo. Neuroscience Letters. 2009;449(3):224–227. doi: 10.1016/j.neulet.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Dayas CV, Aujla H, Baptista MA, Martin-Fardon R, Weiss F. Activation of group II metabotropic glutamate receptors attenuates both stress and cue-induced ethanol-seeking and modulates c-fos expression in the hippocampus and amygdala. The Journal of Neuroscience. 2006;26(39):9967–9974. doi: 10.1523/JNEUROSCI.2384-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Karlsson C, Liang T, Xiong W, Kimura M, Tapocik JD, et al. Loss of metabotropic glutamate receptor 2 escalates alcohol consumption. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(42):16963–16968. doi: 10.1073/pnas.1309839110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Yuan Q, Mash DC, Goldman D. Substance-specific and shared transcription and epigenetic changes in the human hippocampus chronically exposed to cocaine and alcohol. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(16):6626–6631. doi: 10.1073/pnas.1018514108. [DOI] [PMC free article] [PubMed] [Google Scholar]